Abstract
Spinocerebellar ataxia type 1 (SCA1) is a neurodegenerative disease caused by the expansion of a CAG repeat encoding a polyglutamine tract in Ataxin-1 (ATXN1). Both WT and mutant ATXN1 interact with 14-3-3 proteins, and 14-3-3 overexpression stabilizes ATXN1 levels in cells and increases ATXN1 toxicity in flies. To determine whether reducing 14-3-3 levels might mitigate SCA1 pathogenesis, we bred Sca1154Q/+ mice to mice lacking one allele of 14-3-3ε. 14-3-3ε haploinsufficiency rescued cerebellar pathology and motor phenotypes but, surprisingly, not weight loss, respiratory dysfunction, or premature lethality. Biochemical studies revealed that reducing 14-3-3ε levels exerted different effects in two brain regions especially vulnerable in SCA1: Although diminishing levels of both WT and mutant ATXN1 in the cerebellum, 14-3-3ε haploinsufficiency did not alter ATXN1 levels in the brainstem. Furthermore, 14-3-3ε haploinsufficiency decreased the incorporation of expanded ATXN1 into its large toxic complexes in the cerebellum but not in the brainstem, and the distribution of ATXN1’s small and large native complexes differed significantly between the two regions. These data suggest that distinct pathogenic mechanisms operate in different vulnerable brain regions, adding another level of complexity to SCA1 pathogenesis.
Keywords: Ywhae, Purkinje, Capicua, RBM17, ataxin1-like
Neurodegenerative “proteinopathies,” such as Parkinson, Alzheimer's, and Huntington disease, often involve disruptions of protein homeostasis resulting from altered protein interactions, functions, or degradation (1–3). This may be most readily appreciated in the polyglutamine diseases, a group that includes nine different conditions caused by an expansion of a translated CAG tract (polyQ) in distinct genes (4). Early investigations sought to understand the toxicity mediated specifically by the polyQ peptides (5), and they do indeed exert widespread neuronal toxicity when expressed on their own. Research during the past decade, however, has demonstrated that other domains in the polyQ-containing proteins are necessary for the disease-specific patterns of neurodegeneration. For example, serine 13 and 16 are important for huntingtin pathogenesis (6), and mutations in caspase cleavage sites block nuclear localization and toxicity of the polyQ-expanded huntingtin and androgen receptor (7, 8). In the case of expanded Ataxin-1 (ATXN1), either mutation of the nuclear localization signal (9), deletion of the AXH domain (10, 11), or a serine-to-alanine substitution at residue 776 prevents the appearance of spinocerebellar ataxia type 1 (SCA1)-like phenotypes in mice (12).
SCA1 usually causes onset of slowly progressive gait ataxia in midlife to late life, eventually impairing overall motor coordination and producing dysarthria, hypometric saccades, weight loss, respiratory dysfunction, and premature death (13, 14). Cerebellar Purkinje cells are the first to be affected, which accounts for the presenting ataxia, but the pons and brainstem are also prominently involved. Sca1 knock-in mice, which bear a CAG expansion of 154Q at the endogenous locus (Sca1154Q/+) (15), recapitulate the features of human SCA1, from ataxia and motor dysfunction to weight loss, progressive Purkinje cell degeneration, and premature death (15). Using these mice, we have shown that the pathogenicity of mutant ATXN1 derives not from interactions with novel proteins but from alterations in its interactions with various native protein partners (16–19). In particular, the toxicity of mutant ATXN1 derives from its incorporation into its large (1.5–3 MDa) native complexes: the S776A substitution, which does not exert toxicity, prevents mutant ATXN1 from incorporating into its large complexes (12, 19), and displacing mutant ATXN1 from its large native complexes by overexpressing its paralogue, ATXN1-like, suppresses the phenotypes of the Sca1154Q/+ mice (17). These findings highlight the importance of ATXN1 large native complexes in SCA1 pathophysiology in vivo.
One group of protein interactors is of particular interest for pathogenesis studies: those whose interaction with ATXN1 requires phosphorylation at S776 and is enhanced by the expansion of the polyglutamine tract (18). The 14-3-3 proteins, among the earliest identified ATXN1 interactors, fall into this category. Widely expressed in eukaryotic cells (20), these proteins are highly conserved from yeast to mammals (21); they bind to phosphopeptide motifs in a variety of cellular proteins and can protect their target proteins from proteolysis and dephosphorylation (22). Consistent with this function, we previously found that overexpression of 14-3-3ε stabilizes ATXN1 in cells and increases the toxicity of ATXN1[82Q] in the fly model (18). We therefore hypothesized that removing one copy of 14-3-3 might mitigate the disease phenotype. Selecting a particular isoform of 14-3-3 for genetic interaction studies was not exactly straightforward: Several of the seven 14-3-3 isoforms found in mammals (23), including ε, ζ, η, β, and γ, interact with ATXN1 (18). The interaction of the ε and ζ isoforms with ATXN1 was identified primarily by immunoprecipitation of ATXN1[82Q], whereas the β and ε isoforms were the most frequently identified partners in a yeast two-hybrid screen for ATXN1 interactors (18). Sequence similarity among 14-3-3 isoforms suggests that some of them are probably functionally redundant (21); thus, the effects of deleting one copy of one isoform might be masked by the presence of other isoforms. The fact that 14-3-3ε−/− mice die at birth, however, indicates that this isoform has distinct nonredundant functions (24). Indeed, 14-3-3ε is the most highly conserved member of the 14-3-3 family (25): Mammalian 14-3-3ε is more similar in sequence to the plant and yeast homologs than it is to other mammalian isoforms (26). We therefore chose 14-3-3ε (also known as Ywhae) to breed with the Sca1154Q/+ mice.
To our surprise, we found that 14-3-3ε haploinsufficiency strongly rescued motor phenotypes but not other aspects of SCA1 that appear unrelated to the cerebellum. This finding led us to investigate the biochemical correlation of such rescue to reveal region-specific differences in ATXN1 complexes. The findings from this study demonstrate that different pathogenic processes can take place in different vulnerable brain regions, highlighting the complexity of mechanisms underlying neurodegeneration in a single disorder.
Results
14-3-3ε Stabilizes ATXN1 Protein in Mouse Cerebellum.
First, we confirmed the in vivo interaction between ATXN1 and 14-3-3ε by immunoprecipitation using WT cerebellar protein extract; ATXN1 was pulled down by 14-3-3ε–specific antibody but not by the IgG control (Fig. S1A). Furthermore, without any alteration in ATXN1 mRNA levels (Fig. S1B), ATXN1 protein level was clearly reduced in crude cerebellar extracts from 14-3-3ε+/− mice (Fig. S1 C and D). We conclude that 14-3-3ε stabilizes ATXN1 protein in vivo in mouse cerebellum.
Given the importance of ATXN1 large protein complexes in SCA1 pathogenesis, we sought to determine whether 14-3-3ε interacts with WT ATXN1 in its large native complexes. We analyzed cerebellar extracts from WT mice using size exclusion chromatography and identified a gradient of 14-3-3ε–containing complexes, ranging from large- to lower molecular-weight proteins, which likely represent dimeric (60 kDa) (27) and monomeric (30 kDa) forms of 14-3-3ε (Fig. S2A). This was not unexpected, because 14-3-3 proteins have diverse functions and bind a large number of partners (28).
We then performed immunoprecipitation on various fractions from HEK293T cells that overexpress FLAG-tagged ATXN1[82Q], which revealed two elution peaks enriched for ATXN1 in fractions similar to those found in WT mouse cerebellum (16). We immunoblotted for the endogenous 14-3-3ε and found that 14-3-3ε is indeed present in the large ATXN1 complexes (Fig. S2B).
Diminishing 14-3-3ε Rescues the SCA1 Motor Phenotype and Cerebellar Neuropathology.
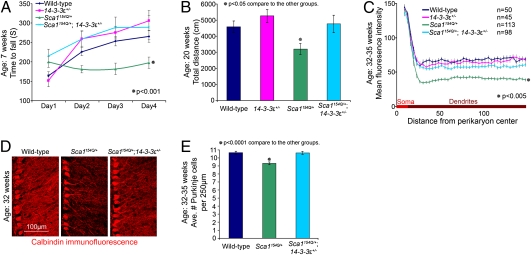
To determine whether 14-3-3ε gene dosage affects the SCA1 phenotype, we crossed 14-3-3ε+/− animals with the Sca1154Q/+ mice and assessed their motor coordination on the accelerating rotarod at 7 wk of age. The Sca1154Q/+; 14-3-3ε+/+ animals (from here on referred to as Sca1154Q/+ mice), as expected, showed significantly worse performance on the rotarod than their WT littermates (P < 0.001) (Fig. 1A). Removing one copy of 14-3-3ε in Sca1154Q/+ mice, however, rescued the rotarod phenotype to WT and 14-3-3ε+/− levels (Fig. 1A).
Fig. 1.
Haploinsufficiency of 14-3-3ε rescues SCA1 cerebellar phenotype. (A) Accelerating rotarod; the average of four trials per day per animal (±SEM) (n > 19 animals per group; ANOVA); 14-3-3ε haploinsufficiency rescues incoordination of Sca1154Q/+ mice (P < 0.001) to WT and 14-3-3ε+/− littermates (P = 0.27 and P = 0.47, respectively). (B) Open-field test; the average total distance traveled by each animal in 30 min (±SEM) (n > 19 animals per group; ANOVA); 14-3-3ε heterozygosity restores activity level of Sca1154Q/+ mice to level of WT or 14-3-3ε+/− littermates (P = 0.78 and P = 0.46 respectively). (C) Quantitative calbindin immunofluorescence of cerebellar Purkinje cell dendrites. Mean fluorescence intensity of the indicated number of optical rectangular subsections from the crusI/II folia of 32- to 35-wk-old animals (n = 3) was plotted as the distance from the perikaryon center (±SEM) (ANOVA); 14-3-3ε heterozygosity rescues the Sca1154Q/+ loss of dendritic arborization phenotype (P < 0.005); there were no significant differences between double mutants and their WT or 14-3-3ε+/− littermates (P = 0.18 and P = 0.47, respectively). (D) Representative confocal images showing Purkinje cell morphology from the crusI/II folia of littermate-matched 32-wk-old animals. (E) Quantification of Purkinje cell rescue, graphed as the average number of soma per 250-μm length (±SEM) along the Purkinje cell layer in individual confocal optical sections (P < 0.0001). (WT, 807 neurons along 19,000 μm in n = 18 optical sections; Sca1154Q/+, 867 neurons along 23,250 μm in n = 23 optical sections; Sca1154Q/+; 14-3-3ε+/−, 858 neurons along 20,250 μm in n = 20 optical sections).
To assess general motor function in older animals (which should have more severe motor degeneration), we used the open-field activity test on 20-wk-old mice, measuring the total distance traveled by animals to evaluate their level of general activity. ANOVA showed that the Sca1154Q/+; 14-3-3ε+/− animals were more active than Sca1154Q/+ mice (P < 0.05) and, again, not significantly different from either their WT or 14-3-3ε+/− littermates (Fig. 1B).
Would 14-3-3ε haploinsufficiency noticeably affect the cerebellar neuropathology of Sca1154Q/+ mice? Although Purkinje cell loss is prominent in human patients, in adult Sca1154Q/+ mice, only about 10% of Purkinje cells are lost; dendritic arborization is the most notable pathological sign as Sca1154Q/+ mice get older (15, 17). At 32–35 wk of age, 14-3-3ε haploinsufficiency dramatically rescued the Purkinje cell dendritic phenotype (P < 0.005) (Fig. 1C). The complexity of fine dendritic arbors and Purkinje cell soma in the Sca1154Q/+; 14-3-3ε+/− mice, as examined by immunofluorescent confocal imaging, resembled that of WT littermates (Fig. 1D), and there was no Purkinje cell loss normally observed in adult Sca1154Q/+ animals (P < 0.0001) (Fig. 1 D and E). Reducing 14-3-3ε dosage thus completely rescued the ataxic phenotype and cerebellar pathology caused by polyglutamine-expanded ATXN1.
14-3-3ε Hapolinsufficiency Reduces Incorporation of Mutant ATXN1 into Its Large Native Complexes in the Cerebellum.
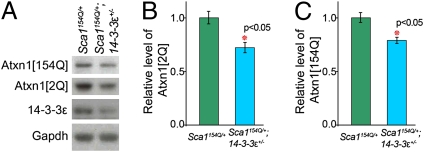
The pathogenicity of mutant ATXN1 correlates directly with its level: Sca1154Q/154Q and homozygote transgenic (B05) mice show significantly more severe phenotypes than their heterozygote littermates (15, 29). Our in vivo data strongly suggest that the stability of the WT ATXN1 depends, at least in part, on the presence of 14-3-3ε at its physiological levels (Fig. S1C). Therefore, 14-3-3ε haploinsufficiency might rescue the SCA1 cerebellar phenotypes by diminishing the levels of mutant ATXN1. Western blot analysis on crude cerebellar extracts showed that the Sca1154Q/+; 14-3-3ε+/− mice had significantly lower levels of both expanded ATXN1 and WT ATXN1 compared with their Sca1154Q/+ littermates (20% and 30%, respectively; P < 0.05) (Fig. 2 A–C).
Fig. 2.
Haploinsufficiency of 14-3-3ε decreases the levels of both WT and expanded ATXN1 in Sca1154Q/+ mice. (A–C) Western blot analysis of mouse cerebellar extracts (genotypes indicated) using antisera to 14-3-3ε, ATXN1 (2Q and 154Q), and Gadph (control). The average level (±95% confidence interval) of ATXN1[2Q] and ATXN1[154Q] proteins in Sca1154Q/+; 14-3-3ε+/− extracts relative to levels in Sca1154Q/+ extracts is illustrated in B and C, respectively (n = 6).
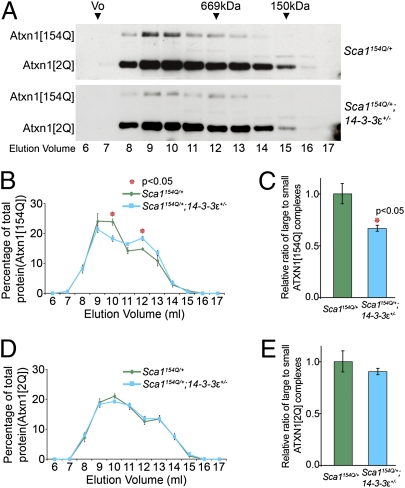
Given the importance of the ATXN1 large complexes in SCA1 pathogenesis, we next compared the formation of small and large ATXN1 complexes in Sca1154Q/+; 14-3-3ε+/− mice and their Sca1154Q/+ littermates by analyzing the elution profiles of ATXN1[2Q] and ATXN1[154Q] in size exclusion chromatography fractions of mouse cerebellar protein extracts (Fig. 3A). Quantification of the ATXN1 elution profiles showed that the ratio of large to small ATXN1[154Q] (but not ATXN1[2Q]) protein complexes in the Sca1154Q/+; 14-3-3ε+/− extracts was significantly lower than in the Sca1154Q/+ extracts (P = 0.011) (Fig. 3 B–E). These data suggest that in addition to reducing ATXN1 levels, heterozygosity for 14-3-3ε decreases the incorporation of glutamine-expanded ATXN1 into the large complexes, some of which are known to be toxic in the cerebellum.
Fig. 3.
Haploinsufficiency of 14-3-3ε shifts mutant ATXN1 from its large to small complexes. (A) Representative Western blots of size exclusion chromatography fractions from mouse cerebellar extracts analyzed for ATXN1[2Q] and ATXN1[154Q]. The column void volume (Vo) and elution volume (mL) of each collected fraction are indicated. (B) ATXN1[154Q] gel filtration elution profiles of cerebellar extracts were plotted as the percentage of ATXN1[154Q] (mean ± SEM) in each fraction (amount per fraction compared with total amount of ATXN1[154Q] in all fractions) from independent extracts. (C) Large (in the peak elution fractions, 9 and 10 mL) to small (in the peak elution fractions, 12 and 13 mL) ATXN1[154Q] complexes (mean ± SEM). (D) ATXN1[2Q] gel filtration elution profiles of cerebellar extract were plotted (n = 4). (E) Large to small ATXN1[2Q] complexes (mean ± SEM) (n = 4).
Heterozygosity for the 14-3-3ε Null Allele Does Not Rescue All SCA1 Phenotypes.
Despite the dramatic rescue of the motor phenotypes and cerebellar pathology, 14-3-3ε haploinsufficiency did not mitigate premature death or weight loss, phenotypes that are less likely to derive from cerebellar dysfunction (Fig. S3 A and B). In fact, the precise cause of death in Sca1154Q/+ mice has never been established. Close observation of several Sca1154Q/+ mice showed that they are able to eat even a few hours before their death; postmortem studies of several Sca1154Q/+ mice found food in the stomach and feces in the colon; thus, the animals do not starve to death. Aside from overall brain atrophy, however, we were unable to detect specific neuropathology in any brain region other than the cerebellum (15).
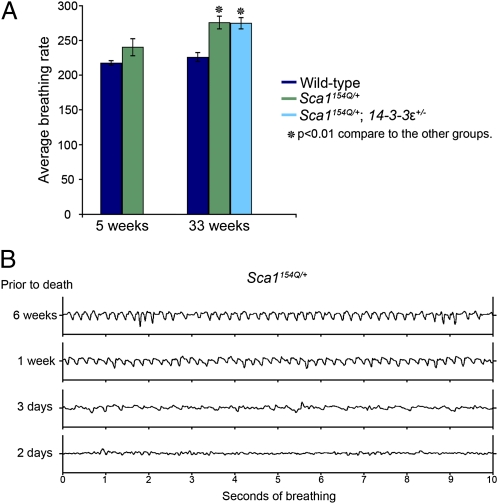
Patients with SCA1 have subclinical pulmonary dysfunction that worsens over time and likely contributes to their death. Using multiple spirometric tests, Sriranjini et al. (14) found evidence of restrictive lung dysfunction, upper airway obstruction, and reduced muscle strength that could arise from a combination of factors, including pulmonary dormancy, poor respiratory muscle coordination, and bulbar dysfunction. We therefore wondered if Sca1154Q/+ mice, which replicate so many other aspects of the human disease, might also experience respiratory dysregulation. Using unrestrained whole-body plethysmography, we assessed the basic respiratory pattern in Sca1154Q/+ mice. At 5 wk of age, the Sca1154Q/+ mice did not differ significantly from their WT littermates (P = 0.16); however, by 33 wk of age, both Sca1154Q/+ and Sca1154Q/+; 14-3-3ε+/− mice showed significantly more shallow and rapid respiration than their WT littermates (P < 0.01) (Fig. 4A). The respiratory phenotype of these mice, which has not been reported before, worsens with time, becoming most severe just before death; Fig. 4B shows a representative pattern from one Sca1154Q/+ mouse 6 wk, 1 wk, 3 d, and 2 d before death. Suffice it to say, 14-3-3ε haploinsufficiency does not appear to rescue phenotypes arising from parts of the nervous system outside of the cerebellum. To ensure that the lack of rescue is not attributable to region-specific changes in CAG repeat length, we assayed the size of the mutant expanded allele in the brainstem, cerebellum, and cortex of Sca1154Q/+ and Sca1154Q/+; 14-3-3ε+/− mice. We found that the expanded allele was stable and similar in all these tissues (Fig. S4). The selective rescue of the SCA1 phenotypes in our genetic interaction raised the possibility of a distinct mechanism limited to the cerebellum.
Fig. 4.
Sca1154Q/+ mice develop respiratory abnormalities. (A) Average (±SEM) rate of respiration in Sca1154Q/+ mice and their WT littermates at 5 and 33 wk of age. At 5 wk, there was no significant difference between the Sca1154Q/+ mice and their WT littermates (n = 4). At 33 wk, both Sca1154Q/+ (n = 5) and Sca1154Q/+; 14-3-3ε+/− (n = 5) mice showed more rapid respiration than their WT littermates (P < 0.01). There was no significant difference between WT animals at the age of 33 and 5 wk, but 33-wk-old Sca1154Q/+ mice had significantly faster respiration than 5-wk-old Sca1154Q/+ mice. Genotypes are indicated by color. (B) Representative respiration at 6 wk, 1 wk, 3 d, and 2 d before death for a Sca1154Q/+ mouse. We recorded this pattern in four mice (two Sca1154Q/+ and two Sca1154Q/+; 14-3-3ε+/− mice). At the time of death, the animals were between 37 and 39 wk old.
ATXN1 Complex Formation Differs Between the Cerebellum and the Brainstem.
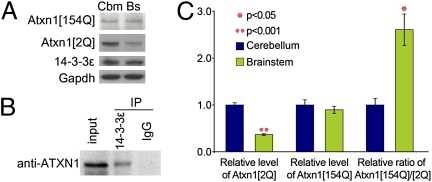
To investigate whether the biochemical changes that we discovered are as specific to the cerebellum as the phenotypic rescue, we examined the brainstem. Not only is this brain region severely affected in SCA1, but bulbar dysfunction may well be involved in the nonrescued respiratory phenotype described above. To continue our biochemical studies on the brainstem and compare them with those on the cerebellum, we had to answer two important questions: (i) whether 14-3-3ε is expressed in the brainstem and (ii) if it is expressed, whether 14-3-3ε and ATXN1 interact in the brainstem. Immunoblot analysis of cerebellar and brainstem lysates showed that the level of 14-3-3ε is the same in both regions (Fig. 5A). This is consistent with previous reports that 14-3-3ε is ubiquitous throughout the brain (30, 31). We then confirmed that ATXN1 coimmunoprecipitated with 14-3-3ε in the brainstem (Fig. 5B). These results show that the reason for not rescuing any phenotype potentially arising from the brainstem is neither lack of 14-3-3ε expression nor lack of its interaction with ATXN1 in the brainstem.
Fig. 5.
Level of WT ATXN1 (but not of 14-3-3ε) and expanded ATXN1 is less in the brainstem. (A) Western blot analysis of Sca1154Q/+ mouse cerebellar and brainstem extracts, using antisera to 14-3-3ε, ATXN1 (2Q and 154Q), and Gadph (control). (B) Coimmunoprecipitation of ATXN1 with 14-3-3ε from WT mouse brainstem extract. (C) Average levels of ATXN1[2Q] and ATXN1[154Q] proteins in Sca1154Q/+; 14-3-3ε+/− relative to levels in Sca1154Q/+ extracts and relative level of ATXN[154Q] to ATXN1[2Q] in the cerebellum and brainstem are illustrated (±95% confidence interval) (n = 6).
A closer look at the protein levels in the extracts from the cerebellum and brainstem of Sca1154Q/+ mice revealed that the level of WT ATXN1 in the brainstem of the Sca1154Q/+ mice was noticeably lower than in the cerebellum but that the level of expanded ATXN1[154Q] was the same in both regions (Fig. 5A). The ratio of mutant to WT ATXN1 in the brainstem was thus two- to threefold greater than in the cerebellum (Fig. 5C), which would likely make 14-3-3 heterozygosity less effective in mitigating brainstem degeneration.
We next quantified ATXN1 levels in crude brainstem extracts from the Sca1154Q/+; 14-3-3ε+/− mice and their Sca1154Q/+ littermates. Despite a decreased level of 14-3-3ε in Sca1154Q/+; 14-3-3ε+/− mice, both WT and mutant ATXN1 levels remained unaltered (Fig. S5 A–C).
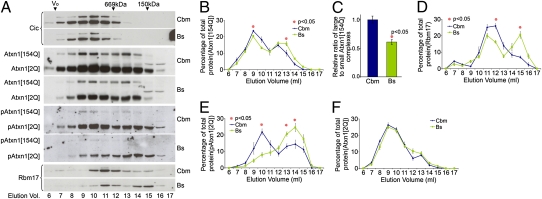
Why does 14-3-3ε haploinsufficiency fail to reduce ATXN1 levels in the brainstem of the Sca1154Q/+; 14-3-3ε+/− mice? The answer cannot involve regional differences in levels of either 14-3-3ε or expanded ATXN1 (they are the same in both the cerebellum and brainstem; Fig. 5A) or any lack of ATXN1/14-3-3ε interaction in the brainstem (Fig. 5B). We concluded that the mechanism by which 14-3-3ε stabilizes ATXN1 must take place in the cerebellum but not in the brainstem. We analyzed the size exclusion chromatography fractions of brainstem protein extracts from Sca1154Q/+; 14-3-3ε+/− mice and their Sca1154Q/+ littermates. Unlike the cerebellum, the elution profiles of ATXN1[2Q] and ATXN1[154Q] in the brainstem of these two genotypes showed very similar patterns (Fig. S5 D–F): thus, 14-3-3ε haploinsufficiency affected the elution profile of mutant ATXN1 in the cerebellum differently from that in the brainstem, suggesting that ATXN1 actually forms different native complexes in these two brain regions. To test this hypothesis, we analyzed the elution profiles of ATXN1, Capicua (a prominent native partner of ATXN1), and RBM17 (a protein that interacts in an S776-dependent manner) (16) in size exclusion chromatography fractions of cerebellar and brainstem extracts from WT mice (Fig. S6A). The elution profiles of total ATXN1, phospho-ATXN1 (pATXN1), and Capicua were the same (Fig. S6 A–C). The distribution of RBM17 in the brainstem, however, differed from that in the cerebellum (Fig. S6 A and D).
In our Sca1154Q/+mouse model, the elution profiles of Capicua and ATXN1[2Q] in the cerebellum and brainstem were similar to those of the WT mice (Fig. 6A), but the ratio of large to small ATXN1[154Q] protein complexes was much lower (P < 0.05) in the brainstem than in the cerebellum (Fig. 6 A–C). This suggests that expanded ATXN1 might have different partners that contribute to different toxic complexes in the two brain regions. In agreement with this notion, the elution profile of RBM17 in Sca1154Q/+ mice differed from the cerebellum to the brainstem (Fig. 6 A and D).
Fig. 6.
Brainstem and cerebellum of Sca1154Q/+ mice show different distributions of ATXN1[154Q], pATXN1, and RBM17. (A) Representative Western blots of size exclusion chromatography fractions from Sca1154Q/+ cerebellar (Cbm) and brainstem (Bs) extracts analyzed for Capicua, ATXN1, pATXN1, and RBM17. (B) ATXN1[154Q] gel filtration elution profiles of Cbm and Bs extracts from Sca1154Q/+ mice were plotted (mean ± SEM) (n = 4). (C) Large to small ATXN1[154Q] complexes (mean ± SEM). Significantly less ATXN1[154Q] is incorporated into the large complexes in the Bs than in the Cbm. Gel filtration elution profiles of RBM17 (D), pATXN1[2Q] (E), and ATXN1[2Q] (F) from Cbm and Bs extracts from Sca1154Q/+ mice were plotted (mean ± SEM) (n = 4).
Phosphorylation of ATXN1 at Serine 776 is critical for SCA1 pathogenesis (12, 18). The distribution of both pATXN1[2Q] and pATXN1[154Q] in Sca1154Q/+cerebellum followed the pattern of total ATXN1, with higher levels in the large complexes than in the small ones (Fig. 6A). In Sca1154Q/+ brainstem, however, the majority of the endogenous WT pATXN1 was in small ATXN1 complexes and very little was present in the large ATXN1 complexes, even though they were enriched for total ATXN1[2Q] (Fig. 6 A, E, and F).
The very low levels of pATXN1[154Q] in the brainstem prevented us from evaluating its elution profile (Fig. 6A). Given that the WT brainstem and cerebellum showed no difference in the distribution of pATXN1 (Fig. S6 A and C), the lack of pATXN1 in the large complexes in the brainstem of Sca1154Q/+ mice must result from a specific effect of mutant ATXN1 in the brainstem. These data highlight major differences in the distribution pattern of mutant ATXN1 complexes in these two brain regions and suggest that pATXN1 does not form the same complexes in the brainstem as it does in the cerebellum.
Discussion
A growing body of literature is clarifying the mechanism of SCA1 pathogenesis in the cerebellum. The cerebellar symptoms in SCA1 are the first to appear and the easiest to measure, but the disease progresses to other serious symptoms that do not arise from the cerebellum. In the present study, we show that 14-3-3ε haploinsufficiency in Sca1154Q/+ mice rescues SCA1 cerebellar pathology and motor phenotypes. Our biochemical studies indicate that 14-3-3ε stabilizes ATXN1; because the level of mutant ATXN1 directly correlates with its pathogenicity (15, 29, 32), it seems likely that the reduction in 14-3-3ε alleviates cerebellar pathology, at least in part, by decreasing levels of mutant ATXN1.
We were surprised to find that 14-3-3ε haploinsufficiency did not rescue all aspects of the phenotype, however: The double-mutant mice still lost weight and died prematurely, just like their Sca1154Q/+ littermates. Toyo-oka et al. (24) showed that 14-3-3ε+/− mice have mildly disorganized hippocampus and cortical thinning. These abnormalities, however, did not interfere with functions we assayed for in this study. We found that 14-3-3ε+/− mice have normal motor coordination and Purkinje cell morphology as well as a normal weight, breathing pattern, and life span. Hence, the mild cortical and hippocampal developmental defects in the 14-3-3ε+/− mice are unlikely to be responsible for the nonrescued life span phenotype in Sca1154Q/+; 14-3-3ε+/− mice. In the course of trying to understand the cause of death, we uncovered a respiratory dysfunction that had not been reported previously in the Sca1 knock-in mice but is documented in human patients (14). While we were investigating this phenotype, four mice (two Sca1154Q/+ and two Sca1154Q/+; 14-3-3ε+/− mice) died within 2 to 4 d of our last recording. Whether respiratory dysfunction was the immediate cause of death is impossible to ascertain; for that matter, the precise cause of death in human patients has not been firmly established. Aspiration pneumonia is a common (and frequently fatal) complication of end-stage SCA1 (33, 34). This unique quantifiable phenotype in Sca1154Q/+ mice could be used as an outcome measure in future studies of therapeutical interventions.
Notwithstanding the uncertain origins of the nonrescued phenotypes, we decided that examining ATXN1 complex formation in the brainstem, a brain region known to be highly vulnerable in SCA1, might provide insight into the apparently cerebellum-specific rescue. We found that, as previously reported (30), 14-3-3ε is expressed equally in the brainstem and in the cerebellum and that ATXN1 and 14-3-3ε interact in both regions. The haploinsufficiency of 14-3-3ε, however, decreased ATXN1 levels only in the cerebellum; brainstem levels of both WT and mutant ATXN1 were unaltered. In exploring the biochemical differences between the cerebellum and the brainstem, we found that some proteins (e.g., RBM17) have completely different patterns in the two regions. In addition, the composition of the large ATXN1 complexes differs between the two regions: pATXN1, which is necessary for 14-3-3/ATXN1 interaction and for the toxicity of mutant ATXN1 in the large complexes in the cerebellum, is not present in the large ATXN1 complexes in the brainstem of Sca1154Q/+ mice.
We also observed that the brainstem of Sca1154Q/+ mice has lower levels of WT ATXN1 relative to the expanded protein. It is interesting to note in this context that previous work has shown WT ATXN1 to exert a protective effect. We have postulated that WT ATXN1 competes with mutant ATXN1 in binding some protein partners, and thus reduces the formation of mutant ATXN1 complexes (16). The lower levels of WT ATXN1 (and thus the greater availability of mutant ATXN1 for complex formation in the brainstem) could explain why genetic suppression by 14-3-3ε heterozygosity is ineffective in the brainstem.
Selective neurodegeneration in the context of ubiquitous expression of the disease protein is a common feature of many neurodegenerative diseases (35); it is a reasonable conjecture that differential vulnerability would be mediated by different protein interactions. What is striking in this study, however, is that we have examined two regions that are both targets of SCA1 and find evidence that pathogenesis in each region differs. In other words, the molecular pathogenesis of neuronal dysfunction within a single disorder can differ among affected brain regions. The finding that the distribution of the mutant protein in its native complexes can vary from one brain region to the next, and that such differences have region-specific effects, underscores the need for in-depth studies on molecular pathogenesis in the multiple cellular contexts typically affected by the disease, with a focus on native complexes. Our data also caution that potential therapeutical modalities should be examined in different brain regions and not generalized to the whole brain from one neuronal group.
Experimental Procedures
Animals and Behavioral Analysis.
The generation and genotyping of Sca1154Q/+ mice have been described (15). Both Sca1154Q/+ and 14-3-3ε+/− mice (24) have been backcrossed to C57BL/6J mice for more than 10 generations. We crossed male Sca1154Q/+ mice to female 14-3-3ε+/− mice to generate double-mutant animals and controls. Rotarod analysis was performed as previously described (36), using 7-wk-old naive animals. The open-field activity test was performed as previously described (36), using 20-wk-old animals.
Analysis of Cerebellar Pathology and Purkinje Cell Loss.
Immunofluorescence staining of cerebellar sections for calbindin (1:1,000 monoclonal antibody to calbindin; Sigma) and indirect quantitative analysis of Purkinje cell dendritic arborization were performed as previously described (15). Optical sections were collected with a Zeiss LSM 510 confocal microscope using ImageJ software (National Institutes of Health) (17).
Immunoprecipitation.
Protein (0.5 mg) from cerebellar or brainstem extracts in 100 μL of TST buffer [50 mM Tris (pH 8), 50 mM NaCl, 0.5% Triton 100X)] was diluted with 400 μL of cold PBS (Input). One microgram of anti–14-3-3ε antibody serum or rabbit nonspecific IgG was added and incubated for 4 h at 4 °C. Immunoprecipitation was performed with BSA-blocked protein G Sepharose beads, and the pellet (immunoprecipitation pellet) was resuspended in sample buffer following three to four washes with cold PBS. Input and immunoprecipitation pellets were analyzed by SDS/PAGE and Western blotting for ATXN1 with mouse monoclonal anti-ATXN1 antibody (Neuromab).
Column Fractionation, Antibodies, and Protein Blot Analysis.
The protocols for protein extraction and gel filtration chromatography have been described (17). The antibodies used for protein blotting were commercially available or are described elsewhere (17): rabbit polyclonal antibody to 14-3-3ε (Cell Signaling), guinea pig polyclonal antiserum to Capicua (CIC) (17), mouse monoclonal antibody to Flag M2 (Sigma), rabbit polyclonal antibody to ATXN1 (11750-VII) (17), mouse monoclonal anti-ATXN1 antibody, rabbit polyclonal antibody to pATXN1 (37), and mouse monoclonal antibody to GAPDH (Advanced ImmunoChemical). Anti-FLAG M2 affinity gel freezer-safe (A2220; Sigma) was used for immunoprecipitation after column fractionation. SDS/PAGE protein blotting and immunohistochemistry followed standard protocols (15, 18). Quantification of proteins in extracts has been described (17).
Breathing Measurements.
Mice were placed within unrestrained whole-body plethysmography chambers (Buxco), ≈500 mL in volume, with a continuous flow rate of 1 L/min flushing the chambers with fresh air. Breathing was recorded for 50 min, and only the minutes during which mice were calm were used for subsequent analysis. Breath waveforms were identified with Biosystem XA software (Buxco) and used to calculate instantaneous breathing frequency and other breathing parameters. The average of the instantaneous frequency was determined for each animal and compared across genotypes and age.
Statistical Analysis.
We performed t tests (two-tailed, not assuming equal variances) and calculated SEMs and 95% confidence intervals using Microsoft Excel. We analyzed rotarod performance and results of the open-field activity test and quantified Purkinje cell dendritic arborization by repeated-measures ANOVA.
Supplementary Material
Acknowledgments
We are grateful to Dr. Anthony Wynshaw-Boris for providing 14-3-3ε+/− mice; the assistance of the mouse phenotyping core facility at Baylor College of Medicine; Sukeshi Vaishnav for mouse genotyping; Dr. Hamed Jafar-Nejad, Dr. Herman Dierick, and the members of the H.Y.Z laboratories for comments on the manuscript; and V. L. Brandt for editorial input. This work was supported by the National Institutes of Health: Grant NS27699 (to H.Y.Z.), Grant NS022920 (to H.T.O.), and Grant HD024064 (to Baylor College of Medicine Intellectual and Developmental Disabilities Research Center).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018748108/-/DCSupplemental.
References
- 1.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell. 2002;111:209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 2.Yamin G, Ono K, Inayathullah M, Teplow DB. Amyloid beta-protein assembly as a therapeutic target of Alzheimer's disease. Curr Pharm Des. 2008;14:3231–3246. doi: 10.2174/138161208786404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giorgini F, Muchowski PJ. Connecting the dots in Huntington's disease with protein interaction networks. Genome Biol. 2005 doi: 10.1186/gb-2005-6-3-210. 10.1186/gb-2005-6-3-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: Mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 5.Nagai Y, Onodera O, Strittmatter WJ, Burke JR. Polyglutamine domain proteins with expanded repeats bind neurofilament, altering the neurofilament network. Ann NY Acad Sci. 1999;893:192–202. doi: 10.1111/j.1749-6632.1999.tb07826.x. [DOI] [PubMed] [Google Scholar]
- 6.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham RK, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 8.Ellerby LM, et al. Kennedy's disease: Caspase cleavage of the androgen receptor is a crucial event in cytotoxicity. J Neurochem. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- 9.Klement IA, et al. Ataxin-1 nuclear localization and aggregation: Role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 10.Tsuda H, et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 11.de Chiara C, Menon RP, Dal Piaz F, Calder L, Pastore A. Polyglutamine is not all: The functional role of the AXH domain in the ataxin-1 protein. J Mol Biol. 2005;354:883–893. doi: 10.1016/j.jmb.2005.09.083. [DOI] [PubMed] [Google Scholar]
- 12.Emamian ES, et al. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 13.Zoghbi HY, Orr HT. Spinocerebellar ataxia type 1. Semin Cell Biol. 1995;6:29–35. doi: 10.1016/1043-4682(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 14.Sriranjini SJ, Pal PK, Krishna N, Sathyaprabha TN. Subclinical pulmonary dysfunction in spinocerebellar ataxias 1, 2 and 3. Acta Neurol Scand. 2010;122:323–328. doi: 10.1111/j.1600-0404.2009.01306.x. [DOI] [PubMed] [Google Scholar]
- 15.Watase K, et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–919. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 16.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman AB, et al. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet. 2007;39:373–379. doi: 10.1038/ng1977. [DOI] [PubMed] [Google Scholar]
- 18.Chen HK, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 19.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi Y. The 14-3-3 proteins: Gene, gene expression, and function. Neurochem Res. 2003;28:1265–1273. doi: 10.1023/a:1024296932670. [DOI] [PubMed] [Google Scholar]
- 21.Aitken A, et al. 14-3-3 proteins: A highly conserved, widespread family of eukaryotic proteins. Trends Biochem Sci. 1992;17:498–501. doi: 10.1016/0968-0004(92)90339-b. [DOI] [PubMed] [Google Scholar]
- 22.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 23.Tzivion G, Avruch J. 14-3-3 proteins: Active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 24.Toyo-oka K, et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: A molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 25.Aitken A. Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol Biol. 2002;50:993–1010. doi: 10.1023/a:1021261931561. [DOI] [PubMed] [Google Scholar]
- 26.Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: Implications for function as adapter proteins. FEBS Lett. 1995;368:55–58. doi: 10.1016/0014-5793(95)00598-4. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhri M, Scarabel M, Aitken A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem Biophys Res Commun. 2003;300:679–685. doi: 10.1016/s0006-291x(02)02902-9. [DOI] [PubMed] [Google Scholar]
- 28.Bridges D, Moorhead GB. 14-3-3 proteins: A number of functions for a numbered protein. Sci STKE. 2005 doi: 10.1126/stke.2962005re10. 10.1126/stke.2962005re10. [DOI] [PubMed] [Google Scholar]
- 29.Burright EN, et al. SCA1 transgenic mice: A model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–948. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 30.Baxter HC, Liu WG, Forster JL, Aitken A, Fraser JR. Immunolocalisation of 14-3-3 isoforms in normal and scrapie-infected murine brain. Neuroscience. 2002;109:5–14. doi: 10.1016/s0306-4522(01)00492-4. [DOI] [PubMed] [Google Scholar]
- 31.Umahara T, et al. Intranuclear immunolocalization of 14-3-3 protein isoforms in brains with spinocerebellar ataxia type 1. Neurosci Lett. 2007;414:130–135. doi: 10.1016/j.neulet.2006.12.026. [DOI] [PubMed] [Google Scholar]
- 32.Lorenzetti D, et al. Repeat instability and motor incoordination in mice with a targeted expanded CAG repeat in the Sca1 locus. Hum Mol Genet. 2000;9:779–785. doi: 10.1093/hmg/9.5.779. [DOI] [PubMed] [Google Scholar]
- 33.Ramio-Torrentia L, Gomez E, Genis D. Swallowing in degenerative ataxias. J Neurol. 2006;253:875–881. doi: 10.1007/s00415-006-0122-2. [DOI] [PubMed] [Google Scholar]
- 34.Shiojiri T, et al. Vocal cord abductor paralysis in spinocerebellar ataxia type 1. J Neurol Neurosurg Psychiatry. 1999;67:695. doi: 10.1136/jnnp.67.5.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 36.Spencer CM, et al. Exaggerated behavioral phenotypes in Fmr1/Fxr2 double knockout mice reveal a functional genetic interaction between Fragile X-related proteins. Hum Mol Genet. 2006;15:1984–1994. doi: 10.1093/hmg/ddl121. [DOI] [PubMed] [Google Scholar]
- 37.Duvick L, et al. SCA1-like disease in mice expressing wild type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.