Abstract
We recently reported that mutations in the widely expressed nuclear protein TOPORS (topoisomerase I-binding arginine/serine rich) are associated with autosomal dominant retinal degeneration. However, the precise localization and a functional role of TOPORS in the retina remain unknown. Here, we demonstrate that TOPORS is a novel component of the photoreceptor sensory cilium, which is a modified primary cilium involved with polarized trafficking of proteins. In photoreceptors, TOPORS localizes primarily to the basal bodies of connecting cilium and in the centrosomes of cultured cells. Morpholino-mediated silencing of topors in zebrafish embryos demonstrates in another species a comparable retinal problem as seen in humans, resulting in defective retinal development and failure to form outer segments. These defects can be rescued by mRNA encoding human TOPORS. Taken together, our data suggest that TOPORS may play a key role in regulating primary cilia-dependent photoreceptor development and function. Additionally, it is well known that mutations in other ciliary proteins cause retinal degeneration, which may explain why mutations in TOPORS result in the same phenotype.
INTRODUCTION
Progressive dysfunction and consequent death of cone and rod photoreceptors in retinal degenerative diseases constitute a major cause of adult blindness. The retina provides extremely sensitive light detection, which is based on the integrity of a highly vulnerable cell—the photoreceptor. Retinal degeneration is genetically the most heterogeneous disease known, with mutations in 146 genes identified to date with an additional 38 loci genetically mapped but the respective genes not yet found. It is arguable that mutations in genes affecting almost every aspect of photoreceptor structure or function can trigger retinal degeneration (1,2).
Among the retinal degenerations, retinitis pigmentosa (RP) is clinically and genetically the most heterogeneous, with an incidence of almost 1 in 3500 people worldwide. Affected individuals suffer from a progressive degeneration of the photoreceptors, eventually resulting in severe visual impairment. The mode of inheritance of RP can be autosomal dominant (adRP), recessive, X-linked or digenic (3). To date, 20 causative genes have been identified for adRP (RetNet). The products of these genes are associated with diverse functions, including photoreceptor structure, ciliary transport, phototransduction and gene expression, e.g. transcription and mRNA splicing.
Recently, we identified a novel gene mapping to chromosome 9p21.1 causing adRP (RP31) (4). The gene encoding topoisomerase I–RS protein (TOPORS, MIM 609923, NM_005802) was found to be mutated in two families of French-Canadian and German origin. Both mutations, a heterozygous 1 bp insertion (c.2474_2475insA) and a 2 bp deletion (c.2552_2553delGA) in exon 3 lead to premature stop codons, p.Tyr825fs and p.Arg851fs, respectively (4). Two additional TOPORS mutations were later found to cause frameshift and termination of the protein prematurely—c.2569delA (p.Arg857GlyfsX9) and c.G2422T (p.Glu808X) (5). A nucleotide substitution c.1205A>C, leading to missense change (p.Q402P), has also been identified in a family with autosomal dominant pericentral retinal dystrophy (6). Prevalence studies indicate that mutations in TOPORS is a rare cause of retinal degeneration and likely to account for 1–2% of adRP.
TOPORS was originally identified in a screen for proteins that bind to the N-terminus of topoisomerase I (7), and it interacts with p53 (8). TOPORS is a tumor-suppressor protein and functions as an E3 ligase for both ubiquitin and small ubiquitin-like modifier 1 (SUMO-1) (9,10). It was recently reported that DNA-damage-inducible SUMOylation of IKK-related kinase—IKK3ε—depends on TOPORS (11). Cumulatively, these studies indicate that TOPORS is a ubiquitously expressed gene, encoding essentially a nuclear protein showing multi-functional character with function that varies according to cell type. The multi-functional nature of this protein may be also related to the different protein domains of TOPORS (4). Although extensive studies have helped define the function of TOPORS in cultured cells, and despite being widely expressed, mutations in TOPORS are only known to cause adRP. Mutations in tissue-specific genes leading to tissue-specific phenotypes define common mechanisms in understanding human genetic diseases. However, TOPORS mutations are beginning to highlight a small group of unique genes that are widely expressed and yet cause a tissue-specific phenotype (such as splicing factor mutations and adRP) (12). This could be due to a unique role for TOPORS in the retina and therefore we may see an altered subcellular localization in the retina, away from the photoreceptor nucleus. Conversely, an altered localization may confer a novel role for TOPORS in the retina.
In the absence of information regarding subcellular localization of TOPORS in the retina of any organism, it is critical to perform a comprehensive study, which will formally establish localization of TOPORS in the retina. For this reason, we undertook this study to delineate the role of TOPORS in the vertebrate retina and to understand why mutations cause only RP. This may help elucidate its specific role in the retina and define general cellular pathways implicated in retinal degeneration, which may have wider consequences for the development of future therapies for this group of patients. The immunolocalization studies presented here show TOPORS localization to the connecting cilium (CC), between inner (IS) and outer segments (OS) of the photoreceptor cell. Mutations in other proteins such as RP1 (13) and RPGR (14) that also localize to the CC are associated with retinal degeneration, thereby providing a basis for a similar explanation for TOPORS mutations causing only RP. All previously published work involving non-retinal cell types demonstrated nuclear localization supporting a nuclear role for TOPORS.
RESULTS
TOPORS localizes to photoreceptor cilia
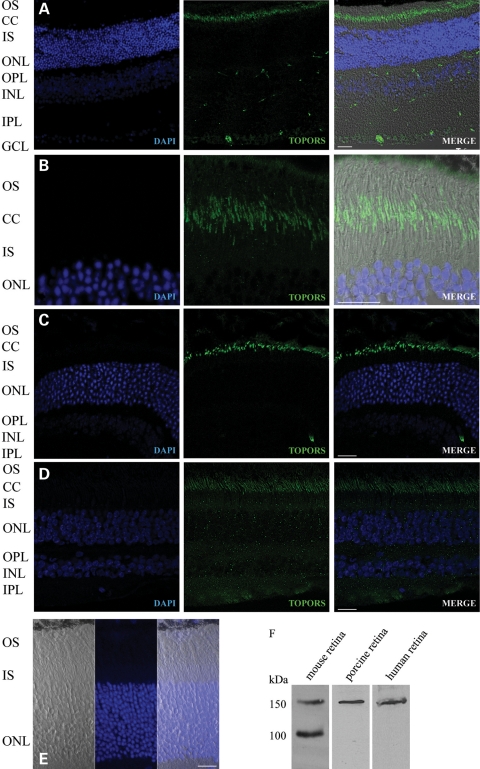
To determine the cellular localization of TOPORS in mammalian retina, we performed indirect immunofluorescence staining of cryosections of adult mouse, porcine and human retinas using an anti-TOPORS antibody. The specificity of the anti-TOPORS antibody was extensively checked in these mammalian species and as expected yielded a consistent band of 150 kDa (Fig. 1F). In all species, we observed TOPORS immunoreactivity predominantly at the junction between photoreceptor IS and OS, called the connecting cilium (Fig. 1A–D). In addition, TOPORS immunostaining was detected in the nuclei of retinal ganglion cells.
Figure 1.
Localization of TOPORS in the retina. (A–D) Indirect immunofluorescence analysis of mouse, porcine and human retina sections using anti-TOPORS antibody (green). (A) TOPORS is localized only in the ciliary region of the photoreceptors and in the nuclei of the ganglion cells in the mouse retina. (B) Higher magnification only of the photoreceptor cell layer of the mouse retina. (C) Immunostaining of anti-TOPORS antibody in the porcine retina. (D) Immunostaining of anti-TOPORS antibody in the human retina. (E) As a negative control, the mouse section was probed with TOPORS antibody blocked with TOPORS recombinant protein (206 amino acid), where no signal could be observed. (F) Immunoblot analysis of the specificity of mouse anti-TOPORS antibody shows a 150 kDa signal detected in mouse, porcine and human retina lysates, where, in the mouse, two bands could be seen with the molecular weight of 100 and 150 kDa due to a specific smaller isoform in this species lacking exon 2 of the gene. OS, outer segment; CC, connecting cilium; IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Nuclei are stained with DAPI (blue). Scale bar: 10 μm.
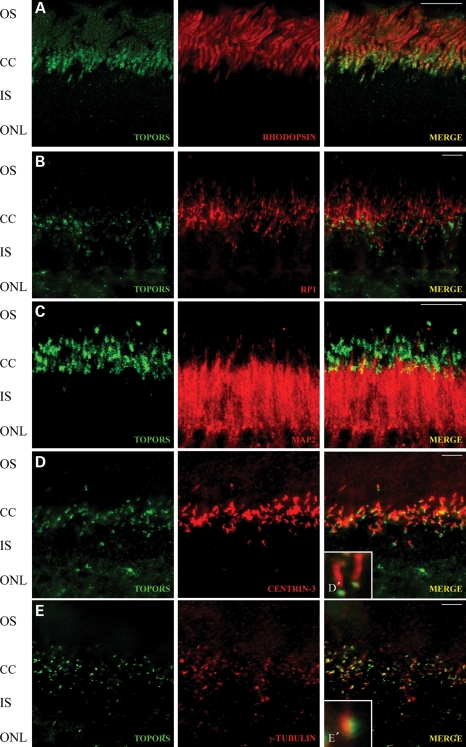
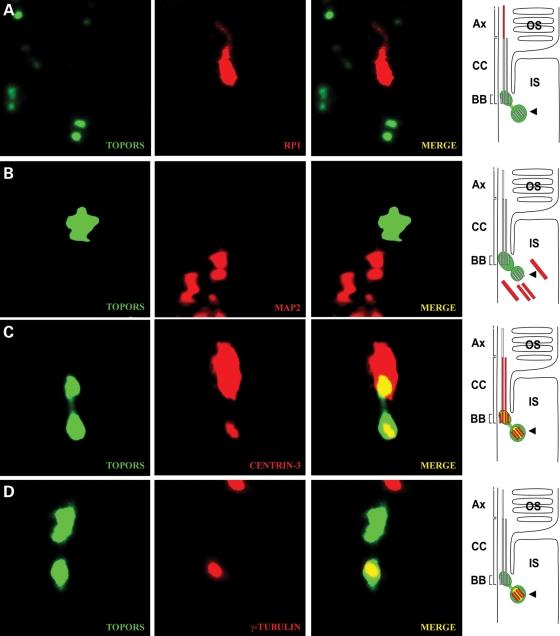
We next sought to define more precisely the spatial distribution of TOPORS along the photoreceptor cilium. We employed double-immunostaining of retinal cryosections with anti-TOPORS antibody and molecular markers for the ciliary subcompartment: anti-RP1 as a marker for the ciliary axoneme extending from the transition region (13); anti-rhodopsin and anti-MAP2, which stain the OSs and the microtubules in the ISs (15); anti-centrin-3 to visualize the transition zone, basal body and adjacent centriole (16); and anti-γ-tubulin as a marker for basal body. Detailed examination of the photoreceptor cilium by high-resolution immunofluorescence microscopy (17,18) revealed that anti-TOPORS antibody stains a pattern of two adjacent dot-like structures (Figs. 2 and 3A). This TOPORS staining did not overlap the rhodopsin, RP1 and MAP2 staining (Fig. 2A–C), thus excluding its presence at the OS, axoneme and IS of photoreceptors. Proximal TOPORS staining co-localized with centrin-3 (Fig. 2D) and γ-tubulin (Fig. 2E), demonstrating the localization of TOPORS to the basal body of the CC of the photoreceptor cells. In comparison with the centriole markers, anti-TOPORS stained a broader dot indicating additional presence of TOPORS in the pericentriolar region (Fig. 3A–D). In conclusion, our detailed inspection of the subciliary molecular distribution in retinal photoreceptor cells revealed specific localization of TOPORS in the basal body of the CC and its daughter centriole.
Figure 2.
Subcellular localization of TOPORS at the ciliary region of the photoreceptor cell. (A–E) Indirect immunofluorescence analysis of mouse retina sections stained with anti-TOPORS antibody (green) with ciliary proteins and proteins of the outer and inner photoreceptor segments (red) in the retina. (A) Photoreceptor localization of TOPORS with the most abundant protein in these cells in the area of the outer segment—rhodopsin. (B) Subcellular localization of TOPORS and RP1 as a marker for photoreceptor ciliary axoneme. TOPORS is localized at the region of the connecting cilium (CC) of the photoreceptor cells. (C) Double-labeling with anti-TOPORS and anti-MAP2. Antibodies against TOPORS stain the CC region. MAP2 is present in the inner segment (IS) of the photoreceptor cell and slightly at the CC region. (D) Indirect immunofluorescence double-labeling with antibodies against TOPORS and the marker of the CC, basal body (BB) and centriole centrin (Centrin-3) in cryosections through mouse retinas. (D′) Higher magnification of merged signal (TOPORS and Centrin-3) from the CC of a single photoreceptor cell. (E) Double-staining with anti-TOPORS and anti-γ-tubulin. TOPORS is localized at the cilium of the photoreceptor cell. Anti-γ-tubulin stains the basal body. (E′) Higher magnification of merged signal of TOPORS and γ-tubulin, which partly co-localizes in the basal body of the CC of a single photoreceptor cell. Scale bar: 10 μm. OS, outer segment; CC, connecting cilia; IS, inner segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer.
Figure 3.
TOPORS is localized at the basal body and centriole of the photoreceptor cell. (A–D) Indirect immunofluorescence double-staining with antibodies against TOPORS and marker proteins for the ciliary region of cryosections in mouse retinas. High-magnification images of double-immunofluorescences of the ciliary region of photoreceptor cells and schematic presentations (on the left) illustrating subciliary localization of TOPORS (green) and marker molecules (red). (A) Anti-TOPORS and anti-RP1 immunolabeling. RP1 is used as a marker indicating the ciliary axoneme (Ax). TOPORS and RP1 do not co-localize. (B) Anti-TOPORS and anti-MAP2 immunolabeling. MAP2 stains the microtubules in the inner segment (IS) of the photoreceptor cells. (C) Centrin-3 labels connecting cilium (CC), basal body (BB) and centriole (arrowhead). According to this centrin-3 labeling, TOPORS is present at the BB and at the centriole (arrowhead). (D) Anti-TOPORS and anti-γ-tubulin immunostaining. γ-Tubulin stains the centriole (arrowhead). TOPORS partly co-localize with γ-tubulin at the BB. OS, outer segment of the photoreceptor cell; IS, inner segment of the photoreceptor cell.
TOPORS co-immunoprecipitates with ciliary-centrosomal proteins in retina
We next examined the interaction of TOPORS with cilia-centrosomal proteins by co-immunoprecipitation in bovine retina extracts. First, we looked at proteins implicated in retinal degeneration and known to localize to the connecting cilia. TOPORS antibody did not immunoprecipitate RPGR–ORF15 (14,19), CEP290/NPHP6 (20), RP1 (13) (Supplementary Material, Fig. S1A–C), pericentrin (21) or centrin (16,22), which are part of the ciliary apparatus of the photoreceptor cells. We found co-immunoprecipitation with centrosomal/basal body protein γ-tubulin (Supplementary Material, Fig. S1D–F). This prompted co-immunoprecipitation experiments with proteins involved in intraflagellar transport (IFT). We consistently detected the association of TOPORS with dynein–dynactin complex proteins (p150glued, p50-dynamitin and cytoplasmic dynein intermediate chain, DIC) that are part of the retrograde transport mechanism (23). Little or no interaction of TOPORS was observed with anterograde motor kinesin-II complex (KIF3A, KAP3 and IFT88) (Supplementary Material, Fig. S1G–L). Reverse co-immunoprecipitation experiments did not reveal TOPORS immunoreactive bands, probably due to its lower abundance in these complexes (data not shown).
TOPORS localizes to primary cilia in cell lines
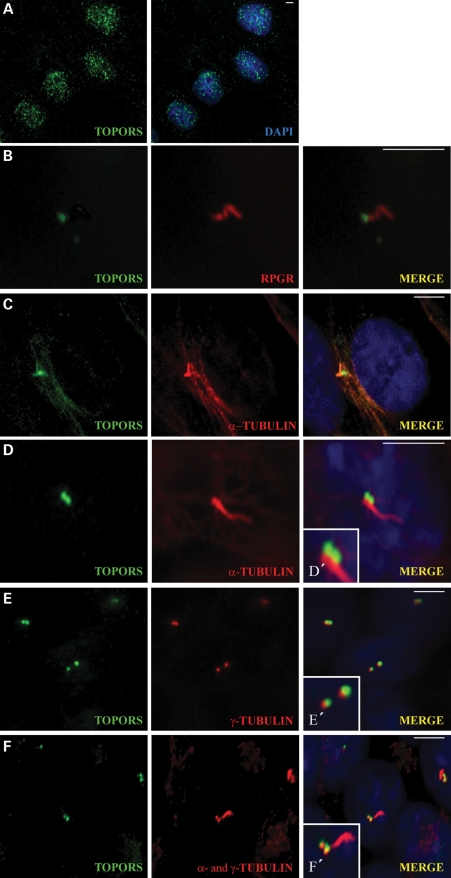
It is well known that many of the connecting cilia proteins are localized in the cilia of non-dividing cells and in the centrosomes of mitotic cells (24). For these reasons, we undertook detailed investigations of TOPORS localization in cell lines. In dividing MDCK cells, TOPORS exhibits the nuclear localization in small speckles (Fig. 4A) as reported previously (7,25). However, in non-dividing ciliated MDCK cells, the staining of endogenous TOPORS is detected only in the basal body, at the base of the cilium (Fig. 4B). The immunostaining was performed using anti-RPGR antibody. Although both proteins appeared in the ciliary region, TOPORS is proximal to RPGR but shares a small region of overlap at the basal body in this particular cell type. Localization of TOPORS to the base of the primary cilium was also detected in ARPE-19 (Fig. 4C) and IMCD3 (Fig. 4D) cell lines.
Figure 4.
TOPORS localizes to the basal body of the primary cilia and associates with microtubule proteins in ciliated cells. (A) Endogenous TOPORS (green) localized in the nucleus stained with DAPI (blue) of mitotic MDCK cells. (B) TOPORS (green) localizes to the base of the ciliary axoneme stained with RPGR (red) in ciliated MDCK cells. (C) In ciliated ARPE-19 cells, TOPORS (green) localizes to the base of the cilium; the ciliary axoneme is stained with α-tubulin (red). (D) In ciliated IMCD3 cells, TOPORS (green) localizes to the base of the cilium; the ciliary axoneme is stained with α-tubulin (red). (D′) Higher magnification of the ciliary structure. (E) Double-staining of ciliated IMCD3 cells with TOPORS (green) and γ-tubulin (red) used as a marker for the basal bodies. Both proteins localize in the same area with some overlap but do not co-localize entirely (see also Supplementary Material, Fig. S3). (E′) Higher magnification of the basal body. (F) Ciliated IMCD3 cells stained with TOPORS (green), a- and γ-tubulin (both in red), highlighting the basal bodies and ciliary axoneme. (F′) Higher magnification of the ciliary apparatus. Nuclei are stained with DAPI. Scale bar: 10 μm.
To confirm the localization of TOPORS to the basal body, we double-immunostained ciliated IMCD3 cells with anti-TOPORS and anti-γ-tubulin antibodies (Fig. 4E). As expected, γ-tubulin was found at the basal body. TOPORS localized to the basal bodies but did not overlap entirely with γ-tubulin. Triple immunostaining using anti-TOPORS, anti-α-tubulin and anti-γ-tubulin antibodies (Fig. 4F) showed TOPORS at the base of the cilia (with γ-tubulin), and α-tubulin along the length of the cilia axoneme.
Cell-cycle-dependent localization of TOPORS to the centrosomes
As TOPORS was previously reported to be a nuclear protein (7,25) and our results with ciliated cells show a predominant cilia staining of the ciliary apparatus, namely the basal body and the adjacent centriole, we hypothesized that TOPORS might exhibit dynamic association with cilia and the nucleus. When expressed as a GFP fusion in RPE-1 cells, TOPORS showed localization to the nucleus and also to the centrosome (Fig. 5A). This allowed independent verification of TOPORS localization using the anti-GFP antibody. The control experiment using only the GFP vector showed diffused staining throughout the cell cytoplasm, whereas TOPORS-GFP localization was restricted to nuclei and centrosomes.
Figure 5.
Localization of TOPORS during cell cycle. (A) RPE-1 cells were transfected with plasmid encoding for TOPORS-GFP and GFP alone, fixed and stained for the centrosomal marker γ-tubulin. (B and C) Synchronized RPE-1 cells were fixed and stained for endogenous TOPORS and γ-tubulin. G0 was stained using anti-acetylated α-tubulin antibody. DNA was stained with DAPI. Insets show higher magnifications of centrosome-containing regions. Representative images of each step of the cell cycle are shown. In colored images, TOPORS staining is in green, centrosomes in red, nuclei in blue and the arrow shows the mid-body. Scale bar: 10 μm.
We performed a detailed analysis of TOPORS localization during different stages of cell division using synchronized RPE-1 cells. Immunocytochemical studies demonstrated that endogenous TOPORS exhibits cell-cycle-stage-dependent localization in the nucleus, centrosomes and mid-body. During G1, G0, S and G2 phases, TOPORS localization is predominantly in the nucleus (associated with PML bodies) and in the centrioles of the centrosomes (co-localized with γ-tubulin) (Fig. 5B). During mitosis, TOPORS is enriched in the centrioles of the spindle poles (Fig. 5C). In addition to centrosome-associated staining, we detected TOPORS at the mid-body, the central part of the cytokinetic bridge in telophase, a feature shared by several other centrosomal proteins (24). Altogether, these results show that TOPORS is associated with the centrosome throughout the cell cycle.
Knockdown of topors results in developmental anomalies in zebrafish
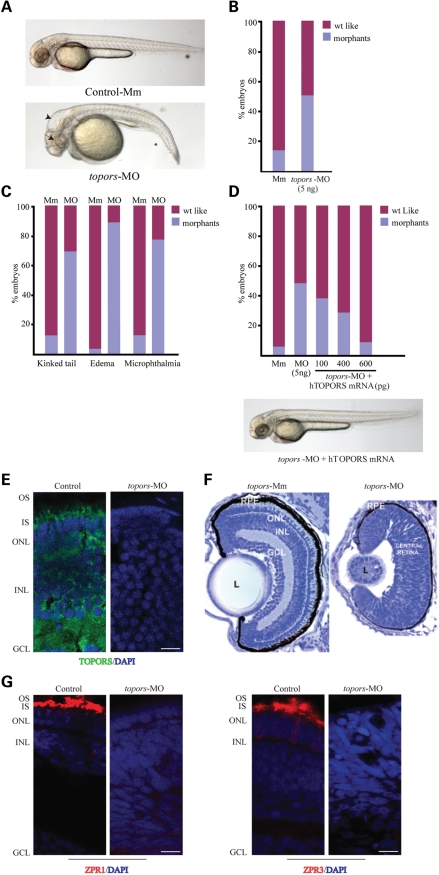
To delineate the function of TOPORS, we knocked down topors expression in zebrafish embryos. Using BLAST, we identified the human orthologue of topors in zebrafish (Accession no. XM_682803). At the protein level, zebrafish TOPORS displays 38% identity and 67% homology to human TOPORS (Supplementary Material, Fig. S2). To assess the function of TOPORS, we injected a translation-blocking (AUG) morpholino (MO) against topors at the one to eight cell stage in zebrafish embryos. Approximately 45% of embryos exhibited developmental anomalies consistent with the effect of depletion of ciliary proteins, such as kinked tail, microphthalmia, hydrocephaly and pericardial effusion at 4 days post fertilization (dpf) (Fig. 6A and B; Supplementary Material, Fig. S3). As a control, injection of a mismatch MO resulted in only ∼15% defective embryos. Further investigation of the specific MO-treated embryos revealed that microphthalmia was present in ∼80% of the morphants, whereas kinked tail and edema were observed in 70 and 85% of morphants, respectively. Similar results were obtained when a splice-blocking MO was injected in fish embryos (Fig. 6C and data not shown). These phenotypes are specific to the silencing of topors, as co-injection of the topors-MO with mRNA encoding human TOPORS was able to rescue the topors-knockdown phenotypes (Fig. 6D).
Figure 6.
Knockdown of zebrafish topors causes developmental anomalies and the effect of the inhibition of topors on zebrafish retina. (A) Injection of anti-sense morpholino (topors-MO) into zebrafish embryos results in developmental disorders, including shortened body axis and hydrocephaly. Embryos injected with the 5 base mismatch (Mm) control are also shown. Asterisk depicts kinked tail; long arrow shows microphthalmia; arrowhead indicates hydrocephaly. (B and C) Histograms showing the frequency of the occurrence of morphants. (B) Incidence of the different phenotypes observed in the MO-treated embryos compared with Mm-treated controls. (C) Results are representative of at least three independent experiments. (D) The topors-knockdown phenotype can be rescued by injecting indicated doses of human TOPORS (hTOPORS) mRNA. Data are representative of three independent experiments (n > 100). Bottom panel shows representative image of an embryo co-injected with the specific MO against topors and mRNA encoding hTOPORS. Majority of the phenotypes seem to be rescued by human TOPORS. (E) Immunohistochemistry of cryosections of 4 dpf zebrafish retina was performed using anti-TOPORS antibody (green). DAPI was used to stain the nuclei (blue). IS, inner segment; OS, outer segment; ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. (F) Retinal histology of control (Mm) and topors-MO injected (MO) embryos at 4 dpf (×40). Plastic sections from 4 dpf zebrafish embryo were stained with toludine blue. (G) Cryosections of 4 dpf zebrafish were immunostained with Zpr1 or Zpr3 antibody (red) for marking cone and rod photoreceptors, respectively. Nuclear layers are stained with DAPI (blue). Scale bar: 10 μm.
TOPORS regulates retinal development in zebrafish
We then investigated the effect of knockdown of topors on retinal development. To this end, we first assessed the extent of knockdown of TOPORS. In contrast to the staining observed in the mouse, porcine and human retinas, TOPORS immunoreactivity in the retina of zebrafish embryos treated with mismatch control was detected in all cellular layers; however, in photoreceptors, the staining was predominantly detected in the IS. No detectable signal was observed in the defective embryos, which were injected with the specific translation-blocking MO against topors (Fig. 6E). We then investigated retinal development in the defective embryos. Consistent with broad developmental defects in embryonic development, histological analysis of retinas from controls and topors-knockdown embryos revealed that depletion of topors results in lamination defects in the retina (the different layers of the retina could not be detected) as well as failure to form OSs (Fig. 6F). We then analyzed the expression of photoreceptor proteins in the topors-knockdown embryos. Cone (Zpr1) and rod (Zpr3) photoreceptor staining were absent in the defective embryos compared with robust expression in the control-treated group (Fig. 6G).
We then examined whether knockdown of topors results in cell death in the retina, by TUNEL staining of control and topors-MO-treated embryos at 3 and 4 dpf. We found that lack of topors resulted in 28–30% TUNEL-positive cells, indicating increase in apoptotic cell death compared with control-injected embryo retina (6–8% TUNEL-positive cells) (Supplementary Material, Fig. S3).
DISCUSSION
Primary (or sensory) cilia are near-ubiquitous microtubule-based organelles that regulate diverse intracellular processes. Mutations in ciliary proteins are associated with severe neurodegenerative disorders, including isolated and syndromic forms of RP, such as Bardet–Biedl syndrome (BBS), Senior–Loken syndrome and Joubert syndrome (20,26–30) and Usher syndrome (17,31,32). Photoreceptors are post-mitotic sensory neurons, which display a unique polarized structure. They are divided into a biosynthetically active IS and a photosensitive OS connected via a bridge-like structure, the connecting (or sensory) cilium, which represents the transition zone of a prototypical cilium (33,34). The OS of the photoreceptor cell is analogous to the primary ciliary structure; here in particular its highly specialized function pertains to phototransduction reliant on a number of proteins that are shuttled in and out of the OS via the CC. Owing to high trafficking demands, defects in cilia-associated proteins are associated with forms of severe retinal degeneration and blindness (13,14).
The RP associated with TOPORS is inherited as an autosomal dominant trait and likely to be due to haploinsufficiency (4). Although TOPORS is a widely expressed protein, patients with mutations in TOPORS do not exhibit a wide range of developmental anomalies, unlike the phenotype detected in topors-knockdown zebrafish embryos. We believe that the residual protein expressed in patients is able to rescue extra-retinal defects. However, as photoreceptors are highly metabolically active neurons, slight perturbations in protein trafficking pathways result in degeneration and eventually blindness. Support for this hypothesis comes from our previous observations of analysis of the function of other ciliary proteins, such as RPGR, CEP290 and RPGRIP1L (20,30,35).
So far, TOPORS was known as a multifunctional protein and all previously published work involving non-retinal cell types demonstrated nuclear localization supporting a nuclear role for TOPORS. It also localizes to the centrioles of centrosomes and spindle poles as well as basal bodies in dividing and quiescent cells, respectively. The association of TOPORS with the centrioles and mid-body indicates that it may also be involved in the regulation of cell division. Its co-localization and close association with γ-tubulin lead us to suggest a possible role for TOPORS in microtubule nucleation, establishment of a bipolar mitotic spindle or normal cell division. It will be important to understand whether TOPORS has a role in the centrosome cycle as a structural component for the centriole/centrosome assembly or is involved in the maintenance of centrosomal adherence.
Defects in the protein transport system responsible for the organization and maintenance of the cilia is implicated as a primary cause for some human diseases also known as ciliopathies. Taken together, we propose that TOPORS dysfunction should be included in the broad category of these diseases. As in other ciliated cells, in photoreceptors, the transport of ciliary components to their destination in the cilium is characterized by different transport modules. In photoreceptor cells, newly synthesized OS components are transported from the biosynthetically active organelles through the cytoplasm of the IS and across the CC to the OS (33,34). At the base of the CC, the cargo which is destined for the OS is transferred from a microtubule-based transport system mediated by minus-end-directed cytoplasmic dynein motor complexes (36) to the ciliary transport systems—the IFT system mediated by kinesin motors (18). Our present data on TOPORS localization indicate that TOPORS is associated with the specialized periciliary machinery essential for the cargo handover from the dynein-mediated IS transport to the IFT complexes (17,34,37). In this periciliary compartment, the ciliary transport complexes are made up of individual IFT molecules (18) and the ciliary cargo delivery into and export from the cilium are mediated by the BBSome (38). Defects of molecules associated with this periciliary reloading complex are common primary causes of retinal degenerations and syndromic ciliopathies (17,26,39,40). Association of TOPORS predominantly with dynactin subunits further supports a possible role for TOPORS in regulating protein trafficking. The dynactin subunits p150glued and p50-dynamitin are responsible for tethering the cargo to the dynein motor and regulate microtubule-associated transport (41). Although direct evidence of a role of retrograde transport in photoreceptor is lacking, the cytoplasmic dynein 2 motor has been identified in the axoneme of photoreceptor OSs (42). Additionally, retrograde transport via the photoreceptor cilium may be involved in the renewal of kinesin motor subunits or for IFT-mediated signaling (43–45). Investigations are underway to validate whether TOPORS is involved in dynein-dependent trafficking.
Knockdown of topors in zebrafish results in defective development, including perturbed retinal lamination and photoreceptor development. This defect is consistent with the phenotype observed in knock out mouse mutants of transcription factors involved in retinal differentiation (46) and in the mutant of Ift88/Polaris (39). We hypothesize that microphthalmia could be the result of defective retinal development (dysplasia) and degeneration. The mechanism of such a severe retinal developmental defect associated with TOPORS may be defective Hedgehog or Wnt signaling cascades, as both are at least partly regulated by ciliary function (47,48). Further evidence for potential involvement of TOPORS in cilia-dependent function comes from our observation that TOPORS associates with ciliary and centrosomal proteins in the mammalian retina. TOPORS has been shown to function in proteasomal degradation pathway by acting as an E3 ubiquitin ligase for p53. As proteasomes are concentrated around the pericentriolar region and are involved in regulating Wnt signaling (49,50), our studies indicate that TOPORS may regulate proteasomal targeting of signaling proteins or cargo molecules. Analysis of the animal models of TOPORS mutations should further our understanding of this phenomenon and delineate the mechanism of pathogenesis of associated retinal degeneration.
MATERIALS AND METHODS
Antibodies
Mouse monoclonal anti-TOPORS antibody (diluted 1:50) and TOPORS recombinant protein were obtained from Abnova Corporation (Taiwan). Rabbit anti-γ-tubulin (1:1000) was obtained from Chemicon (Temeculla, CA, USA), and anti-KIF3A (1:1000) and anti-KAP3 (1:1000) were from Sigma. Anti-RP1 (1:500) and anti-IFT88 (1:1000) antibodies were a generous gift of Dr Eric A. Pierce (University of Pennsylvania, PA, USA) and Dr Bradley K. Yoder (University of Alabama at Birmingham, Birmingham, AL, USA), respectively. Antibody against p150glued (1:500) was purchased from BD Transduction Labs (San Jose, CA, USA). Characterization of the anti-RPGRORF15 (ORF15CP) and anti-CEP290 antibody has been described previously (16,19,20,22). Rat anti-α-tubulin (1:500) was obtained from Abcam, UK; rabbit anti-centrin 3 (1:400) and goat-anti-MAP2 (1:100) from Santa Cruz Biotechnology, Inc., USA; anti-p50 (1:1000) and mouse-anti-DIC (1:500) from Chemicon; rabbit-anti-pericentrin (1:500) from Sigma; and rabbit polyclonal anti-rhodopsin antibody (1:250) from Abcam. Secondary antibodies were FITC-conjugated donkey anti-mouse IgG (1:300), Cy ™3-conjugated donkey anti-rat IgG (1:300), Cy ™3-conjugated donkey anti-goat IgG (1:300), Cy ™3-conjugated donkey anti-rabbit IgG (1:300); horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse IgG from Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA.
Preparation of tissue sections and immunostaining
Eyes of adult mice were dissected after cardiac perfusion with 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.4) and were fixed for overnight at 4°C. The cornea and the lens were removed followed by sucrose gradient infiltration, embedded in optimal cutting temperature mounting medium and cryosectioned at 10 μm. For immunostaining, retinal sections were pretreated with blocking solution (5% normal goat serum and 0.1% Triton X-100 in PBS) for 1 h at room temperature, followed by incubation with primary antibody. The sections were washed and treated with appropriate secondary antibodies. Slides were then visualized using a laser scanning confocal microscope (ZEISS LSM 510, Carl Zeiss, Welwyn Garden City, Herts, UK). Images were processed using LSM5 Image Browser and Adobe Photoshop CS2 (Adobe Systems, USA). Cell nuclei were counterstained in blue with bisbenzimide.
Immunoblot analysis
Retinal bovine lysates were subjected to SDS–PAGE followed by immunoblotting with antibodies. Equal amounts of protein were run on 7.5% gel and transferred to nitrocellulose membrane (BioRad). Membranes were blocked in 5% non-fat dried milk in PBS. Primary and secondary antibodies were diluted in concentrations described above. For visualization, ECL (Amersham Biosciences) was used.
Immunocytochemistry
MDCK and IMCD3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; GIBCO) and ARPE-19 and hTERT-RPE-1 cells in DMEM/F-12+GlutaMAX (GIBCO) supplemented with 10% FCS, penicillin–streptomycin (1000 µg/ml) at 37°C in an atmosphere of 5% CO2. Cells were grown at confluence for at least 10 days (to allow them to form primary cilium) and then subjected to fixation and antibody staining. Slides were treated with ice-cold methanol (5 min at −20°C), dehydrated in PBS (5 min at room temperature) and blocked with PBS–0.5% BSA–20 mm glycine–0.1% NaN3 for 15 min before staining. Cover slips were incubated with the primary antibodies, in dilutions described above, for 1 h at room temperature. Cells were washed in blocking solution and incubated with secondary antibodies for 45 min at room temperature in the dark. Cells were again washed in blocking solution and mounted in ProLong Gold antifade reagent (Invitrogen). The hTERT-RPE-1 cells were synchronized by serum starvation for 24 h followed by addition of 10% FCS and methanol fixation every hour.
Zebrafish studies
A translation-blocking MO (Gene Tools) (5′<GCT CCT TAT CTG TGG TGA TGC CAT>3′) and its 5 base mismatch (Mm) were diluted in Danieau's solution (5 mm HEPES, pH 7.6, 58 mm NaCl, 0.7 mm KCl, 0.4 mm MgSO4, 0.6 mm Ca(NO3)2 and injected into wt zebrafish embryos at the one to eight blastomere stage at different concentrations as described. Embryos were assessed for tail extension anomalies, microphthalmia and edema essentially as described (35). The splice-blocking MO used in this study is 5′<GCC ATG ACC TGA CCT GAT AGA AGA AAC ATT>3′. The splice-blocked morphants were screened with zebrafish topors primers (F: 5′<GTC AGATAATGG CAC CCT CTA AGATGA A>3′ and R: 5′<TCG CTG CAG CTC GTG ATT CTC CG>3′).
For retinal immunohistochemistry, eye cryosections of zebrafish embryos at 4 dpf were stained with different antibodies. Fluorescent images were acquired using Olympus FV500 confocal microscope. Evaluation of cell death in retinal cryosections was performed using Apoptosis Detection Kit (Promega G3250), according to manufacturer's instructions. Briefly, sections were permeabilized with PBS + 0.1% Triton X-100, followed by incubation with equilibration buffer (200 mm potassium cacodylate, pH 6.6, 25 mm Tris–HCl, pH 6.6, 0.2 mm DTT, 0.25 mg/ml BSA, 2.5 mm cobalt chloride) for 10 min at room temperature. Slides were then replaced with 50 μl of rTdT incubation buffer [90 μl of equilibration buffer, 10 μl of nucleotide mix (50 μm fluorescein-12-dUTP, 100 μm dATP, 10 mm Tris–HCl, pH 7.6, 1 mm EDTA, 2 μl of rTdT enzyme)] at 37°C for 60 min in a humidified chamber. At the end of incubation, the slides were washed with 2X SSC buffer, pH 7.6 (87.7 g NaCl, 44.1 g sodium citrate, pH 7.6) and observed for Fluorescein-12-dUTP incorporation (520 nm, green fluorescence). Green fluorescent nuclei were counted in a given area of the section.
For eye measurements, the 5 μm thick plastic sections were examined under the microscope and were imaged. The diameter of the eye was measured as number of pixels on the images using ImageJ/Olympus software. They were then converted to millimeters by multiplying with the calibration factor obtained using a stage micrometer. The data are presented as percent change in diameter compared with the mismatch controls.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by grants from the Foundation Fighting Blindness, EVI-GENORET (LSHG-CT-2005-512036), The Special Trustees of Moorfields Eye Hospital London, National Institute of Health Research (UK) Biomedical Research Centre for Ophthalmology, Fight for Sight (UK), Midwest Eye Banks and Transplantation Center, National Institutes of Health (EY007961), Foundation Fighting Blindness, Biotechnology and Biological Sciences Research Council, Research to Prevent Blindness, Foundation Fighting Blindness-Canada and Fonds de la Recherche en Santee du Quebec, the Deutsche Forschungsgemeinschaft (DFG), the FAUN-Stiftung, Nuermberg, EU FP7 ‘SYSCILIA’ and a scholarship of the Gradiertenförderung of the Johannes Gutenberg University of Mainz, Germany.
ACKNOWLEDGEMENTS
We thank Stephen Lentz, Amiya Ghosh, Lourdes Valdés Sánchez and Paloma Domínguez Giménez for their technical assistance; Drs Mark Lewis and Bret Hughes for providing human retina sections.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hartong D.T., Berson E.L., Dryja T.P. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. doi:10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.Wright A.F., Chakarova C.F., Abd El-Aziz M.M., Bhattacharya S.S. Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat. Rev. Genet. 2010;11:273–284. doi: 10.1038/nrg2717. doi:10.1038/nrg2717. [DOI] [PubMed] [Google Scholar]
- 3.Rivolta C., Sharon D., DeAngelis M.M., Dryja T.P. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum. Mol. Genet. 2002;11:1219–1227. doi: 10.1093/hmg/11.10.1219. doi:10.1093/hmg/11.10.1219. [DOI] [PubMed] [Google Scholar]
- 4.Chakarova C.F., Papaioannou M.G., Khanna H., Lopez I., Waseem N., Shah A., Theis T., Friedman J., Maubaret C., Bujakowska K., et al. Mutations in TOPORS cause autosomal dominant retinitis pigmentosa with perivascular RPE atrophy. Am. J. Hum. Genet. 2007;81:1098–1103. doi: 10.1086/521953. doi:10.1086/521953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowne S.J., Sullivan L.S., Gire A.I., Birch D.G., Hughbanks-Wheaton D., Heckenlively J.R., Daiger S.P. Mutations in the TOPORS gene cause 1% of autosomal dominant retinitis pigmentosa. Mol. Vis. 2008;14:922–927. [PMC free article] [PubMed] [Google Scholar]
- 6.Selmer K.K., Grøndahl J., Riise R., Brandal K., Braaten O., Bragadottir R., Undlien D.E. Autosomal dominant pericentral retinal dystrophy caused by a novel missense mutation in the TOPORS gene. Acta Ophthalmol. 2009;88:323–328. doi: 10.1111/j.1755-3768.2008.01465.x. doi:10.1111/j.1755-3768.2008.01465.x. [DOI] [PubMed] [Google Scholar]
- 7.Haluska P., Jr, Saleem A., Rasheed Z., Ahmed F., Su E.W., Liu L.F., Rubin E.H. Interaction between human topoisomerase I and a novel RING finger/arginine–serine protein. Nucleic Acids Res. 1999;27:2538–2544. doi: 10.1093/nar/27.12.2538. doi:10.1093/nar/27.12.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou R., Wen H., Ao S.Z. Identification of a novel gene encoding a p53-associated protein. Gene. 1999;235:93–101. doi: 10.1016/s0378-1119(99)00203-6. doi:10.1016/S0378-1119(99)00203-6. [DOI] [PubMed] [Google Scholar]
- 9.Rajendra R., Malegaonkar D., Pungaliya P., Marshall H., Rasheed Z., Brownell J., Liu L.F., Lutzker S., Saleem A., Rubin E.H. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 2004;279:36440–36444. doi: 10.1074/jbc.C400300200. doi:10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 10.Weger S., Hammer E., Engstler M. The DNA topoisomerase I binding protein topors as a novel cellular target for SUMO-1 modification: characterization of domains necessary for subcellular localization and sumolation. Exp. Cell Res. 2003;290:13–27. doi: 10.1016/s0014-4827(03)00292-1. doi:10.1016/S0014-4827(03)00292-1. [DOI] [PubMed] [Google Scholar]
- 11.Renner F., Moreno R., Schmitz M.L. SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol. Cell. 2010;37:503–515. doi: 10.1016/j.molcel.2010.01.018. doi:10.1016/j.molcel.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T., et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol. Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. doi:10.1016/S1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q., Zhou J., Daiger S.P., Farber D.B., Heckenlively J.R., Smith J.E., Sullivan L.S., Zuo J., Milam A.H., Pierce E.A. Identification and subcellular localization of the RP1 protein in human and mouse photoreceptors. IOVS. 2002;43:22–32. [PMC free article] [PubMed] [Google Scholar]
- 14.Hong D.H., Pawlyk B., Sokolov M., Strissel K.J., Yang J., Tulloch B., Wright A.F., Arshavsky V.Y., Li T. RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. IOVS. 2003;44:2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 15.Okabe S., Shiomura Y., Hirokawa N. Immunocytochemical localization of microtubule-associated proteins 1A and 2 in the rat retina. Brain Res. 1989;483:335–346. doi: 10.1016/0006-8993(89)90178-9. doi:10.1016/0006-8993(89)90178-9. [DOI] [PubMed] [Google Scholar]
- 16.Trojan P., Krauss N., Choe H.W., Giessl A., Pulvermüller A., Wolfrum U. Centrins in retinal photoreceptor cells: regulators in the connecting cilium. Prog. Retin. Eye Res. 2008;27:237–259. doi: 10.1016/j.preteyeres.2008.01.003. doi:10.1016/j.preteyeres.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Maerker T., van Wijk E., Overlack N., Kersten F.F., McGee J., Goldmann T., Sehn E., Roepman R., Walsh E.J., Kremer H., Wolfrum U. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. doi:10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- 18.Sedmak T., Wolfrum U. Intraflagellar transport molecules in ciliary and nonciliary cells of the retina. J. Cell Biol. 2010;189:171–186. doi: 10.1083/jcb.200911095. doi:10.1083/jcb.200911095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna H., Hurd T.W., Lillo C., Shu X., Parapuram S.K., He S., Akimoto M., Wright A.F., Margolis B., Williams D.S., Swaroop A. RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J. Biol. Chem. 2005;280:33580–33587. doi: 10.1074/jbc.M505827200. doi:10.1074/jbc.M505827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E., et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. doi:10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jurczyk A., Gromley A., Redick S., San Agustin J., Witman G., Pazour G.J., Peters D.J., Doxsey S. Pericentrin forms a complex with intraflagellar transport proteins and polycystin-2 and is required for primary cilia assembly. J. Cell Biol. 2004;166:637–643. doi: 10.1083/jcb.200405023. doi:10.1083/jcb.200405023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giessl A., Pulvermüller A., Trojan P., Park J.H., Choe H.W., Ernst O.P., Hofmann K.P., Wolfrum U. Differential expression and interaction with the visual G-protein transducin of centrin isoforms in mammalian photoreceptor cells. J. Biol. Chem. 2004;279:51472–51481. doi: 10.1074/jbc.M406770200. doi:10.1074/jbc.M406770200. [DOI] [PubMed] [Google Scholar]
- 23.Besharse J.C., Baker S.A., Luby-Phelps K., Pazour G.J. Photoreceptor intersegmental transport and retinal degeneration: a conserved pathway common to motile and sensory cilia. Adv. Exp. Med. Biol. 2003;533:157–164. [PubMed] [Google Scholar]
- 24.Tsvetkov L., Xu X., Li J., Stern D.F. Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. JBC. 2003;278:8468–8475. doi: 10.1074/jbc.M211202200. doi:10.1074/jbc.M211202200. [DOI] [PubMed] [Google Scholar]
- 25.Rasheed Z.A., Saleem A., Ravee Y., Pandolfi P.P., Rubin E.H. The topoisomerase I-binding RING protein, topors, is associated with promyelocyticleukemia nuclear bodies. Exp. Cell Res. 2002;277:152–160. doi: 10.1006/excr.2002.5550. doi:10.1006/excr.2002.5550. [DOI] [PubMed] [Google Scholar]
- 26.Badano J.L., Mitsuma N., Beales P.L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Ann. Rev. Genomics Hum. Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. doi:10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 27.Otto E.A., Loeys B., Khanna H., Hellemans J., Sudbrak R., Fan S., Muerb U., O'Toole J.F., Helou J., Attanasio M., et al. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior–Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 2005;37:282–288. doi: 10.1038/ng1520. doi:10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 28.Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. doi:10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 29.Smith U.M., Consugar M., Tee L.J., McKee B.M., Maina E.N., Whelan S., Morgan N.V., Goranson E., Gissen P., Lilliquist S., et al. The transmembrane protein meckelin (MKS3) is mutated in Meckel–Gruber syndrome and the wpk rat. Nat. Genet. 2006;38:191–196. doi: 10.1038/ng1713. doi:10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- 30.Khanna H., Davis E.E., Murga-Zamalloa C.A., Estrada-Cuzcano A., Lopez I., den Hollander A.I., Zonneveld M.N., Othman M.I., Waseem N., Chakarova C.F., et al. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet. 2009;41:739–745. doi: 10.1038/ng.366. doi:10.1038/ng.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reiners J., Nagel-Wolfrum K., Jürgens K., Märker T., Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. doi:10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Kremer H., van Wijk E., Märker T., Wolfrum U., Roepman R. Usher syndrome: molecular links of pathogenesis, proteins and pathways. Hum. Mol. Genet. 2006;15(Suppl. 2):R262–R270. doi: 10.1093/hmg/ddl205. doi:10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- 33.Horst C.J., Johnson L.V., Besharse J.C. Transmembrane assemblage of the photoreceptor connecting cilium and motile cilium transition zone contain a common immunologic epitope. Cell Motil. Cytoskeleton. 1990;17:329–344. doi: 10.1002/cm.970170408. doi:10.1002/cm.970170408. [DOI] [PubMed] [Google Scholar]
- 34.Roepman R., Wolfrum U. Protein networks and complexes in photoreceptor cilia. Subcell. Biochem. 2007;43:209–235. doi: 10.1007/978-1-4020-5943-8_10. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh A.K., Murga-Zamalloa C.A., Chan L., Hitchcock P.F., Swaroop A., Khanna H. Human retinopathy-associated ciliary protein retinitis pigmentosa GTPase regulator mediates cilia-dependent vertebrate development. Hum. Mol. Genet. 2010;19:90–98. doi: 10.1093/hmg/ddp469. doi:10.1093/hmg/ddp469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tai A.W., Chuang J.-Z., Bode C., Wolfrum U., Sung C.-H. Rhodopsińs carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. doi:10.1016/S0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- 37.Papermaster D.S. The birth and death of photoreceptors: the Friedenwald Lecture. IOVS. 2002;43:1300–1309. [PubMed] [Google Scholar]
- 38.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., Jackson P.K. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. doi:10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 39.Pazour G.J., Baker S.A., Deane J.A., Cole D.G., Dickert B.L., Rosenbaum J.L., Witman G.B., Besharse J.C. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. doi:10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfrum U. Protein networks related to the Usher syndrome gain insights in the molecular basis of the disease. In: Ahuja S., editor. Usher Syndrome: Pathogenesis, Diagnosis and Therapy. 2010. Nova Science Publishers, (in press) [Google Scholar]
- 41.Schroer T.A. Dynactin. Ann. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. doi:10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 42.Mikami A., Tynan S.H., Hama T., Luby-Phelps K., Saito T., Crandall J.E., Besharse J.C., Vallee R.B. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. doi:10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 43.Signor D., Wedaman K.P., Orozco J.T., Dwyer N.D., Bargmann C.I., Rose L.S., Scholey J.M. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. doi:10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Pan J., Snell W.J. Intraflagellar transport particles participate directly in cilium-generated signaling in Chlamydomonas. Cell. 2006;125:549–562. doi: 10.1016/j.cell.2006.02.044. doi:10.1016/j.cell.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 45.Insinna C., Besharse J.C. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev. Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. doi:10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furukawa T., Morrow E.M., Li T., Davis F.C., Cepko C.L. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 1999;23:466–470. doi: 10.1038/70591. doi:10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 47.Neumann C.J., Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. doi:10.1126/science.289.5487.2137. [DOI] [PubMed] [Google Scholar]
- 48.Berbari N.F., O'Connor A.K., Haycraft C.J., Yoder B.K. The primary cilium as a complex signaling center. Curr. Biol. 2009;19:R526–R535. doi: 10.1016/j.cub.2009.05.025. doi:10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wigley W.C., Fabunmi R.P., Lee M.G., Marino C.R., Muallem S., DeMartino G.N., Thomas P.J. Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. doi:10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerdes J.M., Liu Y., Zaghloul N.A., Leitch C.C., Lawson S.S., Kato M., Beachy P.A., Beales P.L., DeMartino G.N., Fisher S., et al. Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 2007;39:1350–1360. doi: 10.1038/ng.2007.12. doi:10.1038/ng.2007.12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.