Abstract
Mutant superoxide dismutase type 1 (MTSOD1) is thought to cause ∼20% of cases of familial amyotrophic lateral sclerosis (FALS) because it misfolds and aggregates. Previous studies have shown that MTSOD1 accumulates inside the endoplasmic reticulum (ER) and activates the unfolded protein response (UPR), suggesting that ER stress is involved in the pathogenesis of FALS. We used a genetic approach to investigate the role of the UPR in FALS. We crossed G85RSOD1 transgenic mice with pancreatic ER kinase haploinsufficient (PERK+/−) mice to obtain G85R/PERK+/− mice. PERK+/− mice carry a loss of function mutation of PERK, which is the most rapidly activated UPR pathway, but have no abnormal phenotype. Compared with G85R transgenic mice, G85R/PERK+/− mice had a dramatically accelerated disease onset as well as shortened disease duration and lifespan. There was also acceleration of the pathology and earlier MTSOD1 aggregation. A diminished PERK response accelerated disease and pathology in G85R transgenic mice presumably because the mice had a reduced capacity to turn down synthesis of misfolded SOD1, leading to an early overloading of the UPR. The results indicate that the UPR has a significant influence on FALS, and suggest that enhancing the UPR may be effective in treating ALS.
INTRODUCTION
Familial amyotrophic lateral sclerosis (FALS) cases comprise 5–10% of the total cases of ALS, and mutant superoxide dismutase type 1 (MTSOD1) is a cause of ∼20% of FALS cases. Although MTSOD1 is an infrequent cause of the total number of cases of ALS, MTSOD1 transgenic mice have been used as a model system in order to clarify the reasons that motor neurons (MNs) die and to identify new therapeutic directions in FALS and sporadic ALS. Despite more than a decade of investigations of this model system, however, the pathogenesis of MTSOD1-induced FALS is still unclear and the prognosis remains grim.
Compelling evidence suggests that MTSOD1 causes FALS through a toxic gain in function (reviewed in 1); however, the nature of the toxicity remains poorly defined. The presence of MTSOD1 aggregates as a characteristic feature of the neuropathology of FALS and the importance of misfolded proteins in the pathogenesis of many neurodegenerative diseases have suggested that the accumulation of misfolded SOD1 is fundamental to the mutant protein's toxicity and leads to the death of MNs. In addition, a number of cellular processes are disturbed during FALS, including axonal flow, mitochondrial function and oxidative stress. It is now known that varied neural cell types contribute to MN death, perhaps through different mechanisms (2–4).
The presence of misfolded proteins in neurodegenerative diseases has prompted interest in the unfolded protein response (UPR) and its potential role in their pathogenesis (reviewed in 5). The accumulation of misfolded or unfolded proteins in the endoplasmic reticulum (ER) leads to protective transduction pathways such as the UPR. The UPR involves activation of three ER-resident stress sensors: pancreatic ER kinase (PERK), activating transcription factor 6 (ATF6) and inositol-requiring kinase-1 (IRE1) (6). These proteins are normally associated with binding immunoglobulin protein (BiP), an ER chaperone, which prevents their signaling; however, in the presence of misfolded or unfolded proteins in the ER, BiP binds and is sequestered by the misfolded and unfolded proteins, releasing PERK, ATF6 and IRE1, with the following consequences: (i) PERK, the most rapidly activated pathway, leads to phosphorylation of eukaryotic initiation factor 2 on the α subunit (eIF2α) with subsequent translational repression. Although PERK represses most translation by phosphorylating eIF2α, it also promotes translation of selected genes as well as transcription factors that can enhance protein folding or can lead to ER-assisted degradation of the misfolded protein. For example, PERK induces translation of activating transcription factor 4 (ATF4), a transcription factor which activates transcription of CCAAT/enhancer-binding protein-homologous protein (CHOP) and growth arrest and DNA damage 34 (GADD34) protein. CHOP can lead to cellular apoptosis and autophagy if the UPR fails to compensate for the misfolding, e.g. if the UPR is prolonged with sustained excessive ER stress. In addition, misfolded proteins in the ER can lead to oxidative stress, which can act in concert with the misfolded proteins, further activating the UPR and potentially leading to apoptosis (7). (ii) ATF6 and IRE1α activation by the UPR upregulates the transcription of multiple genes. When activated, ATF6 is processed and acts as a transcriptional activator, controlling many UPR genes. Following activation, IRE1 initiates splicing of transcriptional factor X-box-binding protein 1 (XBP-1), which activates transcription of many UPR target genes and thereby controls genes important in protein quality. One of the two isoforms of IRE1, IRE1α, has been implicated in neurological disorders (8,9).
Although SOD1 is primarily cytosolic, MTSOD1 and, to a lesser extent, wild-type (WT) SOD1 are also present in the secretory pathway (10–13). These observations have stimulated interest in the possible role of ER stress in FALS. Recent studies have now suggested a role for the UPR and ER stress in SOD1-induced cell death both in vitro (7,14–17) and in tissues from FALS transgenic mice (13,18,19) and ALS patients (20). ER stress has also been implicated in the pathogenesis of another autosomal dominant MN disease caused by vesicle-associated membrane protein-associated protein B (21,22) as well as a number of other neurodegenerative diseases (23).
Although studies have suggested that the UPR plays a role in protecting the central nervous system in FALS, a recent study showed that XBP-1 deficiency protects against MTSOD1-induced damage and disease by increasing autophagy (9). XBP-1 knockdown in an MN and neuroblastoma cell line expressing MTSOD1 led to a decrease in MTSOD1 aggregation and inclusions, with an associated increase in cell survival that was thought to be due to enhanced clearance of the MTSOD1 aggregates by macroautophagy. Knockdown of XBP-1 in the central nervous system of G86R FALS transgenic mice ameliorated disease, decreased MTSOD1 aggregates and increased autophagy in MNs.
The recent observation demonstrating the protective effect on FALS disease of XBP-1 deficiency raised questions about the effect of other pathways of the UPR and identified a need to directly address their involvement. In the present study, we used a genetic approach to investigate the impact of the PERK pathway in FALS by crossing a mouse with PERK haploinsufficiency with a FALS transgenic mouse. These studies showed that interfering with the PERK UPR pathway in FALS significantly accelerates the onset of disease, supporting a role for ER stress and highlighting a protective activity of PERK in FALS pathogenesis.
RESULTS
G85R/Perk+/− transgenic mice have an accelerated onset and shorter early phase of disease compared with G85R transgenic mice
In order to determine the effect of modulating ER stress on MTSOD1-induced FALS, we crossed G85R transgenic mice, which carry G85R genomic sequence flanked by LoxP sites (and were prepared for other planned Cre/LoxP studies) (24), with Perk+/− mice (25), which carry only one copy of Perk. As previously described, there was no clinical phenotype seen with the Perk+/− mice although the mice have decreased phosphorylation of eIF2α with ER stress (25). The G85R/Perk+/− transgenic mice were compared with G85R littermate mice with respect to the following previously defined clinical parameters (2): onset of disease, defined as peak weight before a decline; early phase of disease, defined as the period from peak weight until loss of 10% of maximal weight; late phase of disease, defined as the time from 10% loss in weight until death (when a mouse is unable to right itself within 20 s after being put on its back).
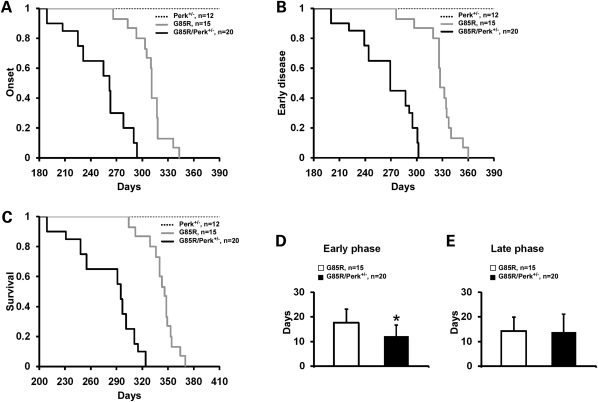
The mean onset of disease for the G85R/Perk+/− transgenic mice was significantly earlier than for G85R mice (253.7 ± 32.9 versus 308.6 ± 18.1 days, P < 0.001) (Fig. 1A). In addition, there was a shortening in the duration of the early phase of disease in G85R/Perk+/− mice versus G85R mice (12.3 ± 4.6 versus 17.6 ± 5.5 days, P = 0.006) (Fig. 1B and D). There was no statistically significant difference in the duration of the late phase of disease (13.8 ± 7.4 versus 14.3 ± 5.6 days, P = 0.809) (Fig. 1E). As expected, survival of G85R/Perk+/− mice was significantly shortened compared with G85R mice (280.0 ± 34.8 versus 340.4 ± 17.9 days, P < 0.001) (Fig. 1C).
Figure 1.
Plots of disease onset (A), the end of early disease (B) and survival (C) as well as diagrams of the duration (with standard deviation) of the early phase (D) and the late phase (E) of disease in G85R/Perk+/− versus G85R transgenic mice. In the Y-axis of (A)–(C), 1 refers to the total number of animals, which is detailed in the upper right. The asterisk indicates P < 0.05.
G85R/Perk+/− transgenic mice have an accelerated pathology and earlier SOD1 aggregation compared with G85R transgenic mice
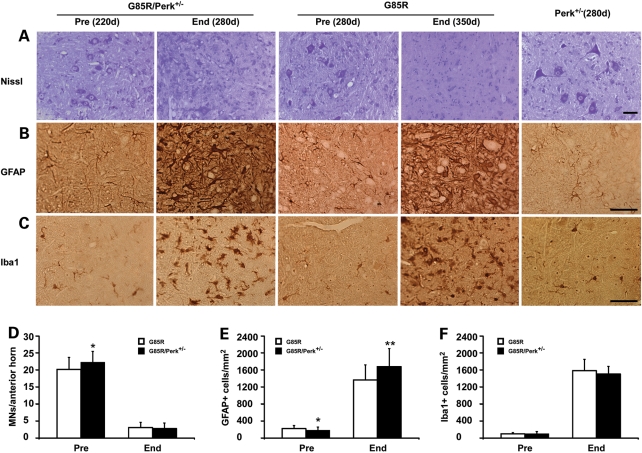
In order to compare the pathology of the different groups of mice, we examined the anterior horn of the lumbar spinal cord from G85R/Perk+/−, G85R and Perk+/− mice that were of different ages (Fig. 2). Subtle pathology was seen in the presymptomatic state (220 days) in the case of G85R/Perk+/− mice (Fig. 2A–C), which was not apparent at this same time point in the case of G85R mice (data not shown). Quantitative studies of G85R/Perk+/− mice at 220 days showed that there was a statistically significant difference in the number of MNs and astrocytes compared with findings in G85R mice at 280 days (Fig. 2D and E); this difference is probably not a relevant one since it presumably results from arbitrarily deciding on a particular number of days as representative of the presymptomatic stage of individual mice in these groups. At the end stage of disease at ∼280 days, G85R/Perk+/− mice demonstrated very significant loss of MNs by Nissl staining (Fig. 2A), vigorous astrocytosis by glial fibrillary acid protein (GFAP) staining (Fig. 2B) and extensive microgliosis by Iba1 staining (Fig. 2C). This pathology was not as prominent in G85R mice of the same age; however, similar findings were observed at the end stage of disease at ∼350 days (Fig. 2). Quantitative studies of moribund mice showed that there was a similar loss of MNs and microgliosis at the end stage of disease in G85R/Perk+/− and G85R mice, but an increase in astrocytes at the end stage in G85R/Perk+/− mice compared with the end stage in G85R mice. No pathological abnormalities were apparent in Perk+/− mice. These results indicate that the pathological and immunohistochemical hallmarks of disease were significantly accelerated in G85R/Perk+/− mice.
Figure 2.
Neuropathological and immunohistochemical studies of the anterior horn of the lumbar spinal cord of G85R/Perk+/−, G85R and Perk+/− transgenic mice of varying ages. Sections of the anterior horn of the spinal cord were stained with (A) Nissl for MNs, (B) anti-GFAP antibody for astrocytes, (C) anti-Iba1 antibody for microglia. In this figure and others: Pre, presymptomatic; End, end stage. Scale bar: 50 µm. Bar diagrams showing the mean ± standard deviation from quantitation of cell types in the lumbar anterior horn of G85R/Perk+/− versus G85R mice: (D) MNs; (E) astrocytes; (F) microglia; *P < 0.05, **P < 0.01.
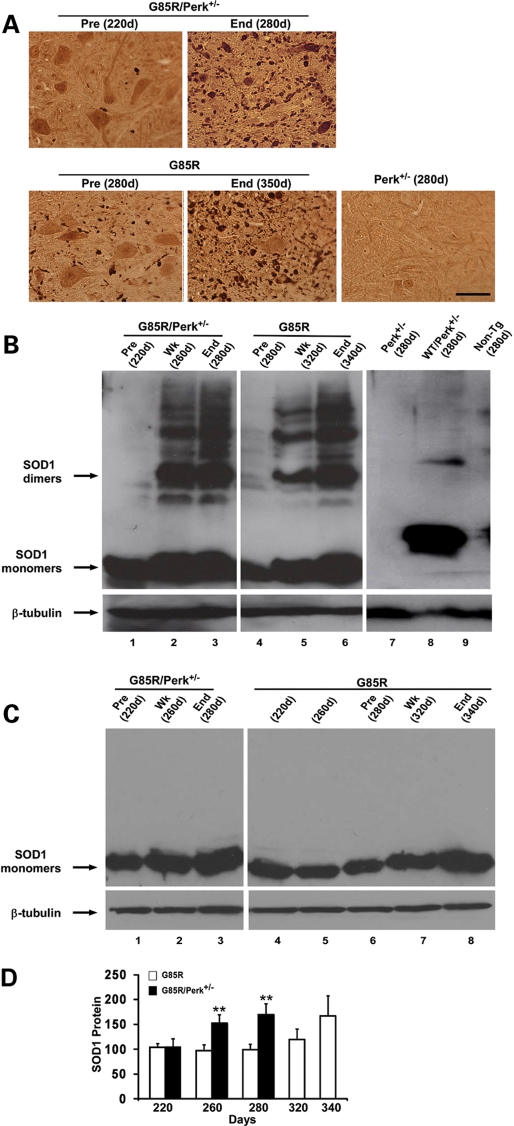
The lumbar spinal cord was examined for SOD1 aggregation by immunohistochemistry and western blots. Abundant MTSOD1 aggregation was detected in cells with MN morphology and in cell processes in G85R/Perk+/− mice at an earlier age than in G85R mice (Fig. 3A). MTSOD1 aggregates were similar in appearance and number at the end stage of disease in G85R/Perk+/− and G85R mice. No aggregates were present in Perk+/− mice. Western blots demonstrated high molecular weight forms of human SOD1 in G85R/Perk+/− and G85R mice (Fig. 3B); these aggregates appeared at an earlier age in G85R/Perk+/− mice than in G85R mice and were not present in controls (Perk+/−, WTSOD1/Perk+/− and non-transgenic littermate mice). There was an increased amount of total MTSOD1 in G85R/Perk+/− mice compared with G85R mice at 260 and 280 days, as shown in Figure 3C and D. The earlier increase in MTSOD1 and earlier aggregate formation in G85R/Perk+/− mice compared with G85R mice are presumably a result of a decrease in eIF2α phosphorylation in the PERK haploinsufficient mice; the decrease in eIF2α phosphorylation would be expected to lead to less suppression of translation of MTSOD1 with a subsequent increase in MTSOD1, including misfolded and aggregated MTSOD1.
Figure 3.
Studies of SOD1 aggregation and accumulation. (A) Immunohistochemical studies of the lumbar anterior horn of G85R/Perk+/−, G85R and Perk+/− transgenic mice of varying ages. (B) Representative western blot of lumbar spinal cord homogenates from varied aged G85R/Perk+/−, G85R and control mice (Perk+/−, WTSOD1/Perk+/− transgenic mice and non-transgenic littermates) subjected to SDS–PAGE under non-reducing conditions. The spinal cord homogenates were immunostained with anti-human SOD1 antibody. The monomeric and dimeric forms of SOD1 are noted with arrows. Note that the monomeric form of G85R MTSOD1 has a different electrophoretic mobility than WTSOD1, and that high molecular weight forms above the dimeric species are present in both G85R/Perk+/− and G85R mice, but they are present at an earlier time in the G85R/Perk+/− mice. Anti-β-tubulin antibody was used as a loading control. In this and subsequent figures: Pre, presymptomatic; Wk, weak; End, end stage. (C) Representative western blot of lumbar spinal cord homogenates from varied aged G85R/Perk+/− and G85R mice subjected to SDS–PAGE under reducing conditions. (D) Quantitation of total MTSOD1 in the spinal cord homogenates from varied aged G85R/Perk+/− and G85R mice as detected in western blots of samples electrophoresed under reducing conditions. The mean amount of MTSOD1 in the spinal cord of G85R mice at ∼220 days was arbitrarily given a value of 100. Note that there is more total MTSOD1 in G85R/Perk+/− compared with G85R mice at 260 and 280 days; no values for G85R/Perk+/− mice are provided after 280 days because they are dead. **P < 0.01.
G85R/Perk+/− transgenic mice have an earlier activation of the UPR and apoptosis pathways than G85R transgenic mice
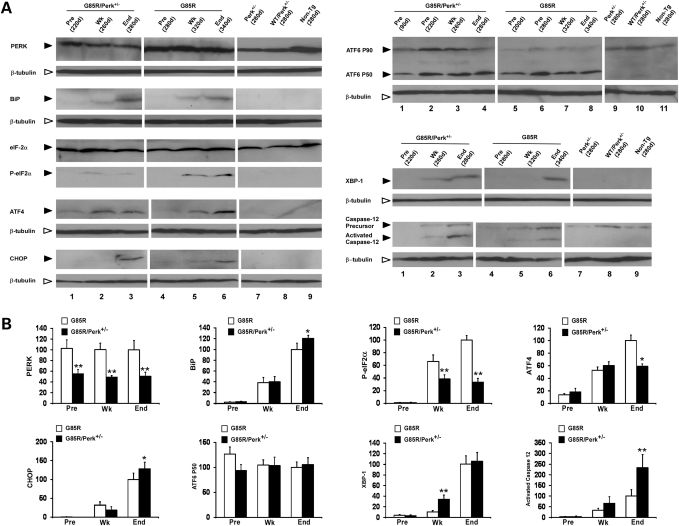
The UPR leads to dissociation of BiP, leading to the activation of PERK, which phosphorylates eIF2α with subsequent translational repression; however, PERK leads to an enhanced translation of ATF4, which subsequently upregulates the expression of CHOP. For this reason, we determined the levels of PERK, BiP (which is upregulated by the UPR), phosphorylated eIF2α, ATF4 and CHOP in the spinal cord of various ages of G85R/Perk+/− and G85R mice as well as controls (Perk+/−, WTSOD1/Perk+/− and non-transgenic littermate mice) (Fig. 4A and B). As expected, the levels of PERK in G85R/Perk+/−, WTSOD1/Perk+/− and Perk+/− mice were significantly less and ∼50% of the levels in G85R and non-transgenic mice. The UPR was induced during disease in both G85R/Perk+/− and G85R mice as demonstrated by increased levels of BiP, phosphorylated eIF2α, ATF4 and CHOP. The markers of activation of the UPR occurred at an earlier age in G85R/Perk+/− mice than in G85R mice, presumably because of the earlier accumulation of misfolded MTSOD1. As expected, considering the PERK haploinsufficiency, the level of phosphorylated eIF2α was lower during disease and at the end stage in G85R/Perk+/− mice. In addition, the level of ATF4 was slightly lower and the level of CHOP was slightly higher at the end stage in G85R/Perk+/− mice compared with G85R mice. The slight decrease in ATF4 may reflect the haploinsufficiency of PERK in G85R/Perk+/− mice. The elevation in CHOP at the end stage in G85R/Perk+/− mice may have occurred because CHOP is induced by a number of pathways in addition to the PERK/eIF2α/ATF4 pathway (26,27). The slight differences in ATF4 and CHOP in dying animals may have also resulted because of the brief duration of disease, in which there is a highly variable and changing number of neural cells undergoing ER stress at the end stage of disease, i.e. MNs are dying whereas astrocytes and microglia are proliferating.
Figure 4.
Quantitation and detection of markers of the UPR and apoptosis in varied aged G85R/Perk+/−, G85R and control mice (Perk+/−, WTSOD1/Perk+/− transgenic mice and non-transgenic littermates) by western blots of lumbar spinal cord homogenates. (A) Representative western blots immunostained with antibodies to the following: PERK, BiP, phosphorylated(P)-eIF2α, ATF4, CHOP, ATF6, XBP-1, activated caspase-12. Anti-β-tubulin antibody was used as a loading control. The level of the individual immunostained protein at the end stage in the case of G85R was arbitrarily given a value of 100. (B) Quantitation of UPR markers on western blots. *P < 0.05, **P < 0.01.
We also examined other pathways of the UPR in lumbar spinal cord homogenates from these different groups of mice. There were increased and similar levels of ATF6 P50 cleavage product in G85R/Perk+/− mice and G85R mice compared with controls (Fig. 4A and B). The enhanced ATF6 cleavage occurred at all of the ages examined, indicating that this UPR pathway was activated relatively early and did not appear to be affected at later times by the haploinsufficiency of PERK. The IRE1 pathway leads to the splicing of XBP1 mRNA, which regulates UPR target genes. The product of the spliced form of XBP-1 was present in the spinal cord homogenates from G85R/Perk+/− mice and G85R mice, but was not detected in the controls; the levels of this product appeared at an earlier age in the G85R/Perk+/− mice, presumably because of the earlier accumulation of misfolded MTSOD1. ER stress activates caspase-12, which can lead to apoptosis. As expected, caspase-12 activation was present in the spinal cord homogenates of G85R/Perk+/− mice and G85R mice, but not in the controls, and activated caspase-12 appeared at an earlier age in the G85R/Perk+/− mice. There were higher levels of activated caspase-12 at the end stage in G85R/Perk+/− versus G85R mice perhaps related to the slightly higher level of CHOP in these mice at this time, which could lead to oxidative stress and thereby promote caspase-12 expression (28); however, the different level of activated caspase-12 may have also been related to the dynamic nature of the brief clinical disease.
DISCUSSION
ALS is a challenging neurodegenerative disease with no effective treatment despite its recognition for more than 100 years, the identification of genes that cause FALS and the availability of FALS transgenic mice. One reason that an effective treatment for FALS is still out of reach is our lack of understanding of why MNs die in FALS and sporadic ALS. It is clear that MTSOD1 kills MNs as a result of a toxicity of the mutant enzyme; however, the nature of the toxicity remains poorly defined.
Although SOD1 was previously considered to be localized to the cytosol, recent studies have demonstrated that MTSOD1 and, to a lesser extent, WTSOD1 are present in the secretory pathways (10–13). MTSOD1 accumulates to a greater extent than WTSOD1 in the ER, perhaps because MTSOD1 binds to BiP (13,19), thereby facilitating its retention in the ER. The misfolding of MTSOD1 and the presence of reactive oxygen species are thought to activate the UPR (7), as demonstrated by the robust expression of UPR target genes in MTSOD1 FALS transgenic mice (15). Overloading of the UPR and oxidative stress can lead to apoptosis and the death of MNs; in addition, stressing the UPR system with translocation of misfolded MTSOD1 into the ER may overload the aggresome system, further contributing to the accumulation of misfolded protein. MNs may be especially sensitive to ER stress because of their large size along with their robust oxidative metabolic activity, protein synthesis and secretory activity—involving the export of large amounts of synaptic vesicles. In addition to the situation with respect to MNs, other cells such as astrocytes, which have aggregates of MTSOD1 (4,29), may undergo toxic effects from ER stress in the G85R/Perk+/− mice. It may be that the greater increase in astrocytes that was seen at the end stage of disease in G85R/Perk+/− mice compared with the end-stage G85R mice (Fig. 2E) was related to the presence of misfolded aggregated MTSOD1 in these cells that like the MNs (and all cell types in G85R/Perk+/− mice) are haploinsufficient for PERK.
A number of investigations have implicated the UPR and ER stress in FALS. MTSOD1 has been shown to aggregate and interact with BiP and other components of the ER stress pathway (13,16,19). Transcription factors associated with the UPR, such as ATF4 (13,18) and activated caspase-12, an ER stress-related cell death effector (13,15), are activated in the spinal cord of FALS transgenic animals. Signs of ER stress, such as phosphorylated eIF2α, are seen in tissues from ALS patients (20). One recent study demonstrated that fast fatiguable MNs in G93A MTSOD1, but not WTSOD, transgenic mice are selectively vulnerable to both degeneration and ER stress (18). Upregulation of the stress-management pathway was found by postnatal day 12 in the G93A mouse; BiP was upregulated in the more vulnerable MNs as early as postnatal day 5, and PERK and ATF4 as early as postnatal day 40. Increased expression of stress-related genes was found at later time periods in the case of less vulnerable MNs. Treatment with salubrinal, which produces an elevation in phophorylated eIF2α and protects cells from ER stress, led to a slower decline in muscle strength and an extension of the lifespan by 25–30 days (18). These results extended previous ones found in vitro, in which salubrinal decreased MTSOD1 aggregation and cell death (14).
The above studies suggested that the UPR is overloaded in FALS and therefore leads to toxicity and apoptosis. A recent study reported markedly increased expression of the spliced form of XBP-1 and ATF4 in the spinal cord of sporadic ALS patients, implicating the UPR in both sporadic and FALS. Surprisingly, these investigators showed that a deficiency of XBP-1 decreased toxicity in cultured cells in vitro and also ameliorated disease in (female) G86R MTSOD1 FALS transgenic mice (9); there was an associated increase in autophagy in the spinal cord of the XBP-1-deficient G86R mice. These results suggest that there may be different effects of the UPR on FALS, depending on the particular UPR pathway that is activated. However, it may be that the effect of the XBP-1 knockdown is at least partly related to the involvement of IRE1/XBP-1 in the innate immune response and inflammation (30).
Previous investigations of the ER stress pathways in FALS have involved the identification of UPR markers in tissues as well as studies of the effect on FALS of drugs and biological interventions that are predicted to perturb the UPR. In contrast, we used a genetic approach involving a cross of the FALS transgenic mouse with a mouse with PERK haploinsufficiency to study the effect of this deficiency on disease of MTSOD1 mice. Compared with G85R mice, G85R/Perk+/− mice had a significantly earlier onset of disease, a shortening of the duration of early disease and a shortened survival. The acceleration of clinical disease was associated with earlier pathological changes and MTSOD1 aggregation. The acceleration of disease seen in G85R/Perk+/− transgenic mice demonstrates the importance of PERK and the UPR as a protective survival response in FALS.
We presume that the accelerated disease in G85R/Perk+/− transgenic mice (with PERK haploinsufficiency) is a result of a progressive increase in misfolded MTSOD1 overloading the UPR. As a result of PERK haploinsufficiency, the G85R/Perk+/− transgenic mice had decreased eIF2α phosphorylation compared with G85R mice, resulting in less suppression of protein synthesis. Therefore, there was an enhanced and accelerated accumulation of misfolded SOD1 in G85R/Perk+/− transgenic mice relative to G85R mice (Fig. 3). The arms of the UPR were activated earlier in G85R/Perk+/− mice than in G85R mice (Fig. 4) as a result of the reduced capacity of G85R/Perk+/− mice to turn down protein synthesis in the face of ER stress. The failure to handle ER stress with the accompanying increase in misfolded MTSOD1 led to the earlier clinical and pathological disease in G85R/Perk+/− transgenic mice. The UPR in G85R mice also became overwhelmed, but at later times. Experiments are in progress using a genetic approach to determine whether decreasing the capacity to dephosphorylate eIF2α can alleviate the overloaded UPR and restore the appropriate homeostatic balance by which the UPR handles misfolded MTSOD1.
MATERIALS AND METHODS
Mice and breeding
WTSOD1 and G85R MTSOD1 transgenic mice carried WT SOD1 and G85R MTSOD1 genomic sequence, respectively, which was flanked by LoxP sites (and were prepared for other studies involving Cre/LoxP) have been previously described (24). These mice were crossed with Perk+/− mice, which are haploinsufficient for Perk (25). All the mice were on a C57BL/6 background. The WTSOD1/Perk+/− and G85R/Perk+/− transgenic mice were identified by PCR using previously published primers for the detection of the SOD1 gene (24) and for Perk (25).
Assessment of disease phenotype
Clinical assessment
Mice were weighed every 2 days and clinically assessed as described previously in studies of FALS transgenic mice (31), including a different line of G85R transgenic mice (32): onset of disease was defined as peak weight before a decline; early phase of disease was the period from peak weight until loss of 10% of maximal weight; late phase of disease was the time from 10% loss in weight until death (when a mouse was unable to right itself within 20 s after being put on its back). In experiments monitoring disease parameters and survival, the G85R/Perk+/− mice were compared with littermate G85R mice.
Pathology
The two groups of mice that were evaluated clinically were also studied and compared with respect to pathology. The pathology of the spinal cord was evaluated as described previously (3,33).
Immunohistochemical evaluation
Nissl staining and GFAP antibody staining have been described (4). SOD1 aggregation was determined by immunohistochemical staining using a rabbit antibody that recognizes the carboxyl end of mouse and human SOD1 (34), as described previously (3). Quantitation of different cell types in the lumbar spinal cord was carried out as described previously (4).
Western blot
For experiments involving UPR markers, the lumbar spinal cord tissue from each mouse was collected and homogenized in 200 µl of homogenizing buffer (20 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 0.5% sodium deoxycholate) containing a protease inhibitor cocktail. The homogenate was then centrifuged 20 000g for 15 min. As described previously (35), 20 µg of total protein from the supernatant was loaded on 10 or 15% SDS–PAGE for western blot analysis. Immunostaining was carried out with the following antibodies: PERK rabbit monoclonal antibody (1:1000, Cell Signaling Technology, Danvers, MA, USA), BiP (1:1000, Enzo Life Sciences International, Inc., Plymouth Meeting, PA, USA), eIF2α (1:1000, Santa Cruz, CA, USA), phosphorylated eIF2α (1:1000, Cell Signaling Technology), CHOP (1:500, Santa Cruz), ATF4 (1:1000, Santa Cruz), caspase-12 (1:1000, Santa Cruz), ATF6 (1:1000, Imgenex, San Diego, CA, USA), XBP-1 (1:500, Biolegend, San Diego, CA, USA) and β-tubulin (1:500, Developmental Studies Hybridoma Bank).
For the detection and quantitation of MTSOD1 aggregates, the soluble and insoluble fractions of the lumbar spinal cord homogenate were subjected to SDS–PAGE electrophoresis, as described (24). In some cases, high molecular weight forms of SOD1 were detected by electrophoresing the samples under non-reducing conditions, as described (24). Immunostaining was carried out with a human-specific anti-SOD1 antibody (34).
Quantitation of the UPR markers and MTSOD1 was carried out from representative western blots as described (24).
FUNDING
This work was supported by the Muscular Dystrophy Association (No. 4346 to R.P.R.), the ALS Association (No. 1211 to R.P.R.) and the NIH (NS34939 to B.P.).
ACKNOWLEDGEMENTS
We thank Heather P. Harding and David Ron for providing us with Perk+/− mice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rothstein J.D. Current hypotheses for the underlying biology of amyotrophic lateral sclerosis. Ann. Neurol. 2009;65:S3–S9. doi: 10.1002/ana.21543. doi:10.1002/ana.21543. [DOI] [PubMed] [Google Scholar]
- 2.Boillee S., Vande Velde C., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. doi:10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Wang L., Sharma K., Grisotti G., Roos R.P. The effect of mutant SOD1 dismutase activity on non-cell autonomous degeneration in familial amyotrophic lateral sclerosis. Neurobiol. Dis. 2009;35:234–240. doi: 10.1016/j.nbd.2009.05.002. doi:10.1016/j.nbd.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L., Gutmann D.H., Roos R.P. Astrocyte loss of mutant SOD1 delays ALS disease onset and progression in G85R transgenic mice. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq463. in press. [DOI] [PubMed] [Google Scholar]
- 5.Zhang K., Kaufman R.J. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–S109. doi: 10.1212/01.wnl.0000192306.98198.ec. doi:10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
- 6.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. doi:10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 7.Malhotra J.D., Miao H., Zhang K., Wolfson A., Pennathur S., Pipe S.W., Kaufman R.J. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl Acad. Sci. USA. 2008;105:18525–18530. doi: 10.1073/pnas.0809677105. doi:10.1073/pnas.0809677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paschen W., Aufenberg C., Hotop S., Mengesdorf T. Transient cerebral ischemia activates processing of xbp1 messenger RNA indicative of endoplasmic reticulum stress. J. Cereb. Blood Flow Metab. 2003;23:449–461. doi: 10.1097/01.WCB.0000054216.21675.AC. doi:10.1097/00004647-200304000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Hetz C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., Martinez G., Cuervo A.M., Brown R.H., Glimcher L.H. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. doi:10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J.P. Chromogranin-mediated secretion of mutant superoxide dismutase proteins linked to amyotrophic lateral sclerosis. Nat. Neurosci. 2006;9:108–118. doi: 10.1038/nn1603. doi:10.1038/nn1603. [DOI] [PubMed] [Google Scholar]
- 11.Turner B.J., Atkin J.D., Farg M.A., Zang D.W., Rembach A., Lopes E.C., Patch J.D., Hill A.F., Cheema S.S. Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J. Neurosci. 2005;25:108–117. doi: 10.1523/JNEUROSCI.4253-04.2005. doi:10.1523/JNEUROSCI.4253-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urushitani M., Ezzi S.A., Matsuo A., Tooyama I., Julien J.P. The endoplasmic reticulum-Golgi pathway is a target for translocation and aggregation of mutant superoxide dismutase linked to ALS. FASEB J. 2008;22:2476–2487. doi: 10.1096/fj.07-092783. doi:10.1096/fj.07-092783. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi H., Almer G., Yamashita S., Guegan C., Nagai M., Xu Z., Sosunov A.A., McKhann G.M., II, Przedborski S. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc. Natl Acad. Sci. USA. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. doi:10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh Y.K., Shin K.S., Yuan J., Kang S.J. Superoxide dismutase 1 mutants related to amyotrophic lateral sclerosis induce endoplasmic stress in neuro2a cells. J. Neurochem. 2008;104:993–1005. doi: 10.1111/j.1471-4159.2007.05053.x. doi:10.1111/j.1471-4159.2007.05053.x. [DOI] [PubMed] [Google Scholar]
- 15.Atkin J.D., Farg M.A., Turner B.J., Tomas D., Lysaght J.A., Nunan J., Rembach A., Nagley P., Beart P.M., Cheema S.S., et al. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J. Biol. Chem. 2006;281:30152–30165. doi: 10.1074/jbc.M603393200. doi:10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- 16.Nishitoh H., Kadowaki H., Nagai A., Maruyama T., Yokota T., Fukutomi H., Noguchi T., Matsuzawa A., Takeda K., Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. doi:10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tobisawa S., Hozumi Y., Arawaka S., Koyama S., Wada M., Nagai M., Aoki M., Itoyama Y., Goto K., Kato T. Mutant SOD1 linked to familial amyotrophic lateral sclerosis, but not wild-type SOD1, induces ER stress in COS7 cells and transgenic mice. Biochem. Biophys. Res. Commun. 2003;303:496–503. doi: 10.1016/s0006-291x(03)00353-x. doi:10.1016/S0006-291X(03)00353-X. [DOI] [PubMed] [Google Scholar]
- 18.Saxena S., Cabuy E., Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat. Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. doi:10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 19.Wate R., Ito H., Zhang J.H., Ohnishi S., Nakano S., Kusaka H. Expression of an endoplasmic reticulum-resident chaperone, glucose-regulated stress protein 78, in the spinal cord of a mouse model of amyotrophic lateral sclerosis. Acta Neuropathol. 2005;110:557–562. doi: 10.1007/s00401-005-1080-y. doi:10.1007/s00401-005-1080-y. [DOI] [PubMed] [Google Scholar]
- 20.Ilieva E.V., Ayala V., Jove M., Dalfo E., Cacabelos D., Povedano M., Bellmunt M.J., Ferrer I., Pamplona R., Portero-Otin M. Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain. 2007;130:3111–3123. doi: 10.1093/brain/awm190. doi:10.1093/brain/awm190. [DOI] [PubMed] [Google Scholar]
- 21.Kanekura K., Suzuki H., Aiso S., Matsuoka M. ER stress and unfolded protein response in amyotrophic lateral sclerosis. Mol. Neurobiol. 2009;39:81–89. doi: 10.1007/s12035-009-8054-3. doi:10.1007/s12035-009-8054-3. [DOI] [PubMed] [Google Scholar]
- 22.Gkogkas C., Middleton S., Kremer A.M., Wardrope C., Hannah M., Gillingwater T.H., Skehel P. VAPB interacts with and modulates the activity of ATF6. Hum. Mol. Genet. 2008;17:1517–1526. doi: 10.1093/hmg/ddn040. doi:10.1093/hmg/ddn040. [DOI] [PubMed] [Google Scholar]
- 23.Uehara T. Accumulation of misfolded protein through nitrosative stress linked to neurodegenerative disorders. Antioxid. Redox. Signal. 2007;9:597–601. doi: 10.1089/ars.2006.1517. doi:10.1089/ars.2006.1517. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Deng H.X., Grisotti G., Zhai H., Siddique T., Roos R.P. Wild-type SOD1 overexpression accelerates disease onset of a G85R SOD1 mouse. Hum. Mol. Genet. 2009;18:1642–1651. doi: 10.1093/hmg/ddp085. doi:10.1093/hmg/ddp085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H., Sabatini D.D., Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. doi:10.1016/S1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 26.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. doi:10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 27.Song B., Scheuner D., Ron D., Pennathur S., Kaufman R.J. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J. Clin. Invest. 2008;118:3378–3389. doi: 10.1172/JCI34587. doi:10.1172/JCI34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brezniceanu M.L., Lau C.J., Godin N., Chenier I., Duclos A., Ethier J., Filep J.G., Ingelfinger J.R., Zhang S.L., Chan J.S. Reactive oxygen species promote caspase-12 expression and tubular apoptosis in diabetic nephropathy. J. Am. Soc. Nephrol. 2010;21:943–954. doi: 10.1681/ASN.2009030242. doi:10.1681/ASN.2009030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruijn L.I., Becher M.W., Lee M.K., Anderson K.L., Jenkins N.A., Copeland N.G., Sisodia S.S., Rothstein J.D., Borchelt D.R., Price D.L., et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. doi:10.1016/S0896-6273(00)80272-X. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman R.J., Cao S. Inositol-requiring 1/X-box-binding protein 1 is a regulatory hub that links endoplasmic reticulum homeostasis with innate immunity and metabolism. EMBO Mol. Med. 2010;2:189–192. doi: 10.1002/emmm.201000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boillee S., Yamanaka K., Lobsiger C.S., Copeland N.G., Jenkins N.A., Kassiotis G., Kollias G., Cleveland D.W. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–1392. doi: 10.1126/science.1123511. doi:10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 32.Lobsiger C.S., Boillee S., Cleveland D.W. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc. Natl Acad. Sci. USA. 2007;104:7319–7326. doi: 10.1073/pnas.0702230104. doi:10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L.J., Lu Y.Y., Muramatsu S., Ikeguchi K., Fujimoto K., Okada T., Mizukami H., Matsushita T., Hanazono Y., Kume A., et al. Neuroprotective effects of glial cell line-derived neurotrophic factor mediated by an adeno-associated virus vector in a transgenic animal model of amyotrophic lateral sclerosis. J. Neurosci. 2002;22:6920–6928. doi: 10.1523/JNEUROSCI.22-16-06920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deng H.X., Shi Y., Furukawa Y., Zhai H., Fu R., Liu E., Gorrie G.H., Khan M.S., Hung W.Y., Bigio E.H., et al. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. doi:10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghadge G.D., Wang L., Sharma K., Monti A.L., Bindokas V., Stevens F.J., Roos R.P. Truncated wild-type SOD1 and FALS-linked mutant SOD1 cause neural cell death in the chick embryo spinal cord. Neurobiol. Dis. 2006;21:194–205. doi: 10.1016/j.nbd.2005.07.006. doi:10.1016/j.nbd.2005.07.006. [DOI] [PubMed] [Google Scholar]