Abstract
Recent studies delineate a pathway involving familial Parkinson's disease (PD)-related proteins PINK1 and Parkin, in which PINK1-dependent mitochondrial accumulation of Parkin targets depolarized mitochondria towards degradation through mitophagy. The pathway has been primarily characterized in cells less dependent on mitochondria for energy production than neurons. Here we report that in neurons, unlike other cells, mitochondrial depolarization by carbonyl cyanide m-chlorophenyl hydrazone did not induce Parkin translocation to mitochondria or mitophagy. PINK1 overexpression increased basal Parkin accumulation on neuronal mitochondria, but did not sensitize them to depolarization-induced Parkin translocation. Our data suggest that bioenergetic differences between neurons and cultured cell lines contribute to these different responses. In HeLa cells utilizing usual glycolytic metabolism, mitochondrial depolarization robustly triggered Parkin–mitochondrial translocation, but this did not occur in HeLa cells forced into dependence on mitochondrial respiration. Declining ATP levels after mitochondrial depolarization correlated with the absence of induced Parkin–mitochondrial translocation in both HeLa cells and neurons. However, intervention allowing neurons to maintain ATP levels after mitochondrial depolarization only modestly increased Parkin recruitment to mitochondria, without evidence of increased mitophagy. These data suggest that changes in ATP levels are not the sole determinant of the different responses between neurons and other cell types, and imply that additional mechanisms regulate mitophagy in neurons. Since the Parkin–mitophagy pathway is heavily dependent on bioenergetic status, the unique metabolic properties of neurons likely influence the function of this pathway in the pathogenesis of PD.
INTRODUCTION
Mitochondrial dysfunction has been heavily implicated in the pathogenesis of Parkinson's disease (PD) (1,2), in which evidence has accumulated of decreased electron transport chain complex function, increased mitochondrially derived reactive oxygen species production and, more recently, dysregulation of mitochondrial dynamics and homeostasis (3–6). Genetic forms of PD have further implicated mitochondrial homeostasis in pathogenesis. Mutations causing loss of function of the proteins PINK1 or Parkin result in early-onset autosomal recessive PD (7,8). Animal models of both PINK1 and Parkin-related PD have demonstrated abnormalities of pathways regulating mitochondrial function and homeostasis (9–13). In addition, genetic complementation studies revealed that PINK1 functions upstream in a pathway with Parkin that appears to regulate mitochondrial fission and/or fusion (10,11,14,15).
More recently, it has been suggested that Parkin regulates mitochondrial degradation through autophagy (mitophagy). In mammalian cell cultures, overexpressed Parkin is recruited to depolarized mitochondria, targeting them for mitophagy (16). Several studies have now confirmed this observation in multiple cell lines, and have revealed a pathway in which PINK1 is required to recruit Parkin to the mitochondria, which subsequently initiates mitophagy (17–20). It has been suggested that this pathway plays an important role in the neurodegeneration of PD, linking mitochondrial quality control to chronic neurodegeneration (3,6,21–23). However, much of the detailed characterization of the Parkin-mediated mitophagy pathway has been completed in immortalized cell lines, both non-neuronal cell lines, such as HeLa cells, and neuronally derived cells, including SH-SY5Y neuroblastoma cells. These ‘neuronal’ and non-neuronal cell types are all much less dependent on mitochondria than neurons, because they preferentially generate ATP through glycolysis and, thus, do not rely on mitochondrial respiration (24–26). There is evidence from yeast studies that this bioenergetic distinction may be critically important in the mitophagy pathway. Kanki and Klionsky (27) found that yeast readily undergo mitophagy under starvation conditions. However, when grown in the presence of a media forcing cells into dependence on mitochondrial respiration for energy production, they exhibited barely detectable levels of mitophagy, even under severe starvation conditions. In view of the unique bioenergetic profile of neurons, which depend heavily on mitochondrial respiration (28), and the potential implications of Parkin-mediated mitophagy in PD neurodegeneration, it is critically important to evaluate this pathway directly in neurons.
We examined the Parkin–mitophagy pathway in neurons. Surprisingly, we found that, unlike in other cell types, rapid cell-wide mitochondrial depolarization in neurons does not cause recruitment of Parkin to mitochondria. Our studies indicate that bioenergetic differences between neurons and other cell types are involved in these different responses, and this may provide a means for tightly controlled regulation of mitochondrial homeostasis in neurons as opposed to other cell types. Our results emphasize that a more thorough understanding of the functions of pathogenesis-related proteins specifically in neurons will help better determine the relevance of proposed pathways to neurodegenerative disease.
RESULTS
Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) treatment does not trigger Parkin recruitment to mitochondria in primary cortical neurons overexpressing human Parkin
In order to evaluate the mitochondrial depolarization-induced recruitment of Parkin to mitochondria in neurons, we utilized primary rat cortical neurons and examined treatments comparable with those previously employed in other cell types (16,17,20). While we could detect endogenous Parkin in cortical neurons by immunocytochemistry (ICC), ICC did not produce a sufficiently robust signal to allow colocalization analysis of endogenous Parkin by microscopy. Consequently, similar to previous studies (16,19,20), we utilized plasmid transfection to overexpress Parkin in cells. We co-transfected primary rat cortical neurons with untagged, full-length human Parkin (hu-Parkin) and mitochondrially targeted DsRed2 protein (mtDsRed2) at days in vitro (DIV) 6. We confirmed that cells were co-transfected with both plasmids (Supplementary Material, Fig. S1), and expressed both hu-Parkin and mtDsRed2 from 72 h to at least 2 weeks post-transfection.
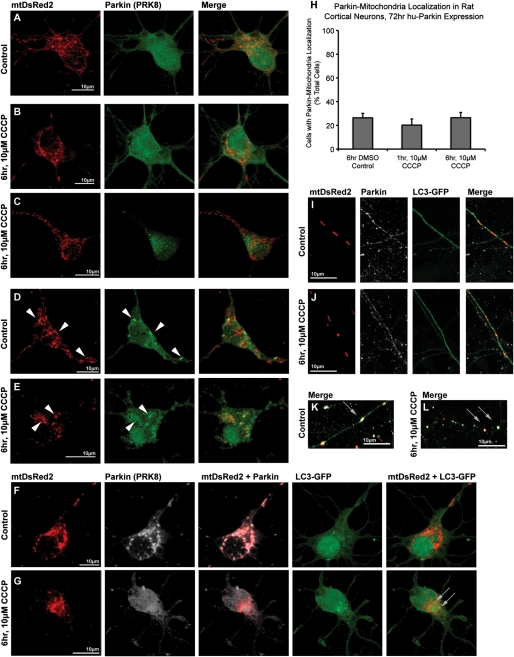
Seventy-two hours after transfection, neurons were treated with 10 µm carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or DMSO vehicle for either 1 or 6 h. Using ICC staining and confocal microscopy to localize hu-Parkin and mtDsRed2, and performing blinded manual analysis of colocalization, we observed that 20–25% of neurons showed evidence of Parkin localized to mitochondria under basal culture conditions (Fig. 1), although some variability was noted between preparations. Neurons observed to have Parkin clustered on mitochondria ranged from those exhibiting lesser than five mitochondria with Parkin colocalization (∼10–15% of total cells, Fig. 1D) to cells exhibiting multiple or all mitochondria with Parkin accumulation (∼10% of total cells, Fig. 1F). Unexpectedly, however, CCCP treatment of neurons did not increase the rate of Parkin–mitochondrial localization (Fig. 1A–C, H). Thus, Parkin was localized to mitochondria in 26 ± 4% (n = 4) of neurons treated with 6 h DMSO control, similar to the number of neurons showing mitochondrial Parkin localization 1 h (20 ± 5%, n = 3) or 6 h (26 ± 4%, n = 4) after application of 10 µm CCCP (P = 0.57, ANOVA). In order to ensure that CCCP treatment depolarized mitochondria in these neuronal cultures, we utilized tetramethyl rhodamine methyl ester fluorescence. We found that 10 µm CCCP rapidly depolarized neuronal mitochondria (Supplementary Material, Fig. S2 and Movies S1 and S2), showing that the absence of Parkin translocation to mitochondria was not due to the inability to depolarize mitochondria.
Figure 1.
Parkin localization in cortical neurons after exposure to CCCP. Rat cortical neurons were co-transfected with hu-Parkin and mitochondrially targeted DsRed2 (mtDsRed2) at DIV6, then treated 72 h later with either DMSO vehicle control (A, D, F, I, K) or 10 µm CCCP (B, C, E, G, J, L) for 1 or 6 h. After 6 h of treatment, a majority of cells in both control and CCCP conditions did not exhibit any translocation of Parkin to the mitochondria (A–C). Most cells exposed to CCCP exhibited minor, if any, mitochondrial morphology changes (b), although a minority of cells exhibited excessive fragmentation (C). A baseline population of Parkin-overexpressing cells exhibited Parkin–mitochondrial accumulation (arrowheads) in both (D) control and (E) CCCP treatment conditions. (F) A neuron co-transfected with mtDsRed2, Parkin and LC3-GFP demonstrates that even with excessive Parkin–mitochondrial localization, no increased mitophagy was observed. (G) Another cell exhibits active mitophagy based on LC3-GFP and mtDsRed2 co-localization (grey arrows), but no Parkin–mitochondrial accumulation. (H) Graph of observed percentage of neurons exhibiting Parkin–mitochondrial localization, with no significance between groups (ANOVA) (30–60 individual cells across three to four independent neuronal preps; ± SEM). Like cell bodies, mitochondria in neuritic projections did not exhibit any differences in Parkin or LC3 localization between (I) DMSO control or (J) CCCP treatments. Definite co-localizations were rare events, and primarily observed in blebbing (K) or fragmented (L) neurites, regardless of treatment.
Mitochondrial depolarization did not lead to increased mitophagy in neurons
To examine mitophagy under these conditions, cortical neurons were co-transfected to express a GFP-tagged form of LC3 (LC3-GFP), which localizes to early autophagic vesicles and is widely used as a marker of autophagy (29), along with mtDsRed2 and hu-Parkin. We defined mitophagic events as areas of colocalization between mtDsRed2 and LC3-GFP fluorescent accumulations. We have previously noted that evidence of mitophagy is rarely observed in primary neurons under control conditions (30), although it has been observed under certain conditions, such as after growth factor withdrawal (31). In our present studies, we rarely noted evidence of mitophagy under control conditions, and we found no observable change following CCCP treatment, indicating that mitochondrial depolarization did not increase mitophagy. Notably, neurons with clear colocalization of Parkin on mitochondria rarely showed evidence of mitophagy, and most exhibited none at all (Figs. 1F). Furthermore, under both control and CCCP conditions, we also observed mitophagic events that were not associated with Parkin localization to mitochondria at all (Figs. 1G). In keeping with the absence of enhanced mitophagy in Parkin-overexpressing neurons, we noted the continued presence of DsRed2-containing mitochondria in Parkin-overexpressing neurons even after several weeks (Supplementary Material, Fig. S1). Further, mitochondria remained present and diffuse even in neurons exposed to CCCP for 24 h, even though, as would be expected, significant toxicity to neurons was apparent (unpublished data). Ultimately, we found no correlation between mitochondrial depolarization with CCCP, Parkin–mitochondrial localization or mitophagy in primary neurons.
The response to CCCP in neuronal axons and dendrites is similar to cell bodies
We next examined Parkin localization in neuritic projections following depolarization, to determine whether regulation of mitochondrial homeostasis differs in the axonal or dendritic compartments. Similar to our findings in the neuronal cell body, we did not observe Parkin translocation to mitochondria in distal axons and dendrites after CCCP treatment for either 1 h (not shown) or 6h (Fig. 1I and J). Parkin expression in axons and dendrites is usually punctate, and Parkin could be observed in proximity to a subset of mitochondria, regardless of whether neurons were treated with DMSO vehicle control media or CCCP. We could not detect increased mitophagy over baseline under any conditions tested, as evaluated by colocalization of mitochondrial fluorescence with GFP-LC3 puncta (Fig. 1I and J). Some distressed neuritic processes, in both CCCP and DMSO control conditions, exhibited significant ‘beading’ or ‘blebbing’, a well-known response to injury (32). In these neurons, the axonal beaded varicosities accumulate proteins, and in some of these there were accumulations of GFP-LC3, Parkin and sometimes mitochondria (Fig. 1K and L). However, in many of these severely injured neurites, there appears to be little or no axonal continuity (Fig. 1L), a feature observed in other injury models and which likely represents degrading neurites with collections of microtubules, proteins and organelles (32).
Endogenous neuronal Parkin does not translocate to mitochondria following CCCP
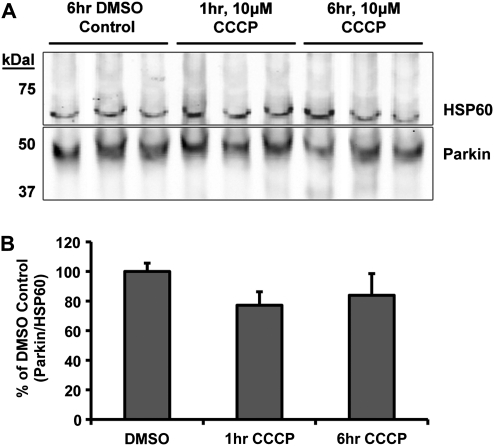
In order to exclude the unlikely possibility that the absence of Parkin localization to mitochondria in neurons is an artifact of overexpression of the human form of the protein, we quantified endogenous Parkin associated with mitochondria before and after depolarization. Since ICC of endogenous Parkin ICC proved difficult to analyze using imaging techniques, we employed a cellular fractionation technique using commercial reagents (see Materials and Methods) to enrich mitochondria from cultured neurons and quantitative western blot analyses to detect Parkin. Cortical neurons (DIV 9) were treated with DMSO or 10 µm CCCP, and harvested 1 or 6 h later. Mitochondrially enriched fractions were prepared from the harvested neurons and subjected to western blot analysis. Blots were probed for Parkin and HSP60, as a mitochondrial loading control (Fig. 2A). The level of endogenous Parkin, relative to mitochondrial loading controls, did not change in the mitochondrial-enriched fraction following CCCP treatment when compared with DMSO control (DMSO 100 ± 6%, 1 h CCCP 77 ± 10%, 6 h CCCP 84 ± 15%, P = 0.075, one-way ANOVA; Fig. 2B). To confirm these results, we repeated the experiments utilizing a modified differential centrifugation procedure using reagents we prepared and ensured were detergent free (33), to guard against any possible dissociation of mitochondrial Parkin during isolation (see Materials and Methods). Western blot analysis from these experiments confirmed the above results, with no significant change in Parkin mitochondrial localization after CCCP treatment (DMSO 100 ± 7%, 1 h CCCP 111 ± 3%, 6 h CCCP 112 ± 21%, P = 0.8, one-way ANOVA). These data further support that conclusion that recruitment of endogenous Parkin onto mitochondria after rapid depolarization did not occur in neurons.
Figure 2.
Western blot analysis of endogenous Parkin localization in cortical neurons after CCCP exposure. Non-transfected cortical neurons were treated and harvested on DIV9 for differential centrifugation isolation of the mitochondrial-enriched fraction. (A) Western blot from the commercial-reagent-based mitochondrial isolation stained for Parkin (PRK8) and mitochondrial loading control HSP60. (B) Quantitative results from western blot analysis presented as the densitometry ratio of Parkin bands over respective mitochondrial HSP60 loading control bands, as percent DMSO control. No significance between results (ANOVA) (n = 3–4 from three independent neuron preps; ± SEM).
Lower concentrations of CCCP did not induce Parkin–mitochondrial translocation in neurons
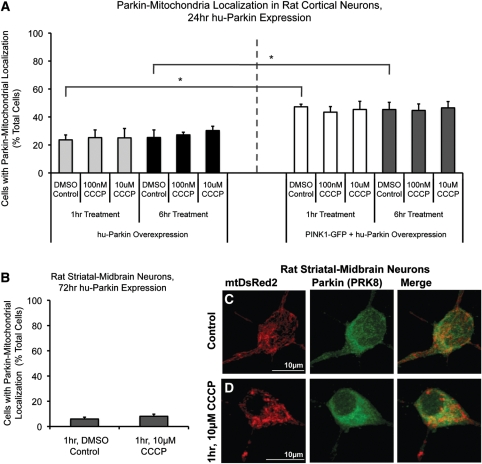
Previous studies employed earlier Parkin-overexpression time points after transfection (24 h) and lower concentrations of CCCP to trigger the Parkin localization effect. Considering that 10 µm CCCP may have been too severe an insult for neurons, and that a previous study reported Parkin–mitochondrial colocalization in primary mouse cortical neurons at 100 nm CCCP (20), we examined both 10 µm and 100 nm CCCP exposure for 1 and 6 h time points after 24 h Parkin expression in DIV 7 cortical neurons. Under these conditions, 23–25% of cells expressing human Parkin showed some colocalization between Parkin and mitochondria (Fig. 3). CCCP exposure at 100 nm did not increase Parkin localization in hu-Parkin overexpressing neurons (25 ± 6% at 1 h, 27 ± 2% at 6 h) compared with vehicle-exposed control (24 ± 4% at 1 h, 25 ± 5% at 6 h; Fig. 3), and neither did 10 μm CCCP (25 ± 7% at 1 h, 30 ± 3% at 6 h; Fig. 3), confirming our previous observations following 72 h of Parkin overexpression (Fig. 1).
Figure 3.
Parkin localization in cortical neurons after 24 h expression, Parkin and PINK1 co-expression, and in striatal/midbrain cultures. (A) Rat cortical neurons were co-transfected with hu-Parkin and DsRed2, or hu-Parkin, PINK1-GFP and mtDsRed2 at DIV6, then treated with either DMSO vehicle control, 100 nm CCCP or 10 µm CCCP for 1 or 6 h on DIV7. Graph represents percentage of observed neurons exhibiting Parkin–mitochondrial localization. No significance between results within Parkin groups or within PINK1 + Parkin groups, respectively (ANOVA). * = P < 0.05, significance between Parkin and Parkin + PINK1 DMSO controls at 1 and 6 h, ANOVA with post hoc Student's t-tests with Bonferroni correction (n = 3–4 from three independent neuron preps; ±SEM). (B) Mixed-culture rat striatal and midbrain neurons were transfected at DIV6 and treated at DIV9. Graph represents percentage of observed neurons exhibiting Parkin–mitochondrial localization. No significance between results; z-test of proportions (n = 20–60 cells/condition from two independent neuron preps; ±SEM). (C and D) Representative images from striatal-midbrain neurons treated with Control vehicle DMSO (C) or 10 μm CCCP (D) for 1 h.
PINK1 overexpression increases Parkin–mitochondria localization, but does not sensitize neurons to depolarization-induced translocation
Multiple studies have identified functional PINK1 expression as an upstream requirement for depolarization-induced Parkin translocation to mitochondria (17–20). Previous studies have shown that cortical neurons express PINK1 (34). However, in order to determine whether the absent depolarization-induced Parkin–mitochondrial translocation response in neurons was attributable to low or absent PINK1 expression in our experimental system, we re-evaluated the response in the presence of over-expressed exogenous PINK1. We co-transfected neurons with GFP-tagged PINK1 and hu-Parkin (confirming co-expression through ICC; Supplementary Material, Fig. S1), and then determined the localization of Parkin after mitochondrial depolarization. Neurons co-expressing both GFP-tagged PINK1 and hu-Parkin exhibited a higher basal rate of Parkin–mitochondrial localization (for 6 h DMSO control, parkin alone: 25 ± 5%; parkin + PINK1: 45 ± 5%; P < 0.05, Student's t-test with Bonferroni correction; Fig. 3). However, the percentage of cells exhibiting Parkin–mitochondria localization did not increase in PINK1-GFP/hu-Parkin expressing cells following 100 nm or 10 µm CCCP-induced depolarization (for 1 h, DMSO: 47 ± 2%; 100 nm CCCP: 43 ± 4%; 10 µm CCCP: 45 ± 6%; for 6 h, DMSO: 45 ± 5%; 100 nm CCCP: 45 ± 5%; 10 μm CCCP: 47 ± 5%; P = 0.99, ANOVA; Fig. 3A). These results suggest that while PINK1 does participate in a pathway directing Parkin to mitochondria in neurons, PINK1 does not sensitize neurons to increased Parkin–mitochondrial localization following rapid depolarization.
Mixed striatal/midbrain neuronal cultures did not undergo Parkin–mitochondrial translocation in response to CCCP-induced mitochondrial depolarization
In order to address whether cortical neurons might be less sensitive than other neuronal populations to mitochondrial depolarization-triggered Parkin translocation, we evaluated a different population of CNS neurons. Mixed striatal/midbrain neuronal cultures were exposed to CCCP under identical conditions to those used for cortical neurons. Striatal/midbrain neurons exhibited a low basal rate of Parkin–mitochondrial accumulation (6% of cells), and CCCP-induced mitochondrial depolarization (1 h, 10 µm CCCP) did not alter Parkin localization in these cells (Fig. 3B–D). These data indicate that the absence of depolarization-induced Parkin translocation to mitochondria is not specific to cortical neurons, but appears to be a general property of cultured primary neurons.
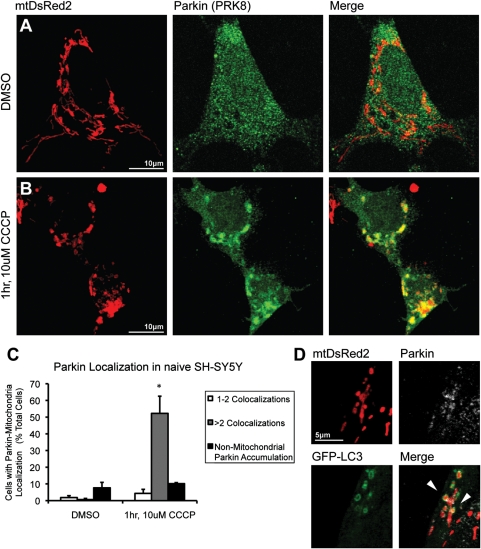
CCCP treatment induces localization of Parkin in both naïve SH-SY5Y cells and neuronally differentiated PC6-3 cells
We then examined CCCP-induced Parkin–mitochondrial translocation in two separate immortalized dopaminergic cell lines, human SH-SY5Y and rat PC6-3 cells, in order to determine whether the absence of a response in primary neuronal cultures was attributable to an artifact of our detection and/or exposure technique. Naïve SH-SY5Y cells were exposed to 10 µm CCCP at 72 h after cotransfection with hu-Parkin and mtDsRed2. Cells overexpressing Parkin under basal conditions exhibited a low (<5%) basal rate of Parkin–mitochondrial colocalization, with most positive cells containing only one to two mitochondria with Parkin accumulation, as well as the presence of non-mitochondrial Parkin accumulations (∼10% of cells; Fig. 4). After 1 h exposure to CCCP, robust recruitment of Parkin to mitochondria was found, with many cells exhibiting a cell-wide association between Parkin accumulation and mitochondria (52.2 ± 10% of cells compared with 0.6% in DMSO control; Fig. 4), similar to previous studies (17,20). Furthermore, co-localization of LC3-GFP positive puncta and Parkin-tagged mitochondria (Fig. 4) indicated upregulation of mitophagy following CCCP treatment.
Figure 4.
Parkin localization in naïve SH-SY5Y cells after mitochondrial depolarization. SH-SY5Y cells co-transfected with mtDsRed2 and hu-Parkin were exposed to (A) 1 h DMSO vehicle control or (B) 1 h, 10 µm CCCP. Cells demonstrate dramatic Parkin–mitochondrial colocalization after CCCP treatment. (C) Graph represents percentage of observed cells exhibiting (white) one to two Parkin–mitochondrial colocalizations per cell, (grey) more than two Parkin–mitochondrial colocalizations per cell and (black) non-mitochondrial cellular Parkin accumulations. *P < 0.05 from DMSO control; ANOVA with post hoc Student's t-tests with Bonferroni correction (n = 3; ±SEM). (D) SH-SY5Y cell co-transfected with mtDsRed2, hu-Parkin and LC3-GFP, demonstrating mitophagy via co-localization of LC3-GFP with mtDsRed2 and Parkin (arrowheads).
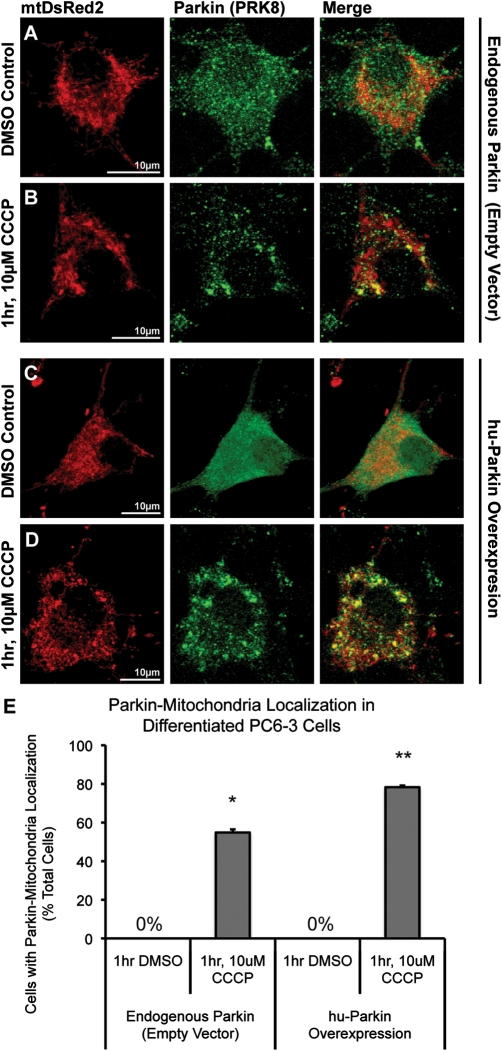
Because our neuronal primary cultures were derived from rat, we wanted to confirm that Parkin translocation could occur in a rat-derived cell line, to exclude the remote possibility that a species-specific difference in Parkin function or localization accounted for the absence of depolarization-induced translocation to mitochondria in neurons. For this, we used PC6-3 cells, a subclone of the dopaminergic rat pheochromocytoma PC12 cell line that can be well-differentiated with NGF to a neuronal phenotype (35). Endogenous Parkin levels in PC6-3 cells were readily detectable by ICC, allowing us to examine localization of both endogenous rat Parkin and overexpressed hu-Parkin. Neuronally differentiated, PC6-3 cells were co-transfected with either mt-DsRed2 and hu-Parkin, or mtDsRed2 and a control empty vector. At 96 h post-transfection, cells were exposed to 10 µm CCCP. Differentiated PC6-3 cells exhibited robust Parkin accumulation on mitochondria following 1 h of CCCP treatment (Fig. 5). This was observed for both endogenous rat Parkin (55 ± 2% of cells compared with 0% in DMSO control) or overexpressed hu-Parkin (78 ± 1% of cells compared with 0% in DMSO control; Fig. 5). Thus, both endogenous rat and overexpressed human Parkin translocate to mitochondria following depolarization in a rat neuronal cell line. To our knowledge, this is the first demonstration of endogenous Parkin translocation in a neuronally differentiated cell line.
Figure 5.
Endogenous and overexpressed Parkin localization in NGF-differentiated PC6-3 cells. PC6-3 cells were neuronally differentiated using NGF, and co-transfected with mtDsRed2 and either (A and B) empty-vector control plasmid or (C and D) hu-Parkin. 96 h after transfection, cells were treated with either (A and C) DMSO vehicle control or (B and D) 1 h, 10 µm CCCP. Both (A and B) endogenous rat Parkin and (C and D) overexpressed hu-Parkin were detected via ICC. Cells demonstrate dramatic Parkin–mitochondrial colocalization after CCCP treatment. (E) Graph represents percentage of observed cells exhibiting Parkin–mitochondrial accumulations. *P < 0.05 and **P < 0.05, significant from respective DMSO controls; z-test of proportions (n = 27–46 cells per condition from two independent platings; ±SEM).
Parkin translocates to depolarized mitochondria in normally glycolytic HeLa cells, but not HeLa cells forced into dependence on mitochondrial respiration
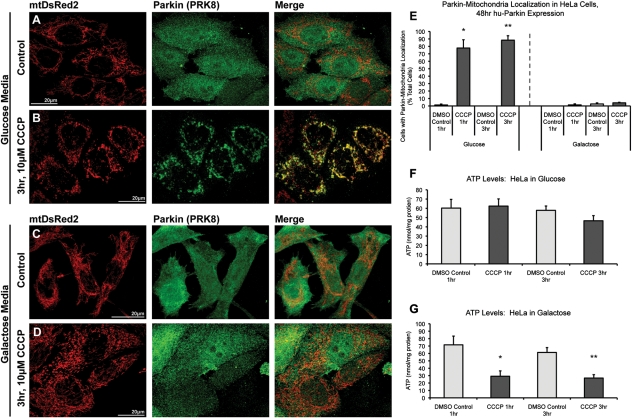
One of the most striking differences between neurons and other cell types (including immortalized cell lines) is the specific dependence of neurons on mitochondrial respiration (28,36). Cancer cells, in particular, obtain most of their energy through glycolysis rather than mitochondrial respiration (25,26). We hypothesized that the differences we observed between neurons and cultured cells might be attributable to bioenergetic differences. To test this hypothesis, we analyzed mitochondrial depolarization-induced Parkin translocation in both normal, glycolytic HeLa cells [where the Parkin–mitochondrial translocation response is robust (16)] and HeLa cells forced into dependence on mitochondrial respiration, mimicking the situation in neurons. HeLa cells were cultured in either standard glucose-based DMEM media or glucose-free DMEM supplemented with galactose (10 mm) and glutamine (2 mm). Under galactose/glutamine culturing conditions, HeLa cells favor utilization of the citric acid cycle and oxidative phosphorylation over lactic acid-generating glycolysis for ATP production (25,26). Cells were cultured without glucose for at least three passages prior to experiments and were then transiently transfected with mtDsRed2 and hu-Parkin and, after 48 h, treated with 10 μm CCCP or vehicle control. As previously reported, HeLa cells cultured in glucose exhibited robust co-localization of Parkin with mitochondria following both 1 and 3 h CCCP (77.8 and 88.4% of cells, respectively), when compared with essentially no co-localization with vehicle-treated control (<2%; Fig. 6). In sharp contrast, however, galactose-cultured HeLa cells, even after 3 h of CCCP treatment, showed no increase in Parkin–mitochondrial translocation (Fig. 6). These data show that the induction of mitochondrial parkin translocation and mitophagy by mitochondrial depolarization is profoundly attenuated by enforcing cellular dependence on mitochondrial respiration.
Figure 6.
Parkin accumulates on depolarized mitochondria in glycolytic, but not oxidative-phosphorylation-dependent HeLa cells. HeLa cells were maintained in either (A and B) glucose-based media or (C and D) glucose-free media supplemented with galactose and glutamine. Cells were co-transfected with mtDsRed2 and hu-Parkin, then treated with either (A and C) DMSO vehicle control or (B and D) 10 µm CCCP for either 1 or 3 h. Cells grown in DMEM containing glucose (B), but not cells grown in galactose/glutamine media (D), exhibit robust Parkin–mitochondrial translocation after 3 h CCCP. (E) Graph represents percentage of observed cells exhibiting Parkin–mitochondrial accumulations. ATP levels were determined in non-transfected HeLa cells after 1 or 3 h of CCCP treatment in both (F) glucose and (G) galactose/glutamine culturing conditions. For all data, *P < 0.05 and **P < 0.05 from respective DMSO controls; ANOVA with post hoc Student's t-tests with Bonferroni correction (n = 3; ±SEM).
Rapid loss of ATP after mitochondrial depolarization correlates with inability to undergo Parkin–mitochondrial translocation
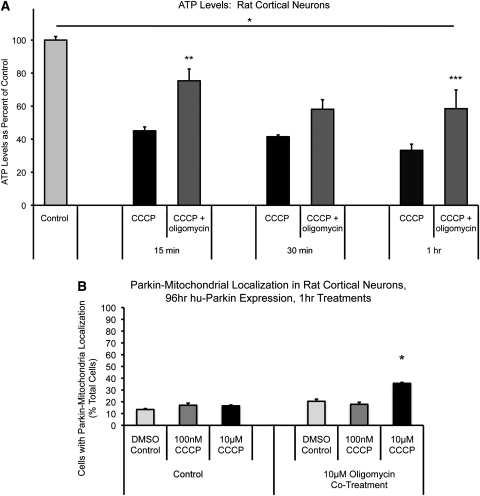
We next measured ATP levels in HeLa cells under both glycolytic and oxidative phosphorylation-dependent conditions. As predicted, mitochondrial depolarization resulted in a dramatic loss of ATP in HeLa cells dependent on oxidative phosphorylation (down to 40% of control after 1 h CCCP), but no ATP loss in normally cultured, glycolytic HeLa cells (Fig. 6C and D). Similarly, ATP loss was dramatic and rapid after CCCP treatment in primary rat cortical neuron cultures as well, with significant loss of ATP observed after 15 min of CCCP (45% of control) and dropping to 33% of control after an hour of CCCP treatment (Fig. 7A).
Figure 7.
Effects of oligomycin co-treatment on CCCP-induced ATP loss and Parkin localization in cortical neurons. (A) Non-transfected neurons were treated on DIV10 and collected at the indicated time points for ATP level analysis. *P < 0.05, DMSO control significant from all treatment groups; **P < 0.05, 15 min CCCP + oligomycin significant from 15 min CCCP; ***P < 0.05, 1 h CCCP + oligomycin significant from 1 h CCCP; ANOVA with post hoc Newman–Keuls (n = 6–15; ±SEM). (B) Cortical neurons were co-transfected with mtDsRed2 and hu-Parkin at DIV6, then treated at DIV10 with either DMSO vehicle control or CCCP, or co-treated with 10 µm oligomycin, for 1 h. Graph represents percentage of observed cells exhibiting Parkin–mitochondrial accumulations. *P < 0.05, significant from respective DMSO control; z-test of proportions (n = 28–157 cells per condition, from three to six separate experiments across three to four independent platings; ±SEM).
Altering neuronal bioenergetics may influence Parkin–mitochondrial translocation in response to mitochondrial depolarization
Our data suggest that bioenergetic differences between neurons and immortalized cell lines dramatically affect the PINK1-Parkin mitophagy pathway. We hypothesized that if this difference was attributable to the loss of ATP in neurons after mitochondrial depolarization, then preventing or delaying the loss of ATP after CCCP exposure should lead to Parkin–mitochondrial translocation.
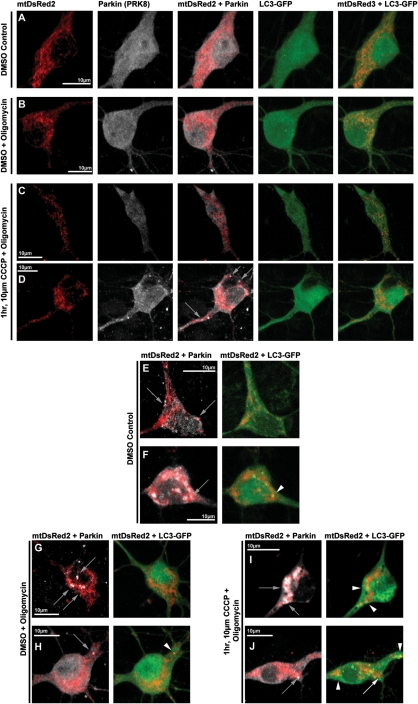
Rapid ATP loss after mitochondrial depolarization in neurons is not only due to the loss of membrane potential-driven oxidative phosphorylation, but also by rapid ATP hydrolysis through reversal of the ATP synthase, which serves as a compensatory mechanism to maintain mitochondrial membrane potential by transporting hydrogen ions from the matrix (37,38). It has been shown that inhibition of the ATP synthase with oligomycin (which also blocks its reverse hydrolysis activity) enables neurons to temporarily maintain a greater pool of ATP in response to rapid mitochondrial depolarization (37). We tested whether the presence of oligomycin in the setting of mitochondrial depolarization would prevent the loss of ATP in response to CCCP, and whether it would alter Parkin localization after CCCP. As noted above, ATP loss occurs rapidly after CCCP exposure in neurons (Fig. 7A). The presence of oligomycin (10 µm) partially prevented the loss of ATP up to an hour after CCCP-induced depolarization compared with DMSO vehicle control (15 min, CCCP: 45 ± 2%, oligomycin + CCCP: 75 ± 7%; 30 min, CCCP: 41 ± 1%, oligomycin + CCCP 58 ± 6%; 1 h, CCCP: 33 ± 4%, oligomycin + CCCP: 58 ± 11%; Fig. 7A). We then evaluated the localization of Parkin in response to mitochondrial depolarization under these conditions. In these experiments, as in our previous studies, there was no significant increase in Parkin–mitochondrial translocation after 1 h of 10 µm CCCP alone, when compared with DMSO control. However, with co-treatment of 10 µm oligomycin and 10 µm CCCP for 1 h, we did observe a modest, but significant increase in the proportion of cells that undergo Parkin–mitochondrial translocation (36 ± 0.3% of total cells) compared with DSMO control (13 ± 0.3% of total cells; P < 0.05, z-test of proportions with Bonferroni correction; Figs 7B and 8). Despite this difference, we did not observe the robust response seen in other cell types, and the majority of neurons displaying Parkin–mitochondrial translocation consisted of one or two mitochondria with Parkin accumulation (Fig. 8D), and not the whole-cell translocation occurring most often in other cell types. In addition, we were unable to detect any correlation between treatment and altered mitophagy by evaluating colocalization of mitochondria with Parkin and LC3-GFP; Parkin-associated mitophagy occurred rarely in neurons, regardless of treatment, and it did not vary by the treatment group (Fig. 8), in contrast to the easily detected association in other cell types (Fig. 4). These data indicate that while partial mitigation of the bioenergetic defect that accompanies rapid depolarization of mitochondria in neurons allowed Parkin–mitochondria translocation to proceed at a low level, we still did not observe this phenomenon in the majority of neurons, it did not occur globally within most individual positive neurons and we did not observe associated mitophagy. These observations suggest that ATP depletion is not the only mechanism underlying the inhibited Parkin localization response in neurons.
Figure 8.
Respresentative images of Parkin and LC3-GFP localization in cortical neurons following oligomycin and CCCP co-treatments. Rat cortical neurons were co-transfected with hu-Parkin, mtDsRed2 and LC3-GFP at DIV6, then treated 96 h later with either DMSO vehicle Control (A, E, F), DMSO with 10 µm oligomycin co-treatment (B, G, H) or 10 µm CCCP with 10 µm oligomycin co-treatment (C, D, I, J) for 1 h. After treatment, a majority of cells in all conditions did not exhibit any translocation of Parkin to the mitochondria (A–C). CCCP-oligomycin co-treatment exhibited a modest but significant increase in the number of cells exhibiting Parkin–mitochondria localization (D), though most positive cells presented with more than five Parkin-positive mitochondria (D, grey arrows). While Parkin–mitochondria localization could also be identified in all conditions (D–J, grey arrows), most cells exhibited no apparent mitophagy, based on LC3-GFP accumulation localization, regardless of Parkin localization (A–E, G). Mitophagy rates did not appear to be affected by oligomycin or CCCP-oligomycin co-treatments, and rare mitophagy events could be identified in all conditions, including Parkin-positive mitophagy (F, H, I; white arrowheads), and mitophagy events independent of Parkin localization (J, white arrow indicates a Parkin-positive mitochondrion without LC3 accumulation; white arrowheads indicate LC3-positive mitochondria without Parkin localization).
Notably, oligomycin inhibition of ATP hydrolysis may also serve to cause more rapid mitochondrial depolarization after CCCP, since mitochondrial membrane potential would not be maintained for as long without this compensatory response. This could also potentially contribute to the modest increase observed in recruitment of Parkin to mitochondria. However, given the rapid depolarization noted after CCCP alone (Supplementary Material, Movie S2), this would seem to be an unlikely explanation for the findings.
DISCUSSION
We observed that rapid mitochondrial depolarization did not induce Parkin translocation to mitochondria in primary rat cortical neurons or striatal/midbrain primary neurons, unlike the response observed in cell lines and other cell types. Neurons are unique in that even in the presence of glucose, much of their energy is derived from mitochondrial oxidative phosphorylation (28,38). Immortalized cell lines such as SH-SY5Y and HeLa cells, as well as cells such as fibroblasts, which also exhibit robust depolarization-induced Parkin–mitophagy (16), instead preferentially utilize glycolysis and lactic acid generation as the primary pathway for ATP generation when cultured in glucose-containing media (24,26). We discovered that when HeLa cells were forced into dependence on mitochondrial oxidative phosphorylation (more similar to neurons), they failed to recruit Parkin to their mitochondria following CCCP depolarization, unlike the robust Parkin–mitochondrial accumulation after depolarization in glycolytic HeLa cells. These results suggest that cellular bioenergetics play a role in dictating Parkin recruitment, and possibly subsequent mitophagy.
As expected, both neurons and HeLa cells dependent on mitochondrial oxidative phosphorylation were unable to fully maintain ATP levels after mitochondrial depolarization with CCCP. Partial protection of the depolarization-induced ATP loss by the addition of oligomycin increased Parkin recruitment to neuronal mitochondria to a modest, though significant degree. However, it still did not lead to the robust Parkin recruitment observed in cell lines, nor was robust evidence of mitophagy observed. This suggests that ATP depletion might partially, but not completely, explain the differences observed in neurons and oxidative phosphorylation-dependent HeLa cells compared with other cells.
The partial influence of ATP levels is interesting, in that it could provide a means in neurons by which low-level individual mitochondrial dysfunction, within the setting of otherwise-functioning, ATP-producing mitochondria, might be regulated through the Parkin–mitophagy pathway. It remains to be seen whether, in fact, individually depolarized mitochondria would undergo Parkin-associated mitophagy in the setting of otherwise normal neuronal mitochondria. On the other hand, even individually depolarized mitochondria might result in very localized changes in ATP levels, particularly in axons, where mitochondria are more sparsely distributed, and where it has been hypothesized that individual docked mitochondria might contribute to localized changes in ATP levels (39). This could be a means for inhibiting Parkin-associated mitophagy in subcellular regions like axons where mitochondria are necessary. Although it is possible that the partial loss of ATP could simply be preventing energy-dependent enzymatic activity in the Parkin–mitophagy pathway from occurring in both neurons and other cells forced to depend on oxidative phosphorylation, our data suggest that this is not a complete explanation.
Other regulatory mechanisms may exist in neurons that tightly regulate this pathway, and which may be linked to bioenergetics. We do observe some evidence of Parkin-associated mitophagy in neurons (Fig. 8F, H, I), suggesting that this pathway does function in these cells. In our studies, however, it is not triggered or upregulated by widespread mitochondrial depolarization. In addition, on an individual basis, many cells exhibited Parkin–mitochondrial accumulation without evidence of colocalization with GFP-LC3, and conversely, some cells with evidence of mitophagy (GFP-LC3 and mitochondrial colocalization) showed no associated Parkin–mitochondrial colocalization (Figs 1G and 8J). While we realize that GFP-LC3 colocalization with mitochondria is only a single measure of mitophagy, and there are caveats to our evaluation, within our measures and time points of detection we were unable to detect an association of mitophagy with Parkin translocation in neurons, whereas we can readily detect this association in other cell types. These data suggest additional mechanisms of mitophagy may be independent of Parkin translocation to mitochondria, consistent with the ability of Parkin to promote compensatory mitophagy in PINK1-deficient cells (22,40).
It remains to be determined what other regulatory mechanisms might exist for mitophagy, but it will be important to understand this more fully specifically in neurons to better elucidate the role of Parkin-regulated mitochondrial homeostasis in neurodegeneration. Such unique regulation would not be unexpected, as it has been suggested that high levels of mitophagy may contribute to cell death in neurons (31). We propose that this bioenergetic regulation is a protective mechanism—that unlike HeLa cells, which are able to eliminate their entire complement of mitochondria following extended CCCP exposure (16,17), neurons are completely dependent on maintaining their mitochondrial population. Thus, it would not be surprising that mitophagy would be a very tightly controlled and more rare event in neurons, like other aspects of mitochondrial dynamics (41). In fact, there is precedence for this, as it has been shown that yeast mitochondria are prevented from undergoing mitophagy when mitochondria are required for cellular metabolism (27). In neurons, there may be more importance placed on repair of mitochondria, and this could be one of the reasons why mitochondrial fusion and fission are so critical to neuronal health (3,4).
Our studies differ from two previous studies, which, while mainly detailing Parkin-induced mitophagy using cell lines and other cell types, report briefly on neuronal populations. Vives-Bauza et al. (20) report an increase in Parkin–mitochondria localization in primary neurons from PINK1-expressing, but not PINK1 (−/−) mice following depolarization, although this is not well detailed. In addition, Narendra et al. (16) reported immunoblot detection of increased Parkin in the crude mitochondrial fraction of cortical neurons treated with CCCP, but do not otherwise detail neuronal findings. We are not certain as to why our results differ, but we have tested multiple conditions carefully, evaluated different neuronal populations and ensured our observations were not due to species differences or technique. Similar to these and other studies, we did find that overexpression of PINK1 increased Parkin accumulation on mitochondria, suggesting that a mechanistic link between PINK1 and Parkin is present in neurons. We did not find, however, that PINK1 overexpression sensitized neurons to depolarization-induced Parkin translocation or subsequent mitophagy.
It is also possible that Parkin is involved in other aspects of neuronal mitochondrial quality control. Many studies have implicated PINK1 and Parkin in a pathway that regulates mitochondrial fission and fusion (10,11,14,40,42,43), although regulation appears to vary depending on cell type (reviewed in 5). It is not presently clear whether the functions of regulating fission/fusion and initiating mitophagy are separate or interrelated pathways for both PINK1 and Parkin, although some studies suggest they may be related (9,40). Thus, PINK1 and Parkin may be influencing other aspects of mitochondrial dynamics and homeostasis in neurons beyond mitophagy regulation, and not all of these functions may depend upon stable association of Parkin with mitochondria.
The role of PINK1 and Parkin in targeting mitochondria to mitophagy has elucidated a novel pathway of mitochondrial homeostatic control. However, our studies reveal that this pathway may be regulated differently in neurons, due to their dependence on mitochondrial energy production. Our study emphasizes the importance of neuron-specific bioenergetics in studies of mitochondrial function in neurological disease.
MATERIALS AND METHODS
Cortical neuron culture
Primary cortical neurons were derived from E18 Sprague–Dawley rats utilizing previously described methods (44) with minor modifications. Briefly, cortices from E18 rats were dissected on ice in HBSS, then pooled and treated with papain enzyme solution (<200 U) followed by soybean trypsin inhibitor. Cells were washed, triturated and plated at a density of 3 × 105 per ml on glass cover slips or plastic culture dishes coated with poly-d-lysine and mouse laminin. Cells were seeded in neurobasal medium (NBM) containing Penicillin/Streptomycin (Pen/Strep), 2% glutamax, B27 supplement and 5% fetal calf serum and kept overnight. The following day, media were replaced with serum-free NBM (SF-NBM) supplemented with Pen/Strep, 2% glutamax and B27 supplement, and maintained with 1/2 media changes every 3 days.
Cortical neuron transfection and treatment
Transfection was performed using Lipofectamine 2000 according to the manufacturer's instructions with modifications. Briefly, a ratio of 0.8–1.0 µg of total DNA/1 µl lipofectamine pre-incubated in Optimem was combined with transfection media (MEM pH 7.4, supplemented with 2% glutamax, 20 mm HEPES, 33 mm glucose, 1 mm Na-pyruvate) on DIV 6. Cells were incubated at 37°C in a non-CO2 incubator for 2 h, then media were replaced with a mixture of conditioned neuronal media and fresh supplemented SF-NBM, half each. Cells were transfected with plasmids expressing mtDsRed2 (Clontech), hu-Parkin [a gift from Dr Matthew Farrer, Mayo Clinic Jacksonville (45)], GFP-tagged human PINK1 (a gift from Dr Scott Kulich, Veterans Affairs Hospital, University of Pittsburgh; originally purchased from Genecopoeia), GFP-tagged LC3 (a gift from Dr Tamotsu Yoshimori, Research Institute of Microbial Diseases, Osaka University, Japan) and/or empty control vector pcDNA3 plasmid. CCCP or DMSO vehicle control treatments were performed 24 or 72 h following transfection, at 7 or 10 DIV, with the exception of oligomycin and creatine treatments, in which cells were pretreated with creatine (1 mm) or control media for 24 h starting at 72 h following transfection, then treated with oligomycin (10 µm in DMSO), CCCP or control. For treatments, cells received 1/2 media changes with 2× concentrations of CCCP or equivalent DMSO vehicle.
Cell line culture, transfection and treatment
The dopaminergic PC12 cell subline, PC6-3, was generously provided by Dr Bruce Pittman (35). Naïve cells were maintained in DMEM-F12 with 10% horse serum, 5% FBS and Pen/Strep. For differentiation, cells were plated at 12 000 cells/ml on Collagen IV (Trevigen) coated glass cover slips. Differentiation media consisted of RPMI media supplemented with 1% heat-inactivated horse serum; 0.5% FBS; insulin, transferrin and selenium supplement (final concentrations: 5.0 µg/ml insulin, 5.0 µg/ml transferrin, 5.0 ng/ml sodium selenite; Lonza) and 40 ng/ml NGF (BD Biosciences). Cell culture media were replenished by 1/2 every 3 days. Cells were found to be differentiated based on neuronal outgrowth and arrest of proliferation at 6 days of NGF. Cells were transfected with Lipofectamine according to the manufacturer's instructions in Optimem using plasmids and DNA/lipofectamine ratios described above on day 3 of differentiation. Cells were treated with CCCP or vehicle control on day 8 after differentiation plating (96 h after transfection). SH-SY5Y and HeLa cells were maintained in DMEM with 10% FBS and Pen/Strep, and plated at 1.7 × 105 cells/ml and 50.000 cells/ml, respectively. Cells were transfected as described above for PC6-3 cells at 48 h after plating, and treated with CCCP or vehicle control at 72 h (SH-SY5Y) or 48 h (HeLa) after transfection.
Immunocytochemistry
Cells cultured on glass cover slips were fixed using 4% paraformaldehyde/4% sucrose for 20 min at RT, followed by three rinses with PBS. Fixed cells were permeabilized with 0.2% Triton-100X in PBS, then blocked with 5% goat serum. Primary antibodies used included PRK8 mouse-anti-parkin (1:40; Santa Cruz) or mouse-anti-GFP (1:2000; Chemicon), and were detected using AlexaFluor 647-conjugated goat-anti-mouse IgG2 secondary (1:500; Invitrogen). Cover slips were rinsed in PBS and water, then mounted using Fluoromount-G mounting media (SouthernBiotech).
Mitochondrial isolation and western blot
Neurons for western blot were plated on and collected from 10 cm culture dishes. Following treatment, DIV10 neurons were scraped off the plates in cold PBS, then pelleted by centrifugation at 2000g. Cell pellets were washed, cells repelleted and then supernatant removed. The resulting cell pellets then underwent one of two methods to isolate the mitochondrial-enriched fraction. Pellets were subjected to differential centrifugation using the Pierce Mitochondria Isolation Kit for Cultured Cell (following manufacturer's instructions), or using a previously described method that utilizes no detergents (33). Cells were disrupted using Knotes 1 ml glass tissue homogenizers with Teflon pestles. Following the respective isolation procedures, the resulting mitochondrial-enriched pellets were immediately resuspended and lysed in a small volume of urea/CHAPS lysis buffer [9 m urea, 2% CHAPS, in 30 mm Tris, with 1x protease inhibitor cocktail (Sigma)]. Protein concentrations were determined by the Bradford method (46) and samples stored at −80°C until use. Samples were diluted in a reducing sample buffer and boiled prior to use. Protein samples were then subjected to SDS–PAGE using Peirce pre-cast tricine gels and transferred to polyvinylidene fluoride using a BioRad SemiDry Transfer apparatus. Western blots of the gels were then probed using PRK8 mouse-anti-parkin (1:500; Santa Cruz), mouse-anti-HSP60 (1:1000; Stressgen) and/or mouse-anti-cytochrome C oxidase subunit IV (1:25,000; AbCam). LiCor Oddessy compatible IR680- and IR800-conjugated goat-anti-mouse secondaries (LiCor) were used for detection, and blots were imaged and analyzed using a LiCor Odyssey system.
Imaging and quantification
All immunocytochemical images were captured using an Olympus Fluoview 1000 confocal microscope (×60 oil immersion lens, NA:1.42) at room temperature. Images of individual cells were taken as z-stacks (0.5 μm slices, 640 × 640 pixel resolution, 20 µs/pixel) encompassing the entire depth of the cell, using sequential laser imaging and Kalman filter correction. Cells were imaged randomly from across each cover slip, and 10–30 cells/cover slip were imaged. Images were blinded to condition, and cells were analyzed throughout each z-plane using Olympus Fluoview Viewer software. For analysis of parkin–mitochondrial localization, the presence of Parkin accumulation on mitochondria was determined as any cell exhibiting at least one fluorescent mitochondrion with clearly colocalized accumulation of parkin fluorescence. For manuscript images, acquired images were adjusted for brightness and/or contrast only prior to exportation from Olympus Fluoview Viewer software at TIFF images, and then compiled into figures using Adobe Photoshop CS4 software.
ATP measurements
Treated cells were washed and harvested by scraping into ice-cold PBS, pelleted by centrifugation at 2000g for 5 min, and resuspended in urea/CHAPS lysis buffer as described above. Samples were briefly centrifuged to remove insoluble material and supernatants stored at−80°C until analysis. Intracellular ATP levels were determined using an ATP Determination Kit (Molecular Probes) based on the reaction of luciferase with luciferin, using manufacturer procedures. Luminescence was measured using a L Max II Luminometer (Molecular Devices), and data were normalized to individual sample protein concentration determined with the Bradford assay (46).
Statistics
Significance was determined using one-way ANOVA or z-test of proportions, where appropriate, followed by post-hoc analysis (Student's t-tests with Bonferroni correction or Newman–Keuls test) using alpha = 0.05. Both GraphPad PRISM and Microsoft Excel software programs were utilized for data analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by the American Parkinson Disease Association (to S.B.B.); National Institutes of Health (K08NS059576 to S.B.B., R01AG026389 and 1R56NS065789 to C.T.C., and NS07391 to V.S.V.); and the 2010 Ellison Medical Foundation/American Federation for Aging Research (to V.S.V.).
ACKNOWLEDGEMENTS
We would like to thank R. Dagda, J.T. Greenamyre and T.G. Hastings for helpful insight and manuscript comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Beal M.F. Mitochondria and neurodegeneration. Novartis Found. Symp. 2007;287:183–192. doi: 10.1002/9780470725207.ch13. discussion 192-186. [DOI] [PubMed] [Google Scholar]
- 2.Schapira A.H. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet. Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen H., Chan D.C. Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knott A.B., Bossy-Wetzel E. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration. Ann. N. Y. Acad. Sci. 2008;1147:283–292. doi: 10.1196/annals.1427.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Laar V.S., Berman S.B. Mitochondrial dynamics in Parkinson's disease. Exp. Neurol. 2009;218:247–256. doi: 10.1016/j.expneurol.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitworth A.J., Pallanck L.J. The PINK1/Parkin pathway: a mitochondrial quality control system? J. Bioenerg. Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- 7.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 8.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 9.Cherra S.J., 3rd, Dagda R.K., Tandon A., Chu C.T. Mitochondrial autophagy as a compensatory response to PINK1 deficiency. Autophagy. 2009;5:1213–1214. doi: 10.4161/auto.5.8.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poole A.C., Thomas R.E., Andrews L.A., McBride H.M., Whitworth A.J., Pallanck L.J. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lutz A.K., Exner N., Fett M.E., Schlehe J.S., Kloos K., Lammermann K., Brunner B., Kurz-Drexler A., Vogel F., Reichert A.S., et al. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J. Biol. Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palacino J.J., Sagi D., Goldberg M.S., Krauss S., Motz C., Wacker M., Klose J., Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 13.Flinn L., Mortiboys H., Volkmann K., Koster R.W., Ingham P.W., Bandmann O. Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio) Brain. 2009;132:1613–1623. doi: 10.1093/brain/awp108. [DOI] [PubMed] [Google Scholar]
- 14.Deng H., Dodson M.W., Huang H., Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exner N., Treske B., Paquet D., Holmstrom K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H.H., Gasser T., et al. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 2007;27:12413–12418. doi: 10.1523/JNEUROSCI.0719-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 18.Kawajiri S., Saiki S., Sato S., Sato F., Hatano T., Eguchi H., Hattori N. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas K.J., Cookson M.R. The role of PTEN-induced kinase 1 in mitochondrial dysfunction and dynamics. Int. J. Biochem. Cell Biol. 2009;41:2025–2035. doi: 10.1016/j.biocel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dagda R.K., Chu C.T. Mitochondrial quality control: insights on how Parkinson's disease related genes PINK1, parkin, and Omi/HtrA2 interact to maintain mitochondrial homeostasis. J. Bioenerg. Biomembr. 2009;41:473–479. doi: 10.1007/s10863-009-9255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu C.T. Tickled PINK1: mitochondrial homeostasis and autophagy in recessive Parkinsonism . Biochim. Biophys. Acta. 2010;1802:20–28. doi: 10.1016/j.bbadis.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gogvadze V., Zhivotovsky B., Orrenius S. The Warburg effect and mitochondrial stability in cancer cells. Mol. Aspects Med. 2010;31:60–74. doi: 10.1016/j.mam.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Reitzer L.J., Wice B.M., Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J. Biol. Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 26.Rossignol R., Gilkerson R., Aggeler R., Yamagata K., Remington S.J., Capaldi R.A. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 27.Kanki T., Klionsky D.J. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolanos J.P., Almeida A., Moncada S. Glycolysis: a bioenergetic or a survival pathway? Trends Biochem. Sci. 2010;35:145–149. doi: 10.1016/j.tibs.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold B., Cassady S.J., Vanlaar V.S., Berman S.B. Integrating multiple aspects of mitochondrial dynamics in neurons: age-related differences and dynamic changes in a chronic rotenone model. Neurobiol. Dis. 2011;41:189–200. doi: 10.1016/j.nbd.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue L., Fletcher G.C., Tolkovsky A.M. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol. Cell. Neurosci. 1999;14:180–198. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 32.Emery D.G., Lucas J.H. Ultrastructural damage and neuritic beading in cold-stressed spinal neurons with comparisons to NMDA and A23187 toxicity. Brain Res. 1995;692:161–173. doi: 10.1016/0006-8993(95)00726-7. [DOI] [PubMed] [Google Scholar]
- 33.Dukes A.A., Van Laar V.S., Cascio M., Hastings T.G. Changes in endoplasmic reticulum stress proteins and aldolase A in cells exposed to dopamine. J. Neurochem. 2008;106:333–346. doi: 10.1111/j.1471-4159.2008.05392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taymans J.M., Van den Haute C., Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. J. Neurochem. 2006;98:951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- 35.Pittman R.N., Wang S., DiBenedetto A.J., Mills J.C. A system for characterizing cellular and molecular events in programmed neuronal cell death. J. Neurosci. 1993;13:3669–3680. doi: 10.1523/JNEUROSCI.13-09-03669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gusdon A.M., Chu C.T. To eat or not to eat: neuronal metabolism, mitophagy and Parkinson's disease. Antioxid. Redox Signal. 2010 doi: 10.1089/ars.2010.3763. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budd S.L., Nicholls D.G. A reevaluation of the role of mitochondria in neuronal Ca2+ homeostasis. J. Neurochem. 1996;66:403–411. doi: 10.1046/j.1471-4159.1996.66010403.x. [DOI] [PubMed] [Google Scholar]
- 38.Nicholls D.G., Johnson-Cadwell L., Vesce S., Jekabsons M., Yadava N. Bioenergetics of mitochondria in cultured neurons and their role in glutamate excitotoxicity. J. Neurosci. Res. 2007;85:3206–3212. doi: 10.1002/jnr.21290. [DOI] [PubMed] [Google Scholar]
- 39.Cai Q., Sheng Z.H. Mitochondrial transport and docking in axons. Exp. Neurol. 2009;218:257–267. doi: 10.1016/j.expneurol.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dagda R.K., Cherra S.J., 3rd, Kulich S.M., Tandon A., Park D., Chu C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berman S.B., Chen Y.B., Qi B., McCaffery J.M., Rucker E.B., 3rd, Goebbels S., Nave K.A., Arnold B.A., Jonas E.A., Pineda F.J., et al. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J. Cell Biol. 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui M., Tang X., Christian W.V., Yoon Y., Tieu K. Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J. Biol. Chem. 2010;285:11740–11752. doi: 10.1074/jbc.M109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park J., Lee G., Chung J. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem. Biophys. Res. Commun. 2008;378:518–523. doi: 10.1016/j.bbrc.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 44.Ghosh A., Greenberg M.E. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- 45.Petrucelli L., O'Farrell C., Lockhart P.J., Baptista M., Kehoe K., Vink L., Choi P., Wolozin B., Farrer M., Hardy J., et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36:1007–1019. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- 46.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.