Abstract
Mutations in PTEN-induced kinase 1 (PINK1) cause early onset autosomal recessive Parkinson's disease (PD). PINK1 is a 63 kDa protein kinase, which exerts a neuroprotective function and is known to localize to mitochondria. Upon entry into the organelle, PINK1 is cleaved to produce a ∼53 kDa protein (ΔN-PINK1). In this paper, we show that PINK1 is cleaved between amino acids Ala-103 and Phe-104 to generate ΔN-PINK1. We demonstrate that a reduced ability to cleave PINK1, and the consequent accumulation of full-length protein, results in mitochondrial abnormalities reminiscent of those observed in PINK1 knockout cells, including disruption of the mitochondrial network and a reduction in mitochondrial mass. Notably, we assessed three N-terminal PD-associated PINK1 mutations located close to the cleavage site and, while these do not prevent PINK1 cleavage, they alter the ratio of full-length to ΔN-PINK1 protein in cells, resulting in an altered mitochondrial phenotype. Finally, we show that PINK1 interacts with the mitochondrial protease presenilin-associated rhomboid-like protein (PARL) and that loss of PARL results in aberrant PINK1 cleavage in mammalian cells. These combined results suggest that PINK1 cleavage is important for basal mitochondrial health and that PARL cleaves PINK1 to produce the ΔN-PINK1 fragment.
INTRODUCTION
Mutations in the (PTEN-induced kinase 1) Pink1 gene (PARK6) are an important cause of autosomal recessive Parkinson's disease (PD) (1). PINK1 is synthesized as a 581 amino acid protein containing a mitochondrial localization sequence, a predicted transmembrane (TM) domain and a serine/threonine kinase domain (2). Mutations within the kinase domain and C-terminus of the protein reduce PINK1 kinase activity, and this loss of kinase function is thought to be responsible for PD pathogenesis (3). Consistent with this hypothesis, a truncated fragment of the PINK1 protein containing the active kinase domain was shown to be sufficient to provide cells with protection against MPTP toxicity, but this effect was abrogated by the presence of kinase-inactivating mutations (4). However, a series of PD-associated mutations lie within the N-terminal region of the PINK1 protein and it is mechanistically unclear how these mutations would account for loss of kinase function (5–7).
Interestingly, an accumulation of the cleaved ΔN-PINK1 protein, but not full-length PINK1 (FL-PINK1), has been reported in the brains of both idiopathic and heterozygous mutant PINK1 (Y431H, N451S, C575R) PD patients. The authors suggest that this accumulation is due to an upregulation of ΔN-PINK1 expression in response to PD-related stress (8). Given that truncated fragments of PINK1 can confer protection, these combined data suggest that cleavage of PINK1 plays a role in protecting neurons during pathogenesis. More recently, accumulation of the FL-PINK1 protein at the mitochondria has been linked to the induction of mitophagy where damaged or impaired mitochondria are removed from the cell (9,10). This recent discovery suggests that prevention of PINK1 cleavage, due to mitochondrial depolarization, can drive mitophagy, but a consensus on whether this effect is protective or detrimental has yet to be reached.
Several studies have demonstrated that the FL-PINK1 protein is cleaved to produce a predominant product of ∼53 kDa (ΔN-PINK1) and a minor one of ∼45 kDa (ΔN2-PINK1) (8,11–14). Conversion of the full-length protein to these two cleavage products is rapid and occurs within 3 min of full-length protein synthesis (11). Interestingly, ΔN-PINK1 is highly unstable and is rapidly degraded by the ubiquitin proteosome system (UPS) (8,11). In contrast, the ΔN2-PINK1 protein species appears to be relatively stable due to association with the Hsp90 chaperone (11). PINK1 cleavage occurs within mitochondria (15) and is dependent on mitochondrial integrity (10,11,16). However, investigations to determine the biological significance of the full-length and PINK1 cleavage products have been significantly hindered because the cleavage site of PINK1 was unknown. This has additionally prevented the elucidation of whether the ΔN2-PINK1 protein is produced through sequential cleavage of ΔN-PINK1.
Here we report the identification of the PINK1 cleavage site at position A103 to produce the 53 kDa ΔN-PINK1 protein. Through mutational analysis of this site and the surrounding region, we identified two key residues which, when mutated, can reduce or enhance PINK1 cleavage, respectively. Utilizing these mutants, we demonstrate that a reduction in PINK1 cleavage, causing an accumulation of full-length protein, results in a significant decrease in mitochondrial membrane potential (Δψm), increased reactive oxygen species (ROS) production and an altered mitochondrial network—a phenotype previously ascribed to PINK1-deficient cells (17–19). Moreover, we report that, in our system, the accumulation of full-length protein results in a reduction in mitochondrial mass, which does not correspond to induction of autophagy. We subsequently show that the PINK1 PD mutations C92F, Q115L and R147H, located close to the cleavage site, cause an accumulation of the FL-PINK1 protein, which results in an intermediate mitochondrial phenotype.
Finally, in an attempt to identify the protease responsible for PINK1 cleavage at A103, we selected two mitochondrial proteases, high temperature requirement protein A2 (HtrA2) and presenilin-associated rhomboid-like protein (PARL), for investigation of their ability to cleave PINK1 at this site. PINK1 is known to interact with the mitochondrial serine protease HtrA2 (20) so this was an obvious candidate to investigate for its ability to cleave PINK1. PARL was selected based on a study in Drosophila suggesting that the PARL homologue, Rhomboid-7, is involved in dPINK1 processing (21). Notably, PINK1, HtrA2 and PARL are all reportedly located in the inner mitochondrial membrane (IMM) (22–24). While PINK1 processing is unaffected by the loss of HtrA2, we show that in the absence of the mitochondrial protease PARL, PINK1 is aberrantly cleaved and the commonly observed ΔN-PINK1 fragment is not produced. We demonstrate that PINK1 interacts with PARL and that re-expression of wt PARL, but not the catalytically inactive PARL mutant, can rescue abnormal PINK1 cleavage in PARL-deficient cells. These combined results suggest that PARL is responsible for PINK1 cleavage at position A103.
RESULTS
Identification of the PINK1 cleavage site
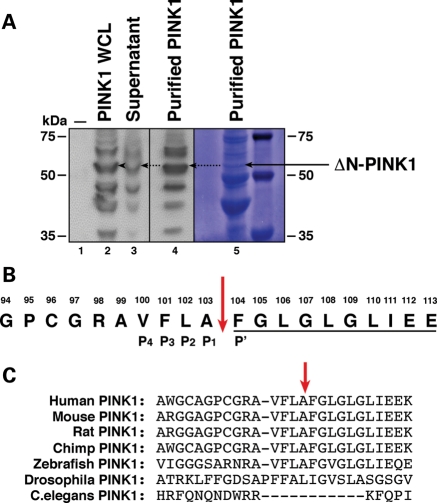
To determine the cleavage site of PINK1, PINK1-3xHA was transiently expressed in HEK293T cells to enable recovery of both full-length and cleaved PINK1 (ΔN-PINK1) from lysates. Since ΔN-PINK1 is unstable and has a half-life of 30 min (11,13), transfected cells were treated with the proteosome inhibitor MG132 prior to lysis in order to increase the amount of cleaved protein in the final sample (8,11). Notably, this was the only experiment which utilized MG132 treatment to enhance ΔN-PINK1 in cells. PINK1-3xHA was then immunoprecipitated using anti-HA-agarose beads and the resulting products were assessed by SDS–PAGE, western blot (WB) analysis and Coomassie staining (Fig. 1A). The 53 kDa band was then analysed by Edman N-terminal sequencing. A full 10 amino acid read was obtained confirming the protein was ΔN-PINK1 and revealing that the PINK1 cleavage site lies within the TM domain between residues A103 and F104 (Fig. 1B). While the cleavage site sequence is not conserved in Drosophila or Caenorhabditis elegans, the site is highly conserved between mammals and zebrafish (Fig. 1C).
Figure 1.
Isolation of ΔN-PINK1 and identification of the cleavage site. (A) WB analysis of vector control-transfected HEK293T cells (lane 1), PINK1-3xHA expression in whole-cell lysate samples before purification with anti-HA beads (lane 2), the presence of residual unbound PINK1-3xHA in supernatant post-purification (lane 3) and enrichment of the ΔN-PINK1 protein in the purified sample (lane 4). WBs were performed using an anti-HA antibody. Lane 5 displays the presence of the ΔN-PINK1 protein assessed by Coomassie staining. This membrane was then sent for N-terminal Edman degradation sequencing analysis. (B) N-terminal Edman sequencing result. The 10 amino acids underlined display the amino acid read obtained during the analysis. The sequence of PINK1 spanning amino acids 94–112 is shown with the cleavage site highlighted by a red arrow. The P4–P1 and P′ sites of the PINK1 cleavage site are also indicated. (C) Sequence alignment of the PINK1 protein surrounding the cleavage site showing conservation between mammalian species. Alignment was performed in Clustal X.
Mutational analysis of the PINK1 cleavage site
To confirm the cleavage site of PINK1, we conducted mutational analysis of residues at and surrounding the cleavage site. We identified one residue which, when mutated, appears to stabilize the ΔN-PINK1 product, leading to an accumulation of ΔN-PINK1 in cells, and one mutation which reduced PINK1 cleavage resulting in cells accumulating FL-PINK1.
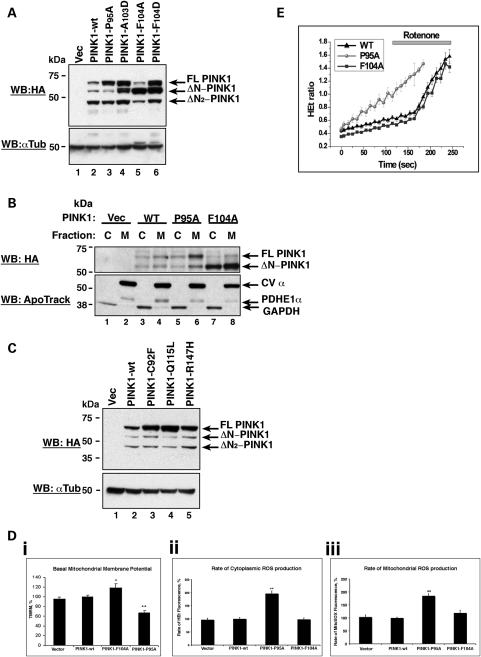
Mutation of the F104 residue at the P′-position to either aspartic acid or alanine had a significant effect on the production of ΔN-PINK1. When residue F104 was converted to an aspartic acid (F104D), we observed an accumulation of both FL- and ΔN-PINK1 protein in cells compared with PINK1-wt (Fig. 2A, lane 4). In contrast, conversion of this residue to alanine (F104A) resulted in a decrease in FL- PINK1 protein expression combined with an accumulation of ΔN-PINK1 (Fig. 2A, lane 5). Subsequent mutagenesis of the area surrounding the cleavage site revealed that mutation of residue P95 to alanine (P95A) greatly reduced PINK1 cleavage, resulting in a significant accumulation of FL-PINK1 and a corresponding reduction in ΔN-PINK1 production (Fig. 2A, lane 3). To ensure that the mutations were affecting the processing of PINK1 rather than altering the subcellular localization of the protein, we investigated whether the reduced cleavage mutant PINK1-P95A and the enhanced cleavage mutant PINK1-F104A still localized to mitochondria. Figure 2B illustrates that both the P95A and F104A PINK1 mutant proteins are predominantly localized to mitochondria suggesting that the altered residues directly affect the cleavage process.
Figure 2.
Efficient PINK1 cleavage is required for mitochondrial health. (A) Cleavage status of the PINK1 cleavage mutants compared with PINK1-wt. Vector control, PINK1-wt-3xHA, PINK1-P95A-3xHA, PINK1-F104A and PINK1-F104D constructs were expressed in SH-SY5Ys. Lysates were then assessed by WB analysis using an anti-HA antibody. (B) PINK1 cleavage mutants still localize to mitochondria. Vector control, PINK1-wt-3xHA, PINK1-P95A-3xHA and PINK1-F104A constructs were expressed in SH-SY5Y cells. Cell lysates were fractionated to isolate cytoplasmic and crude mitochondria. The presence of the PINK1 cleavage mutants in cytosolic and mitochondrial fractions was assessed by WB analysis using an anti-HA antibody. The membrane was subsequently re-probed with anti-ApoTrack cocktail which demonstrates the purity of the fractions and equal loading. (C) Cleavage of the N-terminal PINK1 PD mutant proteins. SH-SY5Y lysates expressing PINK1-C92F, PINK1-Q115L and PINK1-R147H constructs were analysed by WB with an anti-HA antibody. (D) (i) The cleavage status of PINK1 has important effects on basal mitochondrial membrane potential. Failure to cleave the PINK1 protein efficiently, shown through expression of the PINK1-P95A mutant, induces a significant reduction in mitochondrial membrane potential and makes cells more susceptible to stress-mediated death. Vector-transfected control cells were taken to be 100%. *P< 0.05 and **P< 0.005. (ii) A reduced ability to cleave PINK1 results in a significant increase in basal cytoplasmic ROS production. Basal ROS production in vector-transfected control cells was taken as 100%. **P< 0.005. (iii) Impaired PINK1 processing additionally results in a significant increase in basal mitochondrial ROS production. Basal ROS production in vector control-transfected cells was taken as 100%. **P< 0.005. (E) Assessment of ROS production in the PINK1-P95A and PINK1-F104A cleavage mutants compared with PINK1-wt. Basal ROS production increases over time in cells expressing the PINK1-P95A mutant without any stimulus. Stimulation of ROS production using rotenone to block complex 1 function has no effect on ROS production in PINK1-P95A cells but dramatically increases ROS production in both PINK1-wt and PINK1-F104A cells.
The cleavage site of PINK1 lies directly between two of the N-terminal PD reported mutations: C92F and Q115L. We therefore hypothesized that these mutations might affect the cleavage process. Consequently, we assessed the cleavage status of PINK1-C92F, PINK1-Q115L and an additional downstream mutant PINK1-R147H. While the production of the ΔN-PINK1 protein is unaffected by the presence of these PD mutations, an accumulation of the FL-PINK1 protein compared with PINK1-wt was consistently observed (Fig. 2c). Therefore, while these disease-associated mutations do not prevent PINK1 cleavage, our observation indicates that they increase the ratio of FL- to ΔN-PINK1 protein within cells.
PINK1 cleavage is important for mitochondrial health
Utilizing the P95A and F104A cleavage mutants, we investigated whether the cleavage status of PINK1 affected its function at the mitochondria in SH-SY5Y cells.
Mitochondrial membrane potential (Δψm) is a marker of mitochondrial function, and PINK1 deficiency is known to be associated with a reduction in the Δψm (17–19). To assess whether the cleavage status of PINK1 would alter the Δψm, cells were transfected with PINK-wt, PINK1-F104A or PINK1-P95A in a pIRES-GFP vector and loaded with the cationic lipophilic dye, tetramethylrhodamine methylester (TMRM). The fluorescent intensity of the TMRM within the mitochondria is proportional to the Δψm, and the presence of the green fluorescent protein (GFP) marker permitted the identification of transfected cells. Cells expressing the F104A mutant, and thus expressing predominantly ΔN-PINK1, had significantly higher basal Δψm than cells expressing PINK1-wt (an increase in the basal ΔΨm to 123 ± 9.9% of wt, n= 48 cells, P< 0.05) (Fig. 2Di). In contrast, cells expressing the P95A mutant (predominantly FL-PINK1) were found to have consistently and significantly lower Δψm than PINK1-wt-expressing cells (a decrease in the basal ΔΨm to 71.7 ± 4.8% of wt, n= 39 cells, P< 0.001). These data demonstrate that altering the cleavage status of PINK1 induces significant changes in the ΔΨm, and that reduced cleavage of PINK1 is associated with low basal Δψm.
PINK1 is known to be involved in protection from oxidative stress (17,25,26) and we have previously shown that PINK1 deficiency results in an increase in cytosolic and mitochondrial ROS production (18,19). Therefore, to assess the effect of PINK1 cleavage status on the redox balance of the cell, we measured ROS production in both the cytosol and mitochondrial matrix in SH-SY5Y cells stably expressing vector control, PINK1-wt, PINK1-F104A or PINK1-P95A. Cytosolic ROS production was assessed by measuring cytosolic hydroethidium (HEt) fluorescence while mitochondrial ROS production was assessed by measuring MitoSOX fluorescence (the mitochondrially targeted equivalent of HEt). Expression of the PINK1-P95A protein resulted in a significant increase in the basal cytosolic ROS production compared with PINK1-wt or vector (196.4 ± 10.52% compared with control, P< 0.01, n= 98 in wt cells; n= 67 in PINK1-P95A cells) (Fig. 2Dii). In addition, the mitochondrial ROS production was also significantly higher in cells expressing PINK1-P95A compared with PINK1-wt (185.4 ± 7.9% of control value, P < 0.01, n= 116 wt cells; n= 101 PINK1-P95A cells) (Fig. 2Diii). Notably, there was no significant difference in the basal ROS production in cells expressing either vector, PINK1-wt or PINK1-F104A in both the cytosol and mitochondria (Fig. 2Dii and iii). Previously, we demonstrated that the mechanism of mitochondrial ROS production in PINK1-deficient cells was due to the inhibition of complex I (18,19). We therefore examined whether the alteration in mitochondrial ROS production, associated with reduced PINK1 cleavage, was occurring via the same mechanism. As expected, inhibition of complex I by rotenone in cells expressing either PINK1-wt or PINK1-F104A resulted in activation of mitochondrial ROS production (197.8 ± 11.2% of basal rate; n= 98; P> 0.001; Fig. 2E). In contrast, application of rotenone failed to produce any further increase in mitochondrial ROS production in cells expressing PINK1-P95A, suggesting that ROS production is already maximally activated via this mechanism (Fig. 2E). These data show that reduction of PINK1 cleavage and the subsequent accumulation of full-length protein produces a pathophysiological phenotype similar to PINK1 deficiency in our system.
PINK1 cleavage status affects mitochondrial network and mass
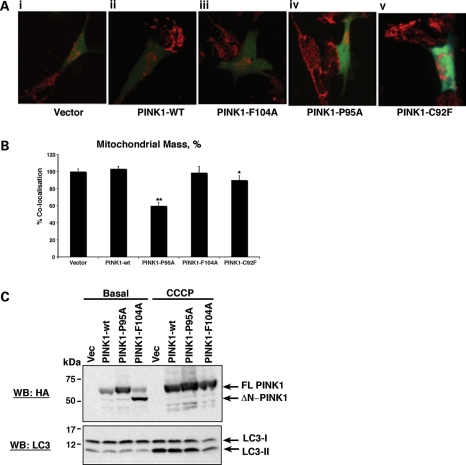
More recently, suppression of PINK1 cleavage through treatment of cells with uncoupling agents has been extensively shown to induce the removal of mitochondria from cells via mitophagy (27,28). Given our observation that the PINK1-P95A mutant protein induces an accumulation of FL-PINK1 protein and that its expression alone is sufficient to reduce the basal Δψm, we examined whether expression of the PINK1-P95A mutant protein had an effect on the mitochondrial network and/or mitochondrial mass within cells. In addition, due to the altered ratio of FL-PINK1 to ΔN-PINK1 in cells expressing the N-terminal PD-associated mutations C92F, Q115L and R147H, we included the PINK1-C92F mutant protein in our analysis.
Utilizing the TMRM dye to visualize mitochondria in non-transfected cells, or those expressing either pIRES vector, PINK1-wt or PINK1-F104A, the mitochondria form an intricate interconnected network that is distributed evenly throughout the cell (Fig. 3Ai, ii, iii and Supplementary Material, Movies S1–S3). In contrast, when cells express the PINK1-P95A protein, the mitochondria are aggregated in one part of the cell, and no clear network is visible (Fig. 3Aiv and Supplementary Material, Video S4). Strikingly, cells expressing the PD-associated mutant PINK1-C92F protein also display abnormally distributed and aggregated mitochondria (Fig. 3Av and Supplementary Material, Video S5). However, retention of the TMRM dye is dependent on Δψm. As exogenous PINK1-P95A expression induces a reduction in Δψm, we repeated these experiments using a membrane potential-independent method. To this end, the PINK1 pIRES constructs were transiently co-transfected with DsRed-Mito to confirm the alteration in mitochondrial distribution. Again we observed the same alterations in the mitochondrial network in cells expressing either PINK1-P95A or PINK1-C92F (data not shown). In order to clarify the basis of this appearance, we subsequently assessed whether there were any differences in mitochondrial mass between the cells using live cell imaging as previously described (29,30). Utilizing the DsRed-Mito and pIRES-GFP constructs, this approach quantifies the degree of co-localization of the mitochondrial (DsRed-Mito) signal with the cytosolic (GFP) signal in z-stacks of cells. This enables a measure of the relative volume of the cell occupied by the mitochondria and reflects the mitochondrial mass within a cell (30). The degree of co-localization for cells expressing vector control was taken as 100%. As shown in Figure 3B, expression of either vector control, PINK1-wt or PINK1-F104A showed no significant alteration in co-localization index. In contrast, the expression of PINK1-P95A resulted in a marked reduction in co-localization (from 100 to 66.4%, n= 4 experiments, P < 0.001) suggesting a reduction in mitochondrial mass. Notably, expression of the PD mutant, PINK1-C92F, was also associated with a 12% reduction in the co-localization index. It is noteworthy that cells expressing the N-terminal PD PINK1 mutation are still capable of producing the ΔN-PINK1 protein at levels comparable with PINK1-wt (Fig. 2C). Therefore, our results suggest that the ratio of FL-PINK1 to ΔN-PINK1 is important for mitochondrial function and maintenance of the mitochondrial population. These data complement the reported findings that the accumulation of FL-PINK1 induces mitochondrial removal via mitophagy (9,16,28). Therefore, to assess whether the reduction in mitochondrial mass, caused by expression of our PINK1-P95A mutant protein, was due to activation of macroautophagy/mitophagy, we assessed the basal and CCCP-induced levels of LC3I-II cleavage in cells stably expressing vector, PINK1-wt, PINK1-P95A and PINK1-F104A. Surprisingly, despite the accumulation of FL-PINK1, reduction in Δψm and reduction in mitochondrial mass, we did not detect an increase in LC3I-II cleavage at the basal level in PINK1-P95A cells compared with cells expressing vector, PINK1-wt or PINK1-F104A (Fig. 3C). Treatment of these cell lines with the mitochondrial uncoupler CCCP suppressed PINK1 cleavage in all stable cell lines and additionally resulted in an increase in LC3I-II cleavage (Fig. 3C). Our results therefore suggest that an accumulation of FL-PINK1 is sufficient to reduce the basal Δψm significantly and can induce a reduction in mitochondrial mass independently of autophagy/mitophagy activation.
Figure 3.
PINK1 cleavage status affects the mitochondrial network and mass. (A) Failure to cleave PINK1 results in an abnormal mitochondrial network and altered location. Images display SH-SY5Y cells expressing the indicated PINK1 pIRES constructs (green cells) and neighbouring untransfected control cells where only mitochondria are visible. Image (i) shows the normal mitochondrial network observed in cells expressing vector control; (ii) PINK1-wt; (iii) PINK1-F104A; (iv) the loss of mitochondrial network and presence of aggregated mitochondria in cells expressing PINK1-P95A; (v) the intermediate phenotype between PINK1-wt and PINK1-P95A observed in cells expressing the PD PINK1-C92F mutant protein. Mitochondria were visualized using TMRM dye. 3D movies are provided in Supplementary Material, Figures S1–S5. (B) Mitochondrial mass appears reduced in cells predominantly expressing FL-PINK1. Cells expressing PINK1-P95A or PINK1-C92F PD mutant protein show significantly reduced mitochondrial mass compared with vector-, PINK1-wt- and PINK1-F104A-expressing cells. Vector control-transfected cells were taken as 100%. (C) Loss of mitochondrial mass through expression of PINK1-P95A does not correlate with autophagy induction. Cells stably expressing either vector control, PINK1-wt, PINK1-P95A or PINK1-F104A were exposed to DMSO control or CCCP for 12 h prior to lysis. Lysates were analysed by WB with an anti-HA antibody (PINK1) and LC3 antibody to detect autophagy activation by LC3I-II cleavage. Expression of PINK1-P95A did not induce an increase in LC3I-II cleavage at basal levels compared with vector control, PINK1-wt or PINK1-F104A cells. CCCP treatment of all cell lines induced LC3I-II cleavage and suppressed PINK1 cleavage in all cell lines.
The Rhomboid protease PARL interacts with and cleaves PINK1
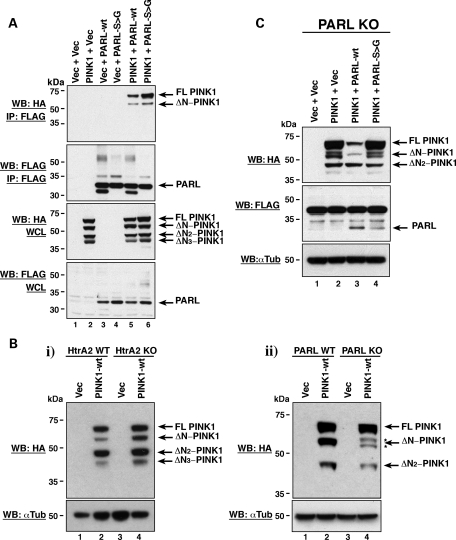
As mentioned in the introduction, the Rhomboid-7 protease is reported to cleave dPINK1 in Drosophila (21). PARL, the Rhomboid-7 human homologue, belongs to a family of proteins, the Rhomboids, with a reported preference for cleaving within TM domains (23). It is, therefore, interesting that the cleavage site of PINK1 lies within its predicted TM domain. However, since there was no published observation of an interaction between PINK1 and PARL in mammals, we initially assessed their ability to interact in HEK293T cells. We find that PINK1 interacts with both wild-type PARL (PARL-wt) and the catalytically inactive PARL-S277G mutant (Fig. 4A). In addition, PARL was able to interact with both the full-length and cleaved PINK1 products, suggesting that amino acids 104–581 of PINK1 are sufficient for the interaction to occur (Fig. 4A). To confirm this exogenous interaction, we assessed whether endogenous PINK1 interacted with PARL in SH-SY5Y cells by mass spectrometry. To control for non-specific interactors, PARL complexes were purified in the presence or absence of peptide competition and all isolated proteins were assessed. In agreement with our exogenous interaction, endogenous PINK1 was readily detected as a PARL interactor (see Supplementary Material, Fig. S2). Consequently, our results indicate that at least two IMM proteases, PARL and HtrA2 (20), can interact with PINK1 in mammalian cells. We therefore examined the cleavage status of the PINK1 protein in mouse embryonic fibroblasts (MEFs) derived from PARL and HtrA2 knockout (KO) mice (31,32). PINK1 was cleaved normally in the absence of HtrA2 (Fig. 4Bi), indicating that HtrA2 is not required for PINK1 cleavage. However, in the absence of PARL, PINK1 was aberrantly cleaved and the 53 kDa ΔN-PINK1 protein was not generated (Fig. 4Bii), strongly suggesting that PARL cleaves PINK1 to produce ΔN-PINK1. However, while the 53 kDa ΔN-PINK1 cleavage product is absent in PARL KO MEFs, two alternative and previously unreported cleavage products are produced. Importantly, normal PINK1 cleavage could be restored in the PARL KO MEFs upon co-transfection with PARL-wt, but not with the inactive PARL-S277G mutant, further demonstrating the specificity of PINK1 cleavage by PARL to generate ΔN-PINK1 (Fig. 4C). Notably production of the ΔN2-PINK1 cleavage product was similar in the PARL KO and WT MEFs, suggesting that PARL is dispensable for this second cleavage event. To confirm the cleavage of PINK1 by PARL, we assessed the cleavage status of our PINK1-P95A and PINK1-F104A mutants in PARL WT and KO MEFs. Figure 5A demonstrates that while the mutant proteins still show reduced and enhanced PINK1 cleavage, respectively, in the WT MEFs both mutant proteins are processed the same as PINK1-wt in the absence of PARL. These results strengthen the specificity of PINK1 cleavage by PARL and suggest that PARL may be the default protease required for ΔN-PINK1 production. While we endeavoured to assess endogenous PINK1 cleavage in the PARL MEFs, the endogenous ΔN-PINK1 protein could not be detected with the antibodies currently available (Supplementary Material, Fig. S3).
Figure 4.
PINK1 interacts with and is cleaved by PARL. (A) Co-immunoprecipitation of PINK1 with both PARL-wt and the catalytically inactive PARL-S277G mutant. Whole-cell lysates (WCL) from HEK293T cells transiently expressing PINK1-wt-3xHA, PARL-wt-FLAG and PARL-S277G-FLAG were immunoprecipitated (IP) with anti-FLAG® M2-agarose affinity gel prior to analysis by WB with the indicated antibodies. (B) (i) PINK1-wt cleavage status in HtrA2 KO and WT MEFs. HtrA2 KO and WT MEFs were transfected with PINK1-wt-3xHA. Lysates were then assessed by WB analysis using an anti-HA antibody. (ii) PINK1-wt cleavage status in PARL KO and WT MEFs. PARL KO and WT MEFs were transfected with PINK1-wt-3xHA. Lysates were then assessed by WB analysis using an anti-HA antibody. (C) PINK1 cleavage can be restored in PARL KO cells overexpressing PARL-wt but not the catalytically inactive PARL-S277G mutant. PARL KO MEFs were transfected with PINK1-wt-3xHA, PARL-wt-FLAG and PARL-S277G-FLAG. Lysates were then analysed by WB with the indicated antibodies.
Figure 5.
The PINK1-P95A mutation does not abrogate the PINK1–PARL interaction. (A) Alterations in PINK1 cleavage induced by P95A and F104A mutations are suppressed by the loss of PARL. Vector control, PINK1-wt, PINK1-P95A and PINK1-F104A were expressed in PARL WT and KO MEFs. Lysates were assessed by WB analysis using an anti-HA antibody. (B) PINK1-P95A still interacts with PARL-wt or PARL-S277G. Lysates from HEK293T cells expressing PINK1-wt-3xHA, PINK1-P95A-3xHA, PARL-wt-FLAG and PARL-S277G-FLAG were immunoprecipitated (IP) with anti-FLAG® M2-agarose affinity gel prior to analysis by WB with the indicated antibodies. (C) Structural predictions of the PINK1-wt TM domain and alterations made by the P95A mutation. (D) Assessment of ROS production in cells expressing vector control, PINK1-wt or PINK1-P95A in a PARL RNAi or scrambled RNAi control background.
Notably, the cleavage recognition site for PARL remains unknown. However, since the PINK1-P95A mutant protein demonstrates reduced cleavage in WT cells, despite the fact that the cleavage site remains intact, we hypothesized that either the proline residue could be important for the PINK1–PARL interaction (by being required for the correct folding of the PINK1 protein) or this residue may form part of a substrate recognition system for PARL. Interestingly, the presence of a helix-breaking residue, such as proline, has been reported as a requirement for substrate recognition with additional rhomboid proteins (33).
We subsequently assessed whether the presence of the P95A mutation abrogated the interaction between PINK1 and PARL. Our results demonstrate that the P95A mutation does not affect the ability of PINK1 to interact with PARL (Fig. 5B), strongly suggesting that the protein is folded correctly. Proline is a conformationally restricted amino acid and is therefore predicted to induce a ‘kink’ in the α-helical structure of the PINK1 TM domain. Structural modelling of the wt and P95A mutated TM domain using PyMoL (DeLano, W.L. The PyMoL Molecular Graphics System 2002, http://www.pymol.org) suggested that the P95A mutation removes this ‘kink’ and results in PINK1 displaying a conventional α-helix (Fig. 5Ci and ii). We therefore hypothesize that cleavage of PINK1 by PARL may be dependent upon structural recognition of a ‘kinked’ α-helix within the membrane rather than amino acid sequence recognition alone.
Given our observation that PARL is necessary for PINK1 cleavage to generate ΔN-PINK1, we reasoned that if reduced PINK1 cleavage impairs mitochondrial function then cells expressing PINK1-wt in a PARL KO background should also demonstrate increased ROS production. We find that in a PARL RNA interference (RNAi) background, SH-SY5Y cells expressing PINK1-wt or vector control show a significant increase in ROS production compared with scrambled RNAi control cells expressing the same constructs (2.1-fold increase for vector control, n= 4 experiments, P< 0.001; 2.2-fold increase for PINK1-wt, n= 4 experiments, P< 0.001) (Fig. 5D). An increase in ROS production in cells expressing the PINK1-P95A protein in a PARL knockout background was also observed (2.1-fold increase, n= 4 experiments, P< 0.001). These combined results therefore strengthen our finding that PINK1 cleavage is important for basal mitochondrial health.
DISCUSSION
In PD, mutations within the PINK1 gene have been extensively shown to be responsible for early onset autosomal recessive disease. PINK1 is a mitochondrial protein (22) and increasing evidence suggests that mitochondrial dysfunction is a key player in PD (18,19). The aim of this study was to gain insight into the biological function of the PINK1 protein by determining the N-terminal cleavage site to generate ΔN-PINK1, assessing the cellular consequences of impaired PINK1 cleavage and identifying the protease responsible for cleavage at this site.
In this study, we show that PINK1 is cleaved between Ala-103 and Phe-104 to generate the 53 kDa ΔN-PINK1 protein. Through the study of PINK1 cleavage mutants, our results show that a reduced ability to cleave PINK1 results in a significant reduction in basal Δψm and a significant increase in both cytoplasmic and mitochondrial ROS production—a phenotype similar to that described in PINK1 KO cells (17,18). Furthermore, the predominant expression of full-length protein by the PINK1-P95A mutant appears to negate any protective effect provided by the presence of both the combined endogenous and low-level exogenous ΔN-PINK1 product in cells. This observation suggests that accumulation of FL-PINK1 might have a dominant negative effect on PINK1 protein function. Notably, as cells expressing the PINK1-C92F N-terminal PD mutation display an increased ratio of FL- to ΔN-PINK1, in combination with an intermediate mitochondrial phenotype, these results may offer some explanation for the mechanism by which N-terminal mutations contribute towards pathogenesis. In support of this, a similar intermediate mitochondrial phenotype has been reported in fibroblasts of a patient carrying a heterozygous N-terminal mutation (17). These combined data suggest that the ratio of FL-PINK1 to ΔN-PINK1 protein is critical for mitochondrial health.
Our finding that ΔN-PINK1 appears to be the ‘mature’ form of PINK1 responsible for conferring protection against oxidative stress to cells correlates with the previous findings of Haque et al. (4) who demonstrated that a fragment of PINK1 (amino acids 112–581) can rescue cells from toxin-mediated death in vivo. Moreover, our current study demonstrates that overexpression of ΔN-PINK1 in human neuronal cells is not detrimental to mitochondrial function. Evidence that expressing more ΔN-PINK1 may be beneficial is supported by the finding that an increased deposition of ΔN-PINK1 protein has been observed in the surviving neurons in PD brain samples (in both idiopathic and heterozygous mutant PINK1 PD cases) (8). Therefore, finding a means to modulate the PINK1 cleavage process at the endogenous level could prove interesting for future therapeutic applications. Unfortunately, we were unable to assess whether the N-terminal PD mutations C92F, Q115L or R147H cause accumulation of FL-PINK1 in patient tissue due to the absence of material.
It has previously been established that PINK1 is cleaved to produce a secondary, minor product of 45 kDa (ΔN2-PINK1) (11). To date, however, the biological significance of this product has not been assessed. Since production of the ΔN2-PINK1 fragment is unaffected in our PINK1 cleavage mutant cells, our data demonstrate that the ΔN2-PINK1 cannot compensate for the loss of ΔN-PINK1 because mitochondrial function is still impaired in its presence. Consequently, our study suggests that ΔN2-PINK1 is not required for PINK1's reported protective effect in neuronal cells. Interestingly, two previous studies have proposed a role for the Hsp90/cdc37 complex in PD pathogenesis (as both FL-PINK1 and the ΔN2-PINK1 cleavage product are stabilized through association with this complex) (12,34). While our data show that the ΔN2-PINK1 product does not confer protection to cells, it is possible that through stabilization of the FL-PINK1 precursor protein, the Hsp90/cdc37 complex may help regulate the ratio of FL-PINK1 to ΔN-PINK1 protein, which we have shown is crucial for mitochondrial health. In recent years, the application of Hsp90 inhibitors as a PD therapeutic has been proposed based on their ability to clear disease-associated α-synuclein aggregates from cells and through destabilizing the PD-mutated LRRK2 kinase (35–37). If modulation of Hsp90 levels can additionally destabilize the FL-PINK1 protein and subsequently alter the ratio of FL- to ΔN-PINK1 in vivo, it is plausible that this treatment could promote neuronal survival by tipping the balance in favour of ΔN-PINK1.
Notably, a role for PINK1 in mitochondrial fission/fusion dynamics has been demonstrated in HeLa cells and Drosophila model systems (17,38,39). Current evidence suggests that PINK1 promotes fission in Drosophila and therefore the loss of PINK1 results in mitochondrial aggregates and swollen mitochondria (17,18,38,39). In mammalian cells, loss of PINK1 function results in fragmented mitochondria (17). More recently, a series of studies have reported that accumulation of FL-PINK1 is required to recruit parkin to the mitochondria and drive mitophagy (10,16). In these studies, mitophagy is induced by exposing cells to uncoupling agents such as CCCP, which reduce the Δψm, and this prevents PINK1 cleavage (10,16) presumably by blocking mitochondrial import of PINK1 (15). In this study, we have demonstrated that the cleavage status of PINK1 and therefore the ratio of FL-PINK1 to ΔN-PINK1 affects the mitochondrial network within the cell. Moreover, expression of the reduced cleavage PINK1-P95A mutant, in the absence of any uncoupling agents, was consistently observed to reduce the basal Δψm significantly and induce a reduction in mitochondrial mass. In addition, expression of the PINK1-C92F PD mutant protein, which results in more FL-PINK1 expression, also induced a reduction in Δψm and mitochondrial mass. It is therefore possible that N-terminal PD mutations within the PINK1 protein distort the cell's natural balance between full-length and cleaved PINK1 protein. This in turn could impose an effect on the basal rates of mitophagy and mitochondrial biogenesis and contribute to pathogenesis. However, while these findings are in agreement with the recent observations on PINK1's role in mitophagy (28), we did not observe an increase in macroautophagy activation when assessed with LC3I-II cleavage. This result is somewhat surprising, given that mitophagy is currently the only identified mechanism by which mitochondria are removed from the cell. However, reports examining autophagy in healthy neurons have suggested the possibility of an alternative autophagic pathway in these cells (40). Whether this hypothesized pathway would permit the specific removal of whole organelles such as mitochondria is currently unknown. At present, therefore, we are unable to explain the mechanism behind the mitochondrial loss in our model, but it is intriguing that damaged mitochondria could be removed from neuronal cells via an as yet unidentified pathway.
Notably, the reduction in the mitochondrial mass of cells predominantly expressing the FL-PINK1 protein may underlie the differences observed in the mitochondrial network. While it remains a possibility that the altered mitochondrial appearances are linked to the dynamic processes of fission/fusion, loss of mitochondrial mass has never been reported through their alteration (41).
The accumulation of FL-PINK1, and reduction of ΔN-PINK1, results in mitochondrial dysfunction, characterized by reduced mitochondrial membrane potential, increased oxidative stress and abnormal mitochondrial appearances. Such effects may render neurons susceptible to stress, and are known to contribute to the reduced neuronal viability seen in PINK1 knockdown/KO mammalian neurons (18). Therefore, a reduced ability to cleave PINK1 produces a phenotype similar to PINK1 deficiency.
In our study, we additionally show that PARL is a novel PINK1-interacting protein in mammals by co-immunoprecipitation and mass spectrometry and reveal that PARL is responsible for cleavage of PINK1 at position A103 to generate DN-PINK1. However, in the absence of PARL, PINK1 is aberrantly cleaved by a secondary, as yet unidentified, protease to generate two previously unreported cleavage products. The observation that these aberrant cleavage products are not normally seen under physiological conditions (8) suggests that in the absence of PARL, compensation mechanisms are initiated to ensure FL-PINK1 cleavage. Notably, these compensation mechanisms are absent in Drosophila because deletion of the PARL homologue, Rhomboid-7, is sufficient to completely block dPINK1 cleavage. Further studies to reveal the compensatory protease(s) responsible for PINK1 cleavage in the absence of PARL in mammals are therefore required. PARL is a serine protease located within the IMM that has been reported to perform an anti-apoptotic role (32). We have shown that the proline residue within the PINK1 TM domain appears to be required for optimal recognition of PINK1 by PARL as a substrate. While the necessity for a helix-breaking residue, such as proline, for substrate recognition is not unprecedented with rhomboid proteins (33), this is the first observation that PARL may utilize this mechanism. While we were able to confirm that endogenous PINK1 interacts with PARL in human neuroblastoma cells by mass spectrometry, we are currently unable to assess aberrant PINK1 cleavage in the PARL KO MEFs due to the absence of reliable antibodies capable of detecting both full-length and cleaved PINK1 at the endogenous level (42,43). The inability to examine PINK1 cleavage events using the current PINK1 antibodies is highlighted by the study of Narendra et al. (10). In this study, the group investigated PINK1 cleavage in PARL WT and KO MEFs using an anti-PINK1 antibody, which recognizes only FL-PINK1. The group subsequently reported that failure of PARL shRNA to augment FL-PINK1 accumulation indicated that PARL was not required for PINK1 processing. While the conclusion drawn by Narendra et al. is not incorrect—PINK1 is indeed still processed in PARL's absence—our results demonstrate that PINK1 is aberrantly cleaved resulting in the loss of the ΔN-PINK1 fragment. Nevertheless, these findings prompted us to sequence the PARL gene in our familial PD cases. Upon sequence analysis of the entire coding region of PARL in 82 unrelated familial PD cases, no mutations were identified. Consequently, there is currently no evidence to suggest that PARL directly contributes to Mendelian forms of PD.
We therefore propose that the ΔN-PINK1 product is the functionally active form of the protein conferring cell protection against stress. Moreover, Haque et al. (4) have shown that this protective effect is achieved through its kinase activity. With the combination of these results, we postulate that the full-length precursor protein has weak intrinsic kinase activity and that cleavage of PINK1 may stimulate this activity. If this is the case, it may explain why previous attempts to identify PINK1 kinase substrates, using predominantly the full-length protein, have been hindered. It will be important therefore to assess whether utilizing the ΔN-PINK1 product, or the PINK1-F104A mutant, one will be able to observe a more robust kinase activity of PINK1, thus facilitating the identification of bona fide PINK1 kinase substrates and potential new therapeutic targets for PD.
MATERIALS AND METHODS
Antibodies
Anti-HA monoclonal antibody was obtained from Roche. Anti-HA agarose affinity gel, M2 anti-FLAG antibody and M2 anti-FLAG agarose affinity gel were obtained from Sigma. Anti-LC3 antibody was obtained from Cell Signalling Technology, while the anti-ApoTrack cocktail was obtained from Mitosciences. Anti-mouse, anti-rabbit and anti-rat secondary antibodies coupled with horseradish peroxidase were from Santa Cruz. All WBs were visualized on X-ray film using ECL reagent (PIERCE) according to the manufacturer's instructions.
Mammalian constructs
Mammalian expression constructs encoding PARL-wt-FLAG and PARL-S277G-FLAG were obtained from Dr L. Pellegrini through the Addgene site (Addgene plasmids 13639 and 13615, respectively). The PINK1-3xHA construct was generated by subcloning human PINK1 cDNA into the pcDNA3-3xHA vector (this vector is under an MTA to the Ashworth Laboratory, Breakthrough Breast Cancer Research Center, ICR, London, UK). PINK1 cleavage and selected PD mutants were all generated by site-directed mutagenesis of the PINK1-wt-3xHA construct using the QuickChange site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. For mitochondrial membrane, ROS production and mitochondrial co-localization assessments, PINK1-wt and PINK1 mutants were isolated from pcDNA3 with the triple HA tag attached and inserted into pIRES2-EGFP to enable simultaneous GFP labelling of all PINK1-transfected cells for analysis. All constructs are available subject to MTA. The pIRES2-EGFP and DsRed-Mito constructs were purchased from Clontech.
PARL siRNA
PARL (PSARL siGENOME SMARTpool) and control scrambled (siGENOME Non-Targeting siRNA Pool 1) siRNAs were purchased from Dharmacon and used as per the manufacturer's instructions.
Cell culture
SH-SY5Y, HEK293T cells (ECACC) and MEF lines were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum gold (PAA Laboratories) and 1×penicillin/streptomycin (Sigma). MEFs derived from WT or HtrA2 KO mice were obtained through collaboration with Dr L. M. Martins (31). MEFs derived from WT and PARL KO mice were obtained through collaboration with Dr B. De Strooper (32).
HEK293T, SH-SY5Y cells and MEFs were transiently transfected using Effectene (Qiagen) according to the manufacturer's guidelines. Stable SH-SY5Y cell lines were generated by selecting transfected cells with 700 µg/ml G418 24 h post-transfection.
Protein biochemistry
General cell lysis
All cell lines were lysed on ice in 50 mm Tris–HCl pH 8, 150 mm NaCl, 1 mm EDTA, 0.5% NP40 (v/v) and 1× protease inhibitor cocktail (Roche). Lysates were homogenized using 30 passes of a hand-held homogenizer then snap frozen in liquid nitrogen. All lysates were homogenized and frozen three times before the insoluble material was pelleted by centrifugation. The resulting supernatants were retained and their protein concentrations were assayed using the BIORAD DC Protein Assay Kit. Unless otherwise stated, 100 µg of protein was loaded for each sample.
Co-purifications
Cells were lysed as described above except the supernatants were not snap frozen. Control (whole-cell lysate) samples of 100 µl were taken from each lysate to confirm protein expression prior to the purification and to determine total protein concentrations to enable equal loading for WB analysis. The rest of the lysate was incubated with the respective antibody-coupled agarose beads. Samples were incubated with the beads overnight, the beads were pelleted and washed in wash buffer three times before complexes were eluted with either acidic glycine in the case of anti-HA beads or FLAG peptide in the case of anti-FLAG beads. Wash buffer consisted of 10 mm Tris–HCl pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.2% NP40 and 1× protease inhibitors. Complexes were then subjected to SDS–PAGE. All gels were transferred onto Immobilon™ PVDF membranes (Millipore). The membranes were subsequently incubated with the indicated primary and appropriate secondary antibody. Where indicated, membranes were stripped before re-blotting.
PINK1 N-terminal Edman degradation sequencing
For isolation of the ΔN-PINK1 protein, two separate samples of purified material were resolved by SDS–PAGE and transferred onto an Immobilon™ PVDF membrane. The membrane was then cut in half—one half was assessed by WB using the anti-HA antibody, while the other half was stained with colloidal Coomassie Blue (Sigma). The band corresponding to cleaved PINK1 on the Coomassie-stained membrane was marked with an asterisk before the membrane was desiccated using silica gel overnight. The dried membrane was sent to Dr J. Keen for N-terminal Edman degradation sequencing (Dr J. N. Keen, School of Biochemistry and Molecular Biology, University of Leeds, Leeds, UK).
Mass spectrometry analysis of PARL–PINK1 interaction
Clonal SH-SY5Y cells expressing PARL-wt FLAG were lysed as described above. Control (whole-cell lysate) samples of 100 µl were taken from the lysate to confirm protein expression prior to the purification and to determine total protein concentration to enable equal loading of proteins onto two purification columns. The remaining lysate was split equally between the purification columns, one containing FLAG agarose and the other containing FLAG agarose pre-incubated with FLAG peptide to prevent FLAG-conjugated proteins from binding to the resin. The lysates were incubated with the antibody-coupled agarose beads ± peptide competition for 1 h, washed in wash buffer three times and then complexes were eluted with FLAG peptide. Samples were then concentrated using viva-spin columns to 25 µl samples and prepared for mass spectrometry analysis by SDS–PAGE and tryptic digest. Samples were assessed by LC-MS-MS and scored using emPAI. All proteins present within the eluted samples were analysed.
To assess basal and induced macroautophagy levels, stable cell lines expressing either 3xHA vector control, PINK1-wt-3xHA, PINK1-P95A-3xHA or PINK1-F104A-3xHA were seeded into six-well plates and either treated with DMSO control or 10 µm CCCP for 12 h. Cells were then harvested and protein expression was assessed by WB.
Mitochondrial fractionations were performed as described previously (44).
Structural modelling of the PINK1 TM domain
The PINK1 TM domain was modelled using PyMoL and the automated building software using the wt as well as the PINK1-P95A sequence as input.
Mitochondrial membrane potential (ΔΨm) assessment
For measurements of ΔΨm, SH-SY5Y cells were loaded with 25 nm TMRM for 30 min at room temperature and the dye was present during the experiment. In these experiments, TMRM is used in the redistribution mode to assess ΔΨm, and therefore a reduction in TMRM fluorescence represents mitochondrial depolarization. Confocal images were obtained using a Zeiss 510 uv-vis CLSM equipped with a META detection system and a 40× oil immersion objective. Illumination intensity was kept to a minimum (at 0.1–0.2% of laser output) to avoid phototoxicity and the pinhole set to give an optical slice of ∼2 µm. TMRM was excited using the 543 nm laser line and fluorescence measured using a 560 nm longpass filter. Z-stacks of 20–30 cells per cover slip were acquired, and the mean maximal fluorescence intensity was measured for each cell group. The differences in ΔΨm are expressed as a percentage compared with cells expressing PINK1-wt (taken as 100%). Three independent experiments were performed on transiently transfected SH-SY5Y cells. The results were further confirmed in triplicate using SH-SY5Y cell lines that stably express the PINK1-wt or PINK1-P95A pIRES constructs.
Assessment of ROS production
For measurement of mitochondrial ROS production, cells were pre-incubated with MitoSOX (5 µm, Molecular Probes, Eugene, OR, USA) for 10 min at room temperature. For measurement of cytosolic ROS production, dihydroethidium (2 µm) was present in the solution during the experiment. No pre-incubation (‘loading’) was used for dihydroethidium to limit the intracellular accumulation of oxidized products. Both HEt and MitoSOX were excited using the 543 and 360 nm laser lines and a ratiometric measurement was used.
Mitochondrial network and mass assessment
Mitochondrial localization was assessed by two methods. (i) Cells were transiently transfected with vector, PINK1-wt, PINK1-P95A or the PD mutation PINK1-C92F. Transfected cells were loaded with 25 nm TMRM for 30 min at room temperature. High-resolution z-stacks were obtained of ∼10 transfected cells. (ii) Cells were co-transfected with either vector, PINK1-wt or PINK1-P95A together with DsRed-Mito. Doubly transfected cells were easily identified by the presence of green cytosolic fluorescence and red mitochondrial fluorescence. High-resolution z-stacks were acquired for ∼10 cells per group. Both methods enabled adequate visualization of the mitochondrial network. However, we wanted to ensure that the alterations in mitochondrial localization measured were independent of any alterations in ΔΨm. In order to quantify the changes in the mitochondrial network, the percentage co-localization of the green (cytosolic) signal and the red (mitochondrial) signal was attained. This ratio represents the volume of the cell that is occupied by mitochondria. The co-localization of these signals was set as 100% in vector-transfected cells, to enable a comparison to be made between different cell groups. The histogram reflects the co-localization attained through both methods in independent experiments.
Statistical analysis
Statistical analysis and exponential curve fitting were performed using Origin 7 (Microcal Software Inc., Northampton, MA, USA) software. Data were analysed by parametric Student's t-tests and significance is expressed as follows: *P< 0.05, **P< 0.005, unless otherwise stated. For all graphs bars represent mean ± SEM.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by Parkinson's Disease Society (grant number G-0612); Medical Research Council (Programme grant number G0400000); the Brain Research Trust (grant number PRO09101) and by the Wellcome/MRC Parkinson's Disease Consortium grant to UCL/IoN, the University of Sheffield and the MRC Protein Phosphorylation Unit at the University of Dundee (grant number WT089698). Funding to pay the Open Access publication charges for this article was provided by the Wellcome/MRC Parkinson's Disease Consortium grant to UCL/IoN, the University of Sheffield and the MRC Protein Phosphorylation Unit at the University of Dundee.
ACKNOWLEDGEMENTS
We would like to thank Dr J. Keen for performing the N-terminal Edman degradation sequencing service and Dr R. Elliott for technical assistance. We would additionally like to thank Dr B. De Strooper for the PARL KO MEFs (32), L. Scorrano for useful discussions and Dr L. Pellegrini for the PARL constructs (45,46).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Ibanez P., Lesage S., Lohmann E., Thobois S., De Michele G., Borg M., Agid Y., Durr A., Brice A. Mutational analysis of the PINK1 gene in early-onset parkinsonism in Europe and North Africa. Brain. 2006;129:686–694. doi: 10.1093/brain/awl005. [DOI] [PubMed] [Google Scholar]
- 2.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 3.Sim C.H., Lio D.S., Mok S.S., Masters C.L., Hill A.F., Culvenor J.G., Cheng H.C. C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum. Mol. Genet. 2006;15:3251–3262. doi: 10.1093/hmg/ddl398. [DOI] [PubMed] [Google Scholar]
- 4.Haque M.E., Thomas K.J., D'Souza C., Callaghan S., Kitada T., Slack R.S., Fraser P., Cookson M.R., Tandon A., Park D.S. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc. Natl Acad. Sci. USA. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valente E.M., Salvi S., Ialongo T., Marongiu R., Elia A.E., Caputo V., Romito L., Albanese A., Dallapiccola B., Bentivoglio A.R. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 2004;56:336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 6.Bonifati V., Rohe C.F., Breedveld G.J., Fabrizio E., De Mari M., Tassorelli C., Tavella A., Marconi R., Nicholl D.J., Chien H.F., et al. Early-onset parkinsonism associated with PINK1 mutations: frequency, genotypes, and phenotypes. Neurology. 2005;65:87–95. doi: 10.1212/01.wnl.0000167546.39375.82. [DOI] [PubMed] [Google Scholar]
- 7.Healy D.G., Abou-Sleiman P.M., Gibson J.M., Ross O.A., Jain S., Gandhi S., Gosal D., Muqit M.M., Wood N.W., Lynch T. PINK1 (PARK6) associated Parkinson disease in Ireland. Neurology. 2004;63:1486–1488. doi: 10.1212/01.wnl.0000142089.38301.8e. [DOI] [PubMed] [Google Scholar]
- 8.Muqit M.M., Abou-Sleiman P.M., Saurin A.T., Harvey K., Gandhi S., Deas E., Eaton S., Payne Smith M.D., Venner K., Matilla A., et al. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J. Neurochem. 2006;98:156–169. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- 9.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin W., Kang U.J. Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weihofen A., Ostaszewski B., Minami Y., Selkoe D.J. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum. Mol. Genet. 2008;17:602–616. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 13.Takatori S., Ito G., Iwatsubo T. Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of PINK1. Neurosci. Lett. 2008;430:13–17. doi: 10.1016/j.neulet.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Petit A., Kawarai T., Paitel E., Sanjo N., Maj M., Scheid M., Chen F., Gu Y., Hasegawa H., Salehi-Rad S., et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J. Biol. Chem. 2005;280:34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 15.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E.M., Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Exner N., Treske B., Paquet D., Holmstrom K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H.H., Gasser T., et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A.S., Miljan E.A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandhi S., Wood-Kaczmar A., Yao Z., Plun-Favreau H., Deas E., Klupsch K., Downward J., Latchman D.S., Tabrizi S.J., Wood N.W., et al. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol. Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R.J., McDonald N., et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat. Cell Biol. 2007;9:1243–1252. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 21.Whitworth A.J., Lee J.R., Ho V.M.-W., Flick R., Chowdhury R., McQuibban G.A. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis. Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gandhi S., Muqit M.M., Stanyer L., Healy D.G., Abou-Sleiman P.M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A.J., et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini L., Scorrano L. A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 2007;14:1275–1284. doi: 10.1038/sj.cdd.4402145. [DOI] [PubMed] [Google Scholar]
- 24.Kadomatsu T., Mori M., Terada K. Mitochondrial import of Omi: the definitive role of the putative transmembrane region and multiple processing sites in the amino-terminal segment. Biochem. Biophys. Res. Commun. 2007;361:516–521. doi: 10.1016/j.bbrc.2007.07.053. [DOI] [PubMed] [Google Scholar]
- 25.Pridgeon J.W., Olzmann J.A., Chin L.S., Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoepken H.H., Gispert S., Morales B., Wingerter O., Del Turco D., Mulsch A., Nussbaum R.L., Muller K., Drose S., Brandt U., et al. Mitochondrial dysfunction, peroxidation damage and changes in glutathione metabolism in PARK6. Neurobiol. Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Deas E., Wood N.W., Plun-Favreau H. Mitophagy and Parkinson's disease: the PINK1–parkin link. Biochim. Biophys. Acta. 2010 doi: 10.1016/j.bbamcr.2010.08.007. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu C.T. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum. Mol. Genet. 2010;19:R28–R37. doi: 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agnello M., Morici G., Rinaldi A.M. A method for measuring mitochondrial mass and activity. Cytotechnology. 2008;56:145–149. doi: 10.1007/s10616-008-9143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Addabbo F., Ratliff B., Park H.C., Kuo M.C., Ungvari Z., Csiszar A., Krasnikov B., Sodhi K., Zhang F., Nasjletti A., et al. The Krebs cycle and mitochondrial mass are early victims of endothelial dysfunction: proteomic approach. Am. J. Pathol. 2009;174:34–43. doi: 10.2353/ajpath.2009.080650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martins L.M., Morrison A., Klupsch K., Fedele V., Moisoi N., Teismann P., Abuin A., Grau E., Geppert M., Livi G.P., et al. Neuroprotective role of the reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol. Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 33.Urban S., Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 34.Moriwaki Y., Kim Y.J., Ido Y., Misawa H., Kawashima K., Endo S., Takahashi R. L347P PINK1 mutant that fails to bind to Hsp90/Cdc37 chaperones is rapidly degraded in a proteasome-dependent manner. Neurosci. Res. 2008;61:43–48. doi: 10.1016/j.neures.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Wang L., Xie C., Greggio E., Parisiadou L., Shim H., Sun L., Chandran J., Lin X., Lai C., Yang W.J., et al. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J. Neurosci. 2008;28:3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Putcha P., Danzer K.M., Kranich L.R., Scott A., Silinski M., Mabbett S., Hicks C.D., Veal J.M., Steed P.M., Hyman B.T., et al. Brain-permeable small-molecule inhibitors of Hsp90 prevent alpha-synuclein oligomer formation and rescue alpha-synuclein-induced toxicity. J. Pharmacol. Exp. Ther. 2010;332:849–857. doi: 10.1124/jpet.109.158436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riedel M., Goldbaum O., Schwarz L., Schmitt S., Richter-Landsberg C. 17-AAG induces cytoplasmic alpha-synuclein aggregate clearance by induction of autophagy. PLoS ONE. 2010;5:e8753. doi: 10.1371/journal.pone.0008753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Ouyang Y., Yang L., Beal M.F., McQuibban A., Vogel H., Lu B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl Acad. Sci. USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poole A.C., Thomas R.E., Andrews L.A., McBride H.M., Whitworth A.J., Pallanck L.J. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yue Z., Friedman L., Komatsu M., Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim. Biophys. Acta. 2009;1793:1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan D.C. Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 42.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E.A., Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl Acad. Sci. USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siddall H.K., Warrell C.E., Davidson S.M., Mocanu M.M., Yellon D.M. Mitochondrial PINK1—a novel cardioprotective kinase? Cardiovasc. Drugs Ther. 2008;22:507–508. doi: 10.1007/s10557-008-6136-5. [DOI] [PubMed] [Google Scholar]
- 44.Samali A., Cai J., Zhivotovsky B., Jones D.P., Orrenius S. Presence of a pre-apoptotic complex of pro-caspase-3, Hsp60 and Hsp10 in the mitochondrial fraction of jurkat cells. EMBO J. 1999;18:2040–2048. doi: 10.1093/emboj/18.8.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeyaraju D.V., Xu L., Letellier M.C., Bandaru S., Zunino R., Berg E.A., McBride H.M., Pellegrini L. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc. Natl Acad. Sci. USA. 2006;103:18562–18567. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sik A., Passer B.J., Koonin E.V., Pellegrini L. Self-regulated cleavage of the mitochondrial intramembrane-cleaving protease PARL yields Pbeta, a nuclear-targeted peptide. J. Biol. Chem. 2004;279:15323–15329. doi: 10.1074/jbc.M313756200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.