Abstract
Viruses of the genus Flavivirus are responsible for significant human disease and mortality. The N-terminal domain of the flaviviral nonstructural (NS)3 protein codes for the serine, chymotrypsin-fold proteinase (NS3pro). The presence of the nonstructural (NS)2B cofactor, which is encoded by the upstream gene in the flaviviral genome, is necessary for NS3pro to exhibit its proteolytic activity. The two-component NS2B-NS3pro functional activity is essential for the viral polyprotein processing and replication. Both the structure and the function of NS2B-NS3pro are conserved in the Flavivirus family. Because of its essential function in the posttranslational processing of the viral polyprotein precursor, NS2B-NS3pro is a promising target for anti-flavivirus drugs. To identify selective inhibitors with the reduced cross-reactivity and off-target effects, we focused our strategy on the allosteric inhibitors capable of targeting the NS2B-NS3pro interface rather than the NS3pro active site. Using virtual ligand screening of the diverse, ∼275,000-compound library and the catalytic domain of the two-component West Nile virus (WNV) NS2B-NS3pro as a receptor, we identified a limited subset of the novel inhibitory scaffolds. Several of the discovered compounds performed as allosteric inhibitors and exhibited a nanomolar range potency in the in vitro cleavage assays. The inhibitors were also potent in cell-based assays employing the sub-genomic, luciferase-tagged WNV and Dengue viral replicons. The selectivity of the inhibitors was confirmed using the in vitro cleavage assays with furin, a human serine proteinase, the substrate preferences of which are similar to those of WNV NS2B-NS3pro. Conceptually, the similar in silico drug discovery strategy may be readily employed for the identification of inhibitors of other flaviviruses.
Introduction
West nile virus (WNV) and Dengue virus (DV) are transmitted to humans by the bites of infected female Aedes mosquitoes. According to WHO, there were >890,000 reported cases of Dengue in the Americas in 2007 alone, of which 26,000 cases were Dengue hemorrhagic fever. Anti-flaviviral therapies and vaccines are currently unavailable.
After flavivirus entry into the host cell, its ∼11-kb positive-sense RNA genome is uncoated and serves as a template for the translation of a single C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5 polyprotein precursor (for reviews see1 and references herein). The nascent polyprotein should be inserted into the endoplasmic reticulum membrane for its expression and processing by the host and viral proteinases. This processing results into the generation of the three structural proteins (C, prM, and E) and seven nonstructural (NS) proteins (NS1-NS5). The structural proteins are components of mature virus particles, whereas the NS proteins are not packaged into mature particles. The flaviviral full-length NS3 protein sequence represents a multifunctional protein in which the N-terminal ∼180-residue portion encodes serine proteinase (NS3pro) and the C-terminal ∼440-residue portion codes for an RNA helicase. The presence of the NS2B cofactor is necessary for NS3pro to exhibit its proteolytic activity.2,3 NS3pro is responsible for the cleavage of the capsid protein C, and also at the NS2A/NS2B, NS2B/NS3, NS3/NS4A, and NS4B/NS5 boundaries and, in addition, at the junction of NS4A/2K peptide. Inactivating mutations of the NS3pro cleavage sites in the polyprotein abolish replication of the virus.4 Therefore, NS2B-NS3pro is a promising anti-flaviviral drug target.5,6
There were several, though modestly successful, high-throughput screening (HTS) attempts to identify inhibitors of WNV NS2B-NS3pro.7–12 Our previous HTS studies suggested that the 5-amino-1-(phenyl)sulfonyl-pyrazol-3-yl class inhibitors interacted with the NS2B-binding cavity in the NS3pro domain and that they interfered with the unique feature of the flaviviral proteinases such as the productive interactions of the NS2B cofactor with the NS3pro domain.7,8 In turn, the enzyme active site is largely conserved in the human and viral serine proteinases, and it lacks the structural features, which could be readily exploited to achieving both the specificity and the potency of the inhibitors. Thus, it is likely that the small molecule interference with the productive conformation of the NS2B cofactor is a superior drug discovery strategy when compared with targeting of the active site of the viral proteinase. To validate this hypothesis, we employed a focused structure-based approach to identify de novo the allosteric small molecule inhibitors of NS2B-NS3pro using virtual ligand screening (VLS) technology.
Materials and Methods
Proteinase Expression and Purification
E. coli BL21 CodonPlus (DE3)-RIPL cells (Stratagene) were transformed with the individual recombinant pET101/DTOPO vectors encoding the WNV and the DV type 2 NS2B-NS3pro proteins.13–15 Transformed cells were grown in LB broth at 37°C to reach A600 = 0.6. The protein expression was induced at 37°C using 1 mM isopropyl β-D-thiogalactoside for an additional 6 h. The cells were collected by centrifugation, re-suspended in 20 mM Tris-HCl, pH 8.0, containing 1 M NaCl and 1 mg/mL lysozyme, and disrupted by sonication. Cell debris was removed by centrifugation. The WNV and DV proteins were purified from the supernatant fraction using HiTrap Co2+-chelating chromatography. The 6xHis-tagged NS2B-NS3pro constructs were eluted using a 0–500 mM gradient of imidazole concentrations. The fractions were analyzed using sodium dodecyl sulfate gel electrophoresis followed by Coomassie staining, and also by Western blotting with a 6xHis antibody (Clontech).
Proteinase Assays with Fluorescent Peptide
The assay was performed in 0.2 mL 20 mM Tris-HCl buffer, pH 8.0, containing 20% glycerol and 0.005% Brij 35. The cleavage peptide pyroglutamic acid Pyr-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin (Pyr-RTKR-AMC) and the enzyme concentrations were 25 μM and 10 nM, respectively. Reaction velocity was monitored continuously at λex = 360 nm and λem = 465 nm on a Spectramax Gemini EM fluorescence spectrophotometer (Molecular Devices). All assays were performed in triplicate in wells of a 96-well plate. Before the enzymatic studies, the concentrations of catalytically active NS2B-NS3pro were quantified by active site titration of the purified samples.16 Active site titration was performed with aprotinin (ki = 20 nM). NS2B-NS3pro (10 nM) was incubated with increasing concentrations of aprotinin. Residual activity of NS2B-NS3pro was measured by determining the rate of cleavage of Pyr-RTKR-AMC. The data were plotted versus the amounts of aprotinin and a line was fitted through the data points. The intercept on the x-axis equals to the concentration of active enzyme. The concentration of active NS2B-NS3pro was close to 100% when compared to the protein concentration. Furin activity assays with Pyr-RTKR-AMC were performed as described previously.17
Determination of the IC50 Values of the Compounds
NS2B-NS3pro (10 nM) was preincubated for 30 min at 20°C with increasing concentrations of the individual compounds in 0.1 mL 20 mM Tris-HCl buffer, pH 8.0, containing 20% glycerol and 0.005% Brij 35. The Pyr-RTKR-AMC substrate (25 μM) was added in 0.1 mL of the same buffer. All assays were performed in triplicate in wells of a 96-well plate. IC50 values were calculated by determining the concentrations of the compounds needed to inhibit 50% of the NS2B-NS3pro activity against Pyr-RTKR-AMC. GraphPad Prism was used as fitting software. Routine enzyme kinetics and inhibition assays have been described in our previous publication.16
Inhibition of WNV and DV RNA Replication
BHK21 cells stably expressing the WNV (New York 99 strain) and the DV type 2 (New Guinea C strain) sub-genomic replicons tagged with Renilla luciferase18,19 were a kind gift from Dr. R. Pad Padmanabhan (Georgetown University, Washington, DC) and Dr. Michael Diamond (Washington University, St. Louis, MO), respectively. Assays were performed in triplicate in wells of a 96-well, flat-bottom, white-wall plates (E&K Scientific). Cells (6,000/well) were incubated for 24 h at 37°C in 5% CO2 incubator. The increasing concentrations of the compounds dissolved in Dulbecco's modified Eagle's medium were added to the samples and incubation was continued for an additional 24 h. The cellular luciferase activity was measured using the EnduRen Live Cell Substrate for Renilla luciferase (Promega) according to the manufacturer's instructions. The IC50 values of the compounds were calculated using the GraphPad Prism software.
Cell Toxicity Assays
Assays were performed in triplicate in wells of a 96-well flat-bottom, white-wall plates. BHK21 cells (6,000/well) were incubated for 24 h at 37°C in 5% CO2 incubator. The increasing concentrations of the compounds dissolved in Dulbecco's modified Eagle's medium were then added to the samples and incubation was continued for an additional 24 h. The viable cells were counted using the ATP-Lite assay kit (PerkinElmer) according to the manufacturer's instructions. The TC50 values of the compounds were calculated using GraphPad Prism software.
Virtual Ligand Screening
The in silico experiments were performed using Q-MOL™ molecular modeling package (Q-MOL L.L.C., San Diego, CA; www.q-mol.com). The Optimized Potential for Liquid Simulations (OPLS) all atom force field20 is uniformly utilized within the Q-MOL program. The ligand docking simulations were conducted using the NS3pro crystal structure coordinates from PDB 2IJO (chain B). The protein molecule preparation included adding of hydrogen atoms and the assignment of the standard OPLS atom types. The ligand binding site was defined as a 10Å sphere centered at the Phe116, which is at the interface between the NS3pro domain and the NS2B cofactor. Thus, the distance between Phe116 and the Ser135 residue of the catalytic triad is 13.4Å (PDB 2IJO). The NS2B cofactor was then deleted from the structure. To increase the speed of calculations and to incorporate implicitly the flexibility of the NS3pro, the protein molecule was treated as a set of grid-based potentials accounting for the relevant NS3pro protein–ligand interactions. The ligands were docked into the grid-based potentials using the Monte Carlo simulation in the internal coordinate space as implemented in the Q-MOL program. The preparation of each ligand for docking simulation initially included an automatic OPLS atom-type assignment and conversion of the two-dimensional sketch-like models (input as the MDL MOL format) into the three-dimensional molecular models. The full-atom ligand structure was then minimized using the Q-MOL small molecule minimization protocol. The protocol combines minimization in both internal and Cartesian coordinates to properly optimize the rotable bonds of a small molecule. The complete NCI database (∼275,000 compounds) was used as a ligand source. The compounds were minimally filtered by applying lower molecular mass cut-off of 200 Da, a polyphenols sub-structure filter, and a filter detecting chlorine atoms attached to aliphatic carbons. Because of the stochastic nature of the Q-MOL docking protocol, each ligand was docked at least three times. The best energy conformers with the lowest binding energy were then selected. To differentiate between the true and false binders, Q-MOL VLS uses a proprietary protein–ligand binding energy evaluation function. This function is based on the re-parameterized OPLS force field, and, in addition to the protein–ligand interactions, it accounts for the internal energy change of the docked ligand. The exact implementation of the protocols will be described elsewhere.
Preparation of the NCI Compound Library for the VLS Process
To increase the overall performance of VLS, the compound database was converted into a binary space partitioning tree-like structure. The cluster tree is constructed by the iterative use of the modified k-value clustering algorithm (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertonline.com/adt). For this purpose, the chemical fingerprint keys are computed for each library compound. The precomputed keys are then used for a pair-wise, binary, grouping of the compounds. Only one compound from each clustered pair as well as all of the unclustered compounds are allowed to reach the next level. The chemical similarity threshold parameter controls the clustering within the individual tree levels. At level 1, which corresponds to the original compound library, the chemical similarity threshold was set at 95%. The clustering routine keeps track of the minimal and maximal similarities, which are computed at each of the individual levels of the tree. The compound relationship information is saved for each of the individual levels. For the subsequent tree levels, the minimal and maximal similarity threshold values are adjusted based on those of the preceding level. This algorithm generates a smooth distribution of the compounds within the clustering tree (Supplementary Fig. 1B). Because of the random nature of the k-value clustering, the cluster tree is generated for each individual VLS process with a unique random seed number. The cluster level numbers and the compound distribution for each level may vary, though insignificantly, among the VLS processes.
The binary space partitioning cluster tree is used in the VLS process as follows. The tree structure is traversed from the root to the upper levels. First, the start tree level is selected. This level must contain 50 ligands or more. These ligands are individually docked to the receptor. The ligands are then sorted according to their binding energy. The top 50 binders with the lowest energy are retained. These binders are used as seeds to compile a new ligand set from the preceding level. The closest analog is then selected for each of the individual ligands according to the chemical similarity parameter. This newly compiled set of 50 ligands is again docked to the receptor. The 2 sets are then combined, sorted according to their binding energy, and the 50 lowest energy binders are retained again. This procedure is repeated until the top level is reached. In the present docking study, 3,000–5,000 individual ligands were docked to the receptor and this number appears to sufficient to identify the top scoring ligands from the 275,000 compound NCI library. The total number of ligands that is required for docking may vary depending on the nature of the receptor, the compound distribution within a clustering tree and other parameters of the VLS process.
Modeling of Protein–Ligand Complexes
The predicted binding modes of the in vitro validated ligands were built using the full-atom flexible protein–ligand docking in the internal coordinates as implemented in the Q-MOL program. Briefly, the initial ligand conformations were taken from the VLS experiments. The protein–ligand complex was then globally optimized in the OPLS force field using the Monte Carlo simulation. The protein Phi, Psi, and Xi angles were allowed to changes. In the case of a ligand molecule, its positional and rotable torsion variables were unfixed.
In Silico Optimization of the Hits
The chemical structures of the initial in vitro validated hits were used as seeds for searching of the NCI database and for building a focused compound library (100 derivatives per each initial hit, ∼1,500 compounds after filtering). The Q-MOL chemical fingerprints were used as a chemical similarity measure during the search. The original validated hits were also included into the library as references. The focused library of derivatives was docked into the NS3pro proteinase using VLS protocol. The 50 best predicted binders with the lowest binding energy were ordered from NCI for the in vitro activity testing.
Ligand Source and Compound Databases
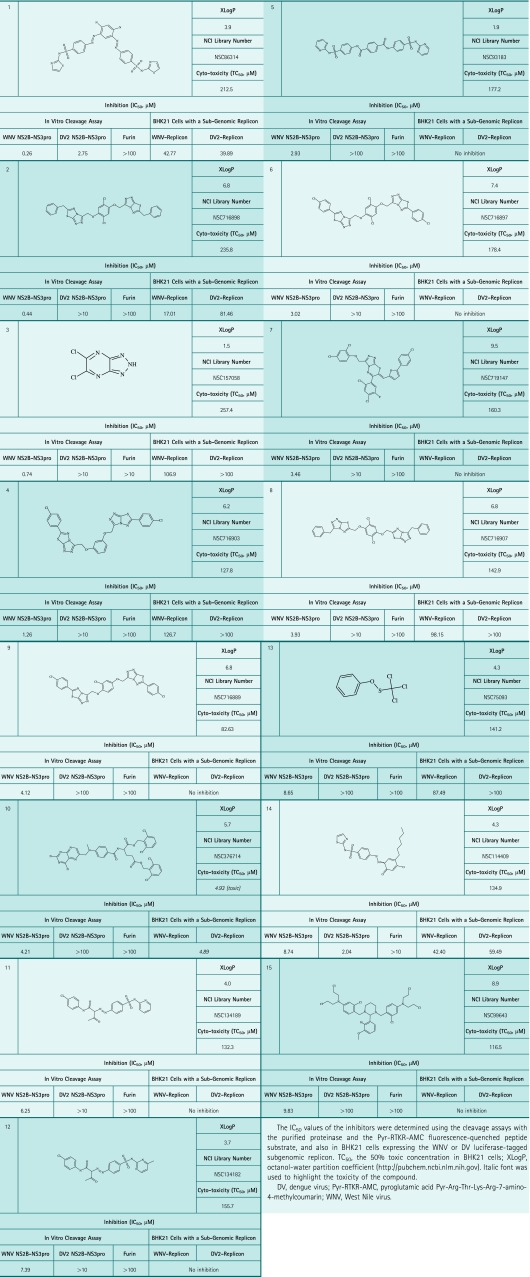
The ligands used in this work and the corresponding databases in SDF format were obtained from “The NCI/DTP Open Chemical Repository” at http://dtp.nci.nih.gov. The compounds were all >95% pure as certified by the supplier (NCI DTP Discovery Services). The ligands NCI/DTP accession numbers (NSC) are provided in Table 1. The ligands were dissolved in 100% dimethyl sulfoxide and stored at −20°C until use.
Table 1.
Inhibitors of West Nile Virus and Dengue Virus
Molecular Modeling Software
The Q-MOL molecular modeling package (Q-MOL L.L.C.; www.q-mol.com) was used in this work to conduct all molecular modeling experiments. The ordinary desktop computer (Intel Core 2 Quad Q6600 2.2 gGz, 4 GB RAM, OpenSUSE 11.0) was used to perform VLS and additional molecular modeling experiments.
Results
VLS and Protein–Ligand Docking
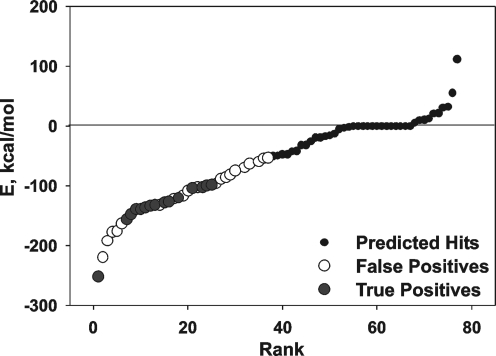
The diverse 275,000-compound NCI database was screened using Q-Mol software. The NS2B cofactor binding site rather than the active site groove of the catalytic NS3pro domain was used as a receptor for the ligand docking. The NBS2B cofactor binding site (the center of which is at a 13.4Å distance from Ser135 of the catalytic triad) was specifically selected for our VLS experiments. We selected this site because the compounds we have previously identified in our HTS studies performed as allosteric inhibitors of the catalytic activity and because they interacted with this particular site in the NS3pro domain.7,8 The application of the VLS protocol, followed by in silico optimization, resulted in the identification of a limited subset of 76 compounds, which predominantly included symmetric bilateral (C2 symmetry) compounds (Fig. 1, Table 1). A number of ligands, including ligands 3, appeared to be fragments of the larger compounds. Several inhibitory scaffolds we identified share common chemical features with the previously identified (phenyl)sulfonyl-pyrazol inhibitory scaffold.7 Despite their symmetry and the resulting substantial size, approximately half of the identified ligands exhibited the calculated octanol–water partition coefficients (XlogP) below 5, which was within the acceptable range for drug-like compounds.
Fig. 1.
Virtual ligand screening of the NCI compound library against WNV NS3pro. The complete NCI database (∼275,000 compounds) was docked into the NS2B cofactor-binding cavity in the NS3pro domain. The initial screening led to identification of the top 80 hits, which were then re-docked under more stringent conditions and ranked according to their relative binding energy E. The top 50 predicted binders were ordered from NCI and their inhibitory potency was tested in the in vitro cleavage assays. A significant fraction of the tested ligands were true-positives (active, gray dots), whereas other ligands were false-positives (inactive, white dots). WNV, West Nile virus.
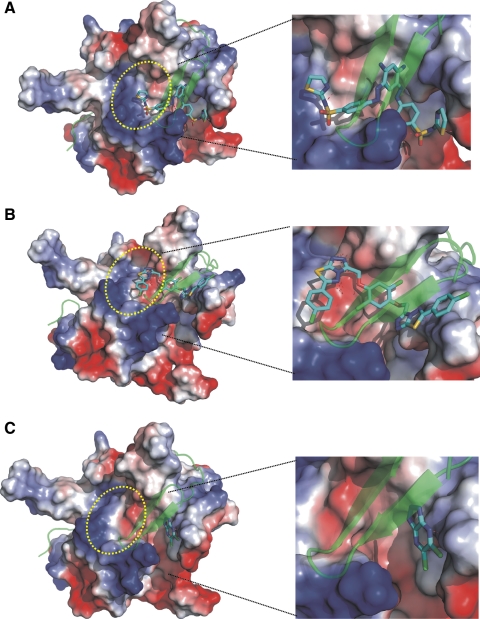
The predicted binding modes of the ligands 1 and 2 suggested their possible interactions with both the active site of the NS3 proteinase and the NS2B cofactor-binding site (Fig. 2). In contrast, ligand 3 was predicted to interact only with the cofactor-binding site. The predicted binding modes of ligands 1 and 2 seem to indicate that bilateral symmetry has certain structural benefits because the equivalent parts of the structure are likely to be equally engaged in the interactions with the protein target. These ligands were favored during the VLS experiments because of the significant entropic gain due to their symmetry.
Fig. 2.
Predicted binding models of the WNV NS2B-NS3pro inhibitors. (A) Ligand 1; (B) ligand 2; (C) ligand 3. The NS3pro is rendered as a molecular surface, colored by its electrostatic potential. NS2B is transparent green. The approximate position of the active site is highlighted by a yellow oval. Hydrogen bonds are shown as red dotted lines. Refer to Table 1 for the ligand structures. The figure was generated using PyMol Molecular Graphics software.
Our protein–ligand complex modeling experiments suggested that the identified small molecule ligands are capable of interfering with the productive positioning of the NS2B cofactor relative to the NS3pro active site. Our earlier structural work has determined that the NS2B cofactor interacts directly with both the active site of the NS3pro and the P3 residue of the substrate.2 In agreement, the deletion of the NS2B sequence or the focused mutagenesis of the NS3 cofactor inactivates the catalytic activity of the two-component NS2B-NS3pro.3,21,22 Our modeling studies, however, could not provide an explicit answer if the identified inhibitors interact only with the NS3 proteinase domain or with both the proteinase and the NS2B cofactor.
Determination of the Inhibitory Efficiency and Specificity of the Ligands
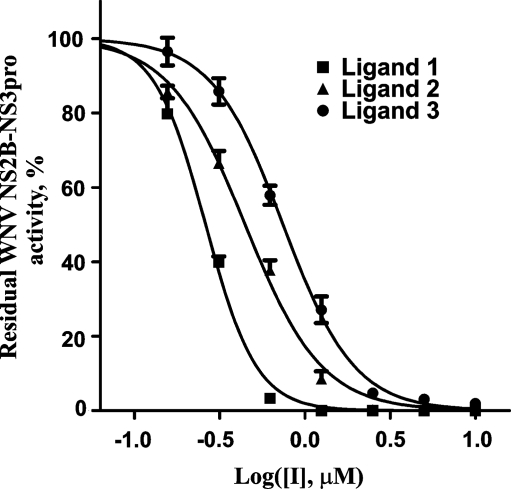
The available compounds that have been predicted by VLS to perform as efficient binders were ordered from the publicly available NCI compound library of the Developmental Therapeutics Program NCI/NIH (http://dtp.nci.nih.gov). The inhibitory potency of the ordered compounds was then directly tested in the cleavage reactions in vitro using the recombinant WNV and DV NS2B-NS3 proteinases and human furin and Pyr-RTKR-AMC as the peptide cleavage substrate (Table 1, Fig. 3).
Fig. 3.
Selected inhibitors efficiently inhibit the catalytic activity of WNV NS2B-NS3pro. Before the addition of the pyroglutamic acid Pyr-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin substrate (25 μM), the purified WNV proteinase (10 nM) was co-incubated for 30 min with increasing concentrations of the inhibitors. The residual activity was then monitored continuously at λex = 360 nm and λem = 465 nm to determine the initial velocity of the reactions. The initial velocity was calculated as a percentage of residual activity versus the untreated proteinase (control). Refer to Table 1 for the ligand structures.
In addition to identifying their inhibitory potency, we have tested the ligands for off-target activity against homologous serine proteases, including furin and DV NS2B-NS3pro. None of the ligands significantly inhibited furin. DV NS2B-NS3pro, which is 70% identical to WNV NS2B-NS3pro at the amino acid sequence level, was only moderately inhibited by ligands 1 and 14. Ligand 1, however, was 10-fold more potent against WNV NS2B-NS3pro (IC50 = 260 nM) as compared with the DV proteinase (IC50 = 2.75 μM). Ligand 14 was a weak WNV inhibitor (IC50 = 8.74 μM).
Determination of the Inhibitory Mechanism
On the basis of our drug discovery strategy, the identified inhibitors should display the noncompetitive inhibition mechanism. To test this suggestion experimentally, we determined the inhibition mechanism using the most potent inhibitors of WNV NS2B-NS3pro. The most efficient compounds 1 and 2 exhibited a mixed inhibitory mechanism, which affected both the KM and the VM parameters. The mixed mechanism also suggested that there are the alternative, opened and closed, conformations of NS2B-NS3pro. These results are consistent with the most recent observations by others.11,23–27 Ligand 3 performed as a noncompetitive inhibitor of NS2B-NS3pro (Table 2). The data obtained for these ligands are consistent with our proteinase–inhibitor complex modeling studies (Fig. 2). Ligands 1 and 2 were predicted to interact with both the active site and the NS2B cofactor-binding site. In contrast, ligand 3 was predicted to interact only with the NS2B cofactor-binding site in the NS3pro domain.
Table 2.
Kinetic Parameters of the Selected Inhibitors
| |
|
|
Inhibitor Concentrations, μM |
||||||
|---|---|---|---|---|---|---|---|---|---|
| ID | NSC | Kinetic Parameters | 0 | 0.156 | 0.3125 | 0.625 | 1.25 | 2.5 | 5 |
| 1 |
NSC86314 |
VM, RFU/s |
35.84 ± 0.26 |
— |
34.65 ± 0.50 |
32.46 ± 0.59 |
28.77 ± 0.49 |
21.87 ± 0.70 |
13.10 ± 0.61 |
| |
|
KM, μM |
57.38 ± 1.02 |
— |
51.79 ± 1.92 |
51.45 ± 2.40 |
46.38 ± 2.08 |
40.95 ± 3.59 |
25.02 ± 3.64 |
| 2 |
NSC716898 |
VM, RFU/s |
34.24 ± 0.28 |
— |
35.25 ± 0.24 |
35.94 ± 0.67 |
32.34 ± 0.26 |
27.27 ± 0.34 |
21.78 ± 0.66 |
| |
|
KM, μM |
48.84 ± 1.02 |
— |
49.37 ± 0.86 |
54.75 ± 2.56 |
43.29 ± 0.92 |
36.83 ± 1.29 |
28.26 ± 2.60 |
| 3 |
NSC157058 |
VM, RFU/s |
32.27 ± 0.59 |
32.87 ± 0.53 |
31.90 ± 0.50 |
30.18 ± 0.50 |
26.46 ± 0.68 |
24.08 ± 0.49 |
— |
| KM, μM | 44.17 ± 2.17 | 45.69 ± 1.97 | 45.93 ± 1.90 | 45.35 ± 2.01 | 40.87 ± 2.86 | 40.55 ± 2.26 | — | ||
The KM and Vmax values were determined using the purified WNV NS2B-NS2pro samples and the pyroglutamic acid-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin fluorescence-quenched peptide substrate in the presence of the indicated concentrations of the inhibitors.
RFU, relative fluorescence unit.
Inhibition of WNV RNA Replication
To support our in vitro data, the inhibition of WNV RNA replication was then measured using baby hamster kidney fibroblast BHK21 cells that expressed the WNV sub-genomic replicon encoding the Renilla luciferase reporter.19,28,29 Because the polyprotein processing followed by the assembly of the viral replication complex is required for RNA replication, the cellular permeability of the inhibitors could be assessed by monitoring the replicon-encoded luciferase activity as a function of inhibitor concentration. These cell-based tests demonstrated that the low micron range concentrations of the selected inhibitors were capable of penetrating into the cell and of repressing the luciferase activity that is indicative of RNA replication. Similarly, the inhibitors were capable of inhibiting, though less efficiently, the cleavage activity in vitro and replication of the DV replicon (Table 1).
Our additional studies in BHK21 cells demonstrated that the inhibitors (except ligand 10) did not exhibit a significant level of cytotoxicity. In addition, the selected compounds did not affect the luciferase activity itself (data not shown). Thus, it is highly likely that the inhibitory ligands, including ligands 1, 2, and 3, directly affected the replicon-encoded NS2B-NS3pro activity.
Discussion
Inactivating mutations of the WNV NS3pro cleavage sites in the polyprotein abolish replication of the virus.4 As a result, NS2B-NS3pro is a promising anti-flaviviral drug target.5,6 Our previous mutagenesis studies demonstrated that both the integrity and the productive conformation of the NS2B cofactor are critical for the activity of the NS3pro.2,7,8,16 According to the studies of others, it is highly likely that NS2B-NS3pro normally exists in two distinct conformations: open and closed. In the open, active conformation, the NS2B cofactor interacts with the active site of the enzyme. In contrast, in the closed, inactive conformation, the NS2B cofactor remains bound to the NS3pro domain, but it is shifted away from the proteinase active site.24,25,30,31
We hypothesized that these conformational transitions could be exploited to abolish the enzyme activity and to help to identify small molecule inhibitory compounds, capable of inducing the nonproductive, closed conformation of NS2B-NS3pro. In agreement, our recent HTS studies suggested that certain class of inhibitors interacted with the interface of the NS2B cofactor and the NS3pro catalytic domain interfering with the formation of the productive cofactor–catalytic domain complex.7,8
The small molecule interference with the productive interaction of the NS2B cofactor with NS3pro could be a superior drug discovery strategy when compared with targeting of the conserved active site of the flaviviral proteinase. To test this assumption, we employed a focused structure-based approach to identify de novo the allosteric small molecule inhibitors of NS2B-NS3pro. We have used high-throughput in silico docking of a diversity set of ∼275,000 molecules from a publicly available NCI compound library of the Developmental Therapeutics Program NCI/NIH (http://dtp.nci.nih.gov). The docking site was set at the interface of the NS2B cofactor and the NS3pro catalytic domain.7 Before the docking simulations the NS2B cofactor was removed from the NS2B-NS3pro structure, and the NS3pro catalytic domain alone was used as a receptor molecule in the VLS experiments.
Protein–ligand docking was followed by extensive experimental in vitro and cell-based tests. These combined studies led us to the identification of the three most promising and novel inhibitory scaffolds. According to our in vitro tests, these scaffolds performed as a nanomolar range, selective inhibitors of WNV NS2B-NS3pro. The inhibitors did not display cross-reactivity with human furin and moderately inhibited highly homologous DV NS2B-NS3pro. These cross-reactivity studies also dismissed the potential promiscuity of the compounds, which could be associated with their aggregation. Poorly soluble ligands could appear as nonspecific inhibitors because they may cause protein sequestration. The distinct specificity profile of our selected inhibitors indicates that they are not prone to the formation of aggregates. In agreement, two compounds we selected inhibited flaviviral replication in cell-based experiments, thus providing additional evidence in support of their specificity. Because of their sub-optimal drug-like parameters, many of the identified structural scaffolds would be difficult to transform into the drug-like hits. Several compounds, including ligands 1, 3, and 5, with the XLogP value below 5 may considered as potential candidates for medicinal chemistry optimization (Table 1). The identification of these scaffolds, however, confirms the efficiency of our VLS approach and also the presence of the allosteric druggable site in the NS3pro catalytic domain that is outside of the active site cavity of the proteinase.
The inhibition mechanism studies and the predicted structures of the NS3pro-inhibitor complexes suggest that the discovered inhibitors induce the inactive, closed conformation of NS2B-NS3pro. The symmetric nature (bilateral C2) seems to significantly contribute to the binding affinity of the compounds. The bilateral symmetry offers entropic advantage to a ligand because productive binding modes are more frequent if compared to nonsymmetric compounds. For example, natural compounds with bilateral symmetry are more frequently enzyme inhibitors (22%) in comparison with nonsymmetrical natural compound (8%).32
The inhibitory characteristics of the scaffolds we identified are comparable with those selected by others. The existing WNV and DV NS2B-NS3 inhibitors are predominantly peptide based and they target directly the active site of the NS3pro domain. The ki values of these peptide-based inhibitors are in a low nanomolar range.16,33–35 The practical use of these peptide inhibitors in vivo, however, is limited. In addition, fragment-based docking led to the small-molecule inhibitors the reported IC50 values of which were in a low micron range.10 Similarly, the inhibitory compounds identified by Mueller et al.12 and Ganesh et al.36 exhibited the ki values above 3 and 14 μM, respectively. The SRI-19093 inhibitor described by Chung et al.37 displayed the 440 nM IC50 value.
Taken together with our earlier data,2,7,8 our proof-of-principle work has demonstrated that disrupting the interactions of the NS2B cofactor with the NS3 protease is the promising strategy for rational structure-based inhibitor development. Further medicinal chemistry work is required to optimize the current inhibitory leads. Conceptually, the drug discovery strategy employed in our study could be readily applied for the identification of inhibitors of other flaviviral proteinases including DV NS2B-NS3pro.
Supplementary Material
Abbreviations
- DV
Dengue virus
- HTS
high-throughput screening
- NS
nonstructural
- NS3pro
NS3 proteinase
- OPLS
Optimized Potential for Liquid Simulations
- Pyr-RTKR-AMC
pyroglutamic acid Pyr-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin
- RFU
relative fluorescence unit
- VLS
virtual ligand screening
- WNV
West Nile virus
Acknowledgments
This work was supported by Public Health Service Grants U01AI078048 and U01AI061139 from the NIAID, NIH.
Disclosure Statement
No competing financial interests exist.
References
- 1.Hanley KA. Weaver SC. Caister Academic Press; Norfolk, UK: 2010. Frontiers in Dengue Virus Research. [Google Scholar]
- 2.Aleshin AE. Shiryaev SA. Strongin AY. Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erbel P. Schiering N. D'Arcy A, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue, West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 4.Chambers TJ. Droll DA. Tang Y, et al. Yellow fever virus NS2B-NS3 protease: characterization of charged-to-alanine mutant and revertant viruses and analysis of polyprotein-cleavage activities. J Gen Virol. 2005;86:1403–1413. doi: 10.1099/vir.0.80427-0. [DOI] [PubMed] [Google Scholar]
- 5.Chappell KJ. Stoermer MJ. Fairlie DP. Young PR. West Nile Virus NS2B/NS3 protease as an antiviral target. Curr Med Chem. 2008;15:2771–2784. doi: 10.2174/092986708786242804. [DOI] [PubMed] [Google Scholar]
- 6.Lescar J. Luo D. Xu T, et al. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Johnston PA. Phillips J. Shun TY, et al. HTS identifies novel, specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 8.Sidique S. Shiryaev SA. Ratnikov BI, et al. Structure-activity relationship, improved hydrolytic stability of pyrazole derivatives that are allosteric inhibitors of West Nile Virus NS2B-NS3 proteinase. Bioorg Med Chem Lett. 2009;19:5773–5777. doi: 10.1016/j.bmcl.2009.07.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller NH. Yon C. Ganesh VK. Padmanabhan R. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol. 2007;39:606–614. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 10.Ekonomiuk D. Su XC. Ozawa K, et al. Flaviviral protease inhibitors identified by fragment-based library docking into a structure generated by molecular dynamics. J Med Chem. 2009;52:4860–4868. doi: 10.1021/jm900448m. [DOI] [PubMed] [Google Scholar]
- 11.Ekonomiuk D. Su XC. Ozawa K, et al. Discovery of a non-peptidic inhibitor of west nile virus NS3 protease by high-throughput docking. PLoS Negl Trop Dis. 2009;3:e356. doi: 10.1371/journal.pntd.0000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller NH. Pattabiraman N. Ansarah-Sobrinho C. Viswanathan P. Pierson TC. Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of west nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52:3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shiryaev SA. Aleshin AE. Ratnikov BI. Smith JW. Liddington RC. Strongin AY. Expression and purification of a two-component flaviviral proteinase resistant to autocleavage at the NS2B-NS3 junction region. Protein Expr Purif. 2007;52:334–339. doi: 10.1016/j.pep.2006.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiryaev SA. Kozlov IA. Ratnikov BI. Smith JW. Lebl M. Strongin AY. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of Dengue and West Nile viruses. Biochem J. 2007;401:743–752. doi: 10.1042/BJ20061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiryaev SA. Ratnikov BI. Aleshin AE, et al. Switching the substrate specificity of the two-component NS2B-NS3 flavivirus proteinase by structure-based mutagenesis. J Virol. 2007;81:4501–4509. doi: 10.1128/JVI.02719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiryaev SA. Ratnikov BI. Chekanov AV, et al. Cleavage targets, the D-arginine-based inhibitors of the West Nile virus NS3 processing proteinase. Biochem J. 2006;393:503–511. doi: 10.1042/BJ20051374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gawlik K. Shiryaev SA. Zhu W, et al. Autocatalytic activation of the furin zymogen requires removal of the emerging enzyme's N-terminus from the active site. PLoS One. 2009;4:e5031. doi: 10.1371/journal.pone.0005031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padmanabhan R. Mueller N. Reichert E, et al. Multiple enzyme activities of flavivirus proteins. Novartis Found Symp. 2006;277:74–84. doi: 10.1002/0470058005.ch6. discussion 84–76, 251–253. [DOI] [PubMed] [Google Scholar]
- 19.Puig-Basagoiti F. Deas TS. Ren P. Tilgner M. Ferguson DM. Shi PY. High-throughput assays using a luciferase-expressing replicon, virus-like particles, and full-length virus for West Nile virus drug discovery. Antimicrob Agents Chemother. 2005;49:4980–4988. doi: 10.1128/AAC.49.12.4980-4988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen WL. Maxwell DS. Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 21.Radichev I. Shiryaev SA. Aleshin AE, et al. Structure-based mutagenesis identifies important novel determinants of the NS2B cofactor of the West Nile virus two-component NS2B-NS3 proteinase. J Gen Virol. 2008;89:636–641. doi: 10.1099/vir.0.83359-0. [DOI] [PubMed] [Google Scholar]
- 22.Yusof R. Clum S. Wetzel M. Murthy HM. Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 23.Robin G. Chappell K. Stoermer MJ, et al. Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation. J Mol Biol. 2009;385:1568–1577. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 24.Su XC. Ozawa K. Qi R. Vasudevan SG. Lim SP. Otting G. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl Trop Dis. 2009;3:e561. doi: 10.1371/journal.pntd.0000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su XC. Ozawa K. Yagi H, et al. NMR study of complexes between low molecular mass inhibitors, the West Nile virus NS2B-NS3 protease. FEBS J. 2009;276:4244–4255. doi: 10.1111/j.1742-4658.2009.07132.x. [DOI] [PubMed] [Google Scholar]
- 26.Tomlinson SM. Malmstrom RD. Russo A. Mueller N. Pang YP. Watowich SJ. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res. 2009;82:110–114. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wichapong K. Pianwanit S. Sippl W. Kokpol S. Homology modeling and molecular dynamics simulations of Dengue virus NS2B/NS3 protease: insight into molecular interaction. J Mol Recognit. 2010;23:283–300. doi: 10.1002/jmr.977. [DOI] [PubMed] [Google Scholar]
- 28.Lo MK. Tilgner M. Shi PY. Potential high-throughput assay for screening inhibitors of West Nile virus replication. J Virol. 2003;77:12901–12906. doi: 10.1128/JVI.77.23.12901-12906.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi PY. Tilgner M. Lo MK. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology. 2002;296:219–233. doi: 10.1006/viro.2002.1453. [DOI] [PubMed] [Google Scholar]
- 30.Tomlinson SM. Watowich SJ. Substrate inhibition kinetic model for West Nile virus NS2B-NS3 protease. Biochemistry. 2008;47:11763–11770. doi: 10.1021/bi801034f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekonomiuk D. Caflisch A. Activation of the West Nile virus NS3 protease: molecular dynamics evidence for a conformational selection mechanism. Protein Sci. 2009;18:1003–1011. doi: 10.1002/pro.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greer A. Wauchope OR. Farina NS. Haberfield P. Liebman JF. Paradigms and paradoxes: mechanisms for possible enhanced biological activity of bilaterally symmetrical chemicals. Struct Chem. 2006;17:347–350. [Google Scholar]
- 33.Stoermer MJ. Chappell KJ. Liebscher S, et al. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability, antiviral activity. J Med Chem. 2008;51:5714–5721. doi: 10.1021/jm800503y. [DOI] [PubMed] [Google Scholar]
- 34.Yin Z. Patel SJ. Wang WL, et al. Peptide inhibitors of dengue virus NS3 protease. Part 2: SAR study of tetrapeptide aldehyde inhibitors. Bioorg Med Chem Lett. 2006;16:40–43. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 35.Yin Z. Patel SJ. Wang WL, et al. Peptide inhibitors of Dengue virus NS3 protease. Part 1: Warhead. Bioorg Med Chem Lett. 2006;16:36–39. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 36.Ganesh VK. Muller N. Judge K. Luan CH. Padmanabhan R. Murthy KH. Identification and characterization of nonsubstrate based inhibitors of the essential dengue and West Nile virus proteases. Bioorg Med Chem. 2005;13:257–264. doi: 10.1016/j.bmc.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 37.Chung DH. Jonsson CB. Maddox C, et al. HTS-driven discovery of new chemotypes with West Nile virus inhibitory activity. Molecules. 2010;15:1690–1704. doi: 10.3390/molecules15031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.