Abstract
Loss-of-function mutations in the genes encoding dystrophin and the associated membrane proteins, the sarcoglycans, produce muscular dystrophy and cardiomyopathy. The dystrophin complex provides stability to the plasma membrane of striated muscle during muscle contraction. Increased SMAD signaling due to activation of the transforming growth factor-β (TGFβ) pathway has been described in muscular dystrophy; however, it is not known whether this canonical TGFβ signaling is pathogenic in the muscle itself. Drosophila deleted for the γ/δ-sarcoglycan gene (Sgcd) develop progressive muscle and heart dysfunction and serve as a model for the human disorder. We used dad-lacZ flies to demonstrate the signature of TGFβ activation in response to exercise-induced injury in Sgcd null flies, finding that those muscle nuclei immediately adjacent to muscle injury demonstrate high-level TGFβ signaling. To determine the pathogenic nature of this signaling, we found that partial reduction of the co-SMAD Medea, homologous to SMAD4, or the r-SMAD, Smox, corrected both heart and muscle dysfunction in Sgcd mutants. Reduction in the r-SMAD, MAD, restored muscle function but interestingly not heart function in Sgcd mutants, consistent with a role for activin but not bone morphogenic protein signaling in cardiac dysfunction. Mammalian sarcoglycan null muscle was also found to exhibit exercise-induced SMAD signaling. These data demonstrate that hyperactivation of SMAD signaling occurs in response to repetitive injury in muscle and heart. Reduction of this pathway is sufficient to restore cardiac and muscle function and is therefore a target for therapeutic reduction.
INTRODUCTION
Mutations in the genes encoding dystrophin and its associated proteins lead to muscular dystrophy and cardiomyopathy in vertebrates and invertebrates. Dystrophin associates with a composite of transmembrane proteins including dystroglycan, sarcospan, syntrophins and the sarcoglycan proteins to mediate stability of the plasma membrane of muscle (1). Dystrophin links to cystoskeletal actin, and dystroglycan binds laminin in the extracellular matrix connecting the cytoskeleton and the matrix (2). This organization supports a role for the dystrophin complex in preserving the mechanical integrity of the plasma membrane. In addition to this mechanical support, additional roles have been proposed, including regulation of nitric oxide, calcium homeostasis and MAP kinase signaling (1,3).
The dystrophin complex contributes to a mechanically strong link between the cytoskeleton and the plasma membrane. In the face of muscle contraction, muscle lacking dystrophin displays more injury than normal muscle (4,5). This is particularly evident when muscle is subject to eccentric contraction, a protocol where lengthened muscle is repetitively subjected to contraction to produce strain on the sarcolemma. In the mdx mouse that lacks dystrophin, the diaphragm muscle displays the greatest degree of disease pathology, nearly on par with what is seen in human muscle (4,6). Loss-of-function mutations in the genes encoding the sarcoglycan subunits also lead to muscular dystrophy and cardiomyopathy similar to what is seen from dystrophin mutations. Interestingly, loss of γ-sarcoglycan does not render muscle more susceptible to eccentric contraction-induced damage, highlighting the non-mechanical aspects of the dystrophin complex (7). The vital tracer, Evans Blue Dye, a small molecule that binds albumin, has been used to document membrane disruption in vivo in the muscular dystrophies. Dye uptake can be readily detected in muscle lacking dystrophin or the sarcoglycan subunits (8–10), indicating that a loss of any of these proteins is sufficient to destabilize the plasma membrane. The fragile muscle membrane coupled with repetitive insult from exercise and contraction leads to progressive myofiber loss.
Dystrophic muscle is characterized by progressive replacement of the muscle fibers by fibrosis. Transforming growth factor-β (TGFβ) is known to contribute to the fibrotic response in a number of pathological processes such as pulmonary fibrosis, liver cirrhosis and renal disease. Increased TGFβ signaling has also been noted in human and murine muscular dystrophy. In dystrophic muscle biopsies taken from patients with Duchenne muscular dystrophy, TGFβ1 is localized near and within injured muscle fibers (11). Moreover, reducing TGFβ signaling in the mdx mouse model, either using TGFβ-neutralizing antibodies or angiotensin receptor blockers, improved muscle regeneration and reduced fibrosis in mdx mice (12). However, others have shown that TGFβ-neutralizing antibodies reduced fibrosis but also produced an unfavorable cytokine profile in mdx diaphragm muscle (13). The role of TGFβ in muscular dystrophy was also reinforced by the recent observation that Ltbp4, a gene encoding a TGFβ-sequestering protein, serves as a genetic modifier of muscular dystrophy where reduced TGFβ signaling was associated with reduced membrane disruption and reduced fibrosis (14). Cumulatively, these data demonstrate that TGFβ signaling, at the level of matrix release and receptor activation, is critical for muscular dystrophy pathogenesis. However, whether and which downstream TGFβ-signaling pathways mediate disease has not been explored.
The utility of Drosophila models of human disease is promoted by the comparatively more rapid genetic analyses that can be conducted. We previously generated a muscular dystrophy model in Drosophila melanogaster using imprecise P element excision to generate deletions in the gene encoding γ/δ-sarcoglycan [Sgcd, Flybase Scgδ (15)]. In Drosophila, there is only a single sequence equally related to mammalian Sgcg and Sgcd, referred to as Sgcd. Sgcd-deleted flies develop normally but with age, muscle tears become apparent, climbing ability declines and the heart tube enlarges and develops reduced contractility. This mirrors what is seen in humans with muscular dystrophy and mouse models of the disease. Drosophila models of muscular dystrophy also include mutations that affect of dystrophin (Dys) or dystroglycan (Dg), altered via RNA interference, overlapping deletions, hypomorphic mutations or mutations in putative dystroglycan glycosylating enzymes (16–19). These perturbations also result in heart and muscle deficits, providing genetic evidence for evolutionary conservation of dystrophin and its associated proteins. Extramuscle features are present in some of these mutants, underscoring the role of the dystrophin complex in other cell types and potentially providing surrogate phenotypes for study (18,20,21). However, a striking difference is that Drosophila Sgcd null muscle does not develop increased fibrosis, and therefore offers the opportunity to examine the role of TGFβ as a direct mediator of muscle disease and dysfunction.
In the canonical TGFβ-signaling pathway, TGFβ family ligands bind to type II TGFβ receptors (22). These receptors bind and phosphorylate type I receptors. This triggers phosphorylation of a signal transduction molecule, the r-SMAD, at its carboxy-terminal SSXS motif. Phosphorylated r-SMADs bind to co-SMADs, translocate to the nucleus and direct changes in gene transcription (23). In Drosophila, there are two r-SMADs. Mothers against dpp (MAD) is homologous to mammalian SMADs 1, 5 and 8 and is phosphorylated in response to bone morphogenic protein (BMP) family signaling. Smad on X (Smox) is homologous to SMADs 2 and 3, and is phosphorylated in response to activin signaling. There is a single co-SMAD, Medea, which is functionally homologous to SMAD4. This lack of redundancy means that mutations of either r-SMAD will affect the entire corresponding family of TGFβ ligands and mutations of Medea will decrease all TGFβ receptor-induced signaling. Significantly, there is not a precise ortholog of TGFβ in Drosophila.
We now used the Sgcd null fly to investigate whether increased TGFβ signaling drives progression of muscular dystrophy. We found that exercise causes increased muscle injury in the Sgcd[840] mutant fly. Using a reporter of TGFβ activity, we find that TGFβ signaling occurs in myonuclei immediately adjacent to the sites of muscle injury in the Sgcd[840] mutant. We show that genetically decreasing SMAD signaling using haploinsufficient alleles was sufficient to rescue skeletal and cardiac muscle dysfunction in this mutant. Finally, we show that exercise increases SMAD activity in the Sgcg null mouse and notably does so in centrally placed nuclei immediately after exercise. These findings identify a central role for SMAD signaling in the progression of muscular dystrophy.
RESULTS
Exercise induces muscle disruption in Sgcd[840] mutants
Deletion mutants in Sgcd were generated using imprecise P element-mediated excision (15). We selected the Sgcd[840] line for analysis since this line has a molecular defined genetic deletion encompassing exons 1–4. Exon 2 encodes the initiator methionine and the complete cytoplasmic and transmembrane domains, and the deletion in the Sgcd[840] line is associated with no detectable sarcoglycan protein expression. In some mammalian models of muscular dystrophy, notably those lacking dystrophin and δ-sarcoglycan, muscle contraction enhances muscle damage (4,6,8). The standard vial used to house Drosophila greatly limits the opportunity to utilize the indirect flight muscles. To determine whether exercise induces damage in the Drosophila Sgcd[840] line, we evaluated mutant and wild-type flies that housed in a 20 × 20 × 20 cm box to permit ad libitum flight from eclosion to 14 days of age. The volume in these boxes was approximately 300 times greater than a vial, providing ample opportunity to exercise. Qualitatively, Sgcd[840] null flies did not spend as much time in flight as their wild-type counterparts, but the amount of flight was sufficient to induce muscle injury (Supplementary Material, Movies S1 and S2).
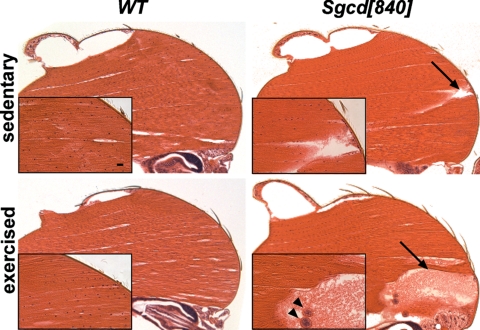
Muscle disruption was occasionally apparent in non-exercised Sgcd[840] flies, as we previously reported [(15); Fig. 1]. Tears were more readily observed in Sgcd[840] flies that had been exercised. Occasionally large muscle tears were associated with an increase in amorphous extracellular material (Fig. 1). Very rarely, we observed infiltration of non-muscle cells that morphologically resemble the macrophage-like hemocytes [Fig. 1; (24)]. The histopathology of the Sgcd[840] muscle represents a simpler model since mammalian muscular dystrophy exhibits effects from non-myocyte lineages including fibrosis, fatty infiltration, inflammatory infiltrate and calcification (Supplementary Material, Fig. S1). Mutant Drosophila muscle only rarely showed the amorphous material in Figure 1 and never displayed the interstitial fibrosis derived from fibroblasts that characterize mammalian dystrophic muscle. Because Drosophila muscle lacks fibrosis, this model is highly useful to study the myogenic component of muscle dysfunction.
Figure 1.
Exercise induces muscle tears in the Sgcd[840] mutants. WT and Sgcd[840] males were aged to 14 days. H&E staining is shown of indirect flight muscles. Muscle disruption was apparent in Sgcd[840] flies (arrows), but not in WT. When the flies were housed in a 20 × 20 × 20 cm box to allow flight, disruptions in the Sgcd[840] mutants were larger and more frequently observed. Infiltrating cells of the Drosophila innate immune system were also rarely noted (arrowheads on the inset panel, lower right). Tears were not apparent in exercised WT flies. Size bar, 10 μm.
Exercise-induced muscle tears in the Sgcd[840] mutant lead to SMAD signaling
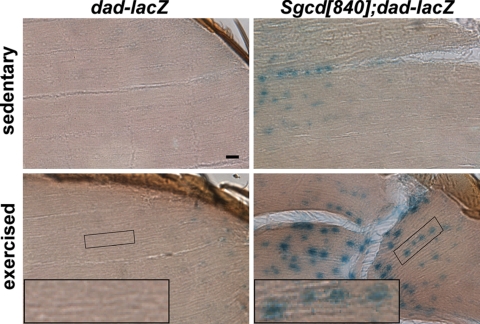
Because enhanced TGFβ signaling has been implicated as pathogenic in human and murine muscular dystrophy, we investigated whether increased SMAD signaling was present in the Sgcg[840] line. We introduced the dad-lacZ reporter into the Sgcd[840] background. dad-lacZ is an insertion of a lacZ enhancer trap element near the daughters against decapentaplegic (dad) gene and is homologous to mammalian SMAD7 (25). In flies bearing the dad-lacZ allele, β-galactosidase (β-gal) is synthesized in response to TGFβ signaling. We generated Sgcd[840] mutant flies that also carried the dad-lacZ reporter, as well as the balancer chromosome TM3 which prevents recombination and loss of the dad-lacZ insertion. Sgcd[+]; dad-lacZ/TM3 and Sgcd[840]; dad-lacZ/TM3 males were aged in either ordinary vials or flight boxes for 14 days to represent sedentary and exercised, respectively. Flies were collected and evaluated for β-gal activity. Extensive β-gal activity was evident in muscle nuclei in exercised Sgcd[840]; dad-lacZ/TM3 flies. Those myonuclei immediately adjacent to muscle disruption showed the greatest increase in β-gal activity (Fig. 2) consistent with a model where injury itself triggers TGFβ signaling. To assess this, we injured wild-type muscle using needle penetration and found a similar pattern of dad-mediated β-gal activity (Supplementary Material, Fig. S2). Therefore, muscle injury itself is characterized by enhanced TGFβ signaling. In Sgcd[840] flies, exercise is sufficient to induce muscle injury and then elicit increased dad activity.
Figure 2.
Exercise-induced muscle disruption in Sgcd[840] is associated with increased SMAD signaling. The reporter dad-lacZ was crossed into Sgcd[840] flies. Sedentary flies refer to those raised in a typical vial. With this housing, very little dad-lacZ activity is apparent in the wild-type flies. Sedentary Sgcd[840]; dad-lacZ flies showed some β-gal activity. Comparably aged Sgcd[+] flies that bear the reporter and were allowed to exercise showed minimal β-gal activity. Exercised Sgcd[840] flies with the dad-lacZ reporter showed high levels of β-gal activity, and this pattern was most evident in myonuclei localized adjacent to sites of muscle disruption. Insets highlight the lined texture of muscle tissue and linear arrangement that identify these as myonuclei. Size bar, 10 μm.
TGFβ signaling is increased in Sgcd[840] mutant muscle
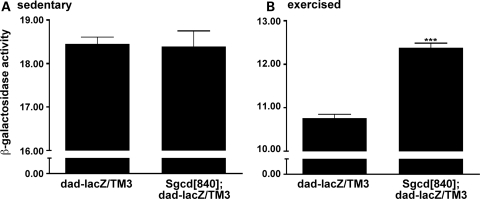
We quantified TGFβ signaling in Sgcd[840] with the dad-lacZ reporter by measuring β-gal activity (26). Since we were primarily interested in signaling within muscle, thoraces were isolated. This portion of the fly body is largely composed of the indirect flight muscles. When comparing sedentary flies, no significant increase in signaling was detected between mutant and wild-type sedentary flies; Sgcd[840]; dad-lacZ/TM3 and Sgcd[+]; dad-lacZ/TM3 flies, 18.45 ± 0.37 versus 18.39x ± 0.95 U/h (P = 0.952). However, when flies were housed in flight boxes for 14 days allowing for exercise, a 15% increase in β-gal activity was detected between Sgcd[840]; dad-lacZ flies compared with controls, 12.38 ± 0.32 versus 10.76 ± 0.28 U/h, (P = 0.0002) (Fig. 3). These data are consistent with a model where increased activity in the presence of the Sgcd mutation produces muscle tearing, which, in turn, triggers increased SMAD signaling. In sedentary flies, where there is less activity and less muscle disruption, SMAD signaling is not activated above background. Wild-type and mutant flies within each group were compared in the same setting. However, due to variability in conditions and the fact that exercised and unexercised flies were assayed at different times, the results cannot be directly compared between sedentary and exercised flies.
Figure 3.
SMAD signaling is increased in Sgcd[840] mutants in response to exercise. The dad-lacZ reporter was crossed into Sgcd[840] and wild-type flies. (A) After 14 days of aging in vials, there was no difference in β-gal activity between Sgcd[840] and dad-lacZ flies. (B) When flies were aged in flight boxes, to allow increased ad libitum activity, Sgcd[840] mutants had increased β-gal activity. Activity is measured as 1000 × AUs at 584 nm/h. ***P < 0.001 versus dad-lacZ/TM3.
To validate the quantitative nature of this assay, we tested the responsiveness of dad-lacZ to mutations in the TGFβ signaling gene Medea. Each of three mutations was independent. We tested Medea[13], Medea[15] or Medea[17] as haploinsufficient alleles in the reporter line and quantified β-gal activity in whole flies after 7 days of aging in vials. Each of the Medea alleles decreased TGFβ activity by an average of 25%. Heterozygous Medea[13] reduced β-gal activity from 55.08 ± 2.53 to 42.27 ± 1.47 U/h (P < 0.001 compared with wild-type with reporter only). Heterozygous Medea[15] or Medea[17] also produced significant decreases (Supplementary Material, Fig. S3).
Reducing TGFβ signaling rescues mobility in sarcoglycan mutant flies
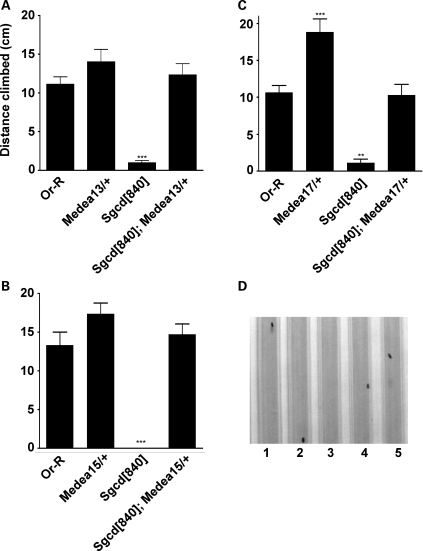
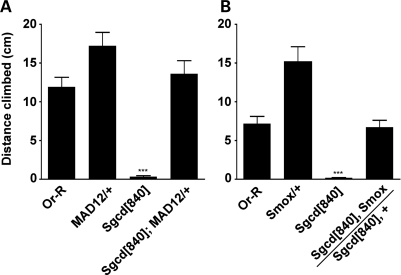
To determine whether the increased SMAD signaling in Sgcd[840] mutant flies is pathogenic, mutations that decrease SMAD signaling were introduced into Sgcd[840]. Outcome was assessed using an assay for negative geotaxis, or the ability to walk upwards against gravity. Age-dependent loss of this climbing ability is a feature not only of Sgcd mutants but also of those lacking dystrophin (15,18). We introduced three different Medea mutations that are known to represent a phenotypic range of TGFβ signaling (Fig. 4), Medea[13], Medea[15] and Medea[17]. The Medea[13] allele results in the mutation of arginine 66 to a stop codon terminating the protein prior to the DNA-binding (MH1) or r-SMAD-interacting (MH2) domains. Medea[15] is a T714K mutation in the MH2 heteromerization domain of Medea, and Medea[17] is a P715T mutation in the MH2 heteromerization domain. Sgcd[840] null flies displayed virtually no climbing response (Supplementary Material, Movie S3). In contrast, Sgcd[840]; Medea[13]/+ flies could ambulate against gravity comparably to wild-type flies (Fig. 4A, n = 19, P < 0.001). We repeated this experiment with Medea[15] and Medea[17] and found that each allele as a heterozygote resulted in similar complete correction of the walking defect. We asked whether these functional improvements correlated to improvement in the morphological abnormalities we previously identified. To address this, we aged Sgcd[840]; Medea[13]/TM3 and Medea[13]/TM3 flies in flight boxes and examined their muscles. Muscle tears were still evident (Supplementary Material, Fig. S4). However qualitatively, the tears appeared smaller, consistent with a role for SMAD signaling in modulating injury and also injury response. Taken together, these data indicate that increased Medea signaling is a direct contributor to the loss of climbing in the Sgcd[840] mutant. Moreover, partial reduction in signaling corrects the deficit.
Figure 4.
Co-SMAD mutants rescue negative geotaxis in Sgcd[840] mutants. We introduced three distinct Medea mutations as heterozygotes into the Sgcd[840] background and measured climbing ability after 14 days. (A) The null mutation Medea[13] and the hypomorphic mutations (B) Medea[15] and (C) Medea[17] restored climbing of Sgcd[840] flies to wild-type levels. n was at least 12 in all groups. ***P < 0.001 versus all other groups. **P < 0.01 versus all other groups. (D) An image of the apparatus used to monitor negative geotaxis. The black dots represent individual Drosophila climbing (Supplementary Material, Movie S3).
r-SMAD mutants rescue negative geotaxis in Sgcd[840] mutants
TGFβ signaling is classically divided into the BMP and activin families of ligands, receptors and r-SMADs. In mammals, BMP signals are transduced by SMADs 1/5/8, while activin signals are transduced by SMADs 2/3. In Drosophila, there is a single r-SMAD for each family, MAD for BMP signaling and Smox for activin signaling. To determine whether the loss of climbing was due to one or the other signaling pathway, we introduced mutations of MAD or Smox into Sgcd[840] mutants. The MAD[12] allele was selected because it is a Q417X mutation that results in protein termination in the MH2 heteromerization domain, but still prior to the carboxy-terminal SSXS phosphorylation site (27). Sgcd[840]; MAD[12]/+ males were indistinguishable from wild-type and were significantly improved from a simultaneously aged cohort of Sgcd[840] flies (Fig. 5A, n = 12, P < 0.001). MAD[12] heterozygotes climbed slightly, but significantly, better than wild-type (P < 0.05).
Figure 5.
r-SMAD mutants rescue negative geotaxis in Sgcd[840] mutants. We introduced SMAD mutations as heterozygotes into the Sgcd[840] background and measured climbing ability after 14 days of aging in vials. (A) Mutations in the BMP r-SMAD, MAD or (B) the activin r-SMAD, Smox restored climbing of Sgcd[840] flies to wild-type levels. n was at least 12. ***P < 0.001 versus all other groups.
We studied the Smox allele Smox[G0348]. In this line, a p{lacW} transposon is inserted into the 5′ UTR of Smox (28). It is hemi- or homozygous lethal, with lethality at the pharate adult stage. The small wings of these individuals are evidence for the lethality being related to a decrease in activin signaling. Sgcd[840], Smox[G0348]/Sgcd[840] flies show normal climbing, significantly better than Sgcd[840]/Sgcd[840] flies (Fig. 5B, n = 20, P < 0.001). Of note, with this assay, there was variation between wild-type (Or-R flies) assayed on different days (compare Fig. 5A and B). We attribute this difference to environmental variability such as temperature and humidity, and for this reason only compared mutants and WT evaluated simultaneously.
Reducing TGFβ signaling rescues heart function in sarcoglycan mutant flies
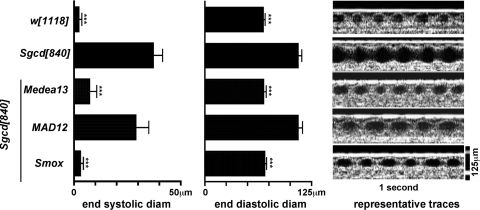
Humans and mice with muscular dystrophy develop cardiomyopathy. Similarly, Sgcd mutant flies have a dilated and poorly contractile heart tube (15). This can be measured using optical coherence tomography (OCT) as increased end-systolic and end-diastolic diameter and reduced measurements of performance (29). To determine whether reducing SMAD signaling would improve heart function, we assayed the heart tube function of Sgcd[840] flies containing the Medea[13], MAD[12] and Smox[G0348] mutations. We analyzed the wild-type stock w[1118] along with Sgcd[840], Sgcd[840]; Medea[13]/TM3, Sgcd[840]; MAD[12]/CyO and Sgcd[840], Smox[G0348]/Sgcd[840] using OCT technique (29) since this method allows quantitative measures of heart function. We measured end-systolic diameter, end-diastolic diameter and heart rate. Heart rate did not vary between any groups (data not shown). Introducing Medea[13] or Smox[G0348] mutations as heterozygotes restored end-systolic and end-diastolic diameters, and thus returned heart function to wild-type levels (Fig. 6). MAD[12] failed to improve heart function, suggesting BMP signaling contributes more to muscle than heart tube dysfunction since BMP disruption was sufficient to correct muscle dysfunction.
Figure 6.
Rescue of Sgcd[840] heart tube function by Medea and Smox heterozygous mutations. The Medea[13], MAD[12] and Smox[G0348] alleles, which each independently improved climbing in the Sgcd[840] flies, were balanced and heart tube function was studied by OCT. OCT allows real-time functional analysis of the heart tube including the end-systolic and end-diastolic diameters. white[1118] was used as the wild-type. Both the end-systolic and end-diastolic diameters were increased in the Sgcd[840] line, providing direct measurements of a dilated heart tube. Heterozygous Medea[13] or Smox[G0348] rescued this dysfunction. However, MAD[12] failed to improve heart function. n was 9–12 for each group. ***P < 0.001 versus Sgcd[840].
Exercise induces TGFβ signaling in Sgcg null mice
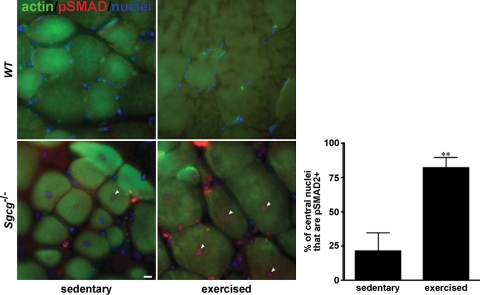
To translate our findings into a mammalian model of muscular dystrophy, we studied mice lacking γ-sarcoglycan (Sgcg null) (9). Drosophila Sgcd is 30–35% identical to both mouse γ- and δ-sarcoglycan, showing equal relationship with the two murine genes. Loss-of-function mutations in human Sgcg or Sgcd cause muscular dystrophy and cardiomyopathy similar to what develops from dystrophin mutations (30,31). Mice null for Sgcg or Sgcd also display a similar phenotype of heart and muscle disease (8,9,32). To determine whether exercise contributes to TGFβ signaling in this model of muscular dystrophy, we exercised Sgcg null and WT mice at 10 m/min for 30 min with a 7° downward angle. Compared with unexercised controls, exercised Sgcg null mice displayed increased nuclear phosphorylated SMAD2/3 (pSMAD, Fig. 7). We quantified the increase by counting the number of central myonuclei that stained for pSMAD2/3. By confining our analysis to central nuclei, we ensured that the nuclei we counted were muscle nuclei, rather than fibroblasts or infiltrating immune cells. In sedentary Sgcg null mice, 21 ± 13% of nuclei were positive, as compared with 82 ± 7% in exercised Sgcg null mutants, (P < 0.01). Of note, there were many extramyofiber nuclei, those found in fibroblasts and infiltrating inflammatory cells were strongly pSMAD2/3 positive. WT mice did not show central nuclei, and had minimal pSMAD2/3, regardless of exercise. The finding that exercise increased the pSMAD signaling in myonuclei indicates a direct effect on myofibers. Therefore, in mammalian muscle, as in Drosophila muscle, the combination of exercise and disruption of the dystrophin–sarcoglycan complex results in increased TGFβ signaling.
Figure 7.
Exercise induces SMAD signaling in Sgcg null mice. Sgcg−/− mice were either exercised by running downhill at 10 m/min for 30 min at a 7° down angle (exercised) or not (sedentary). Nuclear pSMAD2/3 was evident in sedentary Sgcg−/− mice, particularly in the centrally located nuclei of regenerating fibers (arrowheads); however, pSMAD2/3 staining in myonuclei was much more pronounced in the Sgcg−/− muscle that had been subjected to exercise. Nuclear pSMAD2/3 was not exclusive to centrally placed myonuclei since many non-central nuclei showed this staining. To ensure that myonuclei were being quantified, as opposed to infiltrating inflammatory cells or fibroblasts, we compared nuclear pSMAD only in centrally positioned myonuclei. Exercise dramatically increased nuclear pSMAD2/3. Since this analysis was conducted immediately post-exercise, with insufficient time for regeneration, this supports the model where exercise promotes injury and triggers pathogenic pSMAD signaling in muscle nuclei. pSMAD2/3 was not seen in wild-type mice under any of these conditions. **P < 0.01 versus sedentary. Red, pSMAD2/3. Green, Alexa 488 Phalloidin to highlight myofibers. Blue, DAPI to show nuclei. Scale bar, 10 μm.
DISCUSSION
Enhanced TGFβ signaling characterizes muscle injury and disease
TGFβ signaling is mediated through canonical and non-canonical pathways. In mammalian muscle, pSMAD is detected in nuclei, and reduction of nuclear pSMAD was reported in mdx mice treated with TGFβ-neutralizing antibodies (12). In dystrophic muscle, both myonuclei and those nuclei in non-muscle cells display enhanced nuclear pSMAD (12). We now show that the orthologous SMAD-type signaling is increased in a Drosophila model of muscular dystrophy. We utilized the fly model for these studies because dystrophic fly muscle is characterized by a degenerative process. This is in contrast to dystrophic mammalian muscle where there is degeneration alongside regeneration and infiltration by non-muscle elements. Because in mammalian muscle these non-muscle elements, particularly the fibroblasts, have much greater pSMAD signaling, the simpler fly muscle allowed us to examine the myogenic component of injury response. With this approach, we found that exercise enhanced muscle disruption and the degree of β-gal activity from the dad-lacZ indicator allele in Sgcd[840] muscle. The myonuclei most proximal to the region of disruption displayed the greatest amount of β-gal activity, and this is consistent with a model of local TGFβ activity inducing response nearest to the area of injury. Injury alone, in the absence of disease, also elicited this signaling response, if the injury was severe. Apidianakis et al. (33) profiled gene expression of mechanically injured Drosophila thorax and found elevated transcript levels for the BMP-like ligand gbb and the activin ligand daw at 1 and 6h after injury, suggesting that these ligands may be the TGFβ orthologs.
In addition to fibroblast activation and proliferation, mammalian muscular dystrophy is accompanied by an inflammatory response in muscle, in part mediated by mononuclear cells derived from the bone marrow. The equivalent in the fly is a hemocyte-like cell, which we very occasionally observed in sarcoglycan mutant flies. Hemocyte-derived cell lines are known to secrete TGFβ family ligands (34). However, this behavior has not been investigated in Drosophila adult muscle. In mammalian models of muscular dystrophy, immune cells are prominent and a plausible source of TGFβ and other cytokines (35). The use of the dad-lacZ indicator demonstrated that muscle cells clearly possess the capacity to respond to downstream signaling. In mammalian muscle, the finding of myofiber nuclei positive for pSMAD2/3 supports a parallel pathogenesis in mammalian muscle.
TGFβ in regeneration and degeneration in muscular dystrophy
In mammalian muscle affected by muscular dystrophy, degeneration occurs concomitantly with regeneration. In this model reducing TGFβ signaling was thought to primarily mediate its effect by improving muscle regeneration, suggesting that TGFβ signaling has a negative effect on satellite cell function (12). Although the Sgcd[840] model exhibits many features of mammalian muscular dystrophy, we previously did not find evidence of muscle regeneration in this model (15). The absence of obvious regeneration in the fly model suggests that enhanced TGFβ signaling is exerting its effect directly by hastening muscle degeneration either by inhibiting membrane-mediated repair mechanisms or other cellular adaptations. Our findings suggest that the effects of inhibiting TGFβ signaling likely extend beyond the regenerative response, mitigating degeneration and enhancing repair. An additional possibility, raised by the improved climbing performance of normal flies with heterozygous SMAD alleles, is that reducing SMAD signaling developmentally increases muscle function, perhaps by antagonizing the myostatin homolog, myoglianin (36). Further understanding the genes regulated by nuclear pSMADs in injured muscle may help to define the precise pathways that are most critical for eliciting muscle dysfunction. Our results thus far point to the r-SMADs and co-SMADs as targets for therapeutic intervention.
We found that exercising a mouse model of muscular dystrophy increased the pSMAD signaling in those nuclei positioned centrally within myofibers. Centrally positioned nuclei, as opposed to those in the normal peripheral position, are indicative of recent regeneration. However, in our studies, we detected increased pSMAD in central nuclei immediately after exercise without sufficient time for myoblast fusion and regeneration. We interpret these findings to indicate that all myonuclei, including those in the central position, are affected by pathogenic TGFβ signaling. Newly regenerated fibers are particularly important to protect and so targeting this pathway for therapy may offer additional protection.
Sgcd[840] heart and muscle exhibit divergent responses to MAD[12]
We found that heterozygous mutations of Medea, Smox or MAD were dominant suppressors of the loss of walking function in Sgcd[840] mutants. This finding implicates broad downstream signaling, including both the activin and BMP pathways. Curiously, the MAD[12] allele did not improve heart tube function while the Medea[13] and Smox[G0348] alleles did. MAD[12] is the r-SMAD for BMP, and therefore the differential response of the heart tube versus skeletal muscle implies that BMP is more important for muscle versus heart tube function. The Drosophila heart tube is a linear structure. During larval stages, the posterior region is lined with cells expressing cardiac-specific markers (37). Expression of the Hox gene AbdA is sufficient to determine the cardiac fate while the anterior portion is the aorta (37). In the adult, the more anterior segments, A1 through A4, assume cardiac function and are lined with cardiomyocytes derived from the larval cells (38). This developmental paradigm suggests a more intimate relationship between cardiac and vascular structures where activin but not BMP signaling may be essential. Although the SMAD family is more complex in mammals, there still may a differential response to SMAD reduction between heart and skeletal muscles, as inhibition of SMAD signaling may differ between vascular cells and skeletal muscle in mammals.
This study focused on canonical TGFβ signaling in the Sgcd[840] model. Non-canonical TGFβ signaling includes JNK, p38, ERK and Akt signaling (39). Notably, each of these pathways has been shown to be important for muscle regeneration and function. P38 MAP kinase and JNK2 signaling are induced by an exercise regimen in the mdx mouse (40). Akt mediates the muscle proliferative and differentiation functions of insulin-like growth factor-1 (41). In addition, SMADs are not exclusively activated by TGFβ receptors, and the non-transcription factor roles of SMADs should be considered (42) when targeting these pathways for therapeutic intervention.
MATERIALS AND METHODS
Fly stocks
Oregon-R-S (Or-R) was used as the wild-type in all experiments except heart function, which used w[1118]. The Sgcd[840] line was described previously (15). The following strains were used: dad-lacZ, formally PDad[P1883] (25), Medea[13], Medea[15] and Medea[17] (43), MAD[12] (27) and Smox[G0348] (28). Smox[G0348] was from the Bloomington Drosophila Stock Center, number 12246.
Fly husbandry
Flies were raised on standard corn meal, molasses, yeast medium, with dry yeast supplemented for crosses only. Flies were maintained in a 25°C incubator on a 12:12 light:dark cycle, with 20–40% relative humidity. Unless otherwise stated, flies were aged in groups of no more than 10 in 28.5 × 95 mm fly vials and transferred to new vials every 7 days without anesthesia. Flight box-aged flies were aged in groups of 40–60 in 20.5 × 20.5 × 20.5 cm polycarbonate boxes (Eclipse Engineering, Erie CO).
Crosses
To generate Sgcd[840]; Medea[13]/+ flies, Sgcd[840] virgin females were crossed to Medea[13]/TM3, and Sb[+] males were selected as they were Sgcd null and heterozygous Medea[13]. To generate Sgcd[840]; MAD[12]/+ flies, we crossed Sgcd[840] virgin females to MAD[12]/CyO males and selected Cy[+] males. For OCT, the balancers TM3 or CyO were introduced using standard genetic techniques. To introduce heterozygous Smox[G0348] mutations into Sgcd[840] flies, Smox[G0348] and Sgcd[840] were recombined onto a single X-chromosome, generating an Sgcd[840], Smox[G0348]/FM7 stock. Crossing those flies to Sgcd[840] males resulted in Sgcd[840], Smox[G0348]/Sgcd[840] females. The presence of both the Sgcd[840] deletion and the p{lacW} insertion was confirmed by PCR (data not shown). Sgcd[+], SMAD mutant controls were generated by crossing Or-R flies to the SMAD mutant stock (e.g. Medea[13]/TM3).
Quantitative β-gal assay
β-gal was quantified essentially as described (26). Individual flies were briefly anesthetized with CO2, thoraces were dissected and then homogenized in 100 μl assay buffer (50 mM KHPO4, 1 mM MgCl2, pH 7.5) in a 1.5 ml centrifuge tube using a Kontes Pellet Pestle homogenizer and appropriate pestles (Fisher). Whole flies were homogenized in 200 μl assay buffer. Homogenates were centrifuged for 5 min at 14 000g RCF. From each fly, 50μl of supernatant was transferred to a well on a 96-well plate (Costar), one fly or thorax per well. Then, 150 μl of assay buffer plus 1 mM chlorophenol red, β-d-galactopyranoside (CPRG, Roche) was added to each assay well. Plates were incubated at 37°C and the absorbance at 584 nm was read every 2 h for 24 h using a BMG Labtech FLUOstar OPTIMA plate reader. The mean absorbance for Or-R flies was subtracted from each time point and linear regression was performed using Prism 4.0c (GraphPad). The β-gal activity presented is the slope of the regression line ±SD, measured in absorbance units (AUs) at 584 nm per hour. The AUs were multiplied by 1000 and reported as units/hour. Six to eight flies or thoraxes were used per genotype per run. Data presented are representative of at least three runs on different days. Single runs are presented due to day-to-day variability in the assay.
Histology
For hematoxylin and eosin (H&E) staining, flies were fixed and wax embedded according to Fischbach (44), then sectioned and stained using standard protocols. For histochemistry, flies were arranged in freezing medium (TFM, Triangle Biomedical Sciences) and plunged into liquid nitrogen-chilled isopentane, then liquid nitrogen alone. Mouse tissues were collected and quickly frozen. For β-gal activity staining, a published protocol was used with the modification that 2% X-gal was used instead of 10% (45). For immunofluorescence microscopy, 10 μm sections were cut, slides were fixed for 5 min in methanol at −20° and blocked for 1 h using block solution (5% fetal bovine serum in PBS). Primary antibody was incubated for 1 h in block solution. The anti-pSMAD2/3 (Cell Signaling no. 3101) was used at 1:50. After washing in PBS, slides were incubated with goat anti-rabbit IgG conjugated to the Cy3 fluorophore (Jackson ImmunoResearch) and phalloidin conjugated to Alexa Fluor 488. Slides were mounted in Vectashield with DAPI (Vector Labs) and imaged by an observer who was blinded to treatment and exercise status of the mice. For pSMAD2/3+ nucleus counting, five random high-power fields were captured for each condition. pSMAD2/3+ and central nuclei were scored blinded. Images were captured with an Axiophot microscope and iVision software and manipulated within NIH guidelines using Adobe Photoshop 7.0 and ImageJ.
Negative geotaxis
Negative geotaxis was tested at 14 days after eclosion. Individual flies were briefly anesthetized and loaded into 10 ml plastic pipettes affixed to a rigid vertical surface. Flies were given 30 min to recover, and then tapped to the lowest point to engage the negative geotaxis reflex. Flies were allowed to climb for 10 s. Trials were recorded, and the total movement against gravity for each fly was recorded. Groups of flies that were to be compared were assayed simultaneously to eliminate variability attributed to room temperature and room humidity. Each fly was run a total of six times with 1 min between runs. All data presented were from multiple runs on separate days, and flies were compared with flies that were run at the same time. The distance climbed for each fly and each trial was entered into Prism 4.0c. Groups were compared using one-way analysis of variance with a post hoc Tukey test and a significance level of 0.05.
Optical coherence tomography
OCT was performed as described previously (29). Each line was assessed at 7 days after eclosion. Males were studied for all groups except those including Smox[G0348], which is lethal in males. The measured end-systolic and end-diastolic heart tube diameters for each fly were entered into Prism 4.0c. Groups were compared using one-way analysis of variance with a post hoc Tukey test and a significance level of 0.05.
Animal care and use
Sgcg null mice were previously engineered by deleting exon 2, which encodes the initiator methionine and intracellular domain (9). Sgcd null mice were also generated by deleting exon 2 and similarly are a null allele (8). Sgcg null mice were bred through 10 generations into the DBA/2J background as described (46). Exercised mice were subjected to 30 min of running at a rate of 10 m per minute and a 7° downward angle. Post-exercise, animals were sacrificed immediately and muscles were isolated. All experiments were carried out under the approval of the University of Chicago Animal Care and Use Committee.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
Supported by NIH HL61322, NIH T32 GM7261, the American Heart Association 10PRE2600217 and the Muscular Dystrophy Association.
REFERENCES
- 1.Lapidos K.A., Kakkar R., McNally E.M. The dystrophin glycoprotein complex: signaling strength and integrity for the sarcolemma. Circ. Res. 2004;94:1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. doi:10.1161/01.RES.0000126574.61061.25. [DOI] [PubMed] [Google Scholar]
- 2.Ervasti J.M. Costameres: the Achilles' heel of Herculean muscle. J. Biol. Chem. 2003;278:13591–13594. doi: 10.1074/jbc.R200021200. doi:10.1074/jbc.R200021200. [DOI] [PubMed] [Google Scholar]
- 3.Batchelor C.L., Winder S.J. Sparks, signals and shock absorbers: how dystrophin loss causes muscular dystrophy. Trends Cell Biol. 2006;16:198–205. doi: 10.1016/j.tcb.2006.02.001. doi:10.1016/j.tcb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Petrof B.J., Shrager J.B., Stedman H.H., Kelly A.M., Sweeney H.L. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc. Natl Acad. Sci. USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. doi:10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox G.A., Cole N.M., Matsumura K., Phelps S.F., Hauschka S.D., Campbell K.P., Faulkner J.A., Chamberlain J.S. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature. 1993;364:725–729. doi: 10.1038/364725a0. doi:10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- 6.Stedman H.H., Sweeney H.L., Shrager J.B., Maguire H.C., Panettieri R.A., Petrof B., Narusawa M., Leferovich J.M., Sladky J.T., Kelly A.M. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–539. doi: 10.1038/352536a0. doi:10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- 7.Hack A.A., Cordier L., Shoturma D.I., Lam M.Y., Sweeney H.L., McNally E.M. Muscle degeneration without mechanical injury in sarcoglycan deficiency. Proc. Natl Acad. Sci. USA. 1999;96:10723–10728. doi: 10.1073/pnas.96.19.10723. doi:10.1073/pnas.96.19.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hack A.A., Lam M.Y., Cordier L., Shoturma D.I., Ly C.T., Hadhazy M.A., Hadhazy M.R., Sweeney H.L., McNally E.M. Differential requirement for individual sarcoglycans and dystrophin in the assembly and function of the dystrophin–glycoprotein complex. J. Cell Sci. 2000;113(Pt 14):2535–2544. doi: 10.1242/jcs.113.14.2535. [DOI] [PubMed] [Google Scholar]
- 9.Hack A.A., Ly C.T., Jiang F., Clendenin C.J., Sigrist K.S., Wollmann R.L., McNally E.M. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J. Cell Biol. 1998;142:1279–1287. doi: 10.1083/jcb.142.5.1279. doi:10.1083/jcb.142.5.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda R., Nishikawa A., Tanaka H. Visualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J. Biochem. 1995;118:959–964. doi: 10.1093/jb/118.5.959. [DOI] [PubMed] [Google Scholar]
- 11.Bernasconi P., Torchiana E., Confalonieri P., Brugnoni R., Barresi R., Mora M., Cornelio F., Morandi L., Mantegazza R. Expression of transforming growth factor-beta 1 in dystrophic patient muscles correlates with fibrosis. Pathogenetic role of a fibrogenic cytokine. J. Clin. Invest. 1995;96:1137–1144. doi: 10.1172/JCI118101. doi:10.1172/JCI118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohn R.D., van Erp C., Habashi J.P., Soleimani A.A., Klein E.C., Lisi M.T., Gamradt M., ap Rhys C.M., Holm T.M., Loeys B.L., et al. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat. Med. 2007;13:204–210. doi: 10.1038/nm1536. doi:10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreetta F., Bernasconi P., Baggi F., Ferro P., Oliva L., Arnoldi E., Cornelio F., Mantegazza R., Confalonieri P. Immunomodulation of TGF-beta 1 in mdx mouse inhibits connective tissue proliferation in diaphragm but increases inflammatory response: implications for antifibrotic therapy. J. Neuroimmunol. 2006;175:77–86. doi: 10.1016/j.jneuroim.2006.03.005. doi:10.1016/j.jneuroim.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Heydemann A., Ceco E., Lim J.E., Hadhazy M., Ryder P., Moran J.L., Beier D.R., Palmer A.A., McNally E.M. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J. Clin. Invest. 2009;119:3703–3712. doi: 10.1172/JCI39845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allikian M.J., Bhabha G., Dospoy P., Heydemann A., Ryder P., Earley J.U., Wolf M.J., Rockman H.A., McNally E.M. Reduced life span with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum. Mol. Genet. 2007;16:2933–2943. doi: 10.1093/hmg/ddm254. doi:10.1093/hmg/ddm254. [DOI] [PubMed] [Google Scholar]
- 16.Christoforou C.P., Greer C.E., Challoner B.R., Charizanos D., Ray R.P. The detached locus encodes Drosophila dystrophin, which acts with other components of the dystrophin associated protein complex to influence intercellular signalling in developing wing veins. Dev. Biol. 2008;313:519–532. doi: 10.1016/j.ydbio.2007.09.044. doi:10.1016/j.ydbio.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 17.Haines N., Seabrooke S., Stewart B.A. Dystroglycan and protein O-mannosyltransferases 1 and 2 are required to maintain integrity of Drosophila larval muscles. Mol. Biol. Cell. 2007;18:4721–4730. doi: 10.1091/mbc.E07-01-0047. doi:10.1091/mbc.E07-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shcherbata H.R., Yatsenko A.S., Patterson L., Sood V.D., Nudel U., Yaffe D., Baker D., Ruohola-Baker H. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007;26:481–493. doi: 10.1038/sj.emboj.7601503. doi:10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taghli-Lamallem O., Akasaka T., Hogg G., Nudel U., Yaffe D., Chamberlain J.S., Ocorr K., Bodmer R. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. doi:10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kucherenko M.M., Pantoja M., Yatsenko A.S., Shcherbata H.R., Fischer K.A., Maksymiv D.V., Chernyk Y.I., Ruohola-Baker H. Genetic modifier screens reveal new components that interact with the Drosophila dystroglycan–dystrophin complex. PLoS ONE. 2008;3:e2418. doi: 10.1371/journal.pone.0002418. doi:10.1371/journal.pone.0002418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wairkar Y.P., Fradkin L.G., Noordermeer J.N., DiAntonio A. Synaptic defects in a Drosophila model of congenital muscular dystrophy. J. Neurosci. 2008;28:3781–3789. doi: 10.1523/JNEUROSCI.0478-08.2008. doi:10.1523/JNEUROSCI.0478-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. doi:10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 23.Raftery L.A., Sutherland D.J. TGF-beta family signal transduction in Drosophila development: from Mad to Smads. Dev. Biol. 1999;210:251–268. doi: 10.1006/dbio.1999.9282. doi:10.1006/dbio.1999.9282. [DOI] [PubMed] [Google Scholar]
- 24.Rothschild M., Schlein Y., Ito S. A colour atlas of insect tissues via the flea. 1 edn. London, UK: Wolfe Publishing Ltd; 1986. [Google Scholar]
- 25.Tsuneizumi K., Nakayama T., Kamoshida Y., Kornberg T.B., Christian J.L., Tabata T. Daughters against dpp modulates dpp organizing activity in Drosophila wing development. Nature. 1997;389:627–631. doi: 10.1038/39362. doi:10.1038/39362. [DOI] [PubMed] [Google Scholar]
- 26.Hazelrigg T. GFP and other reporters. In: Sullivan W., Ashburner M., Hawley R.S., editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 27.Sekelsky J.J., Newfeld S.J., Raftery L.A., Chartoff E.H., Gelbart W.M. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics. 1995;139:1347–1358. doi: 10.1093/genetics/139.3.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peter A., Schottler P., Werner M., Beinert N., Dowe G., Burkert P., Mourkioti F., Dentzer L., He Y., Deak P., et al. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 2002;3:34–38. doi: 10.1093/embo-reports/kvf012. doi:10.1093/embo-reports/kvf012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf M.J., Amrein H., Izatt J.A., Choma M.A., Reedy M.C., Rockman H.A. Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl Acad. Sci. USA. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. doi:10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nigro V., de Sa Moreira E., Piluso G., Vainzof M., Belsito A., Politano L., Puca A.A., Passos-Bueno M.R., Zatz M. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the delta-sarcoglycan gene. Nat. Genet. 1996;14:195–198. doi: 10.1038/ng1096-195. doi:10.1038/ng1096-195. [DOI] [PubMed] [Google Scholar]
- 31.Noguchi S., McNally E.M., Ben Othmane K., Hagiwara Y., Mizuno Y., Yoshida M., Yamamoto H., Bonnemann C.G., Gussoni E., Denton P.H., et al. Mutations in the dystrophin-associated protein gamma-sarcoglycan in chromosome 13 muscular dystrophy. Science. 1995;270:819–822. doi: 10.1126/science.270.5237.819. doi:10.1126/science.270.5237.819. [DOI] [PubMed] [Google Scholar]
- 32.Coral-Vazquez R., Cohn R.D., Moore S.A., Hill J.A., Weiss R.M., Davisson R.L., Straub V., Barresi R., Bansal D., Hrstka R.F., et al. Disruption of the sarcoglycan–sarcospan complex in vascular smooth muscle: a novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98:465–474. doi: 10.1016/s0092-8674(00)81975-3. doi:10.1016/S0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- 33.Apidianakis Y., Mindrinos M.N., Xiao W., Tegos G.P., Papisov M.I., Hamblin M.R., Davis R.W., Tompkins R.G., Rahme L.G. Involvement of skeletal muscle gene regulatory network in susceptibility to wound infection following trauma. PLoS ONE. 2007;2:e1356. doi: 10.1371/journal.pone.0001356. doi:10.1371/journal.pone.0001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benzakour O., Echalier G., Lawrence D.A. Drosophila cell extracts contain a TGF-beta-like activity. Biochem. Biophys. Res. Commun. 1990;169:1178–1184. doi: 10.1016/0006-291x(90)92020-z. doi:10.1016/0006-291X(90)92020-Z. [DOI] [PubMed] [Google Scholar]
- 35.Spencer M.J., Tidball J.G. Do immune cells promote the pathology of dystrophin-deficient myopathies? Neuromuscul. Disord. 2001;11:556–564. doi: 10.1016/s0960-8966(01)00198-5. [DOI] [PubMed] [Google Scholar]
- 36.Lo P.C., Frasch M. Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-beta superfamily. Mech. Dev. 1999;86:171–175. doi: 10.1016/s0925-4773(99)00108-2. doi:10.1016/S0925-4773(99)00108-2. [DOI] [PubMed] [Google Scholar]
- 37.Lovato T.L., Nguyen T.P., Molina M.R., Cripps R.M. The Hox gene abdominal-A specifies heart cell fate in the Drosophila dorsal vessel. Development. 2002;129:5019–5027. doi: 10.1242/dev.129.21.5019. [DOI] [PubMed] [Google Scholar]
- 38.Monier B., Astier M., Semeriva M., Perrin L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;132:5283–5293. doi: 10.1242/dev.02091. doi:10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- 39.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. doi:10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura A., Yoshida K., Ueda H., Takeda S., Ikeda S. Up-regulation of mitogen activated protein kinases in mdx skeletal muscle following chronic treadmill exercise. Biochim. Biophys. Acta. 2005;1740:326–331. doi: 10.1016/j.bbadis.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Tureckova J., Wilson E.M., Cappalonga J.L., Rotwein P. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J. Biol. Chem. 2001;276:39264–39270. doi: 10.1074/jbc.M104991200. doi:10.1074/jbc.M104991200. [DOI] [PubMed] [Google Scholar]
- 42.Hoover L.L., Kubalak S.W. Holding their own: the noncanonical roles of Smad proteins. Sci Signal. 2008;1:pe48. doi: 10.1126/scisignal.146pe48. doi:10.1126/scisignal.146pe48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hudson J.B., Podos S.D., Keith K., Simpson S.L., Ferguson E.L. The Drosophila Medea gene is required downstream of dpp and encodes a functional homolog of human Smad4. Development. 1998;125:1407–1420. doi: 10.1242/dev.125.8.1407. [DOI] [PubMed] [Google Scholar]
- 44.Fischbach J.A. Mass histology of adult heads. In: Ashburner M., editor. Drosophila, a Laboratory Manual. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 254–259. [Google Scholar]
- 45.Wolff T. Histological techniques for the Drosophila eye, Part II: Adult. In: Sullivan W., Ashburner M., Hawley R.S., editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 46.Heydemann A., Huber J.M., Demonbreun A., Hadhazy M., McNally E.M. Genetic background influences muscular dystrophy. Neuromuscul Disord. 2005;15:601–609. doi: 10.1016/j.nmd.2005.05.004. doi:10.1016/j.nmd.2005.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.