Abstract
Singapore reported its first locally acquired human Plasmodium knowlesi infection in 2007, involving a soldier who had undergone training in a forested area where long-tailed macaques are frequently seen. Comprehensive disease surveillance and monitoring system that was set up after the initial case detected four additional human P. knowlesi cases in 2007 and one in 2008. All involved military personnel who had undergone training in the forested area, and none had traveled out of Singapore 1 month before the onset of symptoms. Screening for malaria parasites on blood obtained from long-tailed macaques revealed that wild monkeys (n = 3) caught from the forested area were infected with P. knowlesi, whereas peri-domestic monkeys (n = 10) caught from a nature reserve park were not infected with any malaria parasites. Phylogenetic analysis of the nonrepeat region of the P. knowlesi csp genes showed that the sequences obtained from the human cases were not distinct from those obtained from wild monkeys. Further, certain genotypes were shared between samples from humans and macaques. Our findings provide evidence that long-tailed macaques are the natural hosts of P. knowlesi in Singapore and the human cases acquired their infection in the same vicinity where these monkeys are found. Further, the risk of acquiring P. knowlesi infection among the general population of Singapore is small as evident from the absence of P. knowlesi in peri-domestic monkeys.
Key Words: Singapore, Plasmodium Knowlesi, long-tailed macaques, Circumsporozoite genes
Introduction
Plasmodium knowlesi was first identified in India in 1931 from a long-tailed macaque (Macaca fascicularis) imported from Singapore (Knowles and Das Gupta 1932). Its ability to infect humans was first described in 1932, when Knowles and Das Gupta successfully transmitted the parasite to two human volunteers by blood passages from infected macaques. However, the first natural human infection of P. knowlesi was only reported in 1965 in an American army surveyor who had acquired the disease while working in the jungle in the state of Pahang, Malaysia (about 300 km north of Singapore) (Chin et al. 1965). This was followed by a presumptive case reported from the state of Johor, Malaysia, which is adjacent to the island of Singapore (Yap et al. 1971). Human infections were thought to be rare until a large focus of humans infected with P. knowlesi were identified by nested polymerase chain reaction (PCR) detection assays in Sarawak, Malaysian Borneo, in 2004 (Singh et al. 2004). Since then, cases of P. knowlesi infections in humans have been reported in other parts of Malaysia, China, Thailand, Singapore, and Philippines (Jongwutiwes et al. 2004, Zhu et al. 2006, Cox-Singh et al. 2008, Luchavez et al. 2008, Ng et al. 2008, Vythilingam et al. 2008), resulting in knowlesi being recognized as the first Plasmodium sp. implicated in zoonotic disease. P. knowlesi infections have also been reported from European travellers returning from endemic countries (Kantele et al. 2008, Bronner et al. 2009).
In Singapore, the first reported locally acquired human P. knowlesi infection occurred in 2007 and involved a soldier in the Singapore military who had no significant travel history and had trained in a restricted-access forested area in Singapore (Ng et al. 2008). Long-tailed macaques, the natural hosts for P. knowlesi, are found in this forested area and also in various nature reserve parks in Singapore that are open to the general public. Comprehensive fever surveillance and monitoring was started among military personnel who had taken part in training exercises in the affected forested area. Five additional human knowlesi malaria cases were identified and confirmed by various laboratories using PCR—four in 2007 and one in 2008.
This study aims to determine whether long-tailed macaques in Singapore are infected with P. knowlesi and whether they are the source of the human knowlesi infections using molecular analysis to determine the epidemiological linkages among the human infections and the macaques.
Methods
Human and macaque samples
To determine the link between human and macaque P. knowlesi, residual blood samples collected in ethylenediaminetetraacetic acid from three out of the five human cases from 2007 and the case from 2008 were sent for further molecular investigation at the Environmental Health Institute (EHI), a national public health laboratory. The samples were denoted SG/EHI/H-001, SG/EHI/H-002, SG/EHI/H-007, and SG/EHI/H-024.
Blood samples in ethylenediaminetetraacetic acid obtained from long-tailed macaques were also received at EHI for analysis. The macaques were caught under an operational surveillance program that has been approved by the Singapore military's medical review committee and by the DSO National Laboratory's Institutional Animal Care and Use Committee. The monkeys caught were turned over to the national veterinary authority for blood sampling, in accordance with ethics practices by the national veterinary authority. Two sets of macaque blood samples were received: Set 1 was sampled from three macaques from the affected restricted training area (SG/EHI/LT-001 and SG/EHI/LT-002 were caught in November 2007, and SG/EHI/LT-017 was caught in June 2009); Set 2 (SG/EHI/LT-003 to SG/EHI/LT-012) was from 10 peri-domestic macaques from a public nature reserve park caught in January 2008.
This study was part of an operational effort by the Singapore military and the National Environmental Agency, Singapore, to assess the risk of P. knowlesi transmission.
DNA extraction and nested PCR assays
DNA was extracted from whole-blood samples using QiaAmp Blood Extraction Kit (Qiagen) according to the manufacturer's protocol. Extracted DNA samples were stored at 4°C until use. Plasmodium sp.–specific nested PCR assays to detect the presence of malaria parasites in the blood of these samples were performed as described by Singh et al. (1999), with slight modifications. Nest 1 PCR amplification was carried out in a 50 μL reaction mixture containing 1× green buffer (Promega), 3 mM MgCl2 (Promega), 200 mM of each dNTPs (Promega), 300 nM of each primers, and 1.25 U of Go Taq DNA polymerase (Promega), and 5 μL of DNA template was used for each reaction. For Nest 2, PCR amplification was carried out in a 20 μL reaction containing 1 × green buffer (Promega), 2 mM MgCl2 (Promega), 200 mM of each dNTPs (Promega), 300 nM of each primers, and 0.5 U of Go Taq DNA polymerase, and 2 μL of the nest 1 PCR product were used as DNA templates. All PCRs were carried out using a T-Gradient thermal cycler (Biometra GmbH). Nest 2 amplicons were analyzed by agarose gel electrophoresis, stained with ethidium bromide, and observed under a ultraviolet transilluminator.
For the detection of P. knowlesi in macaque samples, primers Pmk8 and Pmkr9 were used as described by Singh et al. (2004). The concentration and constituents of the P. knowlesi–specific nested PCR assay were identical to the nest 2 amplification reactions mentioned above.
Cloning and sequencing of the circumsporozoite protein genes of P. knowlesi
The circumsporozoite protein (csp) genes of P. knowlesi from human and monkey samples were amplified with primers PKCSPF2 (5′ TACAAGAACAAGATGARGAAC 3′) and PKCSPR2 (5′ TCAGCTACTTAATTGAATAATGC 3′), respectively. PCR amplification was carried out in a 20 μL reaction mixture containing 1 × HF buffer (Finnzymes), 200 mM of each dNTPs (Finnzymes) and 300 nM of each primer, and 0.02 U of Phusion DNA Polymerase (Finnzymes). The PCR was carried out using a T-Gradient thermal cycler (Biometra). The PCR conditions to amplify the csp gene were as follows: 98°C for 30 s followed by 40 cycles of amplification at 94°C for 7 s, 56°C for 20 s, and 72°C for 20 s followed by a final extension step of 10 min. The expected size of the blunt-ended amplicons for the csp genes is approximately 1.2 kb. PCR products were cloned using Zero Blunt® Topo® PCR cloning kit (Invitrogen) and performed following manufacturer's protocol. Plasmid DNA from clones having the desired DNA fragment was extracted using the Plasmid Miniprep (Qiagen) kit according to the manufacturer's instructions. At least 50 purified plasmids from each monkey sample and 30 from each human sample were used for sequencing. The entire csp gene was sequenced using M13 primers and two internal primers (Singh et al. 2004). Sequencing was performed by a commercial laboratory using the BigDye Terminator Cycle Sequencing kit (Applied Biosystems).
Sequence analysis
The csp gene sequences were analyzed as described previously (McCutchan et al. 1996, Singh et al. 2004). Sequences from the 456 nucleotides that encode the nonrepeat N-terminal (first 195 nucleotides of the coding sequence) and C-terminal (the last 261 nucleotides of the coding sequence) regions of the csp genes were aligned with Clustal W using Megalign software (Lasergene; DNASTAR). The sequences obtained from this study were phylogenetically compared to those in GenBank. The phylogenetic trees were constructed using the neighbor-joining (NJ) method. The NJ tree was constructed using MEGA version 4.0 software (Tamura et al. 2007) and analyzed using the Kimura-2 parameter model including transitions and transversions.
The csp gene sequences obtained from GenBank were the following: Plasmodium falciparum (K02194), Plasmodium vivax (M34697), Plasmodium malariae (J03992), P. knowlesi (M11031, AH013332, AH013334, and AH013337), Plasmodium coatneyi (AY135360), Plasmodium cynomolgi (M15104), Plasmodium berghei (M14135), Plasmodium simiovale (U09765), Plasmodium inui (FJ009512), Plasmodium simium (L05068), and Plasmodium yoelii (J02695).
Results
All six human P. knowlesi cases in Singapore were adult men, with a median age of 20 years (range 18–53), serving in the military. None of the six cases had traveled out of Singapore 1 month before the onset of illness, and none had traveled to areas known to be at risk for P. knowlesi over the previous year. However, all of the servicemen had been training in a forested area in Singapore 1–2 weeks before their onset of illness. Each spent at least one night in the forest, which is the most likely place where the infections were acquired.
Molecular detection of P. knowlesi in macaque samples
Analysis by the nested PCR assays showed that the 3 macaques from the same restricted training areas that were visited by the human cases were positive for P. knowlesi, whereas the 10 sampled at the nature reserve park were negative for Plasmodium DNA.
Molecular investigation of P. knowlesi in human and macaque samples
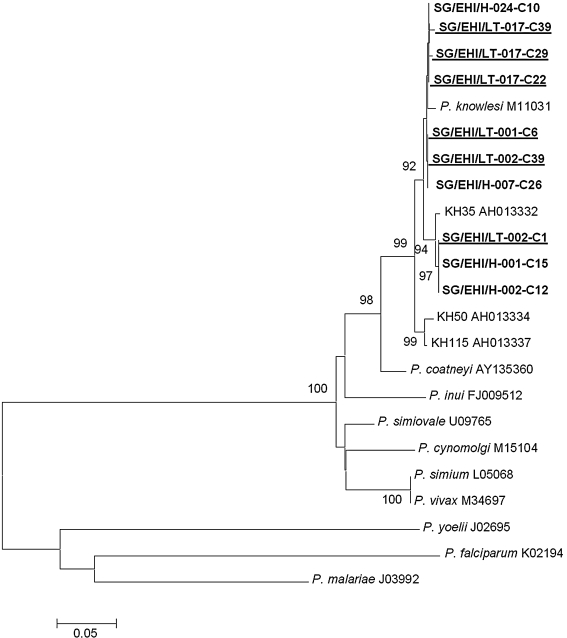
P. knowlesi DNA from four of the six human cases and the three macaques were subjected to PCR amplification, cloning, and sequencing of the Plasmodium sp. csp gene. Phylogenetic analysis inferred from the NJ method showed that the nonrepeat region of the csp genes of the malaria parasites from these human and monkey samples formed a monophyletic clade with other P. knowlesi csp genes obtained from GenBank (Fig. 1). Alignment of the 453-nucleotide residues encoding the nonrepeat N- and C-terminal regions from each clones revealed that each long-tailed macaque was harboring one to three genotypes of P. knowlesi. Analysis of the complete csp gene showed that macaques SG/EHI/LT-017, SG/EHI/LT-002, and SG/EHI/LT-001 had three genotypes, two genotypes, and one genotype, respectively, whereas one genotype was observed from each human isolate. When the 5′ and 3′ flanking regions of the csp genes from the Singapore isolates were compared to that of the reference Nuri strain, the pairwise identity ranges from 97.8% to 99.3% (data not shown). Comparison of P. knowlesi isolated from Singapore with that of Nuri strain revealed 12 polymorphic sites (Table 1), of which 3 and 9 were due to nonsynonymous and synonymous mutations, respectively. However, in general, P. knowlesi sequences from human cases were not phylogenetically distinct from the sequences from macaques.
FIG. 1.
Phylogenetic tree based on the nonrepeat region of the circumsporozoite protein genes of Plasmodium sp. produced by the neighbor-joining method. Clones highlighted in bold are obtained from Singapore isolates and clustered in the Plasmodium knowlesi clade. Clones highlighted are from human isolates, whereas those highlighted and underlined are from monkey isolates. Figures on the branches are bootstrap percentages based on 1000 replicates, and only those above 70% are shown.
For the three human cases from 2007, sequence analysis of the complete csp gene from each sample revealed that two genotypes of P. knowlesi were isolated from the three human cases and the two long-tailed macaques sampled at about the same period. One of the genotypes found in this investigation was shared among the two macaques (SG/EHI/LT-001 and SG/EHI/LT-002) and a human case (SG/EHI/H-007). The second genotype was shared among the other two human cases (SG/EHI/H-001 and SG/EHI/H-002) and one of the macaques (SG/EHI/LT-002).
For the human case (SG/EHI/H-024) detected in December 2008, the complete csp gene sequence analysis revealed that this genotype was also found in the macaque caught in the vicinity in June 2009 (EHI/SG/LT-013). The genotypes found in the human and macaques during 2008–2009 formed a subclade within the P. knowlesi clade and was distinct from those detected in 2007 (Fig. 1).
Discussion
From our study, we have determined that long-tailed macaques from a forested area in Singapore are a natural host of P. knowlesi in Singapore. The six human cases of P. knowlesi infections in Singapore, including the first locally acquired human infection previously reported by Ng and coworkers in 2008, had also visited the same forested area. Molecular analysis suggests that the first three human cases detected in 2007 were epidemiologically linked to the two macaques caught at about the same time, and the most recent human case detected in December 2008 was epidemiologically linked to the macaque caught in 2009. The sharing of identical P. knowlesi csp gene sequences between those found in humans and monkeys and the fact that none of the cases had any significant travel history to known P. knowlesi–endemic areas within 1 month before the onset of symptoms strongly suggest that the human cases had acquired their infections from the affected areas in Singapore where macaques harboring P. knowlesi were found.
As P. knowlesi was first identified in monkeys imported from Singapore in 1931 (Knowles and Das Gupta 1932), it is most likely that continuous sylvatic transmission of P. knowlesi has been occurring in Singapore for a considerable period. However, due to the difficulty in accurately diagnosing P. knowlesi infections based on the parasites' morphology, previous human P. knowlesi cases before 2007 may have been misdiagnosed as P. malariae or P. falciparum since these human malaria parasites share morphological similarities with P. knowlesi (Singh et al. 2004). Four of the cases in this report indeed had initial diagnoses of other human malaria parasites, and this has also been shown to be common in Malaysia (Singh et al. 2004, Vythilingam et al. 2008).
No malaria parasites were detected in the 10 peri-domestic macaques caught from a public nature reserve park in Singapore. Although the number of monkeys tested in these areas was small and may not represent the population of the peri-domestic monkeys present in Singapore, similar results were reported by Vythilingam et al. (2008) in Peninsular Malaysia. In the Malaysian study, all long-tailed monkeys caught in urban areas where the competent vectors are absent were also free from malaria infection; on the other hand, all monkeys caught in the forest were infected with simian malaria parasites of which 97% (n = 73) were found to harbor P. knowlesi. Although peri-domestic long-tailed macaques are frequently seen in fringes of nature reserves and residential areas of Singapore, decades of regular surveillance had not detected Anophelines in most of these areas. The absence or limited presence of known competent vectors of malaria parasites probably explains the absence of malaria parasites in these peri-domestic monkeys.
The vectors of P. knowlesi in Singapore have yet to be identified. Currently, only mosquitoes belonging to the Anopheles leucosphyrus group have been incriminated for transmitting P. knowlesi in nature. These include Anopheles hackeri (Wharton and Eyles 1961) and Anopheles cracens (Vythilingam et al. 2008) in Peninsular Malaysia and Anopheles latens in Sarawak, Malaysian Borneo (Vythilingam et al. 2006, Tan et al. 2008). Several anopheline species of the An. leucosphyrus group have also been found to transmit other simian malaria parasites under natural or experimental conditions (Coatney et al. 1971). The geographic distribution of this group of mosquitoes ranges from Southwestern India, eastward to Southern China, Taiwan, mainland Southeast Asia, Indonesia, and Philippines (Sallum et al. 2005). However, to date, there have not been any reports of mosquitoes belonging to the An. leucosphyrus group in Singapore. During routine entomological surveillance of adult mosquitoes by the Singapore military in 2007 and 2008 at the affected areas under investigation, at least six species of anopheline mosquitoes were caught biting humans. These include Anopheles barbirostris sp. group, Anopheles sinensis, Anopheles tesselatus, Anopheles sundaicus, Anopheles lesteri, and Anopheles kochi (Lam-Phua, SG, Png, AB, Ng, LC, et al., unpublished data). Under experimental conditions, all these anopheline mosquitoes had previously been shown to support the growth of simian malaria parasites, at least to the oocyst stage (Coatney et al. 1971). Of these, An. kochi was found to be the most susceptible to P. knowlesi, P. cynomolgi, Plasmodium eyelsi, Plasmodium fieldi, P. inui, and P. coatneyi (Coatney et al. 1971), and has been shown to feed on monkeys on the ground and at the canopy level (Reid 1968, Vythilingam et al. 2008). In the absence of mosquitoes belonging to the An. leucosphyrus group, An. kochi appears to be a potential vector that might play a role in the transmission of knowlesi malaria in Singapore. However, our vector surveillance data showed that An. kochi represents <1% of the total number of adult anopheline mosquitoes attracted to humans in the affected area. Previous reports have shown that An. kochi is a highly zoophagic mosquito, and have been shown to bite cattle and monkeys more than humans (Reid 1968, Vythilingam et al. 2008). This could explain the small number of personnel acquiring P. knowlesi despite the high frequency of visits by military personnel to the forested areas.
Although only three monkeys were sampled from the restricted forested area, all of them were infected with knowlesi malaria parasites. We are performing additional surveillance to determine the prevalence of infection among macaques in the affected area to understand the transmission dynamics of malaria within the macaque population. Further, a study to determine simioacrodendrophagic mosquitoes will also be conducted to identify potential vectors that transmit the parasite among the monkey population in these areas. Finally, a laboratory-based vector competence study on anopheline mosquitoes identified during our entomological survey will also be conducted to determine potential bridge vectors for P. knowlesi. This is critical for determining potential areas at risk for P. knowlesi transmission so that public health interventions plans can be implemented. Currently, general mosquito control measures through use of Bacilus thuringiensis var. israelensis and environmental management have been intensified. Other preventive measures such as the use of insecticide-treated uniforms and use of repellent have also been implemented. Long-term monitoring to determine how these measures would affect the prevalence of simian malaria parasites in the macaque populations would be essential.
Conclusions
M. fascicularis is a natural host of P. knowlesi in Singapore, and human cases acquired their infections while working in areas where infected monkeys are found. The risk of P. knowlesi is small in the general population of Singapore as the macaques that are close to human dwellings were found to be free of malaria parasites.
Acknowledgments
We thank the Ministry of Finance for the Reinvestment Fund made available for the study. We are also grateful to colleagues at Nparks, Agri-Food, and Veterinary Authority and DSO National Laboratories for their technical assistance.
Disclosure Statement
No competing financial interests exists.
References
- Bronner UP. Divis PC. Färnert A. Singh B. Swedish traveller with Plasmodium knowlesi malaria after visiting Malaysian Borneo. Malar J. 2009;8:15. doi: 10.1186/1475-2875-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin W. Contacos PG. Coatney GR. Kimball HR. A naturally acquired quotidian-type malaria in man transferable to monkey. Science. 1965;149:865. doi: 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- Coatney GR. Collins WE. Warren W. Contacos PG. The Primate Malarias. Washington, DC: U.S. Government Printing Office; 1971. [Google Scholar]
- Cox-Singh J. Davis TME. Lee KS. Shamsul SSG, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S. Putaporntip C. Iwasaki T. Sata T. Kanbara H. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg Infect Dis. 2004;10:2211–2213. doi: 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantele A. Marti H. Felger I. Müller D. Jokiranta TS. Monkey malaria in a European traveler returning from Malaysia. Emerg Infect Dis. 2008;14:1434–1436. doi: 10.3201/eid1409.080170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles R. Das Gupta BM. A study of monkey-malaria and its experimental transmission to man. Indian Med Gaz. 1932;67:301–320. [PMC free article] [PubMed] [Google Scholar]
- Luchavez J. Espino F. Curameng P. Espina R, et al. Human infections with Plasmodium knowlesi, the Philippines. Emerg Infect Dis. 2008;14:811–813. doi: 10.3201/eid1405.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan TF. Kissinger JC. Touray MG. John Rogers M, et al. Comparison of circumsporozoite proteins from avian and mammalian malarias: Biological and phylogenetic implications. Proc Natl Acad Sci USA. 1996;93:11889–11894. doi: 10.1073/pnas.93.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng OT. Ooi EE. Lee CC. Jarrod LP, et al. Naturally acquired human Plasmodium knowlesi infection, Singapore. Emerg Infect Dis. 2008;14:814–816. doi: 10.3201/eid1405.070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JA. Anopheline mosquitoes of Malaya and Borneo. Studies from the Ins Med Res Malaysia: Staples Printing Limited. 1968;31:1–520. [Google Scholar]
- Sallum MAM. Peyton EL. Harrison BA. Wilkerson RC. Revision of the Leucosphyrus group of Anopheles (Cellia) (Diptera, Culicidae) Rev Bras Entomol. 2005;49(Supplement l):1–152. [Google Scholar]
- Singh B. Bobogare A. Cox-Singh J. Snounou G, et al. A genus-and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- Singh B. Lee KS. Matusop A. Radhakrishnan A, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- Tamura K. Dudley J. Nei M. Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan CH. Vythilingam I. Matusop A. Chan ST. Singh B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J. 2008;7:52. doi: 10.1186/1475-2875-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vythilingam I. NoorAzian YM. Tan CH. Jiram AI, et al. Plasmodium knowlesi in humans, macaques and mosquitoes in peninsular Malaysia. Parasites Vectors. 2008;1:26. doi: 10.1186/1756-3305-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vythilingam I. Tan CH. Asmad M. Chan ST, et al. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans R Soc Trop Med Hyg. 2006;100:1087–1088. doi: 10.1016/j.trstmh.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Wharton RH. Eyles DE. Anopheles hackeri, a vector of Plasmodium knowlesi in Malaya. Science. 1961;134:279–280. doi: 10.1126/science.134.3474.279. [DOI] [PubMed] [Google Scholar]
- Yap LF. Cadigan FC. Coatney GR. A presumptive case of naturally occurring Plasmodium knowlesi malaria in man in Malaysia. Trans Royal Soc Trop Med Hyg. 1971;65:839–840. doi: 10.1016/0035-9203(71)90103-9. [DOI] [PubMed] [Google Scholar]
- Zhu HM. Li J. Zheng H. Human natural infection of Plasmodium knowlesi. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2006;24:70–71. (in Chinese). [PubMed] [Google Scholar]