Abstract
The proapoptotic Bcl-2 family protein Bid is cleaved by caspase-8 to release the C-terminal fragment tBid, which translocates to the outer mitochondrial membrane and induces massive cytochrome c release and cell death. In this study, we have characterized the conformation of tBid in lipid membrane environments, using NMR and CD spectroscopy with lipid micelle and lipid bilayer samples. In micelles, tBid adopts a unique helical conformation, and the solution NMR 1H/15N HSQC spectra have a single well resolved resonance for each of the protein amide sites. In lipid bilayers, tBid associates with the membrane with its helices parallel to the membrane surface and without trans-membrane helix insertion, and the solid-state NMR 1H/15N polarization inversion with spin exchange at the magic angle spectrum has all of the amide resonances centered at 15N chemical shift (70–90 ppm) and 1H-15N dipolar coupling (0–5 kHz) frequencies associated with NH bonds parallel to the bilayer surface, with no intensity at frequencies associated with NH bonds in trans-membrane helices. Thus, the cytotoxic activity of tBid at mitochondria may be similar to that observed for antibiotic polypeptides, which bind to the surface of bacterial membranes as amphipathic helices and destabilize the bilayer structure, promoting the leakage of cell contents.
Programmed cell death is initiated when death signals activate the caspases, a family of otherwise dormant cysteine proteases. External stress stimuli trigger the ligation of cell surface death receptors, thereby activating the upstream initiator caspases, which in turn process and activate the downstream cell death executioner caspases (1). In addition, caspases can be activated when stress or developmental cues within the cell induce the release of cytotoxic proteins from mitochondria. This intrinsic mitochondrial pathway for cell death is regulated by the relative ratios of the pro- and antiapoptotic members of the Bcl-2 protein family (2).
Bcl-2 family proteins exert their apoptotic activities through binding with other Bcl-2 family members or other nonhomologous proteins and through the formation of ion-conducting pores that are thought to influence cell fate by regulating mitochondrial physiology. Their functions are also regulated by subcellular location, since they cycle between soluble and membrane-bound forms. The proteins share sequence homology in four evolutionarily conserved domains (BH1–BH4), of which the BH3 domain is highly conserved and essential for both cell killing and oligomerization among Bcl-2 family members. The antiapoptotic family members have all four domains, whereas all of the proapoptotic members lack BH4, and some only have BH3. These BH3-only proteins are activated by upstream death signals, which trigger their transcriptional induction or post-translational modification, providing a key link between the extrinsic death receptor and intrinsic mitochondrial pathways to cell death (3).
One of these BH3-only Bcl-2 family members, Bid, is a 195-residue cytosolic protein that lacks the hydrophobic C-terminal domain often found in other Bcl-2 family members and connects the extrinsic and intrinsic cell death pathways (4). The major mechanism for Bid activation involves its cleavage by caspase-8, after engagement of the Fas or TNFR1 cell surface receptors (5–7). Caspase-8 cleavage of Bid releases the 15-kDa C-terminal fragment tBid, which translocates to the outer mitochondrial membrane, where it induces massive cytochrome c release and cell death. tBid has a 10-fold greater binding affinity for its antagonist Bcl-XL and is 100-fold more efficient in inducing cytochrome-c release from mitochondria than its full-length precursor. Upon translocation from the cytosol to mitochondria, tBid can interact with proapoptotic Bax and Bak to promote mitochondrial apoptosis (8, 9), or it can interact with antiapoptotic Bcl-2 and Bcl-XL and, thus, be sequestered from the cell and held in check (10). Cell-free studies have shown that Bid and Bax are sufficient to form large openings in reconstituted lipid vesicles (11). In addition, tBid can promote the alteration of membrane curvature in artificial lipid bilayers (12), and it can associate directly with mitochondria, inducing a remodeling of mitochondrial structure. tBid-induced mitochondrial remodeling is characterized by the reorganization of inner membrane cristae to form a highly interconnected common intermembrane space and is accompanied by redistribution of the cytochrome c stores from individual cristae to the intermembrane space (13), which may account for the rapid and complete release of mitochondrial cytochrome c that is observed in the presence of tBid.
The Bid BH3 domain, while essential for binding to other Bcl-2 family members, is not required for either translocation to the outer mitochondrial membrane and mitochondrial remodeling or complete cytochrome c release. The specific targeting of tBid to the outer mitochondrial membrane is mediated by the abundant mitochondrial lipid cardiolipin (14, 15), whose metabolism further regulates cytochrome c release and mitochondrion-dependent apoptosis in a Bcl-2- and caspase-independent manner (16). These lines of evidence suggest that Bid induces cell death through two separate mechanisms, a BH3-dependent mechanism that involves binding to other multidomain Bcl-2 family members and a BH3-independent mechanism that involves direct association and interaction with the outer mitochondrial membrane.
The structure of full-length Bid in solution consists of eight α-helices arranged with two central somewhat more hydrophobic helices forming the core of the molecule (17, 18). The third helix, which contains the BH3 domain, is connected to the first two helices by a long flexible loop, which includes the caspase-8 cleavage site, Asp60 (Fig. 1). Despite the lack of sequence homology, the structure of Bid is strikingly similar to those of other anti- and proapoptotic multidomain Bcl-2 family proteins (19–22), and it is also similar to the structure of the pore-forming domains of bacterial toxins. Indeed, like the toxins and other Bcl-2 family proteins, tBid also forms ion-conducting pores in lipid bilayers (23). The structural basis for Bcl-2 pore formation is not known, since the structures that have been determined are for the soluble forms of the proteins, but by analogy to the bacterial toxins, the Bcl-2 pores are thought to form by a rearrangement of their compactly folded helices upon contact with the mitochondrial membrane. One model proposes membrane insertion of the core hydrophobic helical hairpin with the other helices folding up to rest on the membrane surface, whereas an alternative model envisions the helices rearranging to bind the membrane surface without insertion (24, 25).
FIG 1. Amino acid sequence of human Bid (accession number P55957).
The eight helices (H1–H8) are those determined in the solution NMR structure of full-length human Bid (Protein Data Bank number 2BID) (17). The central core helices are H6 and H7. The BH3 domain in H3 and the putative lipid binding motif before H6 are in boldface type and underlined. The arrow marks the caspase-8 cleavage site at Asp60, and the sequences of both wild-type and recombinant tBid start at Gly61. The asterisks mark the four Met residues that were mutated to Leu in recombinant tBid.
In this study, we have examined the conformation and topology of tBid in lipid membrane environments, using solution NMR and CD experiments with micelle samples and solid-state NMR experiments with lipid bilayer samples. In the absence of lipids, tBid is soluble and retains a largely helical conformation, but many of the 1H/15N resonances are missing from the solution NMR heteronuclear single quantum coherence (HSQC)1 spectrum, suggesting that tBid aggregates, adopts multiple conformations, or undergoes dynamic averaging on the NMR time scale. In micelles, tBid adopts a unique helical conformation, reflected in the HSQC spectrum, where each of the 130 amide sites gives rise to a single well resolved resonance. The solid-state NMR polarization inversion with spin exchange at the magic angle (PISEMA) spectrum of tBid in lipid bilayers demonstrates that it binds the membrane with its helices parallel to the membrane surface and without trans-membrane helix insertion. This suggests that the BH3-independent cytotoxic activity of tBid may be similar to that observed for antibiotic polypeptides that bind to the surface of bacterial membranes as amphipathic helices and destabilize the bilayer structure, promoting the leakage of cell contents.
MATERIALS AND METHODS
Protein Expression
The gene encoding human tBid was obtained by PCR amplification of the 405-base pair segment corresponding to amino acids 60–195 of full-length human Bid (accession number P55957). The gene was cloned into the pMMHa vector, which directs the expression of proteins fused to the C terminus of the His9-TrpΔLE polypeptide (26). The plasmid was transformed in Escherichia coli C41(DE3), a mutant strain that was selected for the expression of insoluble or toxic proteins (27). tBid was expressed as an insoluble protein, with the His9-TrpΔLE polypeptide fused to its N terminus. To enable CNBr (cyanogen bromide) cleavage of tBid from the TrpΔLE fusion partner, the Met residues (Met97, Met142, Met148, and Met194) in the tBid sequence were changed to Leu. The His9-TrpΔLE-tBid fusion protein was purified from the inclusion body fraction of the lysate by nickel affinity chromatography (His·Bind Resin (Novagen, Madison, WI) in 6 m guanidine HCl, 20 mm Tris-Cl, pH 8.0, 500 mm NaCl, 500 mm imidazole). After dialysis against water, the fusion protein was dissolved in 0.1 n HCl and then cleaved from the TrpΔLE fusion partner by reaction with a 10-fold molar excess of CNBr (28). tBid was purified by reverse-phase high pressure liquid chromatography (Delta-Pak C4 (Waters, Milford, MA) in 50% water, 50% acetonitrile, 0.1% trifluoroacetic acid). The methods for inclusion bodies isolation and fusion protein purification were as described (26, 29). Cell cultures were in M9 minimal medium as described (29). For the production of uniformly 15N-labeled proteins, (15NH4)2SO4 was supplied as the sole nitrogen source. For selectively labeled protein, individual 15N-labeled and nonlabeled amino acids were provided. All isotopes were from Cambridge Isotope Laboratories (Andover, MA).
Mitochondrial Release Assays
HeLa cells were harvested and then washed and suspended in HM buffer (10 mm HEPES, pH 7.4, 250 mm mannitol, 10 mm KCl, 5 mm MgCl2, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture (Roche Applied Science)). Cells were homogenized with 50 strokes of a Teflon homogenizer with a B-type pestle and centrifuged twice at 600 × g for 5 min at 4 °C. The supernatant was centrifuged at 10,000 × g for 10 min at 4 °C, and the resulting pellet, containing mitochondria, was washed twice in HM buffer and then suspended in HM buffer at a concentration of 1 mg/ml. Isolated HeLa mitochondria (10 µl) were incubated with HM buffer (50 µl) containing increasing amounts of tBid at 30 °C for 1 h. The samples were centrifuged at 10,000 × g for 10 min, and the pellet and supernatant fractions were each resolved by SDS-PAGE and analyzed by immunoblotting using cytochrome c and second mitochondria-derived activator of caspases (SMAC) antibodies.
Bcl-XL Binding Assays
The expression of recombinant His-tagged Bcl-XL(ΔC), without the C-terminal amino acids 212–233, was as described (30). For the binding assays, His6-Bcl-XL(ΔC) protein (15 µg) was incubated at 4 °C for 4 h with 10 µl of Ni2+-NTA resin (Novagen, Madison, WI) in phosphate-buffered saline buffer (10 mm sodium phosphate, 150 mm sodium chloride, pH 7.2), and then the resin was washed three times, tBid (15 µg) was added, and the mixture was further incubated for 4 h. The resin was washed three times with phosphate-buffered saline, and then the bound proteins were eluted with 500 mm imidazole. The samples were resolved by SDS-PAGE stained with Coomassie Blue. As a control, tBid was incubated with the resin in the absence of Bcl-XL.
CD Spectroscopy
The samples were identical to those used for solution NMR experiments but contained 40 µm tBid. The samples were transferred to a quartz cuvette (0.1-mm path length), and far-UV CD spectra were recorded at 25 °C on a model 62A-DS CD spectrometer (Aviv, Lakewood, NJ) equipped with a temperature controller. A 5-s time constant and a 1-nm bandwidth were used during data acquisition over a wavelength range of 180–260 nm. For each sample, three spectra were recorded, averaged, and referenced by subtracting the average of three spectra obtained using the buffer alone. The spectra were analyzed for protein secondary structure with the k2d program (available on the World Wide Web at www.embl-heidelberg.de/~andrade/k2d/) (31).
Solution NMR
The sample of tBid without lipids contained 0.7–1 mm 15N-labeled tBid, in 50 mm sodium phosphate, pH 5. In the absence of lipids, tBid is not soluble in the millimolar concentrations required for NMR at pH values greater than 5. The samples of tBid in micelles contained 1 mm 15N-labeled tBid in 20 mm sodium phosphate, pH 7, with 500 mm SDS (Cambridge, Andover, MA) or 100 mm 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)] (LPPG); Avanti, Alabaster, AL).
Solution NMR experiments were performed on a Bruker AVANCE 600 spectrometer (Billerica, MA) with a 600/54 Magnex magnet (Yarnton, UK), equipped with a triple-resonance 5-mm probe with three-axis field gradients. The two-dimensional 1H/15N HSQC (32) spectra were obtained at 40 °C. The 15N and 1H chemical shifts were referenced to 0 ppm for liquid ammonia and tetramethylsilane, respectively. The NMR data were processed using NMR Pipe and rendered in NMR Draw (33) on a Dell Precision 330 MT Linux work station (Round Rock, TX).
Solid-state NMR
The lipids, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (DOPG) were from Avanti (Alabaster, AL). Samples of tBid in lipid bilayers were prepared by mixing 5 mg of 15N-labeled tBid in water with 100 mg of lipids (DOPC/DOPG, 6:4 molar ratio) that had been sonicated in water to form unilamellar vesicles. The protein and lipid vesicle mixture was incubated on ice for 30 min and then distributed onto the surface of 15 glass slides (11 × 11 × 0.06 mm; Marienfeld, Germany). After allowing excess water to evaporate at 40 °C, the slides were stacked and equilibrated for 12 h at 40 °C and 93% relative humidity, in order to form oriented lipid bilayers. The samples were wrapped in parafilm and then sealed in thin polyethylene film prior to insertion in the NMR probe. Hydrogen-exchanged samples were prepared by exposing the stacked oriented bilayer samples to an atmosphere saturated with D2O.
Solid-state NMR experiments were performed on a Bruker AVANCE 500 spectrometer (Billerica, MA) with a 500/89 AS Magnex magnet (Yarnton, UK). The home-built 1H/15N double resonance probe had a square radiofrequency coil (11 × 11 × 3 mm) wrapped directly around the samples. The one-dimensional 15N chemical shift spectra were obtained with single contact cross-polarization with mismatch-optimized IS polarization transfer (34, 35). The two-dimensional spectra that correlate 15N chemical shift with 1H-15N dipolar coupling were obtained with PISEMA (36). The spectra were obtained with a cross-polarization contact time of 1 ms, a 1H 90° pulse width of 5 µs, and continuous 1H decoupling of 63-kHz radio frequency field strength. The two-dimensional data were acquired with 512 accumulated transients and 256 complex data points, for each of 64 real t1 values incremented by 32.7 µs. The recycle delay was 6 s. The 15N chemical shifts were referenced to 0 ppm for liquid ammonia. The data were processed using NMR Pipe and rendered in NMR Draw (33) on a Dell Precision 330 MT Linux work station (Round Rock, TX).
Calculation of Solid-state NMR Spectra
Two-dimensional 1H/15N PISEMA spectra were calculated on a Linux Dell computer as described previously (37). The 15N chemical shift and 1H-15N dipolar coupling frequencies were calculated for various orientations of an 18-residue α-helix, with 3.6 residues per turn and uniform backbone dihedral angles for all residues (ϕ = −57°; ψ = −47°). The principal values and molecular orientation of the 15N chemical shift tensor (σ11 = 64 ppm; σ22 = 77 ppm; σ33 = 217 ppm; angle (σ11NH) = 17°) and the NH bond distance (1.07 Å) provided the input for the spectrum calculation at each helix orientation.
RESULTS
Protein Expression and Biological Activity
Full-length Bid can be expressed at high levels, in E. coli, as a soluble protein, but tBid overexpression is toxic for bacterial cells. To produce milligram quantities of 15N-labeled tBid for NMR studies, we used the pMMHa fusion protein vector that directs high level protein expression as inclusion bodies, and we transformed the plasmid in E. coli C41(DE3) cells, selected for the expression of insoluble and toxic proteins (27). tBid was separated from the fusion partner by means of CNBr cleavage at the engineered N-terminal Met residue (26). This method yields ~10 mg of purified 15N-labeled tBid from 1 liter of culture. To avoid cleavage within the tBid segment, the four Met residues in the tBid amino acid sequence were mutated to Leu (Fig. 1, asterisks), and therefore, it was important to demonstrate that the recombinant protein retained its biological activity.
We tested the ability of recombinant tBid to induce the release of mitochondrial proteins by incubating it with mitochondria isolated from HeLa cells, assaying the supernatant and pelleted fractions with antibodies to cytochrome c and SMAC. Fig. 2A demonstrates that, in both cases, tBid is fully active, in a dose-dependent manner and at levels similar to wild-type tBid obtained by caspase-8 cleavage (38). Next we tested the ability of recombinant tBid to bind Bcl-XL, since this interaction requires the BH3 domain, and recombinant tBid contains the mutation M97L in this segment. When recombinant tBid, which does not have a His tag, was incubated with Ni2+-NTA without Bcl-XL, it did not bind the resin, as evidenced by the absence of tBid in the imidazole elution (Fig. 2B, lane 4). However, when tBid was incubated with Ni2+-NTA that had been previously incubated with His-tagged Bcl-XL, it also bound to the resin and eluted with Bcl-XL in imidazole (Fig. 2B, lane 5).
FIG 2. Recombinant tBid induces the release of cytochrome-c and SMAC from isolated mitochondria and binds to Bcl-XL in vitro.
A, isolated HeLa mitochondria (10 µl, 1 mg/ml) were incubated with increasing amounts (0–133 nm in 50 µl) of tBid. The supernatant (S) and pellet (P) fractions were separated by centrifugation, resolved on SDS-PAGE, and analyzed by Western blotting with either cytochrome c (top) or SMAC (bottom) antibody. In B, Ni2+-NTA resin (10 µl) was incubated first with His-Bcl-XL(ΔC) (15 µg) and then with tBid (15 µg). The Ni2+-NTA-bound protein was eluted with 500 mm imidazole, and the samples were resolved by SDS-PAGE and stained with Coomassie Blue (lane 5). In the control experiment, tBid was directly incubated with the resin without preincubation with Bcl-XL(ΔC) (lane 4). Individual Bcl-XL(ΔC) monomers (24 kDa) are resolved in lane 2, and individual tBid monomers (15 kDa) are resolved in lane 3. Molecular weight markers are shown in lane 1.
Thus, we conclude that recombinant tBid, isolated from inclusion bodies, is fully active in its ability to induce cytochrome c and SMAC release from isolated mitochondria and that its capacity to bind antiapoptotic Bcl-XL through its BH3 domain is not disrupted by the M97L mutation. The pMMHa vector may be generally useful for the high level expression of other proapoptotic proteins that are difficult to express because of their cytotoxic properties. The use of chemical cleavage eliminates the difficulties, poor specificity, and enzyme inactivation, often encountered with protease treatment of insoluble proteins, and in cases where Met mutation is not feasible, protein cleavage from the fusion partner can be obtained enzymatically by engineering specific protease cleavage sites for the commonly used enzymes thrombin, Fxa, enterokinase, and tobacco etch virus protease. Thrombin and tobacco etch virus retain activity in the presence of detergents, including low millimolar concentrations of SDS.
tBid in Lipid Micelles
The cleavage of Bid by caspase-8 results in a C-terminal product, tBid, which targets mitochondria and induces apoptosis with strikingly enhanced activity (5–7). To characterize the conformation of tBid in lipid environments, we obtained its CD and solution NMR 1H/15N HSQC spectra in the absence or in the presence of lipid micelles (Fig. 3). The HSQC spectra of proteins are the starting point for additional multidimensional NMR experiments that lead to structure determination. In these spectra, each 15N-labeled protein site gives rise to a single peak, characterized by 1H and 15N chemical shift frequencies that reflect the local environment. In addition, the peak line widths and line shapes and their dispersion in the 1H and 15N frequency dimensions reflect protein conformation and aggregation state.
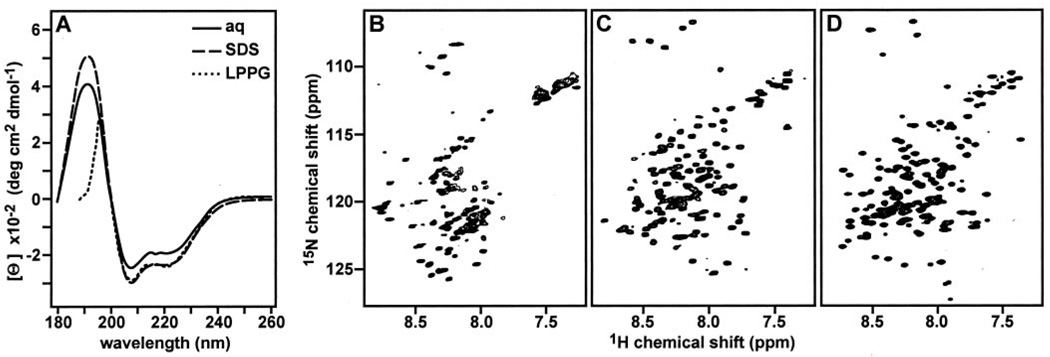
FIG 3. tBid adopts a helical fold in lipid micelles.
The CD spectra in A were obtained at 25 °C for tBid in aqueous solution (solid line), SDS micelles (broken line), or LPPG micelles (dotted line). The 1H/15N HSQC NMR spectra in B–D were obtained at 40 °C for uniformly 15N-labeled tBid, in aqueous solution (B), SDS micelles (C), or LPPG micelles (D). Aqueous samples were in 20 mm sodium phosphate, pH 5; SDS micelle samples were in 20 mm sodium phosphate, pH 7, 500 mm SDS; and LPPG micelle samples were in 20 mm sodium phosphate, pH 7, 100 mm LPPG.
In the absence of lipids, the CD spectrum of tBid displays minima at 202 and 222 nm, characteristic of predominantly helical proteins (Fig. 3A, solid line). The helical content estimated from the CD spectrum is ~65%, which is similar to the 67% helical content determined from the three-dimensional structure of Bid (17) for the 135 C-terminal amino acids that correspond to tBid. However, whereas tBid retains its helical conformation even when it is separated from the 60-residue N-terminal fragment, many of the resonances in its 1H/15N HSQC spectrum cannot be detected (Fig. 3B), suggesting that the protein aggregates in solution, adopts multiple conformations, or undergoes dynamic conformational exchange on the NMR time scale. tBid has very limited solubility at pH values greater than 5, and the addition of KCl up to 200 mm did not improve the appearance of the HSQC spectrum, whereas KCl concentrations above 200 mm yielded a gel. This behavior is consistent with the dramatic changes in the physical properties of the protein that accompany caspase-8 cleavage. Bid has a theoretical pI of 5.3 and a net charge of −7, but caspase-8 cleavage and removal of the 60 N-terminal amino acids shift these parameters to the opposite end of their spectrum, and tBid has a pI of 9.3 and a net charge of +2. In addition, the removal of helix-1 and helix-2 following caspase-8 cleavage exposes hydrophobic residues in the BH3 domain in helix-3 and in the central core helices, helix-6 and helix-7 (17, 18, 23).
When tBid associates with lipid micelles, its HSQC spectrum changes dramatically, and single, well defined 1H/15N resonances are observed for each 15N-labeled NH site in the protein, indicating that tBid adopts a unique conformation in this environment (Fig. 3, C and D). Micelles provide suitable membrane mimetic samples for solution NMR studies of membrane proteins and a realistic alternative to organic solvents, because their small size affords rapid and effectively isotropic reorientation of the protein and because their amphipathic nature simulates that of membranes. The principle goal of micelle sample preparation is to reduce the effective rotational correlation time of the protein so that resonances will have the narrowest achievable line widths while providing an environment that maintains the protein fold.
Several lipids are available for protein solubilization, and we tested both SDS (Fig. 3C) and LPPG (Fig. 3D) for their ability to yield high quality HSQC spectra of tBid for structure determination. Both gave excellent spectra where most of the 130 tBid amide resonances could be resolved. Both SDS and LPPG are negatively charged, but they differ substantially in the lengths of their hydrocarbon chains (C12 for SDS; C16 for LPPG) and their polar headgroups (sulfate for SDS; phosphatidylglycerol for LPPG); thus, the differences in 1H and 15N chemical shifts between the two HSQC spectra most likely reflect the different lipid environments. The spectrum in LPPG has exceptionally well dispersed resonances with homogeneous intensities and line widths. LPPG was recently identified as a superior lipid for NMR studies of several membrane proteins (39) and is particularly interesting for this study because it is a close analog of cardiolipin and monolysocardiolipin, the major components of mitochondrial membranes that bind tBid (14).
In both the SDS and LPPG spectra, the limited chemical shift dispersion is typical of helical proteins in micelles, and this is confirmed by the corresponding CD spectra, which are dominated by minima at 202 and 222 nm, and thus show that tBid retains a predominantly helical fold in both types of micelles (Fig. 3A, broken and dotted lines). We estimated the helical content of tBid in lipid micelles to be ~65%, similar to that of tBid in the absence of lipids and to the value determined for the 135-residue C-terminal segment in the three-dimensional solution structure of Bid (17).
tBid in Lipid Bilayers
By analogy to the structurally homologous colicin and diphtheria bacterial toxins, the mechanism of pore formation by tBid has been thought to involve insertion through the membrane of the two central core helices, helix-6 and helix-7 (23, 25). To examine the conformation of tBid associated with membranes, we obtained one-dimensional 15N chemical shift and two-dimensional 1H/15N PISEMA solid-state NMR spectra of 15N-labeled tBid reconstituted in lipid bilayers (Fig. 4). In these samples, the lipid composition of 60% DOPC and 40% DOPG was chosen to mimic the highly negative charge of mitochondrial membranes. This lipid composition is identical to that of the liposomes used for the measurement of the ion channel activities of Bid and tBid (23), and since the oriented lipid bilayers used in this study were obtained from liposomes prepared in the same way as for the channel studies, the NMR spectra obtained in this work represent the channel-active conformation of tBid.
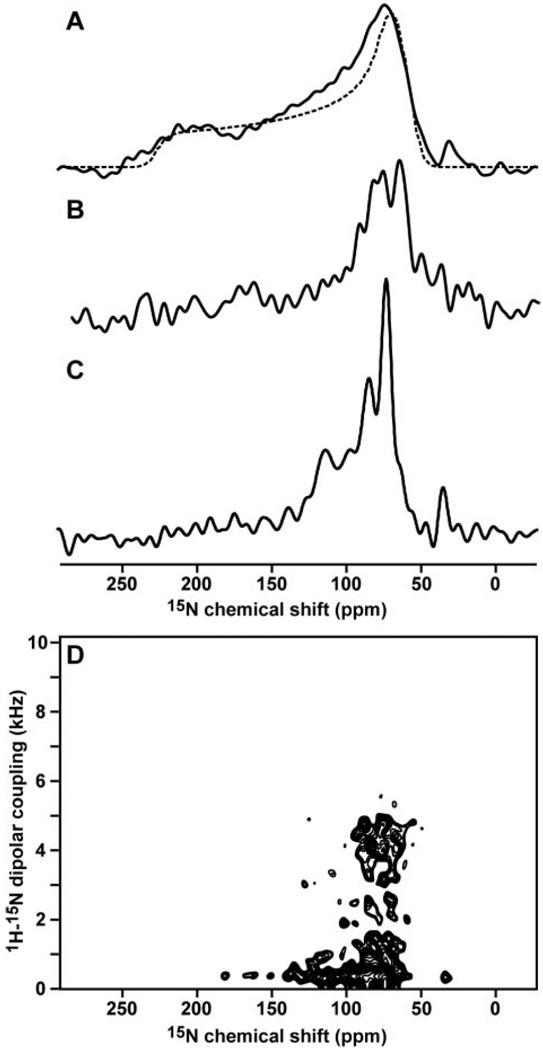
FIG 4. Solid-state NMR spectra of tBid in lipid bilayers show that tBid binds membranes with a unique fold and parallel orientation.
A, one-dimensional 15N chemical shift spectrum of uniformly 15N-labeled tBid in unoriented lipid bilayer vesicles (solid line) and powder pattern calculated for a rigid 15N amide site (dotted line). B, one-dimensional 15N spectrum of selectively 15N-Lys-labeled tBid in oriented lipid bilayers. C, one-dimensional 15N spectrum of uniformly 15N-labeled tBid in oriented lipid bilayers. D, two-dimensional 1H/15N PISEMA spectrum of tBid in oriented lipid bilayers.
When the lipid bilayers are oriented with their surface perpendicular to the magnetic field, the solid-state NMR spectra of the membrane-associated proteins trace out maps of their structure and orientation within the membrane and thus provide very useful structural information prior to complete structure determination. For example, helices give characteristic solid-state NMR spectra where the resonances from amide sites in the protein trace-out helical wheels that contain information about helix tilt and rotation within the membrane (37, 40, 41). Typically, trans-membrane helices have PISEMA spectra with 15N chemical shifts between 150 and 200 ppm, and 1H-15N dipolar couplings between 2 and 10 kHz, whereas helices that bind parallel to the membrane surface have spectra with shifts between 70 and 100 ppm and couplings between 0 and 5 kHz. We refer to these as the trans-membrane and in-plane regions of the PISEMA spectrum, respectively.
The 15N chemical shift spectrum of tBid in unoriented lipid bilayer vesicles is a powder pattern (Fig. 4A, solid line) that spans the full range (60–220 ppm) of the amide 15N chemical shift interaction (Fig. 4A, dotted line). The absence of additional intensity at the isotropic resonance frequencies (100–130 ppm) demonstrates that the majority of amino acid sites are immobile on the time scale of the 15N chemical shift interaction, although it is possible that some mobile unstructured residues cannot be observed by cross-polarization. The peak at 35 ppm is from the amino groups at the N terminus and side chains of the protein.
The spectrum of tBid in planar oriented lipid bilayers is very different (Fig. 4C). The amide resonances are centered at a frequency associated with NH bonds in helices parallel to the membrane surface (80 ppm), whereas no intensity is observed at frequencies associated with NH bonds in trans-membrane helices (200 ppm). The resonances near 120 ppm are unlikely to result from mobile sites, since little or no isotropic intensity is observed in the spectrum from unoriented bilayers; instead, they probably reflect specific orientations of their corresponding sites near the magic angle, which corresponds to 35.3° from the membrane surface. The NMR data show no evidence of conformational exchange on the millisecond to second time scale of the channel opening and closing events, excluding the possibility of transient insertion of tBid in the membrane. Therefore, we conclude that tBid binds strongly to the membrane surface, adopting a unique conformation and orientation in the presence of phospholipids.
Amide hydrogen exchange rates are useful for identifying residues that are involved in hydrogen bonding and that are exposed to water. The amide hydrogens in trans-membrane helices can have very slow exchange rates due to their strong hydrogen bonds in the low dielectric of the lipid bilayer environment, and their 15N chemical shift NMR signals persist for days after exposure to D2O (42). On the other hand, transmembrane helices that are in contact with water because they participate in channel pore formation and other water-exposed helical regions of proteins have faster exchange rates, and their NMR signals disappear on the order of hours (43). To examine the amide hydrogen exchange rates for membrane-bound tBid, we obtained solid-state NMR spectra after exposing the oriented lipid bilayer sample to D2O for 2, 5, and 7 h. The majority of resonances in the 15N chemical shift spectrum of tBid disappeared within 7 h, indicating that the amide hydrogens exchange and, hence, are in contact with the lipid bilayer interstitial water.
The tBid amino acid sequence has four Lys residues (Lys144, Lys146, Lys157, and Lys158), all located in or near helix-6, one of the two helices thought to insert in the membrane and form the tBid ion-conducting pore. The spectrum of 15N-Lys-labeled tBid in bilayers is notable because its amide resonances all have chemical shifts near 80 ppm, in the in-plane region of the spectrum, and this cannot be reconciled with membrane insertion (Fig. 4B). Since tBid maintains a helical fold in lipid micelles and it is reasonable to assume that the helix boundaries are not appreciably changed from those of full-length Bid, the solid-state NMR data demonstrate that helix-6 does not insert through the membrane but associates parallel to its surface. This is also supported by recent EPR data (44).
These findings are confirmed and refined with the two-dimensional 1H/15N PISEMA spectrum of tBid in bilayers (Fig. 4D). Each amide site in the protein contributes one correlation peak, characterized by 1H-15N dipolar coupling and 15N chemical shift frequencies that reflect NH bond orientation relative to the membrane. For tBid, the circular wheel-like pattern of resonances in the spectral region bounded by 0–5 kHz and 70–90 ppm provides definitive evidence that tBid associates with the membrane as surface-bound helices without trans-membrane insertion. The substantial peak overlap reflects a similar orientation of the tBid helices parallel to the membrane, and spectral resolution in this region requires three-dimensional correlation spectroscopy and selective isotopic labeling (45).
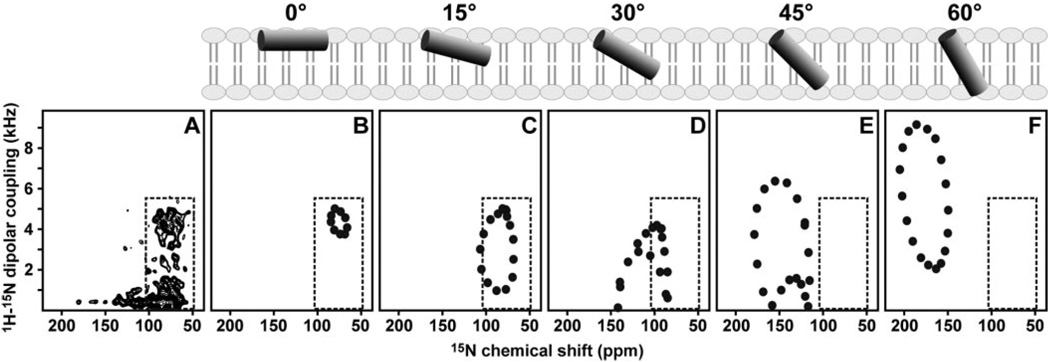
Since the NMR frequencies directly reflect the angles between individual bonds and the direction of the applied magnetic field, it is possible to calculate solid-state NMR spectra for specific models of proteins in oriented samples. For example, the spectra of helices have wheel-like patterns of resonances, called Pisa Wheels, that mirror helix tilt and rotation in the membrane (37, 40, 41), and a comparison of calculated and experimental spectra provides very useful structural information prior to complete structure determination. The spectra calculated for several orientations of an ideal 18-residue helix, with 3.6 residues per turn and identical backbone dihedral angles for all residues (ϕ, ψ = −57°, −47°), are shown in Fig. 5 (B–F) and are compared with the experimental PISEMA spectrum obtained for tBid in oriented bilayers, in Fig. 5A. In each spectrum, the characteristic Pisa Wheel pattern reflects helix tilt, but only the spectra calculated for membrane surface helices, with tilts from 0 to 15°, have intensity in the region of the experimental spectrum of tBid (inset box), whereas trans-membrane helices with tilts ranging from 45 to 90° from the membrane surface, have Pisa Wheels in a completely unpopulated region of the experimental spectrum. Based on this comparative analysis, we estimate that the helices of tBid are nearly parallel to the lipid bilayer plane (0° orientation), with tilts of no more than 20° from the membrane surface.
FIG 5. Two-dimensional solid-state NMR 1H/15N PISEMA spectrum of uniformly 15N-labeleld tBid in oriented lipid bilayers.
The experimental spectrum (A) is compared with the theoretical spectra (B–F) calculated for an 18-residue α-helix, with uniform backbone dihedral angles (ϕ = −57°; ψ = −47°) and different helix tilts (0–60°) relative to the membrane surface, as depicted in the schematic diagrams above the spectra. The 0° orientation is for a helix parallel to the membrane surface.
DISCUSSION
The results of this study demonstrate that tBid adopts a unique helical fold in lipid environments and that it binds the membrane without insertion of its helices. Solid-state NMR studies of the antiapoptotic Bcl-2 family member, Bcl-XL, also indicate that membrane insertion of the Bcl-XL helices is limited (42), and solution NMR studies show that Bcl-XL adopts an extended helical conformation in lipid micelles (46). Both tBid and Bcl-XL form ion-conductive pores that are thought to play a role in apoptosis through their regulation of mitochondrial physiology (23, 25), and since the samples in both the solid-state NMR and ion channel activity studies of tBid were identical in their lipid composition and the manner of sample preparation (23), the membrane surface association of tBid, observed by solid-state NMR in this study, is very likely to represent the channel-active conformation of the protein.
Pore formation by the Bcl-2 family proteins has been thought to involve translocation of the central core helices through the membrane, and the helices of both Bid and Bcl-XL are sufficiently long to span the lipid bilayer. However, their amphipathic character is also compatible with membrane surface association, in a manner that is reminiscent of the antimicrobial polypeptides, where binding of the polypeptide helices to the bacterial membrane surface is thought to transiently destabilize the membrane and change its morphology, inducing leakage of the cell contents, disruption of the electrical potential, and ultimately cell death (45, 47). It is notable that bacterial and mitochondrial membranes have very similar structures and surface charge and that tBid is both capable of altering bilayer curvature (12) and of remodeling the mitochondrial membrane (13), which would be sufficient to cause the release of mitochondrial cytotoxic molecules. The membrane surface association of tBid may also serve to display the BH3 domain on the mitochondrial membrane surface, making it accessible for binding by other Bcl-2 family members.
Although tBid does not insert in DOPC/DOPG lipid bilayers, we cannot exclude the possibility that trans-membrane insertion may be driven by the presence of natural mitochondrial lipids, such as cardiolipin and monolysocardiolipin. These lipids both bind tBid (14) and are concentrated at mitochondrial outer-inner membrane contact sites where tBid localizes; however, a recent EPR study using these lipids also found no evidence of tBid helix insertion through the membrane (44). It is also possible that trans-membrane insertion of tBid requires post-translational N-terminal myristoylation, since this has been shown to enhance Bid-induced cytochrome c release and apoptosis (48), and that the interactions with other Bcl-2 family proteins such as Bak and Bax or with other nonhomologous proteins such as the mitochondrial voltage dependent anion channel may promote insertion of the tBid helices through the mitochondrial membrane.
Wagner and co-workers (17) monitored the solution NMR 1H/15N HSQC spectrum of Bid after the addition of caspase-8, testing the hypothesis that Bid activation is accompanied by a conformational change. They found that the structure of Bid is not changed by caspase-8 cleavage, since tBid remains associated with the N-terminal fragment, but proposed that dissociation occurs at lower physiological concentrations, through a conformational on-off equilibrium. In light of the lipid affinity of Bid and the data in Fig. 3 demonstrating that tBid is well-folded in lipid micelles independently of the N terminus, it is possible that lipids act as a third partner during caspase-8 cleavage of Bid. Thus, whereas in the absence of lipids the two cleavage fragments remain associated, the presence of lipids may enable dissociation by providing an environment where tBid can fold independently.
In this regard, it is interesting to note that the Bid amino acid sequence (141PRDMEKE147) at the beginning of helix-6 is similar to the conserved sequence (95PDVEKE100) that forms a short lipid recognition helix in the lipoprotein apolipophorin-III. In Bid, this sequence forms a short loop that is solvent-exposed and perpendicular to the axis of helix-6, whereas in apolipophorin-III it forms a short helix at one solvent-exposed end of the molecule that is perpendicular to the helix bundle (49). This short motif is conserved in the Bid sequences from various species, suggesting that it plays a role in the protein biological function, and given the documented lipid-binding activity of Bid (14), it may constitute a lipid recognition domain for Bid similar to that of apolipophorin-III. Thus, it is possible that the structure of tBid, destabilized by dissociation from the N-terminal fragment after caspase-8 cleavage, undergoes a conformational change whereby it opens about the flexible loops that connect its helical segments to an extended helical conformation, which binds to the membrane surface. This would be similar to the mechanism proposed for apolipophorin-III, which adopts a marginally stable helix bundle topology that allows for concerted opening of the bundle about hinged loops (49).
Acknowledgment
We thank Jinghua Yu for assistance with the NMR experiments.
Footnotes
This research was supported by Department of the Army Breast Cancer Research Program Grants DAMD17-00-1-0506 and DAMD17-02-1-0313 (to F. M. M.), California Breast Cancer Research Program Grant 8WB0110 (to F. M. M.), and National Institutes of Health Grant R01GM60554 (to J. C. R.). The NMR studies utilized the Burnham Institute NMR Facility, supported by National Institutes of Health Grant P30CA30199 and the Biomedical Technology Resources for Solid-State NMR of Proteins at the University of California San Diego, supported by National Institutes of Health Grant P41EB002031.
The abbreviations used are: HSQC, heteronuclear single quantum coherence; PISEMA, polarization inversion with spin exchange at the magic angle; SMAC, second mitochondria-derived activator of caspases; NTA, nitrilotriacetic acid; DOPC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; DOPG, 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]; LPPG, 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)].
REFERENCES
- 1.Denault JB, Salvesen GS. Chem. Rev. 2002;102:4489–4500. doi: 10.1021/cr010183n. [DOI] [PubMed] [Google Scholar]
- 2.Green DR, Reed JC. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 3.Cory S, Adams JM. Natl. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Zhu H, Xu CJ, Yuan J. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 6.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 7.Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C, Erdjument-Bromage H, Tempst P, Korsmeyer SJ. J. Biol. Chem. 1999;274:1156–1163. doi: 10.1074/jbc.274.2.1156. [DOI] [PubMed] [Google Scholar]
- 8.Eskes R, Desagher S, Antonsson B, Martinou JC. Mol. Cell. Biol. 2000;20:929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. Cell Death Differ. 2000;7:1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 10.Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, Korsmeyer SJ. Mol. Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 11.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 12.Epand RF, Martinou JC, Fornallaz-Mulhauser M, Hughes DW, Epand RM. J. Biol. Chem. 2002;277:32632–32639. doi: 10.1074/jbc.M202396200. [DOI] [PubMed] [Google Scholar]
- 13.Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA, Korsmeyer SJ. Dev. Cell. 2002;2:55–67. doi: 10.1016/s1534-5807(01)00116-2. [DOI] [PubMed] [Google Scholar]
- 14.Degli Esposti M, Cristea IM, Gaskell SJ, Nakao Y, Dive C. Cell Death Differ. 2003;10:1300–1309. doi: 10.1038/sj.cdd.4401306. [DOI] [PubMed] [Google Scholar]
- 15.Lutter M, Fang M, Luo X, Nishijima M, Xie X, Wang X. Nat. Cell Biol. 2000;2:754–761. doi: 10.1038/35036395. [DOI] [PubMed] [Google Scholar]
- 16.Ostrander DB, Sparagna GC, Amoscato AA, McMillin JB, Dowhan W. J. Biol. Chem. 2001;276:38061–38067. doi: 10.1074/jbc.M107067200. [DOI] [PubMed] [Google Scholar]
- 17.Chou JJ, Li H, Salvesen GS, Yuan J, Wagner G. Cell. 1999;96:615–624. doi: 10.1016/s0092-8674(00)80572-3. [DOI] [PubMed] [Google Scholar]
- 18.McDonnell JM, Fushman D, Milliman CL, Korsmeyer SJ, Cowburn D. Cell. 1999;96:625–634. doi: 10.1016/s0092-8674(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 19.Fesik SW. Cell. 2000;103:273–282. doi: 10.1016/s0092-8674(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki M, Youle RJ, Tjandra N. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 21.Denisov AY, Madiraju MS, Chen G, Khadir A, Beauparlant P, Attardo G, Shore GC, Gehring K. J. Biol. Chem. 2003;278:21124–21128. doi: 10.1074/jbc.M301798200. [DOI] [PubMed] [Google Scholar]
- 22.Hinds MG, Lackmann M, Skea GL, Harrison PJ, Huang DC, Day CL. EMBO J. 2003;22:1497–1507. doi: 10.1093/emboj/cdg144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schendel SL, Azimov R, Pawlowski K, Godzik A, Kagan BL, Reed JC. J. Biol. Chem. 1999;274:21932–21936. doi: 10.1074/jbc.274.31.21932. [DOI] [PubMed] [Google Scholar]
- 24.Cramer WA, Heymann JB, Schendel SL, Deriy BN, Cohen FS, Elkins PA, Stauffacher CV. Annu. Rev. Biophys. Biomol. Struct. 1995;24:611–641. doi: 10.1146/annurev.bb.24.060195.003143. [DOI] [PubMed] [Google Scholar]
- 25.Schendel SL, Montal M, Reed JC. Cell Death Differ. 1998;5:372–380. doi: 10.1038/sj.cdd.4400365. [DOI] [PubMed] [Google Scholar]
- 26.Staley JP, Kim PS. Protein Sci. 1994;3:1822–1832. doi: 10.1002/pro.5560031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miroux B, Walker JE. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 28.Gross E, Witkop B. J. Am. Chem. Soc. 1961;83:1510–1511. [Google Scholar]
- 29.Opella SJ, Ma C, Marassi FM. Methods Enzymol. 2001;339:285–313. doi: 10.1016/s0076-6879(01)39319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z, Schendel S, Matsuyama S, Reed JC. Biochemistry. 1998;37:6410–6418. doi: 10.1021/bi973052i. [DOI] [PubMed] [Google Scholar]
- 31.Andrade MA, Chacon P, Merelo JJ, Moran F. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 32.Mori S, Abeygunawardana C, Johnson MO, Vanzijl PCM. J. Magn. Reson. B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Pines A, Gibby MG, Waugh JS. J. Chem. Phys. 1973;59:569–590. [Google Scholar]
- 35.Levitt MH, Suter D, Ernst RR. J. Chem. Phys. 1986;84:4243–4255. [Google Scholar]
- 36.Wu CH, Ramamoorthy A, Opella SJ. J. Magn. Reson. A. 1994;109:270–272. [Google Scholar]
- 37.Marassi FM. Biophys. J. 2001;80:994–1003. doi: 10.1016/S0006-3495(01)76078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 39.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. J. Biomol. NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 40.Marassi FM, Opella SJ. J. Magn. Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Denny J, Tian C, Kim S, Mo Y, Kovacs F, Song Z, Nishimura K, Gan Z, Fu R, Quine JR, Cross TA. J. Magn. Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 42.Franzin CM, Choi J, Zhai D, Reed JC, Marassi FM. Magn. Reson. Chem. 2004;42:172–179. doi: 10.1002/mrc.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh KJ, Barbuto S, Meyer N, Korsmeyer S. The 5th Biennial Structural Biology Symposium; Tallahassee, FL. [Google Scholar]
- 45.Marassi FM, Ma C, Gesell JJ, Opella SJ. J. Magn. Reson. 2000;144:156–161. doi: 10.1006/jmre.2000.2036. [DOI] [PubMed] [Google Scholar]
- 46.Losonczi JA, Olejniczak ET, Betz SF, Harlan JE, Mack J, Fesik SW. Biochemistry. 2000;39:11024–11033. doi: 10.1021/bi000919v. [DOI] [PubMed] [Google Scholar]
- 47.Boman HG. Annu. Rev. Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 48.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Sykes BD, Ryan RO. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]