Abstract
Assembly of the spliceosome onto pre-mRNA is a dynamic process involving the ordered exchange of snRNPs to form the catalytically active spliceosome. These ordered rearrangements have recently been shown to occur cotranscriptionally, while the RNA polymerase is still actively engaged with the chromatin template. We previously demonstrated that the histone acetyltransferase Gcn5 is required for U2 snRNP association with the branchpoint. Here we provide evidence that histone acetylation and deacetylation facilitate proper cotranscriptional association of spliceosomal snRNPs. As with GCN5, mutation or deletion of Gcn5-targeted histone H3 residues leads to synthetic lethality when combined with deletion of the genes encoding the U2 snRNP components Lea1 or Msl1. Gcn5 associates throughout intron-containing genes and, in the absence of the histone deacetylases Hos3 and Hos2, enhanced histone H3 acetylation is observed throughout the body of genes. Deletion of histone deacetylaces also results in persistent association of the U2 snRNP and a severe defect in the association of downstream factors. These studies show that cotranscriptional spliceosome rearrangements are driven by dynamic changes in the acetylation state of histones and provide a model whereby yeast spliceosome assembly is tightly coupled to histone modification.
Keywords: pre-mRNA splicing, Saccharomyces cerevisiae
Removal of noncoding sequences from premessenger RNA is achieved by the activity of a dynamic ribonucleoprotein complex, the spliceosome. As the spliceosomal snRNPs sequentially recognize sequences in the pre-mRNA, the spliceosome undergoes ATP-dependent rearrangement of its RNA and protein components. Recently, it has been recognized that splicing can occur cotranscriptionally, while the RNA polymerase is still actively engaged with the chromatin template.
The monomeric units of chromatin, nucleosomes, are comprised of DNA wrapped around the core histones H3, H4, H2A, and H2B. Histones undergo extensive posttranslational modification on their N-terminal tails that affect compaction of DNA and binding of regulatory factors. Although it is known that splicing occurs cotranscriptionally, it is far less well understood how changes in this chromatin template affect the reaction. Analysis of tiling array data has suggested that nucleosomes and, according to several of these studies, specific histone modifications are enriched in exon sequences, suggesting that there may be specific histone “marks” that are associated with splicing signals (1–7). Additionally, proteins that bind to methylated histones (H3K4me3 and H3K36me3) have been shown to facilitate the recruitment of snRNPs to the nascent transcript and influence efficiency of splicing and alternative splicing (8, 9). The chromatin-bound mammalian SWI/SNF complex associates with components of the spliceosome and affects alternative splicing (10, 11). Although these studies suggest a role for histone modifications in mammalian alternative splicing, there has been little analysis of their roles in constitutive splicing, particularly in Saccharomyces cerevisiae, whose core machinery is shared with higher eukaryotes, and hence, is likely to yield insights into mechanisms that coordinate transcription and splicing.
We recently determined that the yeast histone acetyltransferase, Gcn5, is required for cotranscriptional recruitment of the U2 snRNP to the pre-mRNA. Deletion of Gcn5 or eliminating its catalytic activity, combined with deletion of genes encoding either of the U2 snRNP proteins Msl1 or Lea1, conferred synthetic-lethality to those cells. Deletion of other components of the SAGA complex responsible for targeting Gcn5’s activity to nucleosomes had the same effect. Although there was no evidence of Gcn5-dependent acetylation of U2 snRNP proteins, Gcn5-dependent acetylation in the 5′ region of several intron-containing genes was observed (12).
Histone acetylation and deacetylation are critical for proper gene expression, and hyperacetylation [by histone deacetylase (HDAC) deletion] and hypoacetylation (by removal of histone acetylation activity) can be equally deleterious to the cell (13). Proper cellular function depends on a delicate balance of these two reactions (14). Recent studies also demonstrate that both acetylation and deacetylation occur within the coding region of genes and that rapid removal of acetyl groups from histones within the body of genes is achieved by the activities of multiple HDACs (15, 16).
As the polymerase traverses this complex template to synthesize the RNA, the snRNPs associate with the pre-mRNA in a highly dynamic fashion. Work from several laboratories has demonstrated that the stepwise exchange of factors during cotranscriptional spliceosome assembly can be measured by ChIP (17–19). The 5′ splice site is recognized by the U1 snRNP, followed by U2 snRNP association with the branchpoint. ATP-dependent rearrangements in the spliceosome facilitate release of some U2 snRNP interactions and addition of the U4/U6·U5 triple snRNP. Subsequent rearrangements lead to the formation of the catalytic center and catalysis of the two transesterification steps.
Here we show that, like deletion of GCN5, mutation or deletion of histone H3 lysine residues that are targeted by Gcn5, K9 and K14, leads to synthetic growth defects when combined with LEA1 or MSL1 deletion. Gcn5 is observed throughout intron-containing genes, and although Gcn5-dependent acetylation is observed at the promoter, HDAC deletion reveals acetylation at H3K9 and K14 throughout the body of the gene, indicating that acetylation is masked by the rapid removal of acetyl groups by HDACs. When the HDACs are removed, and acetylation is enriched, the U2 snRNP interactions in the branchpoint region persist and are not efficiently exchanged for downstream snRNPs, suggesting that histone acetylation dynamics are coupled with spliceosome dynamics. These data lead to a model in which acetyl marks within the gene lead to recruitment and rearrangement of splicing factors during cotranscriptional spliceosome assembly.
Results
Mutation of Histone H3 Residues Targeted by Gcn5 Confers Synthetic Lethality with LEA1- and MSL1-Deleted Cells.
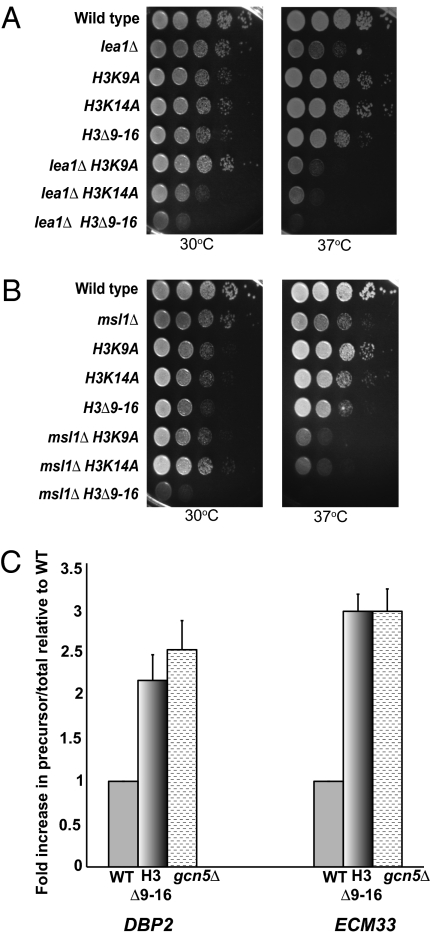
We have previously observed that deletion of either LEA1 or MSL1 in combination with catalytic mutants of GCN5 leads to synthetic lethality (12). To further elucidate whether acetylation of specific N-terminal residues is important for these functional interactions, we analyzed growth in strains deleted of LEA1 or MSL1 with mutations at key residues of histone H3 that are targets of Gcn5; H3K9 or K14 were mutated to A or removed using a short N-terminal deletion Δ9–16 (20). To confirm that amino acids 9 to 16 were deleted, Western blot analysis was performed, and acetylation of these residues was not detected (Fig. S1). At 30 °C, LEA1 deletion combined with H3K14A results in a synthetic growth defect, and the Δ9–16 truncation of histone H3 in combination with deletion of LEA1 results in a severe synthetic growth phenotype (Fig. 1A). At 37 °C, all three double mutants show a severe synthetic growth defect. Truncation of histone H3 in combination with deletion of MSL1 results in a striking growth defect at 30 °C (Fig. 1B), and similar to the lea1Δ double mutants, the msl1Δ double mutants are barely viable at 37 °C. It is unclear why the synthetic growth defect observed with lea1Δ K14A is not observed with the msl1Δ double mutants, but this may hint at slight differences in the cotranscriptional behaviors of the two factors (12). Because the growth defects observed with the double mutants of either LEA1 or MSL1 and a catalytic mutant of Gcn5 are more severe than the histone point mutants (12), we cannot rule out the possibility that acetylation of residues in addition to 9 and 14 contribute to Gcn5’s activity in cotranscriptional splicing. Nonetheless, these results provide evidence that the functional interactions between GCN5 catalytic activity and MSL1/LEA1 are because of Gcn5’s role in acetylating histone substrates.
Fig. 1.
Mutation of histone H3 residues combined with deletion of either MSL1 or LEA1 leads to severe synthetic growth defects. (A) Growth analysis of the double-mutant lea1Δ and histone H3 point mutants or truncation. Cells were grown at 30 °C in YPD medium until the desired OD600 was obtained. Cells were spotted onto YPD plates as a 10-fold serial dilution and grown at 30 °C or 37 °C for 2 d. (B) Dilution series of the double mutant msl1Δ and histone H3 point mutants or truncation. Cells were treated as described in A. (C) Quantitative RT-PCR of DBP2 and ECM33 in the histone H3 truncation mutant histone H3Δ9–16 or gcn5Δ vs. wild-type. Data are represented as a fold-increase in the ratio of precursor (unspliced)/total DBP2 or ECM33 message relative to wild-type. Graph represents three independent experiments and error bars represent SEM.
Because we detect a functional interaction between LEA1/MSL1 and mutations in histone H3, we predicted that splicing would be affected by mutation of specific H3 lysine residues. Therefore, we determined if truncation of the N-terminal tail of histone H3 affects the splicing of the intron-containing genes, DBP2 and ECM33, because deletion of GCN5 results in an increase in DBP2 and ECM33 pre-mRNA (12). Quantitative RT-PCR was performed to determine the ratio of unspliced pre-mRNA to total RNA. The Δ9–16 mutant reproducibly exhibits a 2- to 2.5-fold increase in intron accumulation compared with wild-type, which is comparable to the results observed upon deletion of GCN5 for both intron-containing genes (Fig. 1C). Although the intron accumulation observed for the histone H3 truncation mutant or for cells harboring the deletion of GCN5 is less than that observed upon deletion of core components of the spliceosome, which affects co- and posttranscriptional intron removal (Fig. S2), these data demonstrate that maximal production of these spliced messages is tied to proper histone modification.
Gcn5-Dependent Histone H3 Acetylation Occurs Within the Coding Region of Intron-Containing Genes.
Gcn5 is recruited to actively transcribed genes, leading to histone acetylation and TBP binding at the promoter (21, 22). More recently, it has been shown that Gcn5 can also be found within the body of actively transcribed genes and can affect histone acetylation downstream of the promoter (15, 16). To evaluate the role of Gcn5-dependent acetylation of intron-containing genes, we examined Gcn5 occupancy within DBP2 and observed Gcn5 both in the promoter region and throughout the coding region of DBP2 (Fig. S3 A and B), consistent with our observation of Gcn5-dependent acetylation of this intron-containing gene (12). In these studies, as with those described previously, DBP2 was extended with GFP sequence to facilitate ChIP resolution. We observe similar results with the DBP2 without the extension (12).
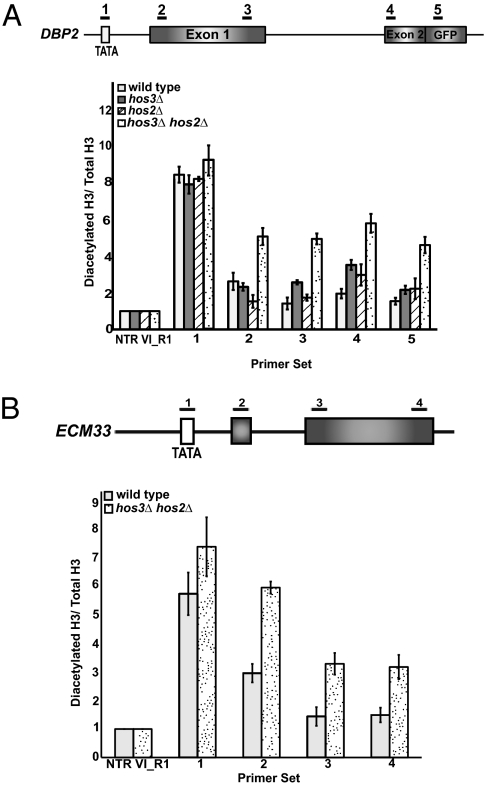
Although our previous studies demonstrated a peak in histone acetylation in the promoter region of the intron-containing genes that we analyzed, studies by others have shown that significant histone acetylation in the coding region can only be detected in the absence of HDACs. Therefore, we considered the possibility that some acetylation marks that are placed by Gcn5 within these genes are removed by HDACs, which could have important effects on cotranscriptional spliceosome assembly. To address this theory, we examined the histone H3 diacetylation profile of DBP2 and ECM33 in cells deleted of multiple HDACs. HOS3 HOS2 deletion has previously been shown to affect H3 acetylation in the coding region (15), as they target the same histone residues as Gcn5 (23, 24). As previously reported, a single deletion of either HDAC does not significantly affect histone H3 diacetylation (12). However, deletion of both HOS3 and HOS2 leads to an increase in H3 acetylation throughout both DBP2 and ECM33 (Fig. 2). For DBP2, H3 acetylation increases within the body of the gene about two- to threefold. However, the overall trend for ECM33 is different from DBP2 in that there is a gradual decrease in acetylation from the 5′ end to the 3′ end, indicating that there are gene-specific effects of HOS3 HOS2 double deletion in the body of intron-containing genes. Nonetheless, these data show that there is acetylation within the region of the gene that contains splicing signals and raise the possibility that the rapid deacetylation mediated by the HDACs affects cotranscriptional spliceosome assembly.
Fig. 2.
Gcn5-dependent histone acetylation in the coding region of DBP2 is masked by histone deacetylation. (A) ChIP analysis of histone H3 K9/14 acetylation of DBP2 in wild-type and histone deacetylase mutants using an antibody that recognizes acetylated histones (06-599; Millipore). Data are represented as diacetylated histone H3 normalized to the total amount of histone H3 (Total H3). (B) ChIP analysis of diacetylation of histone H3 in wild-type and the HOS3 HOS2 double deletion strain for ECM33 (exons are shaded gray). Light gray bars represent wild-type and speckled bars represent the HDAC double mutant. Data are represented as diacetylated H3 normalized to total histone H3. Graphs represent the average of at least three independent experiments, ±SD
Histone Deacetylase Activity Alters Cotranscriptional Recruitment of Lea1 and Msl1 and Alters Spliceosomal Rearrangements.
Previous studies demonstrate that ChIP allows detection of cotranscriptional spliceosome assembly; upon synthesis of the appropriate RNA signal, individual snRNPs appear to associate with nascent RNA in a stepwise manner (17–19, 25). When spliceosomal rearrangements are perturbed, there is an observable lag in the disengagement of snRNPs with pre-mRNA, and factors that are recruited downstream of the lag show diminished association with the pre-mRNA (18, 19, 25). Because deletion of HDACs increases histone acetylation in the coding region, we decided to examine cotranscriptional recruitment of Lea1 and Msl1 to DBP2 in the absence of HOS3 and HOS2. Although single deletion of either gene had little effect on cotranscriptional recruitment of Lea1 (Fig. S3C), in the hos3Δ hos2Δ cells there is an increase in the peak signal at primer set 4, and the typically rapid decrease in signal using primers downstream of primer set 4 is not observed (Fig. 3A). Instead, there is persistent U2 association typical of a defect in snRNP rearrangements. Total protein levels of Lea1 and Msl1 are unchanged (Fig. S4A). Similar results are observed for Msl1 (Fig. 3A and Fig. S3D). To rule out the possibility that the observed changes in U2 snRNP association and dissociation are gene-specific, we examined cotranscriptional recruitment of Lea1 and Msl1 to ECM33 in the HDAC deletion strain. Deletion of HOS3 and HOS2 leads to an increase in association of Lea1 and Msl1 to ECM33, as indicated by enrichment at primer set 3 (Fig. 3B), and as with DBP2, there is an increase in enrichment of both Lea1 and Msl1 at downstream primer sets, suggesting decreased dissociation of the U2 snRNP. Taken together, these results indicate that the activity of multiple HDACs working together to remove acetyl groups from histone H3 is critical for cotranscriptional spliceosomal rearrangements involving the U2 snRNP.
Fig. 3.
Deletion of HDACs Hos3 and Hos2 alters cotranscriptional recruitment of Msl1/Lea1. (A) Graphs depicting the occupancy of Lea1-HA and Msl1-HA at each region of DBP2 in the presence or absence of the HDACs, HOS3 and HOS2, relative to the nontranscribed control. Light gray bars depict the occupancy of Lea1 or Msl1 in the presence of HDACs. Dark gray bars represent Lea1-HA or Msl1-HA occupancy in the absence of HDACs. (B) Graph depicting the occupancy of Lea1-HA or Msl1-HA at each region of ECM33 in the presence and absence of HOS3 and HOS2 relative to the nontranscribed region. Data are represented as in A. Graphs represent the average of three independent experiments, ±SD. (C) Dilution series of the triple deletion mutants, lea1Δ hos3Δ hos2Δ and msl1Δ hos3Δ hos2Δ. Cells were grown at 30 °C in YPD liquid medium until the desired OD600 was obtained. Cells were spotted onto YPD plates as a 10-fold serial dilution, and plates were incubated for 2 d at 30 °C, 3 d at 25 °C, and 5 d at 16 °C.
We have shown that the functional relationship between Gcn5-dependent histone acetylation and Msl1/Lea1 can be revealed by analysis of synthetic genetic interactions. Because HDAC deletion appears to prevent U2 snRNP release, we predicted that conditions under which normally transient interactions are hyperstabilized, such as lowered temperature, would result in a synthetic growth phenotype in the triple mutants lea1Δ hos3Δ hos2Δ and msl1Δ hos3Δ hos2Δ. Although we previously observed no change in viability when the single HDAC deletions were combined with deletion of MSL1or LEA1 (12), strains deleted of either LEA1 or MSL1 and both HOS3 and HOS2 exhibited a severe synthetic growth defect when grown at 16 °C or 25 °C (Fig. 3C). Growth defects at low temperatures often reflect defects in interactions within multiprotein complexes, such as the ribosome (for which this phenotype was initially exploited) (26, 27). These results support a model in which HDACs and MSL1/LEA1 affect a common function in vivo.
Histone Deacetylation Is Necessary for Recruitment of snRNPs Downstream of U2.
Cotranscriptional assembly of snRNPs has been shown to occur in a coordinated, stepwise manner. Based on the defects in U2 snRNP release and the cold sensitivity observed in the triple mutants, we hypothesized that HDAC deletion would adversely affect steps downstream of U2 snRNP rearrangements. For example, deletion of the cap-binding complex, a complex involved in the exchange of factors during splicing, led to persistence of the U1 snRNP as measured by ChIP and prevented proper recruitment of the downstream U snRNPs (18). To address the effect of HDAC deletion on spliceosome assembly, we examined the cotranscriptional recruitment of the triple snRNP, represented by the U5 protein Snu114 (18), in the absence of the HDACs Hos3 and Hos2. Cotranscriptional Snu114 association is observed using primers downstream of the 3′ splice site, a result that is consistent with previous observations (12, 18). However, deletion of both HOS3 and HOS2 leads to a decrease in the recruitment of the U5 snRNP component to DBP2 pre-mRNA (Fig. 4A). Similar but more pronounced results were observed with ECM33 (Fig. 4B).
Fig. 4.
Deletion of the HDACs Hos3 and Hos2 affects spliceosome dynamics downstream of U2 snRNP recruitment. (A) Bar graph depicting the cotranscriptional recruitment of U5 snRNP (Sun114-HA) to DBP2 in the presence and absence of Hos3 and Hos2 (Left). Light gray bars represent the occupancy of Snu114-HA in a wild-type background and the dark gray bars represent Snu114-HA recruitment when HOS3 and HOS2 are deleted. Occupancy is measured as fold-accumulation over the nontranscribed control. Bar graph represents the recruitment of Prp19-HA to DBP2 in the presence and absence of Hos3 and Hos2 (Right). Light gray bars represent wild-type and dark gray bars represent the hos3Δ, hos2Δ double mutant strain. (B) Bar graph depicting the cotranscriptional recruitment of U5 snRNP (Snu114-HA) and Prp19-HA to ECM33 in the presence and absence of Hos3 and Hos2 (Left and Right, respectively). Data are represented as in A. Graphs represent the average of at least three independent experiments.
We next examined the effect of HOS3 and HOS2 double deletion on the recruitment of a factor downstream of U5 snRNP recruitment, Prp19. Prp19 is a component of the nineteen complex that associates after triple snRNP addition and is required for the stable association of the U5 and U6 snRNAs with the spliceosome (28). As with U5 snRNP recruitment, we detect a decrease in recruitment of Prp19 to DBP2 and ECM33 in the absence of both HOS3 and HOS2 (Fig. 4). Significant changes in the levels of expression of these proteins were not observed (Fig. S4B). These results suggest that, although deletion of multiple HDACs increases the association of the U2 snRNP at the branchpoint, this negatively impacts subsequent steps of splicing. Examination of the unspliced (precursor)/Total DBP2 and ECM33 by qRT-PCR in the HDAC double-deletion strain revealed an approximately twofold increase in unspliced DBP2 and ECM33 transcript relative to wild-type (Fig. S5). Hence, as with a gcn5Δ and mutation of histone H3 residues, altering the dynamics of cotranscriptional spliceosome assembly through HDAC deletion affects splicing. Although we cannot unequivocally rule out the possibility that HDACs affect acetylation of some component of the spliceosome, these data together lead to a model in which Gcn5-mediated histone acetylation is required for cotranscriptional recruitment of the U2 snRNP, and deacetylation is required for normal release of U2 snRNP and the subsequent association of snRNPs acting downstream of it.
To determine if RNA polymerase II transcription was altered by deletion of the HDACs, we examined Pol II association with DBP2 and ECM33 by ChIP in the hos3Δ hos2Δ strains. Deletion of HOS3 and HOS2 had a mild effect on Pol II occupancy of DBP2 and ECM33, with a slight increase in Pol II across the gene (Fig. S6). It is possible that HDAC deletion alters elongation properties of polymerase and influences cotranscriptional recruitment of snRNPs, although because the changes in Pol II upon deletion of these two HDACs are relatively small, it seems unlikely that this alone accounts for the strong and differential effects of HDAC deletion on U2 snRNP, triple snRNP, and Prp19 recruitment (Discussion). Nonetheless, these data show that the dynamics of histone acetylation influences snRNP rearrangements during spliceosome assembly.
Discussion
Splicing and transcription are highly dynamic processes that are spatially and temporally coordinated. Here, we provide evidence that histone acetylation and deacetylation affect cotranscriptional splicing by facilitating the dynamic rearrangements of the spliceosome. Not only does Gcn5 show strong functional interactions with specific U2 snRNP components (12), but similar interactions are observed when the Gcn5 targets, H3K9 and H3K14, are mutated or deleted. Removal of Gcn5-targeted residues results in accumulation of unspliced DBP2 and ECM33 pre-mRNA to levels similar to deletion of GCN5 (Fig. 1), suggesting that both Gcn5 and its target histone H3 residues are important for cotranscriptional splicing. Double deletion of the HDACs HOS3 and HOS2 leads to an increase in histone acetylation within intron-containing genes and hampers the ordered exchange of spliceosomal snRNPs and stepwise assembly of the spliceosome (Figs. 2–4). Furthermore, upon HDAC deletion, cells become increasingly dependent on a fully functional spliceosome for viability (Fig. 3), and splicing defects are observed. These data provide evidence in yeast that dynamic modification of histones contributes to the dynamics of spliceosome assembly.
Understanding How Dynamic Histone Acetylation Modulates Spliceosome Assembly.
In situ analyses of cotranscriptionally assembled spliceosomes illustrate that nascent RNPs are found along the chromatin axis, and although there is an extensive exchange of splicing complexes along the nascent transcript, they do not appear to be associated with the polymerase itself (29). Our data suggest that the chromatin may provide signals for exchange of factors that assemble on pre-mRNA. Notably, when U2 snRNP recruitment is eliminated (as when GCN5 is deleted), no recruitment of downstream factors is observed (12). Whereas when the level of U2 snRNP persists, as is the case when HDACs are deleted, recruitment of downstream factors decreases to an extent that correlates with the change in U2. This finding is consistent with there being a tight relationship between U2 association and the association of downstream factors. Although we cannot rule out the possibility that HDACs affect the recruitment of downstream factors independently of their effect on U2, these results support a model in which increased histone acetylation (because of the lack of HDACs) results in altered spliceosome dynamics.
One of the challenges to understanding the mechanisms underlying cotranscriptional spliceosome assembly has been the lack of techniques that directly measure snRNP interactions with nascent RNA, particularly in vivo. ChIP analysis, like that described here, has proven to be a powerful tool for assaying cotranscriptional association of splicing factors with active transcription complexes (12, 17–19). Nonetheless, although the presence of a ChIP signal is a strong indication of association of a particular snRNP with the nascent transcript, it is more difficult to infer meaning from a lack of signal. For example, the absence of a ChIP signal may reflect inaccessibility of the epitope recognized during the immunoprecipitation step rather than the absence of the protein altogether. Hence, we cannot rule out the possibility that the decrease in snRNP association that we observe when HDACs are deleted reflects changes in epitope accessibility because of alterations in spliceosome dynamics. Further analysis will be needed to distinguish between these possibilities. Nonetheless, our results indicate that preventing histone deacetylation significantly alters snRNP dynamics in a manner that is reflected by changes in their ChIP profiles.
Because of the observation that ordered association of splicing factors occurs only upon polymerase synthesis of the appropriate RNA signals, the distance traveled by the polymerase has been used as a proxy for time, suggesting that kinetics of spliceosome assembly can be inferred from ChIP results. For example, our data suggests that increased association of the U2 snRNP leads to a “delay” in the downstream steps. Although this is a model that we find provocative and continue to test further, ChIP is an indirect measure of RNA binding to pre-mRNA, and hence, some aspects of the crosslinked complex, such as epitope accessibility, could complicate the interpretation of results. Ongoing refinement of tools such as ChIP to study cotranscriptional splicing in vivo will continue to elucidate the effects of histone modification on spliceosome assembly kinetics.
A number of studies indicate that the elongation properties of the polymerase can influence splice site recognition in both yeast and mammals (reviewed in ref. 30), and recent reports have indicated that pausing of RNA polymerase at the 3′ splice site or terminal exon is required for cotranscriptional splicing (31, 32). One could imagine that a change in polymerase processivity and possibly a decrease in pausing could contribute to the effects we observe. Nonetheless, it is unlikely that the modest changes in polymerase occupancy with the HDAC deletion are sufficient to explain both the striking increases in U2 snRNP and decreases in downstream snRNPs. Furthermore, it is unlikely that the effects on splicing by Hos3 and Hos2 are exclusively a result of this pausing, as the slight changes in Pol II occupancy in hos3Δ hos2Δ are similar across the length of the genes in this study (Fig. S6).
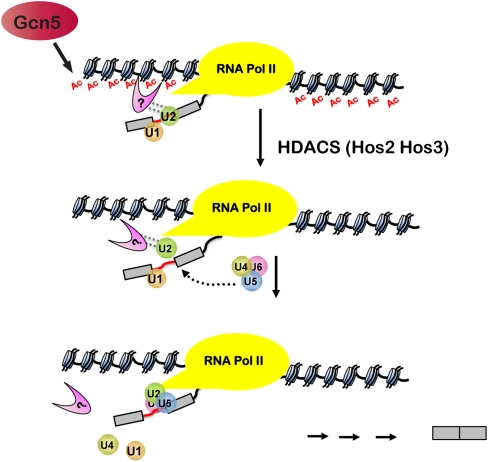
Another intriguing mechanism that would explain our results may be gleaned from studies of mammalian splicing. These results show that histone modifications, such as methylation, create binding sites for factors that facilitate spliceosome assembly and alternative splicing. H3K4me3 provides binding sites for a mammalian protein, Chd1, which associates with snRNPs and facilitates their recruitment (8). Furthermore, alternative splicing can be regulated by splicing factors that are recruited to introns by proteins that bind to H3K36me3 (9). Both of these studies indicate a precedent for “adaptor” proteins that bridge histone marks and the spliceosome. It is possible that acetyl marks create binding sites and that a protein (or proteins) binds to acetylated histone H3 tails to facilitate recruitment of the spliceosome to the nearby RNA. Because neither Msl1 nor Lea1 contains an acetyl-binding domain, it is likely that some other protein that interacts with both histones and the U2 snRNP proteins facilitate cotranscriptional spliceosome assembly. In this model, acetylation provides a binding site for an adaptor that mediates U2 snRNP association, but deacetylation allows for the proper release of the adaptor and the U2 snRNP and facilitates subsequent rearrangements (Fig. 5). This finding is consistent with our results showing that HDAC deletion results in persistent histone acetylation, stabilizes once transient interactions, delays U2 snRNP release, and inhibits the recruitment of downstream snRNPs (Fig. 5). Unlike histone methylation, which is a relatively stable mark, acetylation is sufficiently dynamic that it is easy to imagine that protein binding to acetyl marks could facilitate dynamic rearrangements of the spliceosome, as we observe. Because spliceosomal rearrangements are ATP-dependent, we predict that there are important interactions between the histone modifying machinery and the ATP-dependent RNPases that are central to the rearrangements described here. It will be interesting to identify and characterize these interactions.
Fig. 5.
Histone acetylation and deacetylation play a role in cotranscriptional splicing. Model depicting the role of Gcn5 and HDACs in spliceosomal rearrangements. Gcn5-dependent histone acetylation may create binding sites for a factor that interacts directly or indirectly with U2 snRNP proteins (indicated by dotted lines), to facilitate their cotranscriptional association. Hos3- and Hos2-mediated deacetylation allows for the proper release of the U2 snRNP and assembly of the spliceosome. A testable prediction is that the U2 snRNP may affect recruitment of histone deacetylases.
Materials and Methods
Strains/Viability Assay/Dilution Series.
For growth analysis, strains were grown overnight in YPD media at 30 °C. Cells were diluted to an OD600 of 0.1 in 10 mL of YPD, and incubated at 30 °C until all strains reached an OD600 of 0.5. A 10-fold serial dilution of each strain was spotted onto YPD plates, and incubated 2 d at 30 °C and 37 °C, 3 d at 25 °C, and 5 d at 16 °C. A list of strains used is in Table S1.
ChIP.
Overnight cultures were diluted and grown to an OD600 of 0.5 to 0.6. Cells were fixed and chromatin-isolated, immunoprecipitated, and analyzed by real-time PCR, as described previously (12). Immunoprecipitations were carried out with the following antibodies: Anti-HA 12CA5 (Roche) was used for HA-tagged proteins; Anti-Myc 9E10 (Roche) was used for Myc-tagged proteins; 8WG16 (Covance) was used for RNAPII; and anti-diacetylated H3 (Upstate) and anti-H3 (Abcam) were used to detect histones. Primer sequences are described in ref. 12. The results shown here are based on three or more independent experiments ±SD.
Quantitative PCR.
Total cellular RNA was extracted by hot phenol-chloroform extraction. Before cDNA synthesis, total RNA was treated with DNase I (Promega) according to the manufacturer's instructions. Complementary DNA was synthesized from 1 μg of DNase-treated RNA in a 20 μL reaction mixture containing 1× First Strand Buffer, 2 mM each dNTP, 10 mM DTT, 2 U RNasin (Promega), 1 μM gene-specific primer, and 200 U of SuperScript II Invitrogen). Analysis described in further detail is located in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Lorraine Pillus for reagents and technical advice. This work was supported in part by National Research Service Award Ruth L. Kirchstein Predoctoral Fellowship 5F31GM078745 (to F.Q.G.), National Science Foundation CAREER award MCB-0448010 (to T.L.J.), and National Institutes of Health Grant GM085474.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011982108/-/DCSupplemental.
References
- 1.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol. 2009;10(9):R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 6.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tilgner H, et al. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 8.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batsché E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13(1):22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 11.Tyagi A, Ryme J, Brodin D, Ostlund Farrants AK, Visa N. SWI/SNF associates with nascent pre-mRNPs and regulates alternative pre-mRNA processing. PLoS Genet. 2009;5:e1000470. doi: 10.1371/journal.pgen.1000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YY, et al. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelauer M, Wu J, Suka N, Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- 15.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25(1):31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol Cell Biol. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19(1):53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5′ss base pairing in yeast. Mol Cell. 2005;19(1):65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Dai J, et al. Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampsey M. A SAGA of histone acetylation and gene expression. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- 22.Timmers HT, Tora L. SAGA unveiled. Trends Biochem Sci. 2005;30(1):7–10. doi: 10.1016/j.tibs.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Carmen AA, et al. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc Natl Acad Sci USA. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- 25.Tardiff DF, Rosbash M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guthrie C, Nashimoto H, Nomura M. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc Natl Acad Sci USA. 1969;63:384–391. doi: 10.1073/pnas.63.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 28.Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- 29.Wetterberg I, Zhao J, Masich S, Wieslander L, Skoglund U. In situ transcription and splicing in the Balbiani ring 3 gene. EMBO J. 2001;20:2564–2574. doi: 10.1093/emboj/20.10.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13(1):5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- 31.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.