Abstract
Purpose
To measure T1 and T2 relaxation times of metabolites in glioma patients at 3T and to investigate how these values influence the observed metabolite levels.
Materials and Methods
Twenty-three patients with gliomas and ten volunteers were studied with single voxel 2D J-resolved PRESS using a 3T MR scanner. Voxels were chosen in normal appearing white matter and in regions of tumor. The T1 and T2 of choline containing compounds (Cho), creatine (Cr) and N-acetyl aspartate (NAA) were estimated.
Results
Metabolite T1 relaxation values in gliomas were not significantly different from values in normal white matter. The T2 of Cho and Cr were statistically significantly longer for Grade 4 gliomas than for normal white matter but the T2 of NAA was similar. These differences were large enough to impact the corrections of metabolite levels for relaxation times with tumor grade in terms of metabolite ratios (P<0.001).
Conclusion
The differential increase in T2 for Cho and Cr relative to NAA means that the ratios of Cho/NAA and Cr/NAA are higher in tumor at longer echo times relative to values in normal appearing brain. Having this information may be useful in defining the acquisition parameters for optimizing contrast between tumor and normal tissue in MRSI data, where limited time is available and only one echo time can be used.
Keywords: relaxation times, glioma, 3 T
INTRODUCTION
Gliomas account for the majority of primary brain tumors and vary from benign to malignant. Among all the gliomas, Glioblastoma Multiforme (GBM) is both the most common and the most malignant with a relatively poor prognosis. Proton magnetic resonance spectroscopy (MRS) is a powerful noninvasive tool that has been used for the assessment of metabolites in gliomas and the biochemical profiles of brain tumors have been widely studied (1,2,3,4). A general marker of brain tumors is the elevation of choline containing compounds (Cho), which is thought to be due to increased cell density and membrane turnover in neoplasms, and the reduction of the neural marker N-acetyl aspartate (NAA). The availability of higher field strength MR scanners and multi-channel radiofrequency coils offer the potential of higher signal-to-noise ratio (SNR) and better spectral resolution (5) that can be used to either shorten acquisition time, decrease spatial resolution or improved detection of other brain metabolites, such as glutamate (Glu), a main excitatory neurotransmitter, glutamine (Gln) which acts as a detoxifier, and myo-inositol (mI) that is predominantly located within astrocytes.
In planning the data acquisition parameters for using MR spectroscopic imaging (MRSI) to determine the spatial extent of tumor versus normal brain tissue, it is important to consider how to select values that will emphasize the contrast between metabolites in the different regions. Because of the restrictions on clinical scan time, the repetition time (TR) for acquiring MRSI data is often set at 1~2 s and is usually only possible to acquire data with a single echo time (TE). In deciding which TE is most appropriate for a particular data acquisition it is critical to consider the differences in metabolite intensities caused by the effects of T1 and T2 relaxation times, as well as the changes in metabolite concentrations associated with pathology. Previous studies have reported a difference in T2 relaxation time between tumor and normal tissues in brain at 1.5 T (6,7), but the values reported were variable and there is no literature about the T2 relaxation times in brain tumors at 3 T.
The purpose of this study was to use a single voxel PRESS sequence with an 8-channel phased array coil at 3 T, to measure the longitudinal and transverse relaxation times of Cho, Cr and NAA within anatomic lesions corresponding to grade 3 and grade 4 gliomas and to compare them with white matter (WM) or normal-appearing white matter (NAWM). Because the data acquisition method that was used for this study employed 2D J-resolved spectroscopy, it also allowed the separation of J-coupling information from chemical shift and improved distinction between Glu and Gln in the TE-averaged spectra by a averaging across the different echo times (8). The data were used to compare estimates of metabolite levels in tumor versus NAWM, as well as evaluating the differentials in Cho, Cr and NAA with and without corrections for relaxation times.
MATERIALS AND METHODS
Study Population
Ten volunteers (four males and six females, median age of 28 year) and twenty-three patients (eleven males and twelve females, median age of 44 year) with gliomas at different clinical stages were studied. Tumors were graded by histological examination of tissue samples obtained during biopsy or surgical resection. Of all the patients, there were two Grade 2 oligodendrogliomas, five Grade 3 (two oligodendrogliomas, and three anaplastic astrocytomas), fifteen Grade 4 (GBM) patients and one oligodendroglioma with unknown grade.
Data Acquisition
All the empirical data were acquired using an 8-channel phased array coil on a 3 T GE Signa scanner running the Excite software package (GE Healthcare Technologies, Waukesha, WI). The MR scans included the acquisition of both anatomic and spectral data.
Anatomical MR images comprised T1-weighted sagittal scout images (TR/TE = 70/2 ms), axial FLuid Attenuated Inversion Recovery (FLAIR) (TR/TE/TI = 10002/121/2200 ms) and T1-weighted SPoiled GRadient echo (SPGR) images (TR/TE = 26/3 ms).
All spectra were acquired using 2D J-resolved techniques. For T2 studies and metabolite levels, 2D J-resolved PRESS data were obtained using 64 steps with a time increment of 2.5 ms starting at TE of 35 ms (8). With a TR of 2 s and a number of excitation (NEX) of 2, the total acquisition time was ~5 minutes. For T1 studies, TE-averaged PRESS data was obtained with 16 steps and a time increment of 10 ms at TR = 1s with NEX = 4, TR = 2 s with NEX = 4, and TR = 8 s with NEX = 2, respectively (8,9). The spectral data were acquired with 2048 spectral points, and 5000 Hz spectral width. To obtain estimates of coil sensitivities for the combination of the 8-channel data, unsuppressed water spectra were acquired with an echo time of 35 ms with each of the PRESS spectra.
The 8-cc single voxels were located within WM, NAWM or regions of tumor for participants. The volume of interest (VOI) from volunteers was localized in the parietal white matter. The NAWM in patients was positioned as far as possible from the lesions and included four parietal NAWM voxels and six frontal NAWM voxels. The tumor regions were defined by T2 hyperintensity from T2 FLAIR images and positioned to cover as much of the lesion as possible. The number and regions of interest (ROI) investigated for each patient in the T2 studies were shown as follows: four patients (NAWM only), six patients (NAWM and one tumor voxel) and nine patients (one tumor voxel only). Due to long acquisition times to measure T1 values of metabolite, a relatively small number of patients was involved in the study and these patients varied from low grade gliomas to high grade gliomas.
Data Processing
Post-processing was applied on a Sun workstation (Figure 1). The time domain data were extracted from the raw files using the SAGE software package™ (General Electric, Milwaukee) which is based on the IDL (Research Systems, Inc., Boulder, CO) software package. An internal water reference was used for phase and frequency correction, and 4-Hz Lorentzian apodization was applied in the F2 dimension. 8-channel data were combined in the time-domain using the unsuppressed water signal (10). Spectra were then zero filled and processed with a fast Fourier Transform (FFT). The TE-averaged FID was generated by averaging the different echo times and metabolite quantification was performed without any spectral apodization or zero-filling.
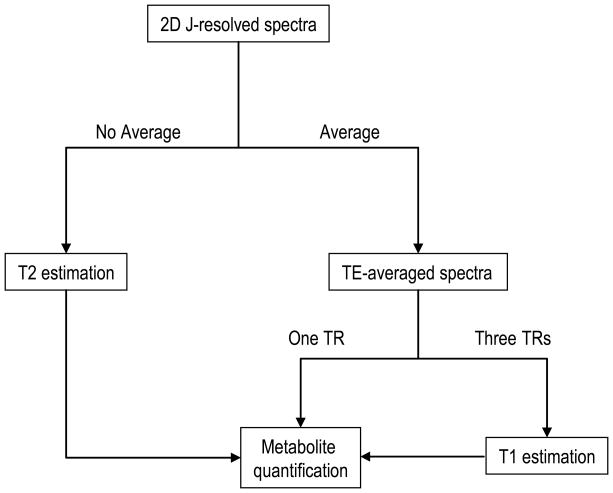
Figure 1.
Scheme of the procedure for data postprocessing.
Metabolite T1 and T2 estimation
To increase the SNR and the accuracy of the T2 calculation, data for each echo-time were averaged with the one before and the one after in the t1 dimension and then Fourier transformed in the t2 dimension. The peak heights of Cho at 3.22 ppm, Cr-CH3 at 3.02 ppm and NAA at 2.02 ppm were extracted from the spectra with TE from 57.5 ms to 190 ms, a total of 54 spectra, to reduce the contamination of macromolecules and/or J-coupled multiplets, and fitted to a single exponential function. Only the T2 fits with variances of the fit residue smaller than 10% were included in the analysis.
T1 relaxation times were calculated from data partial saturation using a two-parameter least-squares fitting routine to the equation S / S0 = 1−2 exp[−(TR −TE / 2) / T1 ] +exp(−TR / T1 ) (11), where S is the signal intensity acquired at three different TRs, S0 is the fully relaxed signal intensity, and the effective TE of the TE-averaged spectra for T1 acquisition is 110 ms. The signal intensities of Cho, Cr and NAA were normalized for the number of acquisitions that were obtained before T1 fitting.
Estimation of Differences in Metabolite Levels
To estimate metabolite levels, the combined data were averaged in the t1 domain before zero filling and then quantified using the LCModel package (12) and QUEST (quantitation based on quantum estimation) quantitation algorithm (13) in the Magnetic Resonance User Interface (MRUI) spectrum analysis program.
An in vitro basis set of individual metabolites, consisting of NAA, Glu, Gln, Cr, Cho and mI, was prepared for LCModel. The fitting was performed between 3.85 ppm and 1.8 ppm to minimize spectral artifact from lipid. Metabolite concentrations included in analysis were those with relative Cramér-Rao lower bounds (CRLB) lower than 5% for Cho, Cr and NAA, and 20% for mI and Glu.
For QUEST, the signals were analyzed in the time domain. To minimize the incorporation of large broad baseline components, the first data point of the free induction decay (FID) was excluded and the first twenty points were weighted by a quarter-wave sinusoid (14). A Hankel Lanczos singular value decomposition (HLSVD) filter was applied to remove the residual water from the spectra (15). Metabolic signals for the basis set were generated using GAMMA simulations (16) with prior knowledge of chemical shift and J-coupling (17). Due to known differences in T2 relaxation times between two singlets of Cr peak, Cr-CH2 and Cr-CH3 are separated into two spectra in the basis set. The selection criteria for QUEST estimates were defined by the output parameters from the QUEST analysis, namely a CRLB smaller than 20% and additional common damping factor smaller than 12 Hz. The calibration factors for both methods were obtained from a standard GE MRS HD phantom (12.5 mM NAA, 10 mM Cr, 3 mM Cho, 12.5 mM Glu, 7.5 mM mI and 5 mM lactate) and 50 mM NAA only phantom (50 mM phosphate buffer, pH 7.2) and were applied in all of the data sets.
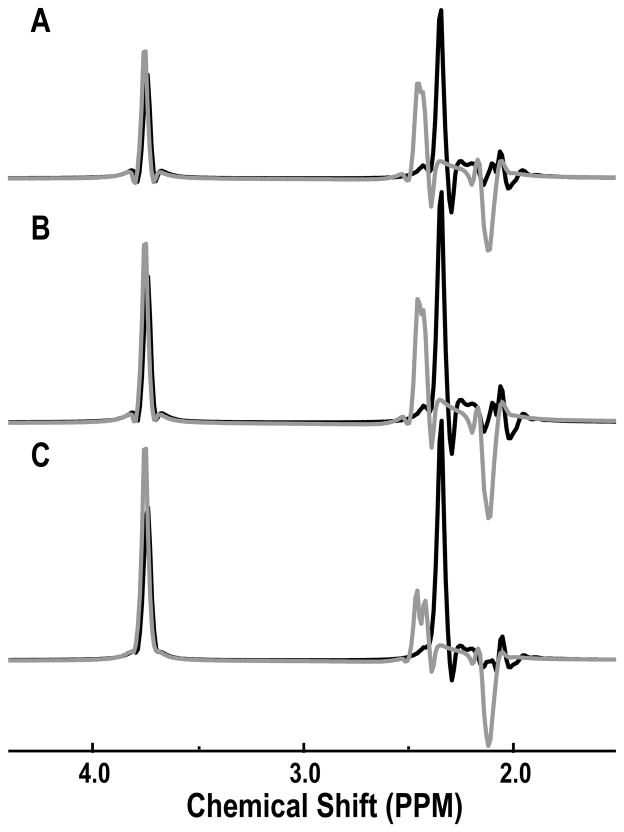
To estimate the T2 values on the effective separation of Glu and Gln in the TE-averaged spectra, the spectra were created with 2048 spectral points and spectral width of 5000 Hz and TE values starting at 35 ms in either 128 steps of 2.5 ms or 64 steps of 2.5 ms. To approximate the effects of transverse relaxation time and inhomogeneities for the in vivo spectra for Glu and Gln, the time data were multiplied by a factor, where t1 is the echo time (TE) and t2 is the total sampling time. was set to 50 ms which was estimated from the mean linewidth of metabolites (5), and a simple monoexponential decay was applied to the data with T2 values of 100 ms, 150 ms and 300 ms, respectively. A 4-Hz Lorentzian apodization, zero filling and Fourier Transform were applied and the spectra were averaged in the t1 domain. Figure 2 shows the TE-averaged spectra of Glu and Gln that were simulated using this approach. The efficacy of separation for C4 protons of Glu and Gln in TE-averaged spectra was the same for different T2 values but the signal intensities of Gln depended on the T2 values. This meant that the signals from the C4 protons of Gln were not fully cancelled in the simulation, but they were clearly separated from the C4 protons of Glu due to the difference in chemical shift being 0.1 ppm.
Figure 2.
Simulation of TE-averaged PRESS spectra of Glu (black) and Gln (grey) with different T2 relaxation times. TE-averaged PRESS spectra were simulated with 64 steps and T2 of 100 ms (A) and T2 of 150 ms (B), and 128 steps and T2 of 300 ms (C).
Corrections of Metabolite Levels for Relaxation Times
The metabolite levels were corrected for transverse and longitudinal relaxation times using the values estimated from the empirical data. T2 correction was applied for levels of Cho, Cr and NAA by multiplying LCModel concentrations and QUEST parameter estimates by a factor fTE =exp[TE·(1/ T2vivo −1/ T2vitro )], respectively, while the effective TE of the TE-averaged spectra for T2 acquisition was 113.75 ms, and T1 correction factor was fTR =[1−exp(−TR / T1vitro )] /[1−exp(−TR / T1vivo )] for Cho, Cr and NAA. Since the metabolite signals used as a basis for QUEST were ideally simulated without any effect of T1 and T2, the calibration factor only required a calculation of the in vivo T1 and T2 values.
Voxel Content Analysis
For the empirical data, the spectral data were referenced to the 3D SPGR image by assuming that there was no movement between the image and spectra acquisition. The FLAIR image was aligned to the corresponding 3D SPGR image. Segmentation of the brain images was performed automatically on the 3D SPGR images using a program that is based on a Markov random field model (18). The segmented white matter mask was then used to identify voxels which had at least 75% normal appearing white matter. The masks of the region of T2 hyperintensity were segmented using region growing segmentation tool (19). The tumor voxels used for the study were restricted to those that had more than 80% within the T2 hyperintensity. These were the voxels used for comparative analysis to the volunteer and patient data.
Statistics
The statistical analysis was performed using the SPSS software package (Chicago, IL). A P-value of <0.05 was regarded as significant for all the tests. For T1 studies, nonparametric Wilcoxon rank sum tests were used to test the difference between lesions and normal tissues.
ANalysis Of VAriance (ANOVA) was utilized to determine whether T2 values and metabolite levels differed between normal, Grade 3 and Grade 4. To check possible confounding effects of age and sex, these variables were added into the model. However, they did not change the effects of tumor grade and they were insignificant. And then Tukey’s honestly significant difference tests were used to test these differences between tumor grades or between T2 hyperintensity lesion and normal white matter.
The Spearman rank correlation coefficients were calculated to determine the association between the estimated metabolite concentrations of LCModel and QUEST for Cho, Cr and NAA after corrections with T1 and T2 relaxation times.
Changes of metabolite ratios by corrections with T1 and T2 relaxation times in each grade and overall was tested using a paired t-test. And then a t-test was used to compare whether the effect of change was dependent on tumor grade.
RESULTS
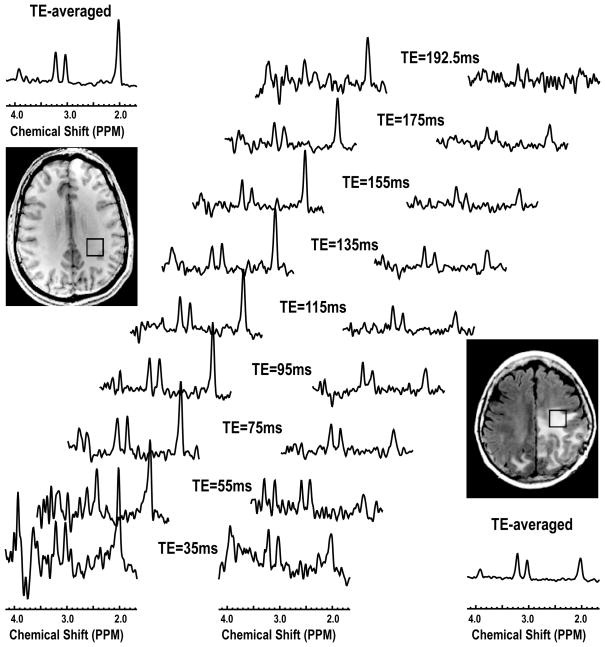
The quality of data for the single voxel data was excellent. Figure 3 shows an example of the 2D J-resolved and TE-averaged spectra from a volunteer and a patient. Note the differences in metabolite levels, with a clear reduction in the level of NAA in the voxel from tumor. At the shorter echo times peaks corresponding to Gln, Gln and mI can be observed.
Figure 3.
Single voxel 2D J-resolved spectra acquired from a volunteer and a patient with a GBM. The TE-averaged spectra were plotted along the individual echo spectra.
Metabolite T1 Relaxation times
The T1 relaxation times of metabolites were available for voxels in 8 volunteers and 8 patients. Values were 1.06±0.11 s (mean ± standard deviation, SD) versus 1.00±0.21 s for Cho, 1.38±0.13 s versus 1.37±0.20 s for Cr, and 1.38±0.13 s versus 1.38±0.21 s for NAA. There was no significant difference between these values.
Metabolite T2 Relaxation times
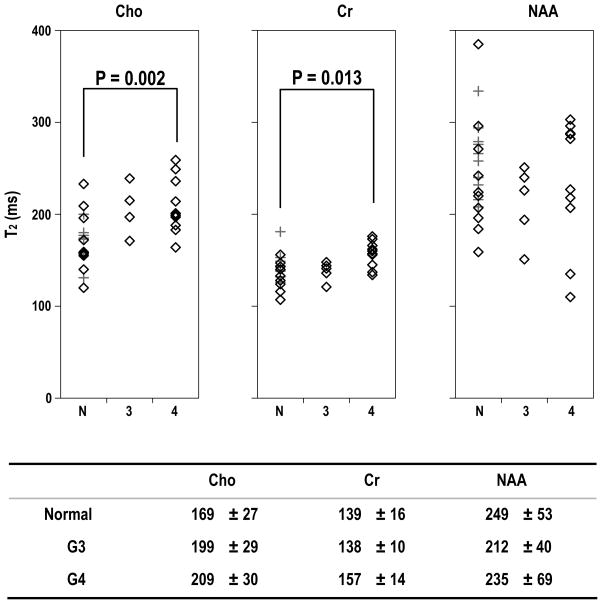
For the estimation of T2 values, there were 10 voxels in WM from volunteers, 3 voxels from NAWM in patients with Grade 3 glioma and 7 voxels from NAWM in patients with Grade 4 glioma. Neither the values in WM and NAWM or their standard deviations were significantly different. The results were therefore pooled as normal white matter VOIs for comparison with tumor voxels. NAWM voxels from patients were treated as independent observations from the values determined in the lesions because they were selected to be clearly distinct from the lesions on T2 weighted images and did not appear to be correlated with tumor values. The pooled white matter values for Cho were 169±27ms, for Cr were 139±16ms and for NAA were 249±53ms. There were tumor voxels from 5 different patients with Grade 3 glioma and 10 different patients with Grade 4 glioma. The variations in T2 relaxation values of metabolites for normal white matter and patients were shown in Figure 4. The range of NAA T2 values was quite large, and they were not significantly different between normal white matter and tumor voxels. For voxels from Grade 3 glioma the T2 values of Cho were 199±29ms but the small sample size meant that this was not significant. For voxels from Grade 4 glioma the T2 values of Cho were 209±30ms and for Cr were 1.57±14ms. These were significantly different from the values in normal white matter with P=0.002 and P=0.013 respectively.
Figure 4.
T2 relaxation times for metabolites (ms). The significance was tested between normal white matter and tumor using Tukey’s honestly significant difference test. The diamond in the figure stands for data from patients, while the cross for volunteers. N, 3 and 4 represent values from WM in volunteers and NAWM in patients, Grade 3 and Grade 4 lesions, respectively.
Metabolite Levels Corrected for relaxation Times
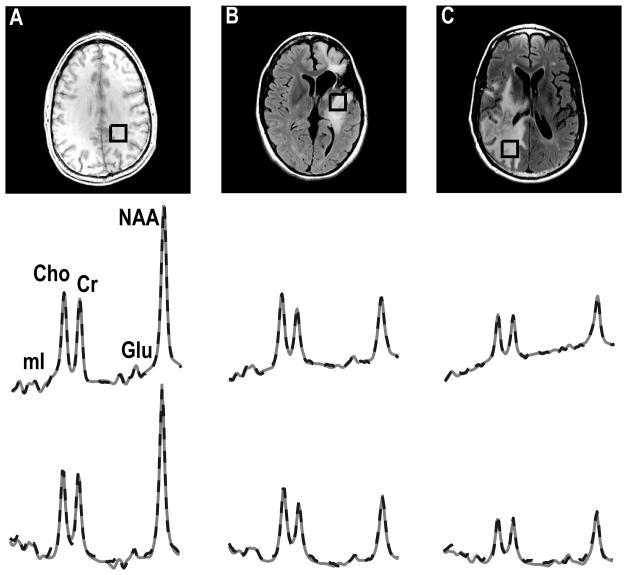
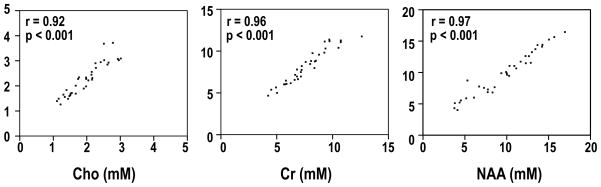
Examples of TE-averaged spectra from normal parietal white matter, Grade 3 glioma and Grade 4 glioma that were quantified by LCModel and QUEST and are shown in Figure 5. Both algorithms fit the data well, with the differences in the plotted spectra being due to the way in which the baseline estimation is performed. Because of the different relaxation times of metabolites in the basis sets for the two methods, due to one being simulated and the other from in vitro phantoms, the comparison between them only considered Cho, Cr and NAA, which were corrected for T1 and T2 values. The fact that the estimates for Cho, Cr and NAA obtained using LCModel and QUEST were consistent is reflected in Figure 6. The Spearman rank correction coefficients were 0.92, 0.96 and 0.97 for Cho, Cr and NAA estimates with P<0.001 for all three metabolites.
Figure 5.
TE-averaged spectra quantified by LCModel (middle row) and QUEST (third row) corresponding to the image in a volunteer (A), Grade 3 (B) and Grade 4 (C) patients. The fitted spectra (straight line) were related to the phased spectra (broken line).
Figure 6.
Scatter plots of Cho, Cr and NAA calculated from LCModel vs. QUEST for all the voxels in the study. Metabolite concentrations on the x-axis represent those determined from LCModel, while the values on the y-axis were obtained from the same spectra using quantification by QUEST. P-value represents the significance test for the correlation coefficient.
The estimates of metabolite levels obtained using QUEST and LCModel are further illustrated in Table 1 and Table 2. Cho and Cr were clearly increased in Grade 3 gliomas but decreased in Grade 4 gliomas relative to normal brain. The differences in Cr between Grade 3 and 4 lesions was statistically significant (P<0.01). The concentrations of NAA were reduced in both tumor grades (P<0.001). Compared with those in normal white matter, the levels of mI estimated using LCModel were increased in Grade 3 gliomas (P=0.002) and statistical significantly different from Grade 4 (P=0.017). No significance was found for the results using QUEST. The levels of Glu were significantly increased in Grade 3 from QUEST (P=0.047), and the difference between Grade 3 and Grade 4 for Gln was marginally significant for LCModel (P=0.05).
Table 1.
Concentrations of Cho, Cr and NAA as corrected using empirical T1 and T2 relaxation times and expressing the (mean±SD) in mM by QUEST and LCMODEL. Statistics were evaluated using Tukey’s honestly significant difference tests.
| Methods | Metabolite | Group | Concentrations | Population | G3 | G4 |
|---|---|---|---|---|---|---|
| QUEST | Cho | Normal | 2.3±0.5 | N=20 | ||
| G3 | 2.9±0.9 | N=5 | P<0.01 | |||
| G4 | 1.9±0.5 | N=10 | P<0.01 | |||
| Cr | Normal | 8.9±1.6 | N=20 | P<0.001 | ||
| G3 | 9.1±2.4 | N=5 | P<0.005 | |||
| G4 | 6.0±0.9 | N=10 | P<0.005 | |||
| NAA | Normal | 12.4±2.1 | N=20 | P<0.001 | P<0.001 | |
| G3 | 6.8±1.5 | N=5 | ||||
| G4 | 6.3±1.6 | N=10 | ||||
| LCModel | Cho | Normal | 2.0±0.5 | N=20 | ||
| G3 | 2.3±0.6 | N=5 | ||||
| G4 | 1.7±0.6 | N=10 | ||||
| Cr | Normal | 8.4±1.5 | N=20 | P=0.001 | ||
| G3 | 8.6±2.6 | N=5 | P<0.001 | |||
| G4 | 5.8±1.1 | N=10 | P<0.001 | |||
| NAA | Normal | 12.5±2.1 | N=20 | P<0.001 | P<0.001 | |
| G3 | 5.5±1.5 | N=5 | ||||
| G4 | 6.6±2.2 | N=10 |
N=number of patients
Table 2.
Levels of mI, mIG, Glu and Gln as quantified by QUEST and LCModel (mean±SD). Statistics were evaluated using Tukey’s honestly significant difference tests.
| Methods | Metabolite | Group | Levels | Population | G3 | G4 |
|---|---|---|---|---|---|---|
| QUEST | mI | Normal | 2.8±0.9 | N=20 | ||
| G3 | 3.8±1.2 | N=5 | ||||
| G4 | 2.8±1.7 | N=8 | ||||
| mIG | Normal | 3.6±1.1 | N=20 | |||
| G3 | 5.1±1.8 | N=5 | ||||
| G4 | 3.3±1.6 | N=8 | ||||
| Glu | Normal | 2.3±0.5 | N=14 | P<0.05 | ||
| G3 | 3.1±0.8 | N=4 | P<0.05 | |||
| G4 | 2.1±0.5 | N=4 | P<0.05 | |||
| Gln | Normal | 2.5±0.9 | N=19 | |||
| G3 | 2.7±0.1 | N=2 | ||||
| G4 | 2.3±0.7 | N=9 | ||||
| LCModel | mI | Normal | 5.9±1.5 | N=14 | P<0.005 | |
| G3 | 10.4±2.4 | N=4 | P<0.05 | |||
| G4 | 6.4±2.7 | N=5 | P<0.05 | |||
| Glu | Normal | 6.3±1.9 | N=14 | n/a | ||
| G3 | 8.4 | N=1 | ||||
| G4 | 5.4±1.5 | N=3 | n/a | |||
| Gln | Normal | 2.2±0.3 | N=3 | |||
| G3 | 2.4±0.6 | N=4 | P=0.05 | |||
| G4 | 1.5±0.5 | N=5 | P=0.05 |
N=number of patients
Differences in Metabolite Ratios With and Without Corrections for T2
Since the time available for data acquisition is relatively short in the case of in vivo MRSI data, the difference in T2 relaxation times may contribute to the distinction between tumor and normal tissue. To investigate the magnitude of this effect we considered the differences in metabolite ratios for our single voxel data with and without correction for relaxation times. The metabolite ratios with and without corrections of relaxation values were shown in Table 3. In all cases the Cho/Cr, NAA/Cr and Cho/NAA ratios in tumors were larger than in normal white matter for the uncorrected compared to the corrected values. This implies that the differential in relaxation times between Cho, Cr and NAA increases the contrast for spectra in tumor relative to normal tissue when there is a higher degree of T2-weighting. Statistical significance was found for the interaction between the correction and tumor grade for all the metabolite ratios with P<0.001.
Table 3.
Relative metabolite ratios (mean±SD). The ratios of Cho/Cr, NAA/Cr and Cho/NAA estimated by LCModel were compared with the same data after the corrections of T1 and T2 values.
| Cho/Cr | NAA/Cr | Cho/NAA | ||||
|---|---|---|---|---|---|---|
| Uncorrected | Corrected | Uncorrected | Corrected | Uncorrected | Corrected | |
| Normal | 0.36±0.05 | 0.24±0.03 | 1.84±0.23 | 1.51±0.19 | 0.20±0.04 | 0.16±0.03 |
| G3 | 0.47±0.13 | 0.28±0.08 | 0.80±0.37 | 0.71±0.33 | 0.70±0.35 | 0.47±0.23 |
| G4 | 0.46±0.09 | 0.29±0.06 | 1.23±0.35 | 1.14±0.33 | 0.42±0.23 | 0.29±0.16 |
DISCUSSION
Gliomas are the most common primary brain tumor and primarily affect white matter. The ability to map out the spatial extent of tumor relative to normal appearing white matter is dependent upon both the variations in absolute concentrations of metabolites and in degree of T1 and T2 weighting in any given acquisition. Designing the most appropriate acquisition parameters for obtaining multi-voxel MRSI data requires the measurement of these variables in localized spectra from normal volunteers and patients with brain tumors.
The methods used in this study to estimate relaxation times and metabolite levels had several benefits over previous approaches. Firstly, the individual spectra that used for T2 fitting were the mean of three spectra with a TE difference of 2.5 ms, which increased the signal-to-noise ratio (SNR), as shown in Figure 3. This can be expected to provide more reliable estimates of the T2 values. Note that J-coupling differences within these three averaged spectra would have been very small and were therefore ignored. The second benefit in using the 2D refocused sequence is that the effective echo time was relatively long would therefore reduce the effects of macromolecules and decrease the contribution of J-coupled resonances of Glu located underneath the NAA peak. Although the last echo in the T2 fit (TE = 190 ms) was not long enough to cover the complete signal decay, more points were involved in our study than in previous publications (20,21,22). Thirdly, since the TE-averaged spectra had a flat baseline, the possibility of overestimating T1 values should be relatively small.
A limitation of the approach used was that the weak signal intensities of Glu and mI translated into a large variation and uncertainty in peak intensities for individual echo times and so the T1 and T2 of these metabolites were not able to be evaluated. To estimate the T2 values of metabolites such as mI, Glu and Gln with complex J coupling patterns would require a metabolite specific spectral editing sequence (23,24).
Several studies have assessed the T1 and T2 relaxation times of normal brain metabolites within different regions of the normal brain and at different field strengths. T1 values were significantly longer at 3 T than at 1.5 T (21), but were not reported as showing major regional variability (22). A distinct correlation was reported between T2 and relative WM/GM composition for NAA and Cr-CH3 (22). This suggests that the T2 of these intracellular metabolites is more affected by the integrity and composition of the tissue, whereas T1 values are influenced by molecular tumbling, reflecting the viscosity of the medium. The T1 relaxation times of metabolites from normal brain that were observed in our study are similar to those previously published (22), and were not significantly different from the T1 values in gliomas. While this is consistent with observations made at 1.5 T (7) it may also be due to the limited number of cases in the study.
The T2 relaxation times of metabolites from normal white matter, including 14 parietal white matter and 6 frontal white matter, in our study, are close to that previously published (9), which also investigated T2 values in the parietal white matter. T2 relaxation times in Cho and Cr were statistically significantly longer in Grade 4 gliomas compared with those in normal white matter. The Cho peak resonates at 3.20 ppm and represents choline, phosphocholine (PC) and glycerophosphocholine (GPC). Previous studies have shown that a switch from GPC to PC is associated with glioma malignancy (25,26). This could be one reason why the observed T2 of Cho increases in tumor. The higher T2 of Cr in Grade 4 glioma may also be associated with the changes of metabolite composition, associated with the increase of phosphocreatine (PCr) to Cr. Note that PCr contains a high energy phosphate bond, which transfers to ADP via the breakdown of PCr to Cr and that reduced PCr has been observed in gliomas (27). This suggests that there is a high energy requirement for maintaining the growth of cells in gliomas. In the previous studies that were performed at 1.5 T, either the T2 relaxation time of NAA and Cr were reported to be shorter in high grade gliomas (6) or the T2 relaxation time of all of the singlets were shorter in the tumor (7). The differences observed in our study may be because of the higher field strength used, the larger number spectra employed in calculating the T2 values or the application of a more effective statistic test, Tukey’s honestly significant difference test, instead of a regular student t-test. Compared to Bonferroni correction, which is known to be overly conservative in the adjustment of multiple comparisons, Tukey’s honestly significant difference test takes into account the correlation structure of the ANOVA model and is more efficient.
The elevation of the Cho peak and the reduction of the neuronal marker NAA are considered to be characteristic of gliomas. Our results suggest that the changes in Cho and Cr peak heights in gliomas are partially caused by T2 effects and partially by changes in absolute concentrations, and that there is a significant interaction exists between corrections for relaxation times and tumor grade. Longer T2 relaxation value of Cho in gliomas means that the ratio of Cho/NAA that is observed in long echo spectra is larger than that in short echo spectra and relatively larger than values in normal white matter at the same echo time. This is consistent with the previous observations at 1.5 T (28).
Two different algorithms were used to quantify the metabolic profiles in our study. LCModel uses a constrained regularization method accounting for the phase, lineshape and baseline (12). QUEST differs from the LCModel in the method used for fitting the baseline, and is sensitive to whether a common extra-damping factor or different metabolite extra-damping are estimated (13). In our case, we used a simulated basis set and found better results when a unique common extra-damping was estimated. Since there is little penalty for the contamination of macromolecules and lipids in the TE-averaged spectra (8,9,29), we used the QUEST quantification algorithm without background adjustment, but weighted the first 20 points with a quarter-wave sinusoid and discarding the first points to reduce the influence of broad resonances underlying metabolites of interest. The results of the estimates of Cho, Cr and NAA showed a very high correlation coefficient between the two methods. Compared with LCModel, QUEST gives more flexibility and reliability but has more subjective interaction, which requires the use of more prior knowledge.
As expected, there was a striking reduction in NAA for all the grades of glioma. The higher metabolite levels in Grade 3 compared to Grade 4 glioma suggested that the levels in the higher grade lesions may be influenced by partially voluming with necrosis (30). The mI is predominantly located within astrocytes and is a precursor for the phosphatidylinositol (PI) second messenger system (31) that is also presumed to act as an osmoregulator. It resolves as an apparent doublet at 3.6 ppm in TE-averaged spectra, which cannot be separated from the Gly peak which resonates at 3.56 ppm. Compared to the simulation, the doublet of mI from the in vitro solution had different effective T2 values, which resulted in the difference of fitting outcomes between LCModel and QUEST. Changes that are associated with mI and reported in the literature show increases in mIG (32) and its ratio to Cr (33) in low-grade astrocytomas but decreased in high grade. In our results, the mI estimated from LCModel was statistically significantly higher in Grade 3 compared to normal white matter.
Although averaged 2D J resolved spectra have less macromolecule contamination compared with short echo acquisitions, macromolecules are always associated with gliomas and result in more uncertainty of small peaks such as Glu/Gln. The changes observed with LCModel and QUEST had a similar pattern of changes. Previous studies have showed that a significant increase in Glx was found in oligodendrogliomas and was used to discriminate them relative to low-grade astrocytomas (34). The increase in Glu that was observed in the Grade 3 glioma considered in our study is consistent with the observation that glioma cells may secrete Glu, resulting in an increase in extracellular Glu (35,36). Increased Glu could be also associated with inflammation in the peritumor tissues since activated microglia and brain macrophages express high affinity glutamate transporters (37) and stimulates tumor cell proliferation. Gln was also slightly increased in Grade 3 but the change was not statistically significant. Because Gln acts as an suppressor for apoptosis, it could contribute to block apoptosis induced by exogenous agents, such as radiation treatment and chemotherapy, and also promote tumor proliferation (38). As predicted by the simulation, the signal intensities of Glu and Gln in the TE-averaged spectra depend on the T2 values. The pattern of changes in Gln for gliomas would be much clearer if estimates of the T2 of Glu and Gln were obtained. Although mobile lipids and lactate are also strongly associated with the higher grade of gliomas, it was not possible to distinguish Lac from lipid in the TE-averaged spectra. It is for this reason that we did not attempt to quantify these metabolites in the current study.
It is well known that gliomas are extremely heterogeneous. The regions of T2 hyperintensity on the T2-weighted image may include edema, gliosis, inflammation, active tumors or treatment effects. While we chose to consider large tumors and required that 80% of the voxel considered was in the tumor, the effect of heterogeneity is a limitation in terms of the results of our study. Due to the large voxel size in the study (8-cc), it was not possible to evaluate the differences between enhancing and non-enhancing lesions. This would require multi-voxel acquisitions (39). Obtaining biopsies from the region with the highest metabolite ratio (Cho/NAA) may further help to understand the underlying biologic characteristics of the tumor. Another factor that may have influenced the data is that there were four different grades of gliomas considered, which arise from different glial cells, such as oligodendrocytes and astrocytes, and could therefore result in different metabolite characterizations (34).
In conclusion, the results of our study demonstrated the differences in metabolite relaxation times and in metabolite concentrations both between normal white matter and tumor and between glioma of Grade 3 and Grade 4. These data suggest that the contrast in metabolite rations between tumor and normal tissue would be greatest at longer echo times and, if the signal to noise is high enough, the use of long echo times may have benefits over short echo times. Our results are encouraging but further studies are required to provide more information about the spatial variations in metabolite concentrations within tumor and peritumor regions using multi-voxel acquisitions in populations of patients with gliomas.
Acknowledgments
Grant Support: NIH RO1 CA59880 and ITL-BIO 04-10148
Footnotes
Presented in part at the 14th Annual Meeting of ISMRM, Seattle, WA, United States, 2006
References
- 1.Gill SS, Thomas DG, Van Bruggen N, Gadian DG, Peden CJ, Bell JD, et al. Proton MR spectroscopy of intracranial tumours: in vivo and in vitro studies. J Comput Assist Tomogr. 1990;14:497–504. doi: 10.1097/00004728-199007000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Fulham MJ, Bizzi A, Dietz MJ, Shih HH, Raman R, Sobering GS, et al. Mapping of brain tumor metabolites with proton MR spectroscopic imaging: clinical relevance. Radiology. 1992;185:675–686. doi: 10.1148/radiology.185.3.1438744. [DOI] [PubMed] [Google Scholar]
- 3.Preul MC, Caramanos Z, Collins DL, Villemure JG, Leblanc R, Olivier A, et al. Accurate, noninvasive diagnosis of human brain tumors by using proton magnetic resonance spectroscopy. Nat Med. 1996;2:323–325. doi: 10.1038/nm0396-323. [DOI] [PubMed] [Google Scholar]
- 4.Tong Z, Yamaki T, Harada K, Houkin K. In vivo quantification of the metabolites in normal brain and brain tumors by proton MR spectroscopy using water as an internal standard. Magn Reson Imaging. 2004;22:1017–1024. doi: 10.1016/j.mri.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Osorio JA, Ozturk-Isik E, Chen AP, Xu D, Crane JC, et al. Considerations in applying 3D PRESS H-1 brain MRSI with an eight-channel phased-array coil at 3 T. Magn Reson Imaging. 2006;24:1295–1302. doi: 10.1016/j.mri.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Isobe T, Matsumura A, Anno I, Yoshizawa T, Nagatomo Y, Itai Y, et al. Quantification of cerebral metabolites in glioma patients with proton MR spectroscopy using T2 relaxation time correction. Magn Reson Imaging. 2002;20:343–349. doi: 10.1016/s0730-725x(02)00500-3. [DOI] [PubMed] [Google Scholar]
- 7.Sijens PE, Oudkerk M. 1H chemical shift imaging characterization of human brain tumor and edema. Eur Radiol. 2002;12:2056–2061. doi: 10.1007/s00330-001-1300-3. [DOI] [PubMed] [Google Scholar]
- 8.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004;51:435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan R, Sailasuta N, Hurd R, Nelson S, Pelletier D. Evidence of elevated glutamate in multiple sclerosis using magnetic resonance spectroscopy at 3 T. Brain. 2005;128:1016–1025. doi: 10.1093/brain/awh467. [DOI] [PubMed] [Google Scholar]
- 10.Wright SM, Wald LL. Theory and application of array coils in MR spectroscopy. NMR Biomed. 1997;10:394–410. doi: 10.1002/(sici)1099-1492(199712)10:8<394::aid-nbm494>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 11.Young IR, Bailes DR, Burl M, Collins AG, Smith DT, McDonnell MJ, et al. Initial clinical evaluation of a whole body nuclear magnetic resonance (NMR) tomograph. J Comput Assist Tomogr. 1982;6:1–18. doi: 10.1097/00004728-198202000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 13.Ratiney H, Sdika M, Coenradie Y, Cavassila S, van Ormondt D, Graveron-Demilly D. Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed. 2005;18:1–13. doi: 10.1002/nbm.895. [DOI] [PubMed] [Google Scholar]
- 14.Knijn A, de Beer R, van Ormondt D. Frequency-selective quantification in the time domain. J Magn Reson. 1992;97:444–450. [Google Scholar]
- 15.Pijnappel WWF, van den Boogaart A, de Beer R, van Ormondt D. SVD-based quantification of magnetic resonance singals. J Magn Reson. 1992;97:122–134. [Google Scholar]
- 16.Smith SA, Levante TO, Meier BH, Ernst RR. Computer Simulations in Magnetic Resonance. An Object-Oriented Programming Approach. J Magn Reson. 1994;106:75–105. [Google Scholar]
- 17.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 19.Saraswathy S, Crawford F, Nelson SJ. Semi-Automated Segmentation of Brain Tumor Lesions in MR Images. Proceedings of the 14th Annual Meeting of ISMRM; Seattle. 2006. (abstract 1609) [Google Scholar]
- 20.Mlynarik V, Gruber S, Moser E. Proton T (1) and T (2) relaxation times of human brain metabolites at 3 Tesla. NMR Biomed. 2001;14:325–331. doi: 10.1002/nbm.713. [DOI] [PubMed] [Google Scholar]
- 21.Ethofer T, Mader I, Seeger U, Helms G, Erb M, Grodd W, et al. Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magn Reson Med. 2003;50:1296–1301. doi: 10.1002/mrm.10640. [DOI] [PubMed] [Google Scholar]
- 22.Traber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging. 2004;19:537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- 23.Choi C, Coupland NJ, Bhardwaj PP, Kalra S, Casault CA, Reid K, et al. T2 measurement and quantification of glutamate in human brain in vivo. Magn Reson Med. 2006;56:971–977. doi: 10.1002/mrm.21055. [DOI] [PubMed] [Google Scholar]
- 24.Choi C, Coupland NJ, Bhardwaj PP, Malykhin N, Gheorghiu D, Allen PS. Measurement of brain glutamate and glutamine by spectrally-selective refocusing at 3 Tesla. Magn Reson Med. 2006;55:997–1005. doi: 10.1002/mrm.20875. [DOI] [PubMed] [Google Scholar]
- 25.Usenius JP, Vainio P, Hernesniemi J, Kauppinen RA. Choline-containing compounds in human astrocytomas studied by 1H NMR spectroscopy in vivo and in vitro. J Neurochem. 1994;63:1538–1543. doi: 10.1046/j.1471-4159.1994.63041538.x. [DOI] [PubMed] [Google Scholar]
- 26.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 27.Negendank W. Studies of human tumors by MRS: a review. NMR Biomed. 1992;5:303–324. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- 28.Kaminogo M, Ishimaru H, Morikawa M, Ochi M, Ushijima R, Tani M, et al. Diagnostic potential of short echo time MR spectroscopy of gliomas with single-voxel and point-resolved spatially localised proton spectroscopy of brain. Neuroradiology. 2001;43:353–363. doi: 10.1007/s002340000473. [DOI] [PubMed] [Google Scholar]
- 29.Bolan PJ, DelaBarre L, Baker EH, Merkle H, Everson LI, Yee D, et al. Eliminating spurious lipid sidebands in 1H MRS of breast lesions. Magn Reson Med. 2002;48:215–222. doi: 10.1002/mrm.10224. [DOI] [PubMed] [Google Scholar]
- 30.Kinoshita Y, Yokota A. Absolute concentrations of metabolites in human brain tumors using in vitro proton magnetic resonance spectroscopy. NMR Biomed. 1997;10:2–12. doi: 10.1002/(sici)1099-1492(199701)10:1<2::aid-nbm442>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 31.Kofman O, Belmaker RH. Ziskind-Somerfeld Research Award 1993. Biochemical, behavioral, and clinical studies of the role of inositol in lithium treatment and depression. Biol Psychiatry. 1993;34:839–852. doi: 10.1016/0006-3223(93)90052-f. [DOI] [PubMed] [Google Scholar]
- 32.Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, et al. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–232. doi: 10.1002/mrm.10367. [DOI] [PubMed] [Google Scholar]
- 33.Castillo M, Smith JK, Kwock L. Correlation of myo-inositol levels and grading of cerebral astrocytomas. AJNR Am J Neuroradiol. 2000;21:1645–1649. [PMC free article] [PubMed] [Google Scholar]
- 34.Rijpkema M, Schuuring J, van der Meulen Y, van der Graaf M, Bernsen H, Boerman R, et al. Characterization of oligodendrogliomas using short echo time 1H MR spectroscopic imaging. NMR Biomed. 2003;16:12–18. doi: 10.1002/nbm.807. [DOI] [PubMed] [Google Scholar]
- 35.Takano T, Lin JH, Arcuino G, Gao Q, Yang J, Nedergaard M. Glutamate release promotes growth of malignant gliomas. Nat Med. 2001;7:1010–1015. doi: 10.1038/nm0901-1010. [DOI] [PubMed] [Google Scholar]
- 36.Ye ZC, Ransom BR, Sontheimer H. (1R,3S)-1-Aminocyclopentane-1,3-dicarboxylic acid (RS-ACPD) reduces intracellular glutamate levels in astrocytes. J Neurochem. 2001;79:756–766. doi: 10.1046/j.1471-4159.2001.00581.x. [DOI] [PubMed] [Google Scholar]
- 37.Piani D, Frei K, Do KQ, Cuenod M, Fontana A. Murine brain macrophages induced NMDA receptor mediated neurotoxicity in vitro by secreting glutamate. Neurosci Lett. 1991;133:159–162. doi: 10.1016/0304-3940(91)90559-c. [DOI] [PubMed] [Google Scholar]
- 38.Mates JM, Perez-Gomez C, Nunez de Castro I, Asenjo M, Marquez J. Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int J Biochem Cell Biol. 2002;34:439–458. doi: 10.1016/s1357-2725(01)00143-1. [DOI] [PubMed] [Google Scholar]
- 39.Li Y, Chen AP, Crane JC, Chang SM, Vigneron DB, Nelson SJ. Three-dimensional J-resolved H-1 magnetic resonance spectroscopic imaging of volunteers and patients with brain tumors at 3T. Magn Reson Med. 2007;58:886–892. doi: 10.1002/mrm.21415. [DOI] [PubMed] [Google Scholar]