Abstract
In this study, we used ischemia-induced retinal neovascularization (NV) as a model to investigate the possible role of microRNAs in a clinically important disease process. Microarray analysis demonstrated seven microRNAs, miR-106a, -146, -181, -199a, -214, -424, and -451, that were substantially increased and three, miR-31, -150, and -184, that were substantially decreased in ischemic retina. Potential targets for the upregulated microRNAs were not identified, but bioinformatic analysis suggested target genes for the downregulated microRNAs that were confirmed by a luciferase reporter assay. Real time RT-PCR confirmed substantial levels of miR-31, -150, and -184 in normal retina that were significantly reduced in ischemic retina. Interestingly, constitutive levels of miR-31 and -184 are high in cornea and lens, two avascular tissues. Intraocular injection of pre-miR-31, -150, or -184 significantly reduced ischemia-induced retinal NV, and injection of pre-miR-31 or -150 also significantly reduced choroidal NV. These data suggest that alteration of microRNA levels contributes to two types of ocular NV and suggest that injection or enhanced expression of microRNAs is a potential therapeutic strategy.
Keywords: age-related macular degeneration, angiogenesis, choroidal neovascularization, diabetic retinopathy, vascular endothelial growth factor
Introduction
Angiogenesis is a complex process in which several gene products participate. There are several stimulators and inhibitors and the balance between them determines whether angiogenesis occurs or is repressed. There are differences among vascular beds in the body, and therefore findings in one tissue cannot be automatically generalized to other tissues, but such findings provide groundwork for testing and confirmation in other tissues.1 With this proviso, retinal and choroidal NV provide models that can be used to search for principles that can be tested for application in other types of angiogenesis, and in addition they are clinically important because they occur in several diseases and are major causes of vision loss (for review, see 2). One theme that retinal and choroidal NV seem to share with several other types of NV in other tissues is that vascular endothelial growth factor (VEGF) is a particularly important stimulator.3–6 Intraocular injections of ranibizumab, an Fab that binds all isoforms of VEGF-A causes significant visual improvement in 35–40% of patients with choroidal NV due to age-related macular degeneration (AMD).7, 8 While this is impressive and has changed the treatment landscape for patients with choroidal NV, there are still 60–65% of these patients who may need something more than blockade of VEGF alone.
While it is clear that transcriptional regulation is important for determining the balance of stimulators and inhibitors of NV, there is mounting evidence suggesting that microRNAs may play a complementary role. MicroRNAs were first identified as the cause of co-suppression in plants.9–14 Genes encoding primary microRNAs are scattered throughout the genomes of eukaryotes and are transcribed to generate RNA species that are cleaved into 60–70 nucleotide stem loop microRNA precursors (pre-microRNAs) by a microprocessor complex containing the nuclear RNaseIII Drosha and DiGeorge syndrome critical region 8 protein.15–17 After export from the nucleus, the pre-microRNAs are cleaved by a cytoplasmic RNase III, Dicer, and following stand selection and separation, mature 22 base pair microRNAs are incorporated into an RNA-induced silencing complex (RISC) to act as target recognition sequences.18 Several mRNAs may contain target sequences and can undergo cleavage or translational repression depending upon whether complementary base pairing with the microRNA is perfect or imperfect.19 The potential role of microRNAs in angiogenesis has been suggested by several observations. Migration and tube formation of cultured vascular endothelial cells are reduced by transfection of miR-221/miR-222 20 or global changes in microRNAs induced by knockdown of Dicer.21 Furthermore, mice with targeted disruption of Dicer die during embryonic life due to disruption of vascular development 22 indicating that microRNAs play a critical role in vasculogenesis, which often shares mechanisms with angiogenesis. In this study, we used microarray analysis to identify microRNAs that are upregulated or downregulated in ischemic retina. Three microRNAs were confirmed to be substantially reduced in ischemic retina by real time RT-PCR and we tested the effect of intraocular injection of these microRNAs on levels of products of target genes and on NV.
Results
Several microRNAs are altered in ischemic retina in which there is NV
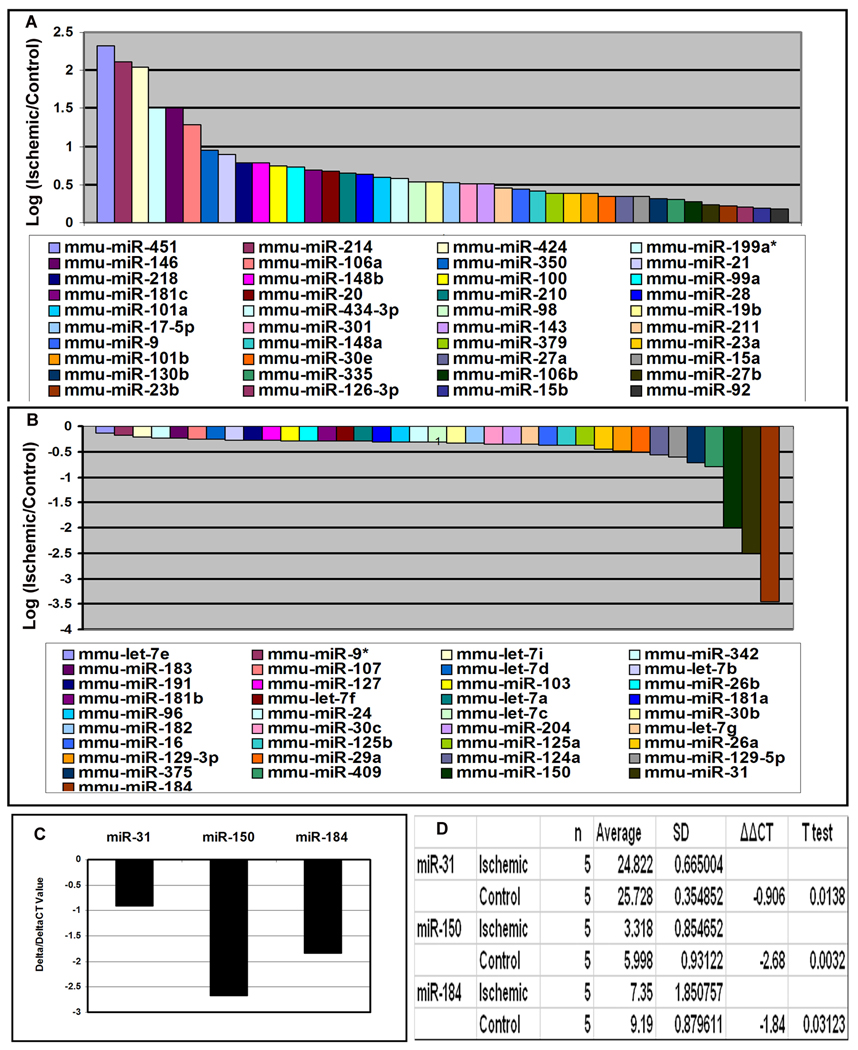
Small RNA species were isolated from the retinas of postnatal (P) 15 mice with ischemic retinopathy and control mice. The RNA species from ischemic retinas were labeled with Cy3 and those from control retinas were labeled with Cy5. The level of several microRNAs was increased in ischemic retina and seven, miR-451, -424, -146, -214, -199a, -181, and -106a were increased above a log2 threshold (Figure 1A). Three microRNAs, miR31, miR150, and miR184, were decreased below a log2 threshold (Figure 1B), and a significant reduction was confirmed by quantitative real time PCR (Figure 1C and D).
Figure 1. Identification of microRNAs that are upregulated or downregulated in ischemic retina.
Six C57BL/6 mice were placed in 75% oxygen at P7 and returned to room air at P12. At P15, mice were euthanized and small RNA species were isolated from the retinas. The RNA samples from ischemic retinas were pooled and labeled with Cy3 and the samples from control retinas were pooled and labeled with Cy5. The RNA probes were hybridized with microarrays as described in Methods. The log2 of the ratio of Cy3/Cy5 signal strength is shown for 40 microRNAs that were upregulated in ischemic retina (A) and 37 were that were downregulated (B). Real time RT-PCR was used to independently determine levels of miR-31, miR-150, and miR-184 relative to levels of 5s rRNA. The mean threshold cycle number (delta CT) for miR-31, miR-150, and miR-184 was determined for RNA from ischemic and control retinas (n=5 for each) and the mean difference (delta/delta CT) ranged from about −1.0 (miR-31) to −2.5 (miR-150, C). Statistical comparisons confirmed that each of the miRs were significantly reduced in ischemic compared to control retinas (D).
Identification of possible target genes for down-regulated microRNAs by bioinformatics
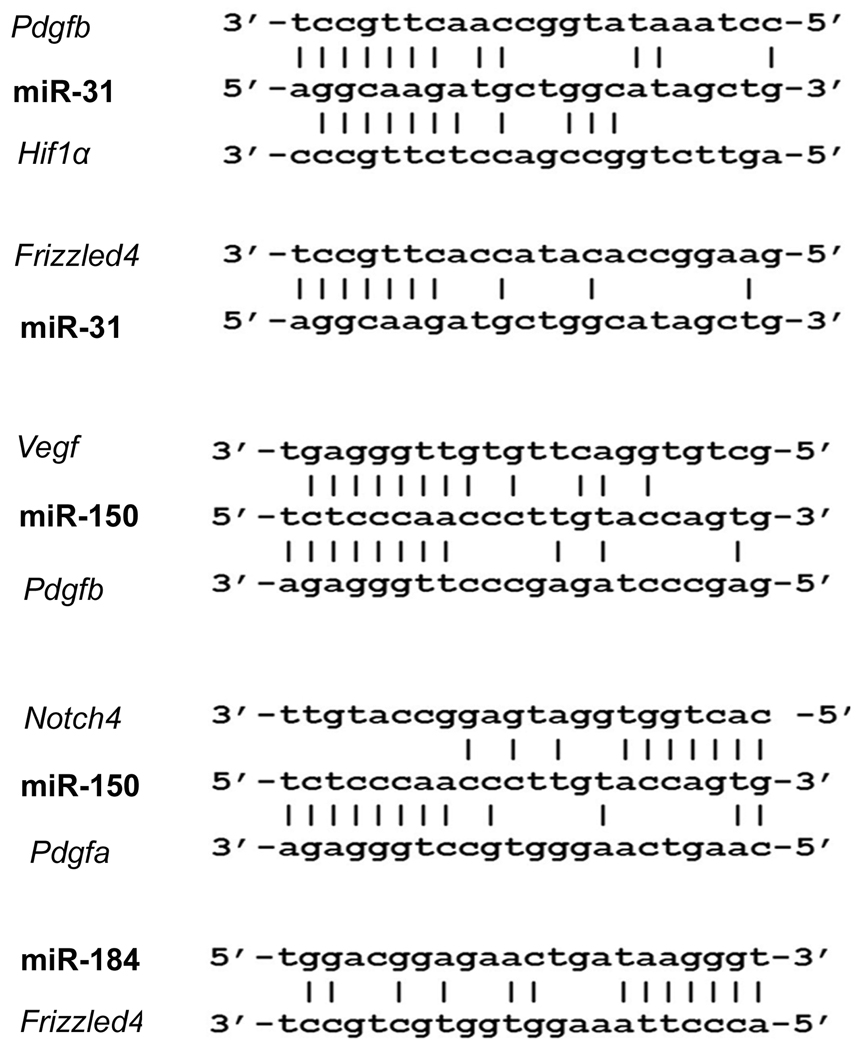
Candidate target genes were identified for the 3 downregulated microRNAs, but none were found for any of the upregulated microRNAs. There were possible target sequences for miR-31 in Pdgfb, Hif1α, and Frizzled4; possible targets for miR-150 in Vegf, Pdfgb, Pdgfa, and Notch4; and possible target sequences for miR-184 in Frizzled4 (Figure 2).
Figure 2. Identification of target genes for down-regulated microRNAs.
The European DNA database was searched for 7–8 consecutive complementary sequences starting 2 bp from the 5’ or 3’ end of miR-31, miR-184, and miR-150. Complementary regions were identified in the 3’UTR of: Pdgfb, Hif1α, and Frizzled4 for miR-31; Vegf, Pdgfb, Pdgfa, and Notch4 for miR-150, and Frizzled 4 for miR-184 (A). Areas of alignment with the 3’UTR of Vegf, Pdfgb, Hif1a and Notch4, but not Frizzled4 were highly conserved among different species.
Confirmation of targets by microRNA reporter assay
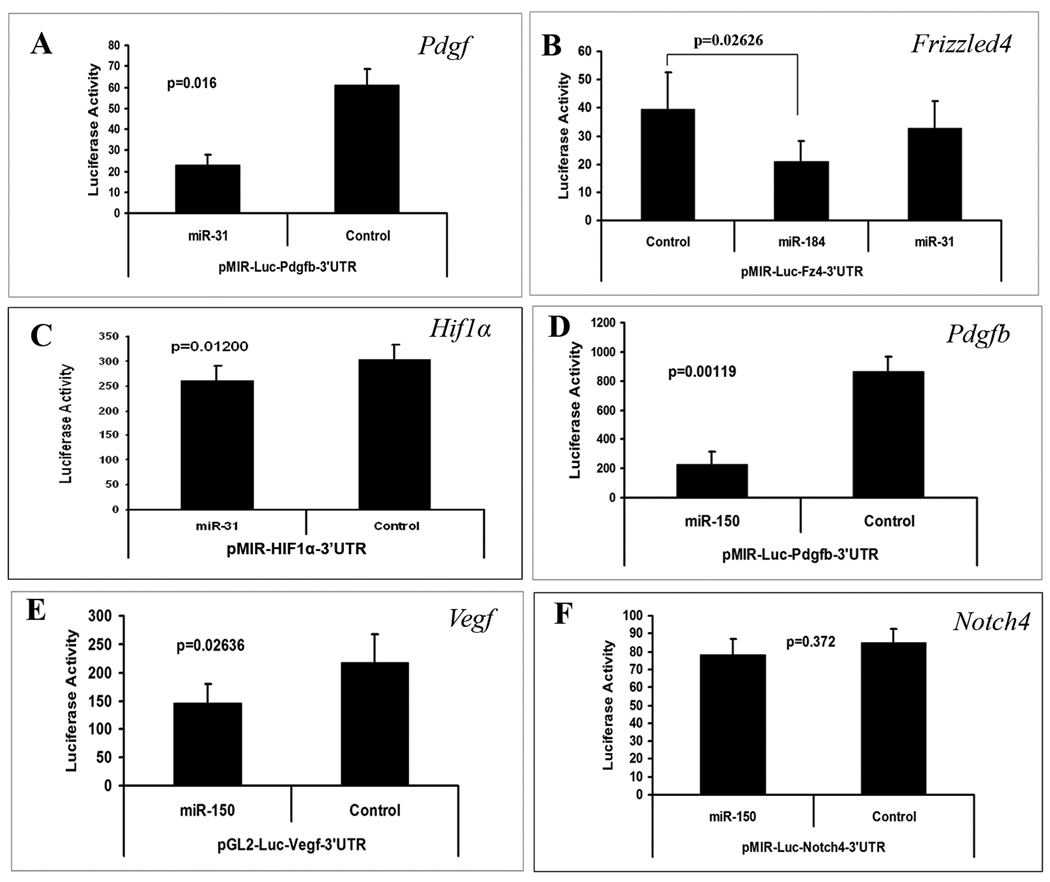
Cultured RPE cells were co-transfected with pre-miR-31, pre-miR-150, or pre-miR-184 and a reporter construct containing one of its possible target sequences. Co-transfection with pre-miR-31 significantly reduced luciferase levels expressed by the Pdgfb and Hif1α reporters, but had no effect on the reporter for Frizzled4 (Figure 3A–C). The reporters for Vegf and Pdgfb, but not Notch4 were down-regulated by pre-miR-150 (Figure 3D–F). The reporter for Frizzled4 was down-regulated by pre-miR-184 (Figure 3B).
Figure 3. The effect of microRNAs on promoter activity of purported target genes.
Luciferase reporter constructs were generated for microRNA reporter assays for each of the potential target sequences listed in Figure 2 except Pdgfa. Each bar represents the mean (±SEM) calculated from three experiments. Compared to a control microRNA with scrambled sequence, miR-31 significantly reduced luciferase expression for pMIR-Luc-Pdgfb-3’UTR (A) and pMIR-Luc-HIF1α-3’UTR (B), but not pMIR-Luc-Fz4-3’UTR (C). Luciferase expression was significantly reduced by miR-184 for pMIR-Luc-Fz4-3’UTR (C). Luciferase expression was reduced by miR-150 for pMIR-Luc-Pdgfb-3’UTR (D) and pGL2-Luc-Vegf-3’UTR (E), but not pMIR-Luc-Notch4-3’UTR (F). Statistical comparisons were made by paired t-test.
Effect of miRNAs on target gene product levels in ischemic retina in vivo
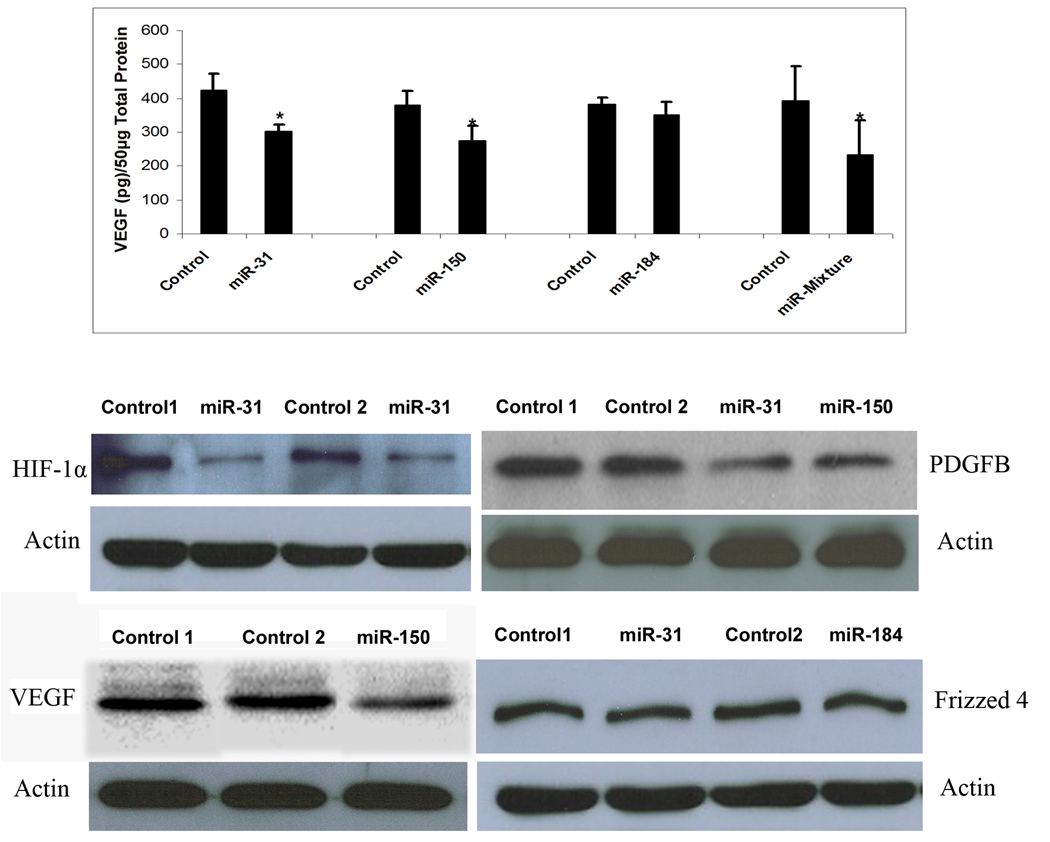
Mice were placed in 75% oxygen at P7 and at P12 they were returned to room air and given a 1 µl intraocular injection of 5 µM experimental pre-microRNA in one eye and 5 µM control pre-microRNA in the other eye. Mice were euthanized at P15 and retinal homogenates were used to assess HIF-1α, PDGF-B and Frizzled 4 by immunoblots and VEGF by both ELISA and immunoblots. Injection of pre-miR-31 or -150, but not pre-miR-184 caused a significant reduction in VEGF levels in ischemic retina (Figure 4 top panel). Since miR-31 does not target Vegf directly, it is likely that its effect occurs indirectly through one of its targets, probably Hif1α. Injection of 1 µl containing 1.67 µM of each of the 3 pre-microRNAs also caused a significant reduction in VEGF. Intraocular injection of pre-miR-31 caused reductions in retinal HIF-1α and PDGF-B (Figure 4, second row). Injection of pre-miR-150 caused reductions in PDGF-B and VEGF (Figure 4, second and third rows). Injections of pre-miR-31 or pre-miR-184 failed to cause reductions in Frizzled 4 protein in the retina.
Figure 4. Effect of microRNAs on target gene product levels in ischemic retina.
C57BL/6 mice were placed in 75% oxygen at P7 and returned to room air at P12 and given an intravitreous injection of 1 µl of 5 µM experimental precursor microRNA (pre-microRNA) in one eye and 1 µl of 5 µM control pre-microRNA in the other eye. At P15, mice were euthanized and VEGF was measured in retinal homogenates by ELISA (top panel). Each bar represents the mean (±SEM) calculated from 3 experiments. VEGF protein was not detectable in retinas from mice in room air at P15 (n=7), but high levels were measured in ischemic retinas (n=10). Compared to injection of control pre-microRNA, injection of pre-miR-31 or -150, but not pre-miR-184 caused significant reduction in VEGF in ischemic retina (top panel). There was also a significant reduction in VEGF in ischemic retinas from eyes injected with 1 µl containing 1.67 µM concentrations of each of the 3 experimental pre-microRNAs compared to fellow eyes injected with 1 µl containing 5 µM control pre-microRNA. Immunoblots of retinal homogenates from eyes injected with pre-miR-31 showed reductions in HIF1α (middle panel, left) and PDGF-B (middle panel, right) compared to eyes injected with control pre-microRNA. Ischemic retinas from eyes injected with pre-miR-150 showed reductions in PDGF-B (middle panel, right) and VEGF (bottom panel, left). Frizzled 4 was not reduced by injection of pre-miR-31 or -184 (bottom panel, right). Each immunoblot was repeated at least once with similar results.
Effect of microRNAs on retinal NV in ischemic retina
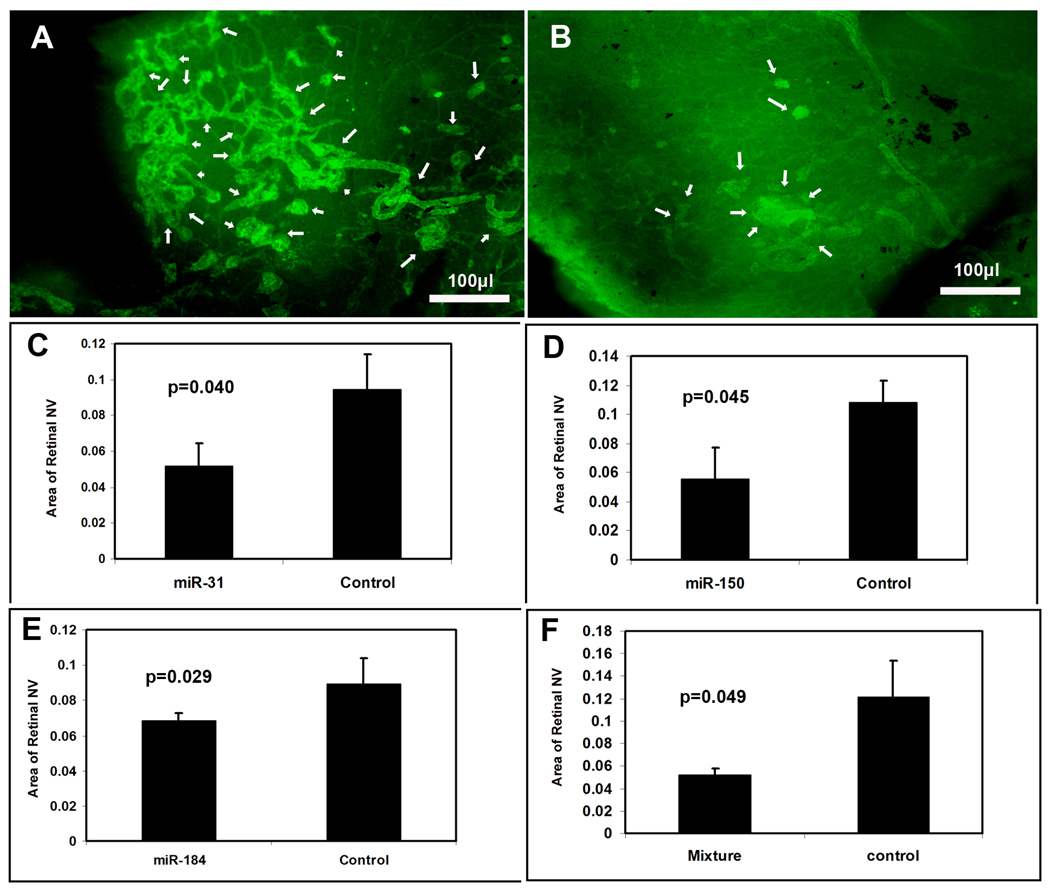
Mice were placed in 75% oxygen at P7 and at P12 they were returned to room air and given a 1 µl intraocular injection of 5 µM experimental pre-microRNA in one eye and 5 µM control pre-microRNA with scrambled sequence in the other eye. At P17, mice were given an intraocular injection of 0.5 µg of rat anti-mouse PECAM1 antibody in each eye and after 12 hours they were euthanized and retinas were incubated with FITC-labeled goat anti-rat IgG, flat mounted, and examined under a fluorescence microscope. This technique provides selective staining of retinal NV on the surface of the retina 23, and retinas from eyes injected with 1 µl containing 5 µM control pre-miR showed extensive NV (Figure 5A). Figure 5B shows a representative retina from an eye injected with 1 µl containing 1.67 µM concentrations of pre-miR-31, -150, and -184; it shows very little NV on the surface of the retina. Retinas from eyes injected with 1 µl containing 5 µM pre-miR-31 or -150, or -184 also appeared to have less NV than eyes injected with control miR (not shown). Measurements by computerized image analysis with the observer masked with regard to treatment confirmed that there was a significant reduction in mean area of NV per retina in eyes injected with pre-miR-31 (C), -150 (D), -184 (E), or a mixture of all 3 pre-miRs compared to fellow eyes injected with control pre-miR.
Figure 5. Effect of microRNAs on ischemia-induced retinal neovascularization (NV).
At P7, litters of C57BL/6 mice were placed in 75% oxygen. At P12, mice (n=8) were given an intraocular injection of experimental precursor microRNA (pre-microRNA) or a mixture of all 3 experimental pre-microRNAs in the right eye and control pre-microRNA in the other eye. At P17, retinal NV was stained as described in Methods and measured on retinal flat mounts by image analysis. Compared to the retinas from mice injected with control pre-microRNA, which showed prominent NV on the surface of the retina (A, arrows), retinas from mice injected with a mixture of pre-miR-31, -150, and -184 showed substantially less NV (B, arrows). Measurements done by image analysis showed that the mean area of NV was significantly less for eyes injected with pre-miR-31 (C), -150 (D), -184 (E), or a mixture of all three (F) compared to fellow eyes injected with control pre-microRNA. Each bar represents the mean (±SD) area of NV calculated from 8 mice. Statistical comparisons were made by paired t-test.
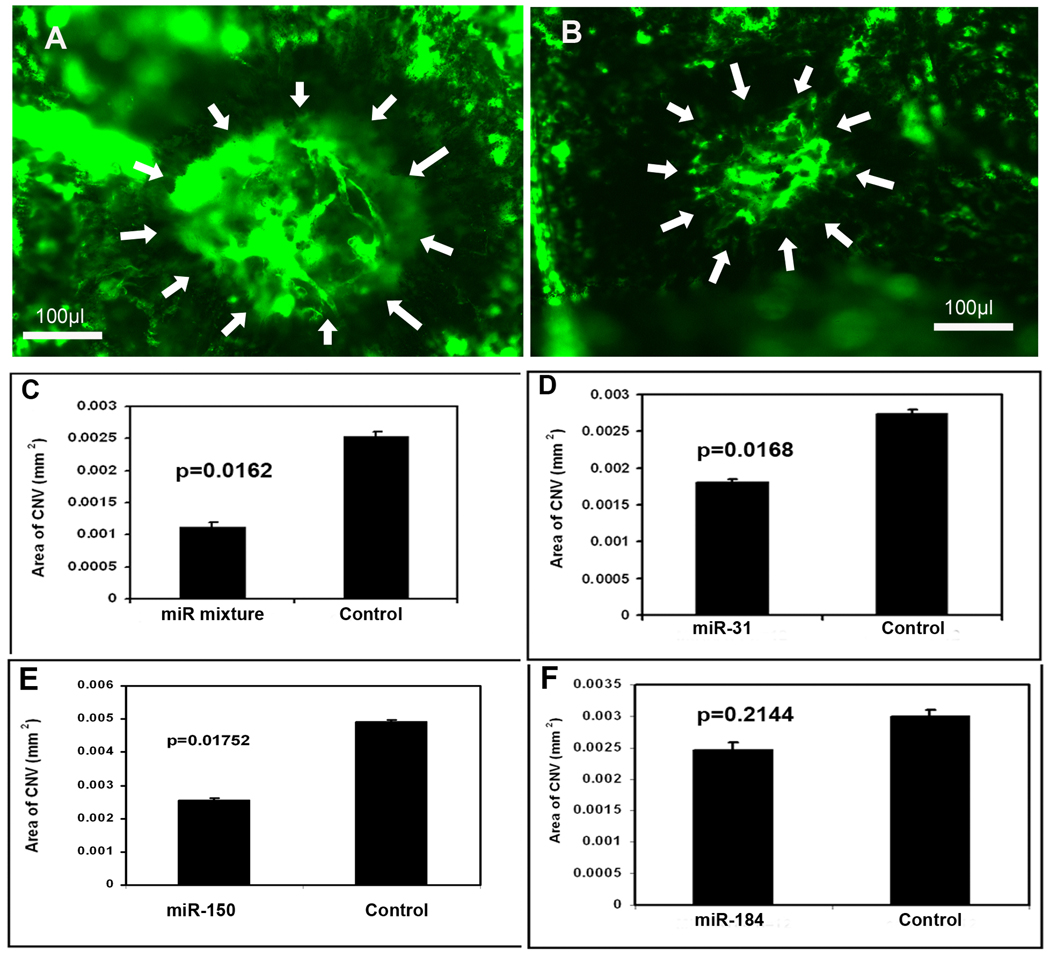
Effect of microRNAs on choroidal NV
Adult C57BL/6 mice had rupture of Bruch’s membrane in 3 locations by laser photocoagulation in each eye followed by a 1 µl intraocular injection of 5 µM experimental pre-microRNA in one eye and 5 µM control pre-microRNA in the fellow eye. After 14 days, mice were perfused with fluorescein-labeled dextran and the area of choroidal NV at Bruch’s membrane rupture sites was measured on choroidal flat mounts. Compared to fellow eyes injected with 1 µl containing 5 µM scrambled pre-microRNA (Figure 6A), those injected with 1 µl containing 1.67 µM concentrations of each of the 3 experimental pre-microRNAs showed smaller choroidal NV lesions (Figure 6B). Measurement of the area of choroidal NV by computerized image analysis with the investigator masked with regard to treatment showed that eyes injected with the mixture of experimental pre-microRNAs had a significant reduction in the mean area of choroidal NV at Bruch’s membrane rupture sites compared to fellow eyes injected with control pre-microRNA (Figure 6C). There was also a reduction in choroidal NV area in eyes injected with 5 µM pre-miR-31 (Figure 6D) or -150 (E) compared to fellow eyes injected with 5 µM control pre-miR, but there was no difference in eyes injected with pre-miR-184 (F).
Figure 6. Effect of microRNAs on choroidal neovascularization (NV).
Adult C57BL/6 mice had rupture of Bruch’s membrane by laser photocoagulation in three locations in each eye and they were given an intraocular injection of 1 µl of 5 µM precursor microRNA (pre-microRNA) in one eye and 1 µl of control pre-microRNA in the fellow eye on day 0 and day 7. On day 14, mice were perfused with fluorescein-labeled dextran and the area of choroidal NV at Bruch’s membrane rupture sites was measured by image analysis. Each bar represents the mean (±SD) area of choroidal NV calculated from at least 12 experimental values. Compared to the area of NV in eyes injected with control pre-microRNA (A), there was a significant decrease in eyes injected with a mixture of all 3 experimental pre-microRNAs (B and C), pre-miR-31 (D), or -150 (E), but not pre-miR-184 (F). Statistical comparisons were made by paired t-test.
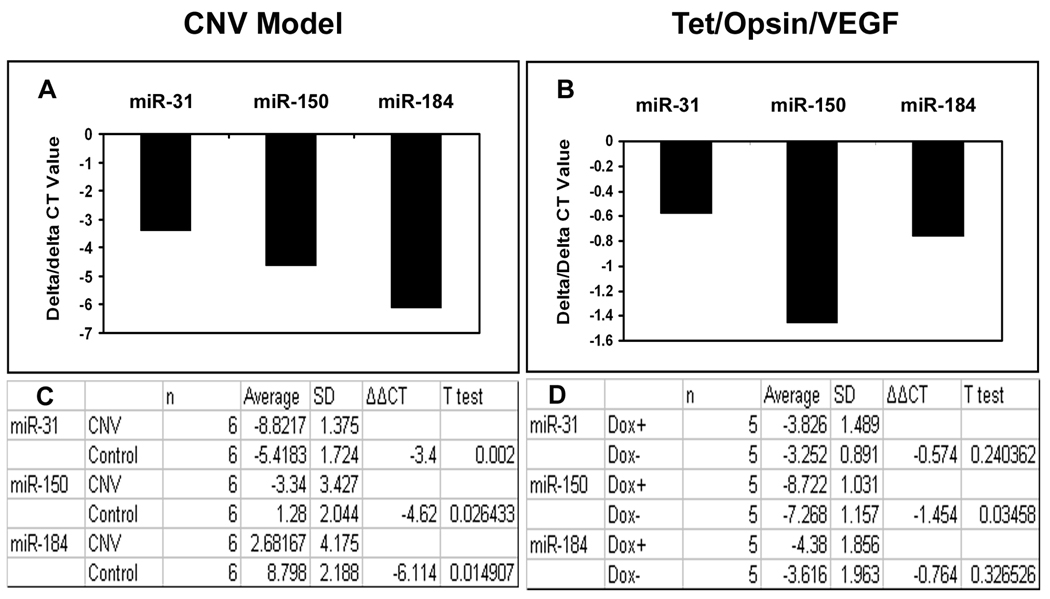
Reduction of miR-31, -150, and -184 in eyes with choroidal NV in the absence of ischemia
In order to determine if ischemia was the immediate cause of down-regulation of miR-31, -150, and -184, their levels were measured in eyes with multiple areas of choroidal NV, which is not associated with ischemia. Compared to contralateral control eyes, those with choroidal NV showed a significant reduction in miR-31, -150, and -184 (Figure 7A and C). In both retinal and choroidal NV, there is a substantial elevation of VEGF levels; to determine if this might be the immediate cause of the reduction of the 3 miRs in association with ocular NV, we utilized Tet/opsin/VEGF double transgenic mice which have doxycycline-inducible expression of VEGF in the retina 24. Three days after initiation of treatment with doxycycline, Tet/opsin/VEGF mice showed a significant reduction of miR-150 in the retina compared to Tet/opsin/VEGF mice that were not treated with doxycycline, but there was no significant difference in miR-31 or -184 (Figure 7B and D).
Figure 7. Expression of miR-31, miR-150 and miR-184 in other models of ocular neovascularization.
C57BL/6 mice (n=6) had laser-induced rupture of Bruch’s membrane at 10 locations in one eye and after 4 days were euthanized, anterior segments were removed from each eye, and small RNAs were isolated from eye cups. Double transgenic Tet/opsin/VEGF mice with doxycycline-inducible expression of VEGF in the retina were given 2 mg/ml of doxycycline in their drinking water (n=5) or maintained on normal water (n=5, controls). After 3 days, the mice were euthanized and small RNAs were isolated from the retinas. Real time RT-PCR for miR-31, -150, -184 and 5S rRNA was done as described in Methods. The mean threshold cycle number (delta CT) for miR-31, miR-150, and miR-184 was determined for RNA isolated from experimental and control eyes and the bars show the mean difference (delta/delta CT). Compared to contralateral control eyes, those with choroidal NV showed substantial reductions in mean threshold number for each of the miRs (A) and statistical comparisons confirmed that the levels of miR-31, -150, and -184 were all significantly reduced in eyes with choroidal NV (C). Compared to untreated Tet/opsin/VEGF mice, those treated with doxycycline resulting in expression of VEGF in the retina showed minimal changes in mean threshold number for miR-31 and -184 that were not statistically significant, but there was a significant reduction in the level of miR-150 (B and D).
Discussion
In this study, we have found microRNAs that are increased and those that are decreased in ischemic retina. Three microRNAs, miR-31, miR-150, and miR-184, which were substantially reduced based upon microarray analysis were confirmed to be reduced in ischemic retinas by quantitative real time RT-PCR. Genes containing complementary sequences that potentially could be targets for these 3 microRNAs were identified and tested for binding in luciferase reporter assays. Using this approach the following purported target genes were identified; Pdgfb and Hif1α for miR-31, Vegf and Pdgfb for miR-150, and Frizzled4 for miR-184. Compared to injection of a control pre-microRNA, injection of pre-miR-31 into the eyes of mice with ischemic retinopathy caused significant reductions in PDGF-B and HIF-1α confirming that they are targets for miR-31. In similar manner VEGF and PDGF-B were confirmed to be targets for miR-150, but we could not confirm that Frizzled4 is definitely a target for miR-184. Interestingly, we found that injection of pre-miR-31 also reduced VEGF in ischemic retina which must occur indirectly, because Vegf is not a target for miR-31. We postulate that the reduction in VEGF may occur through reduction of HIF-1α, which is a transcriptional activator of VEGF that is responsible for the increased VEGF levels in ischemic retina.25, 26 Injection of pre-miR-31, -150, or -184 significantly reduced retinal NV in mice with ischemic retinopathy. These data suggest that the reduction in miR-31, -150 and -184 that occurs in ischemic retina may contribute along with transcriptional regulation mechanisms, to the disease phenotype. The data also suggest that injection or other interventions to ameliorate the reductions in these microRNAs could provide a new approach for therapy. Intraocular injections of pre-miR31 or pre-miR-150 also suppressed the development of choroidal NV indicating that this approach could also apply to this different type of ocular NV.
It does not appear that ischemia is the cause of the reduction of miR-31, -150, and -184, because they were also reduced in eyes with choroidal neovascularization. Induced overexpression of VEGF in the retina caused a significant reduction in miR-150, but not miR-31 or -184. Vegf is a target gene for miR-150 raising the intriguing possibility that products of target genes play a role in regulation of miR levels, a possibility that deserves further study.
The target genes that have been identified for miR-31 and -150 code for proteins that have pro-angiogenic activity, which provides reasonable mechanisms of action for inhibition of NV achieved by injection of these microRNAs. However, microRNAs often have several targets and miR-31 and -150 may have other unidentified targets. At this point, we are not certain of any target genes for miR-184, because while luciferase reporter assays suggested that Frizzled4 may be a target gene, we could not confirm that Frizzled 4 is reduced in ischemic retina by intraocular injection of pre-miR-184. Further work is needed to search for other potential targets for each of the three microRNAs.
We focused on 3 microRNAs that were substantially down-regulated in ischemic retina, because we could not identify any potential targets for the 7 microRNAs that were substantially increased. While results with these microRNAs will be more difficult to interpret, it will still be useful to investigate them in the future. They could target antiangiogenic proteins, the identification of which may provide important new insights. In rat carotid arteries injured by balloon angioplasty, miR-21, -146, -214, and -352 were significantly increased and knockdown of miR-21 substantially reduced neointimal lesion formation by reducing proliferation and promoting apoptosis of vascular smooth muscle cells.27 Thus, either up- or down-regulation of microRNAs may play an important role in disease pathogenesis. It is intriguing that miR-214, which is upregulated in injured carotid arteries, is also upregulated in ischemic retina.
There may be ways that microRNAs contribute to angiogenesis in extraocular sites that do not apply to ocular NV. The proto-oncogene MYC stimulates angiogenesis in one-hit model neoplasms 28, 29 at least in part by upregulating the miR17–92 cluster containing microRNAs that target a 3’-UTR target site found in several members of the thrombospondin type 1 repeat (TSR) superfamily.30 The TSR superfamily members, thrombospondin-1, SPARC and spondin-1 have anti-angiogenic activity 31–33, and another member, connective-tissue growth factor, can promote or suppress angiogenesis depending upon the context.34, 35 The miR17–92 cluster was not upregulated in ischemic retina.
Many microRNAs are expressed in a tissue-specific manner 36 and previous studies have demonstrated that miR-184 and -31 are preferentially expressed in the eye.37–40 Interestingly, both miR-184 and -31 are highly expressed in cornea and lens, two avascular tissues.38 They are also expressed in the retina 40 and it is a reasonable hypothesis that their constitutive expression helps to prevent vascular invasion of cornea, lens, and retina, while when the retina becomes ischemic their down-regulation provides a permissive environment for NV.
Thus it appears that microRNAs may provide a post-transcriptional mechanism that complements transcriptional regulation to alter the balance of proangiogenic and antiangiogenic factors in the eye. Some microRNAs involved in control of ocular NV may also be utilized in other tissues and others may be used primarily or solely in the eye. Additional work is needed to further define targets for all of the microRNAs identified in this study and precisely define the mechanisms by which they regulate ocular NV. The ability of microRNAs to simultaneously alter levels of several gene products in the angiogenesis cascade, and their role as part of an endogenous control system that could potentially be manipulated, suggest that they may be very valuable for development of new treatments.
Materials and Methods
Isolation of small RNA and microRNA array
C57 BL/6 mice were placed in 75% oxygen at postnatal day 7 (P7), returned to room air at P12, and euthanized at P15. Eyes were dissected and retinas were removed. Small RNAs were isolated by Vina Small RNA isolation kit (Ambion, Houston, TX) according to the manufacturer’s instructions. A total of 5 µg of small RNAs pooled from 10 ischemic and 10 control retinas from age-matched mice reared in room air was used for microarray analysis. Hybridizations were performed by the LC Sciences Microarray Service (LC Sciences, Houston, TX). Small RNAs from ischemic retinas labeled with Cy3 and small RNAs from control retinas labeled with Cy5 were hybridized to a chip containing an array of murine microRNA (mmu-miR) probes for six redundant complementary sequences for each of the mmu-miR transcripts in the August 2005 Sanger miRBase Release 7.0 (http://www.sanger.ac.uk/Software/Rfam/mirna/). The chip was scanned with red and green channels and the red and green intensity was determined for each probe. Statistical comparisons were made between the mean difference in intensity for the six probes for each transcript and the intensity differences for control probes, and p < 0.01 was used to identify differentially expressed microRNAs.
Bioinformatics analysis for possible microRNA target genes
A group of angiogenic factors and/or their receptors were selected from www.geneontology.org, including VEGF, HGF, FGF-2, PlGF, IGF-1, TGF-beta, PDGF-A, PDGF-B, VG5q, Delta-like 4, SEMA3a, Sema3F, Notch4, EphA2, Roundabout, HIF-1alpha, Frizzed-4, SDF-1, PEDF, vasohibin-1, thrombspodin-1, Ang-1, Ang-2, Flt-1, KDR, neuropilin-1, neuropilin-2, CXCR4, Tie1, Tie2, angiostatin. The sequences of their 3’-untranslated regions (3’-UTR) were obtained from the European DNA database Mus musculus http://www.ensembl.org/index.html. To predict possible microRNA target sites, 5’ and 3’-UTR sequences were searched using Web-based microRNA target prediction systems, including miRBase (http://microrna.sanger.ac.uk/targets/v4/), miRanda (www.microrna.org), Rnahybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid), and Pic Tar (http://pictar.bio.nyu.edu). Previously reported criteria served as a guide.41, 42 (1) A conserved seed match, i.e., perfect Watson-Crick complementarity between nucleotides 2–7 of the microRNA and a 6 nucleotide section of the 3’UTR of the mRNA. The match is required to occur at corresponding positions for multiple species. (2) A conserved anchoring A, which is an A nucleotide on the 3' UTR just downstream of the seed match. This anchoring A is required to be in the UTRs of the gene in multiple species. (3) A conserved m8-t8 match, which is an A:U or G:C match between the eighth nucleotide of the microRNA and the corresponding position in the 3' UTR in multiple species. Conservation analysis between the mouse sequences and those for humans, rats, and dogs were done using the website www.genome.ucsc.edu.
Validation of microRNA array data by real-time PCR
Small RNA species were isolated from P15 ischemic and control retinas as described above and real time PCR was done to measure microRNA levels as previously described with minor modifications.43 Briefly, small RNAs were reverse-transcribed to cDNA by either microRNA-specific primers with a 36 bp tail from C. elegans genomic sequence or random hexamer priming using Superscript III (Invitrogen, Carlsbad, CA). A 16 bp upstream primer homologous with a particular microRNA and a 19 bp universal primer complementary to the tail sequence were used for amplification of microRNA. For standaridization, primers for 5S RNA were also used (Table 1). Amplifications were performed in Roche Light Cycler using the FastStart Master SYBR Green I reaction mix (Roche Molecular Biochemicals, Indianapolis). For a negative control, cDNA generated from random hexamer priming was used. Data are represented as the difference in threshold cycle number (delta/delta CT). Real time RT-PCR for miR-31, -150, and -184 was also done in mice with laser-induced choroidal NV 44 and mice with doxycycline-inducible expression of VEGF in the retina.24
Table 1.
Primer sequences
| Hif1α-3’UTR |
| Forward: TTGAGCTCTATCAAATAAACATCTTCTGTGGACCAGG |
| Reverse: CCGAGCTCCCTGGTCCACAGAAGATGTTTATTTGATA |
| Pdgfb-3’UTR |
| Forward: TGCATGCTAGGGGCGTCGGCAAGAGTGTGGGCAG |
| Reverse: TATTCGAAACAACTTTAAATACGGAATATAAAT |
| Notch4-3’UTR |
| Forward: CCACTAGTGCAGCTGCGTGGAAGGAAGGA |
| Reverse: TCAAGCTTTAGCACTTCAAATTAGCTTTA |
| Frizzled4-3’UTR |
| Forward: CCACTAGTTTACTTTCTGTGATGGAGCC |
| Reverse: TCAAGCTTAAGTATCATTTCCCCTGCC |
| RT primers for microRNAs |
| miR-31: CGAAATTGCCAGGATCAGGGTTAAAGATATCTCACGTACACGTA |
| miR-150: CGAAATTGCCAGGATCAGGGTTAAAGATATCTCACGTACACTGG |
| miR-184: CGAAATTGCCAGGATCAGGGTTAAAGATATCTCACGTAACCCTT |
| Real-time PCR primers for microRNAs |
| Universal Forward Primer: CGAAATTGCCAGGATC |
| Reverse Primers: miR-31: AGGCAAGATGCTGGCA |
| miR-150: TCTCCCAACCCTTGTA |
| miR-184: TGGACGGAGAACTGAT |
| 5S rRNA |
| Forward: GCCATACCACCCTGAACG |
| Reverse: AGCCTACAGCACCCGGTATT |
microRNA reporter assays
To assess the validity of purported target sequences in Pdfgb, Frizzled4, Hif1α, and Notch4, they were tested in microRNA reporter assays. Purported target sequences were amplified using retinal cDNA as template and primers that included restriction sites for SpeI and HindIII. PCR products were cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA) and sequenced. Once the correct sequence was confirmed, the SpeI and HindIII fragment was released and subcloned into a pMIR-reporter vector (Ambion, Houston, TX). A pGL2-CMV vector containing 1.7kb mouse VEGF 3’UTR coupled to luciferase was provided by Dr. R. C. Nichols, Department of Microbiology and Immunology, Dartmouth School of Medicine, Hanover, NH.
All pre-microRNAs were obtained from Ambion (Houston, TX). ARPE19 cells were cultured in DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS). At 70% confluence, cells were co-transfected with 5 µl of a 5 µMol/L pre-microRNA, 1 µg of pMIR-target-3’UTR, and 2.5 ng of Runillo. For controls, RPE cells were co-transfected with 5 µl of a 5 µMol/L of control pre-microRNA, 1 µg of pMIR-target-3’UTR, and 2.5 ng of Runillo. After 48 hours, cells were harvested and luciferase expression assays were performed using dual–luciferase assay kits (Promega, St. Luis Obispo, CA).
Mice with oxygen-induced ischemic retinopathy
Ischemic retinopathy was generated as previously described.45, 46 At postnatal day (P) 7, mice were placed in 75 ± 3% oxygen for 5 days and then returned to room air at P12 and given a 1 µl intraocular injection of 5 µM experimental pre-microRNA in one eye and 5 µM control pre-microRNA in the fellow eye. The injections were given under a dissecting microscope with a Harvard Pump Microinjection System and pulled glass micropipettes as previously described.47 For measurement of levels of selected target gene products (VEGF, PDGF-B, Frizzled 4, and HIF-1α), mice were euthanized at P15 and retinal homogenates were prepared for ELISAs or immunoblots. To assess the effect of pre-microRNAs on retinal NV, P12 mice with ischemic retinopathy were given a 1 µl intraocular injection of 5 µM experimental pre-microRNA or a mixture of pre-microRNAs in one eye and 5 µM control pre-microRNA in the fellow eye. The area of NV on the surface of the retina was measured at P17 as previously described.48 Briefly, mice were given an intraocular injection of 1 µl of 0.5 µg rat anti-mouse PECAM antibody Pharmingen (San Jose, CA) and after 12 hours they were euthanized and eyes were fixed in 10% formalin for 4 hours. Retinas were dissected out intact, incubated for 40 minutes in 1:500 goat anti-rat IgG conjugated with Alexa488 (Invitrogen, Carlsbad, CA), washed, and whole mounted. An observer masked with respect to treatment group examined the slides with a Nikon Fluorescence microscope and measured the area of NV per retina by computerized image analysis using ImagePro Plus software (Media Cybernetics, Silver Spring, MD).
ELISAs and Immunoblots
The retinas were removed and placed in 200 µl of lysis buffer (50 µl 1M Tris-HCl (pH 7.4), 50 µl 10% sodium dodecyl sulfate, 5 µl 100 nM phenylmethaneculfonyl and 5 ml sterilized, de-ionized water). Retinas were homogenized, sonicated at 4°C for 5 seconds, and centrifuged at 10,000g for 5 minutes at 4°C. The protein concentration of the supernatants was measured with a bicinchoninic acid protein assay kit (BioRad, Hercules, CA). ELISAs were performed using the Quantikine VEGF assay kit (R&D Systems, Minneapolis, MN) using the manufacturer’s instructions. Serial dilutions of recombinant VEGF were assayed to generate a standard curve. The limit of detection was 125 pg/ml of VEGF. For immunoblots, 20 µg of protein were resolved by SDS-PAGE using 8–12% acrylamide gels and transferred to a nitrocellulose membrane. Non-specific binding was blocked by incubation in Tris-buffered saline (TBS) containing 5% skim milk and membranes were hybridized with 0.5 µg/ml rabbit anti-mouse VEGF polyclonal antibody (Abcam, Cambridge, MA), rabbit anti-mouse Frizzled 4 antibody (R&D Systems, Minneapolis, MN), or rabbit anti-mouse PDGF-B antibody (Santa Cruz Biotechnology, Santa Cruz, CA) in TBS containing 0.05% Tween 20 and 2.5% skim milk at 4°C overnight. After three washes with TBS-Tween-20, the membrane was incubated in horseradish peroxidase (HRP)-conjugated goat anti-rabbit polyclonal antibody (1:5000, Amersham-Pharmacia Biotech, Piscataway, NJ). For signal development, membranes were incubated in Enhanced Chemoluminescence-Plus Western blotting detection reagent (Amersham-Pharmacia Biotech, Piscataway, NJ) and exposed to X-ray film (Kodak, Rochester, NY).
Model of choroidal NV
Choroidal NV was generated by modification of a previously described technique.44 Mice were given a 1 µl intraocular injection of 5 µM experimental pre-microRNA or mixture of experimental pre-microRNAs in one eye and 5 µM control pre-microRNA in the fellow eye right after the laser and again on day 7. Fourteen days after laser, mice were perfused with fluorescein-labeled dextran (2×106 average MW, Sigma, St. Louis, MO) and choroidal whole mounts were prepared and examined by fluorescence microscopy. The area of choroidal NV at each Bruch’s membrane rupture site was measured by image analysis by an observer masked with respect to treatment group.46, 48 The 3 CNV areas within each eye were averaged to give one experimental value per eye.
Acknowledgments
Supported by EY12609 from the National Eye Institute and a senior scientist award from Research to Prevent Blindness, New York, NY.
PAC is the George S. and Dolores Dore Eccles Professor of Ophthalmology and Neuroscience.
References
- 1.Campochiaro PA. Ocular versus extraocular neovascularization: mirror images or vague resemblances. Invest. Ophthalmol. Vis. Sci. 2006;47:462–474. doi: 10.1167/iovs.05-1494. [DOI] [PubMed] [Google Scholar]
- 2.Campochiaro PA. Ocular neovascularisation and excessive vascular permeability. Expert. Opin. Biol. Ther. 2004;4:1395–1402. doi: 10.1517/14712598.4.9.1395. [DOI] [PubMed] [Google Scholar]
- 3.Aiello LP, Pierce EA, Foley ED, et al. Suppression of retinal neovascularization in vivo by inhibition of vascular endothelial growth factor (VEGF) using soluble VEGF-receptor chimeric proteins. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10457–10461. doi: 10.1073/pnas.92.23.10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JW, Adamis AP, Shima DT, et al. Vascular endothelial growth factor/vascular permeability factor is temporally and spatially correlated with ocular angiogenesis in a primate model. Am. J. Pathol. 1994;145:574–584. [PMC free article] [PubMed] [Google Scholar]
- 5.Ozaki H, Seo M-S, Ozaki K, et al. Blockade of vascular endothelial cell growth factor receptor signaling is sufficient to completely prevent retinal neovascularization. Am. J. Pathol. 2000;156:679–707. doi: 10.1016/S0002-9440(10)64773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak N, Okamoto N, Wood JM, Campochiaro PA. VEGF is an important stimulator in a model of choroidal neovascularization. Invest. Ophthalmol. Vis. Sci. 2000;41:3158–3164. [PubMed] [Google Scholar]
- 7.Rosenfeld PJ, Brown DM, Heier JS, et al. Ranibizumab for neovascular age-related macular degeneration. N. Eng. J. Med. 2006;355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 8.Brown DM, Kaiser PK, Michels M, et al. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N. Eng. J. Med. 2006;355:1432–1444. doi: 10.1056/NEJMoa062655. [DOI] [PubMed] [Google Scholar]
- 9.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorgensen R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–344. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Lau NC, LIm LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 14.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 15.Denli AM, Tops BBJ, Plasterk RHA, et al. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 16.Gregory RI, Yan KP, Amuthan G, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 17.Han J, Lee Y, Yeom Y-K, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Poliseno L, Tuccoli A, Mariani L, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 21.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 22.Yang WJ, Yang DD, Na S, et al. Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 23.Shen J, Xie B, Dong A, et al. In vivo immunostaining demonstrates macrophages associate with growing and regressing vessels. Invest. Ophthalmol. Vis. Sci. 2007;48:4335–4341. doi: 10.1167/iovs.07-0113. [DOI] [PubMed] [Google Scholar]
- 24.Ohno-Matsui K, Hirose A, Yamamoto S, et al. Inducible expression of vascular endothelial growth factor in photoreceptors of adult mice causes severe proliferative retinopathy and retinal detachment. Am. J. Pathol. 2002;160:711–719. doi: 10.1016/S0002-9440(10)64891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozaki H, Yu A, Della N, et al. Hypoxia inducible factor-1a is increased in ischemic retina: temporal and spatial correlation with VEGF expression. Invest. Ophthalmol. Vis. Sci. 1999;40:182–189. [PubMed] [Google Scholar]
- 26.Kelly BD, Hackett SF, Hirota K, et al. Cell type-specific regulation of angiogenic growth factor gene expression and induction of angiogenesis in nonischemic tissue by a constitutively active form of hypoxia-inducible factor 1. Circ. Res. 2003;93:1074–1081. doi: 10.1161/01.RES.0000102937.50486.1B. [DOI] [PubMed] [Google Scholar]
- 27.Ji R, Cheng Y, Yue J, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ. Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 28.Pelengaris S, Littlewood T, Khan M, et al. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 29.Brandvold KA, Neiman P, Ruddell A. Angiogenesis is an early event in the generation of myc-induced lymphomas. Oncogene. 2000;19:201–210. doi: 10.1038/sj.onc.1203589. [DOI] [PubMed] [Google Scholar]
- 30.Dews M, Homayouni A, Yu D, et al. Aumentation of tumor angiogenesis by a myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1066. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Good DJ, Polverini PJ, Rastinejad F, et al. A tumor supressor-dependent inhibitor of angiogenesis is immunologically and functionally indistinguishable from a fragment of thrombospondin. Proc. Natl. Acad. Sci. USA. 1990;87:6624–6628. doi: 10.1073/pnas.87.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tolsma SS, Volpert OV, Good DJ, et al. Peptides derived from two separate domains of the matrix protein thrombospondin-1 have anti-angiogenic activity. J. Cell Biol. 1993;122:497–511. doi: 10.1083/jcb.122.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tucker RP. The thrombospondin type 1 repeat superfamily. Int. J. Biochem. Cell Biol. 2004;36:969–974. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Inoki I, Shiomi T, Hashimoto G, et al. Connective tissue growth factor binds vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. FASEB J. 2002;16:219–221. doi: 10.1096/fj.01-0332fje. [DOI] [PubMed] [Google Scholar]
- 35.Shimo T, Nakanishi T, Nishida T, et al. Connective tissue growth factor induces the proliferation, migration, and tube formation of vascular endothelial cells in vitro, and angiogenesis in vivo. J. Biochem. (Tokyo) 1999;126:137–145. doi: 10.1093/oxfordjournals.jbchem.a022414. [DOI] [PubMed] [Google Scholar]
- 36.Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002:S97–S107. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 37.Lagos-Quintana M, Rauhut R, Meyer J, et al. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Molec. Vision. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 39.Karali M, Peluso I, Marigo V, Banfi S. Identification and characterization of microRNAs expressed in the mouse eye. Invest. Ophthalmol. Vis. Sci. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- 40.Xu S, Witmer PD, Sumayag S, et al. MicroRNA transcriptome of mouse retina and identification of a sensory organ specific miRNA cluster. J. Biol. Chem. 2007;282:25053–25066. doi: 10.1074/jbc.M700501200. [DOI] [PubMed] [Google Scholar]
- 41.Lewis BP, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 42.Sethupathy P, Megraw M, Hatzigeogiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 43.Raymond CK, Roberts BS, Garrett-Engele P, et al. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobe T, Ortega S, Luna JD, et al. Targeted disruption of the FGF2 gene does not prevent choroidal neovascularization in a murine model. Am. J. Pathol. 1998;153:1641–1646. doi: 10.1016/S0002-9440(10)65753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen J, Samul R, Lima e Silva R, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2005;13:225–234. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- 46.Shen J, Yang XR, Xiao WH, et al. Vasohibin is up-regulated by VEGF in the retina and suppresses VEGF receptor 2 and retinal neovascularization. FASEB J. 2006;20:723–725. doi: 10.1096/fj.05-5046fje. [DOI] [PubMed] [Google Scholar]
- 47.Mori K, Duh E, Gehlbach P, et al. Pigment epithelium-derived factor inhibits retinal and choroidal neovascularization. J. Cell. Physiol. 2001;188:253–263. doi: 10.1002/jcp.1114. [DOI] [PubMed] [Google Scholar]
- 48.Lima e Silva R, Shen J, Hackett SF, et al. The SDF-1/CXCR4 ligand/receptor pair is an important contributor to several types of ocular neovascularization. FASEB J. 2007;21:3219–3230. doi: 10.1096/fj.06-7359com. [DOI] [PubMed] [Google Scholar]