Abstract
Both Foxp3+ regulatory T cells (Tregs) and antigen-expanded Foxp3− Tregs play an important role in regulating immune responses as well as in preventing autoimmune diseases and graft rejection. Molecular mechanisms modulating Treg function remain largely unclear, however. We report here on the expression and function of an inhibitory killer cell Ig-like receptor, KIR3DL1, in a nonobese diabetic (NOD) mouse-derived autoantigen-specific Treg (2D2), which protects from type 1 diabetes (T1D) in adoptive transfer experiments. This gene is not expressed in T1D pathogenic T cells (Tpaths) or non-Tpath T cells. KIR genes are known to play an important role in regulating natural killer (NK) cell function, but their role in Tregs and T1D is unknown. To examine whether KIR3DL1 expression may modulate Treg function, we used shRNA to down-regulate KIR3DL1 expression (2D2-shKIR). We find that KIR3DL1 down-regulation enhances in vitro function, as measured by improved suppression of target cell proliferation. Antibody blockade of IL-10 but not IL-4 partially abrogated suppressive function. In vivo function is also improved. Adoptive transfer of 2D2-shKIR into 10-wk-old NOD mice prevented spontaneous insulitis and T1D, and the inhibitory effect was further improved if the cells were transferred earlier into 6-wk-old NOD mice. These studies indicate that KIR3DL1 expression may negatively regulate Treg function and suggest a previously undescribed target for improving immune tolerance for potential treatment of autoimmune diseases like T1D.
Keywords: autoimmunity, KIR gene family
Regulatory T cells (Tregs) help maintain homeostatic balance in the immune system (1–4). The loss of Tregs, either as a result of failed development or by experimental depletion, leads to multiorgan autoimmune diseases (1–4). Conversely, augmenting Treg populations by adoptive transfer can prevent autoimmune diseases, including type 1 diabetes (T1D) (1–6). Both Foxp3+CD4+ Tregs and antigen-expanded Foxp3−CD4+ Tregs play an important role in regulating immune responses as well as in preventing autoimmune diseases and graft rejection. Antigen-specific Tregs are often more potent than polyclonal Tregs at abrogating disease (5–8), but the molecular mechanisms modulating antigen-specific Treg function remain to be determined.
We previously isolated several CD4+Foxp3− Tregs that are specific for glutamic acid decarboxylase (GAD), a major autoantigen in T1D (9–12). These autoantigen-specific and cytokine-dependent Tregs are able to inhibit proliferation of pathogenic T cells (Tpaths) effectively (9–12). Some of these Tregs required cell contact, but others did not. For example, Treg N206, which is a GAD peptide p206-specific CD4+ Treg line isolated from nonobese diabetic (NOD) mice, does not require cell contact, but it inhibits Tpath proliferation and prevents T1D in adoptive transfer experiments (9). The regulatory function of NR206, another p206-specific Treg line isolated from nonobese diabetes-resistant (NOR) mice, is dependent on both IFN-γ production and cell contact with target cells (11). It is not known how these Tregs acquire or perform their regulatory function.
To identify factors that may be involved in modulating Treg function, we performed genome-wide comparative expression analyses of CD4+ Tregs, Tpaths, and non-Tpaths and noted unique expression of killer cell Ig-like receptor (KIR) 3DL1 in Tregs. KIRs are transmembrane glycoproteins expressed by natural killer (NK) cells. KIR genes have been studied mainly in humans, where they are known to be involved in self-nonself recognition by binding to ligands on target cells (13–19). For some KIRs, the ligand is an HLA class I membrane protein. By recognition of HLA ligands on prospective target cells, KIRs play a critical role in regulating NK cell function (17, 18, 20, 21). One outcome is that NK cells are inactivated as a result of KIRs recognizing self-HLA ligands; thus, they will not attack target cells bearing self-HLA molecules. NK cells express a wide range of both activating and inhibitory KIRs, and the varied expression profile and balance of these receptors can dictate the NK cell function and activities (14, 17, 18). Inhibitory KIRs may play an important role in immune regulation by actively promoting peripheral tolerance, enhancing effector cell survival, or dampening immune responses (16, 20–22). On the other hand, activating KIRs are implicated in conditions including active host defense against infectious organisms (23–25). Normal T cells express very few or no KIRs, but KIR expression can be detected on a small subset of T cells in patients with autoimmune diseases, such as lupus and rheumatoid arthritis (22, 26–31). Based on these studies, it has been proposed that KIR expression may play a role in aberrant T-cell responses (13, 15, 22, 29, 30). Beyond this, little is known, and expression of KIRs in Tregs has not been studied.
Herein, we report results obtained from comparative gene array analyses and further functional studies of mouse KIR3DL1. We find that this KIR is expressed at relatively high levels in Tregs but not in Tpaths or non-Tpaths. Because KIR3DL1 is homologous to the human inhibitory KIR3DL1, we hypothesized that expression of KIR3DL1 in Tregs will inhibit their regulatory functions. To address this hypothesis, we used gene-specific shRNA to down-regulate KIR3DL1 expression in 2D2 cells, a Treg clone derived from N206 cells. Our studies demonstrate that KIR3DL1 expression indeed can negatively regulate their Treg function and suggest that KIR3DL1 is a previously undescribed important target for modulation of Treg function for treatment of autoimmune diseases like T1D.
Results
KIR3DL1 Is Expressed in Tregs but Not in Tpaths.
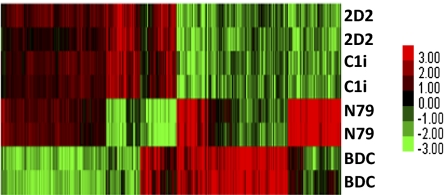
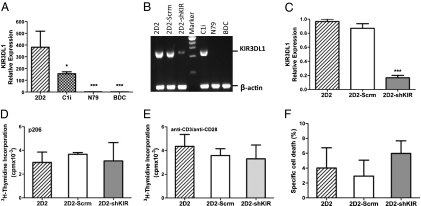
To gain insight into the nature of CD4+ GAD-specific Tregs that suppress T1D in a mouse model, genome-wide expression analysis was done to compare diabetes-causing CD4+ Tpaths (BDC) and non-Tpath neutral T cells (N79) with Tregs. Fig. 1 shows a heat map of genes that differ in expression by twofold or more between the cell lines. The expression patterns of the two Treg clones (2D2 and Cli) are very similar and quite different from the diabetes-causing BDC cells or neutral N79 cells. Examples of genes specifically up-regulated in both Treg clones but not in BDC or N79 cells include Kir3dl1, Adora3, Slamf7, Tnfsf13b, and Havcr2. A full list of genes that differ between the cells is given in Tables S1 and S2. A more complete analysis of expression and other epigenetic features will be published elsewhere. For this paper, we focus on KIR3DL1, which was found to be expressed highly only in Tregs. The initial results obtained by transcription profiling were confirmed by additional studies (Fig. 2 A and B). KIR3DL1 mRNA is easily detectable in Tregs but not in BDC or N79 cells.
Fig. 1.
Heat map of genome-wide analyses of genes expressed in Tregs, Tpaths, and neutral T cells. Expression profiles of genes in Tregs (2D2 cells and C1i cells) vs. Tpaths (BDC cells) and neutral cells (N79 cells) are shown. The data for 2D2 cells (and the other cells) were collected from two biological replicates processed on two separate microarray chips. Affymetrix microarrays were used (details are presented in Materials and Methods).
Fig. 2.
Gene-specific shRNA down-regulates KIR3DL1 in Tregs without affecting cell proliferation and survival in response to stimulation. (A) Relative expression of KIR3DL1 in T cells. Real-time RT-PCR was used to measure the expression of KIR3DL1 in four types of CD4+ T cells: 2D2, C1i, N79, and BDC. The SD for at least three measurements is shown. (B) Gel showing Kir3dl1 gene expression in various cell lines, down-regulation by shRNA (2D2-shKIR), and no down-regulation by scrambled shRNA (2D2-Scrm). (C) Real-time RT-PCR results showing down-regulation of KIR3DL1 in 2D2-shKIR cells but not in 2D2-Scrm cells. Proliferation is not affected. Cells (2D2, 2D2-Scrm, and 2D2-shKIR) were activated with either anti-CD3/CD28 (D) or their antigenic peptide, GAD p206 (E). (F) Cell death is not affected. Cell death was measured by Annexin V staining and analyzed by flow cytometry. Data are mean ± SD of at least four independent experiments. *P < 0.05; ***P < 0.001.
Knockdown of KIR3DL1 Improves Treg Function.
To investigate the function of KIR3DL1 in the 2D2 Tregs, we used a lentiviral vector to introduce KIR3DL1-specific shRNAs. Some of these shRNAs reduced the level of KIR3DL1 mRNA, and stable knocked-down cell lines were obtained. As shown in Fig. 2C, more than 80% reduction was obtained for 2D2-shKIR cells. Sequence-scrambled shRNA, which was used as a control, caused no reduction (Fig. 2C, 2D2-Scrm cells).
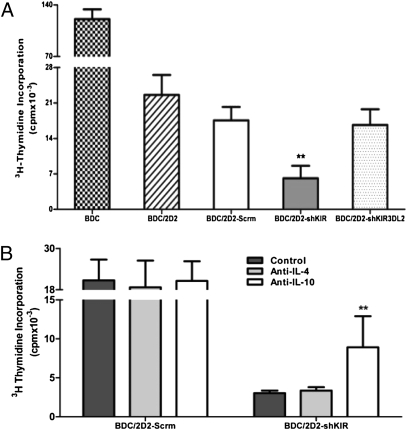
Down-regulation of KIR3DL1 had no significant effect on growth or cell survival (Fig. 2 D–F). As shown in Fig. 3, however, the suppressive function of 2D2 Tregs is affected. In the presence of 2D2 or 2D2-Scrm cells, ∼15% of BDC cells proliferated in response to stimulation when compared with BDC cells cultured without these Tregs (Fig. 3A). In the presence of 2D2-shKIR cells, only ∼5% of BDC cells proliferated (Fig. 3A). Thus, 2D2-shKIR shows an approximately threefold improved suppression, whereas a scrambled shRNA (2D2-Scrm) or shRNA cells directed against another KIR gene, KIR3DL2, had no significant effect.
Fig. 3.
Down-regulation of KIR3DL1 significantly improves the regulatory function of 2D2 Tregs in suppressing proliferation of Tpaths. (A) Suppression of BDC cell proliferation by Tregs in a mixed cell inhibition assay (Materials and Methods). BDC cells were cultured either alone or with various Tregs, including 2D2, 2D2-Scrm, 2D2-shKIR (harboring shRNA against KIR3DL1), and 2D2-shKIR3DL2 (harboring shRNA against KIR3DL2) cells. At least four measurements were made, and the SD is shown. (B) Regulatory function of 2D2-shKIR is partially dependent on IL-10 production. Inhibition assays were performed in which the cells were cultured either alone or with the presence of a saturating amount (24 μg/mL) of either anti–IL-4 or anti–IL-10 antibodies. At least five measurements were made, and the SD is shown. **P < 0.01.
Knockdown of KIR3DL1 Leads to Increased IL-10, Which Contributes to Suppressive Function.
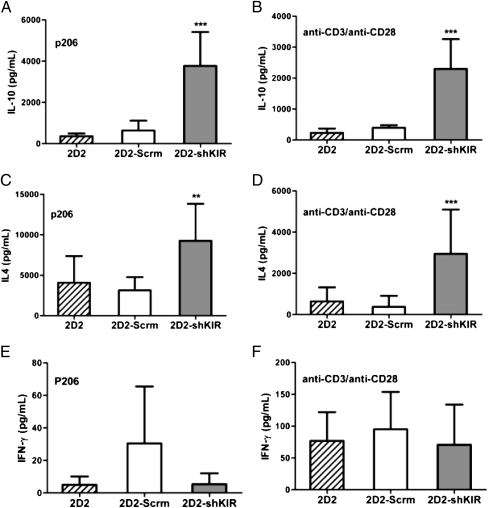
Our previous studies showed that N206 cells, the parent cell line of 2D2, produced IL-10, IL-4, and some IFN-γ (9). Therefore, we examined whether their production is altered in 2D2-shKIR cells. As shown in Fig. 4, all three types of cells produced IL-10 and IL-4 in response to stimulation by either anti-CD3/CD28 or GAD p206 in a mixed-cell assay. Compared with 2D2 and 2D2-Scrm cells, however, 2D2-shKIR cells produced more IL-10 and IL-4. In addition, none of these three types of cells produced a large amount of IFN-γ following stimulation. Based on their cytokine secretion profiles, it appears that the 2D2-shKIR cells are more like IL-10–producing Tregs, such as the Tr1 cells, which produce a larger amount of IL-10 and a smaller amount of other effector cytokines (32, 33).
Fig. 4.
Down-regulation of KIR3DL1 significantly increases IL-10 production. Tregs were activated with either p206 plus irradiated APCs (A, C, and E) or with anti-CD3/anti-CD28 antibodies (B, D, and F) for 24 h. Cell culture supernatant was harvested for ELISA to detect the presence of IL-10 (A and B), IL-4 (C and D), or IFN-γ (E and F). At least three measurements were made, and the SD is shown. **P < 0.01; ***P < 0.001.
We then performed cytokine-blockade assays to determine whether IL-10 produced by 2D2-shKIR cells contributes to their regulatory functions. As shown in Fig. 3B, addition of anti–IL-10 antibody but not anti–IL-4 antibody to the cell cultures increased BDC cell proliferation approximately threefold compared with that of cultures in the absence of the antibody (Fig. 3B). Thus, 2D2-shKIR Tregs exert their enhanced regulatory function, at least in part, by increased secretion of IL-10.
Down-Regulation of KIR3DL1 Significantly Increases the Ability of 2D2-shKIR Cells to Inhibit Spontaneous T1D in Both Early-Preonset and Late-Preonset Models of NOD Mice.
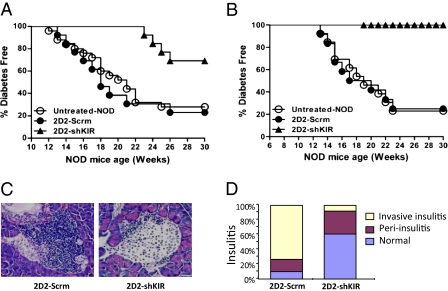
Does down-regulation of KIR3DL1 in 2D2-shKIR cells increase their ability to inhibit T1D in NOD mice? To address this question, we transferred 2D2-Scrm or 2D2-shKIR cells into 10-wk-old late prediabetic NOD mice. At this age, 2 wk before the onset of overt diabetes, it is known that destructive invasive insulitis has developed for several weeks (usually starting from 3–4 wk of age). As shown in Fig. 5A, 2D2-shKIR Treg treatment delayed the onset of spontaneous T1D in the treated NOD mice by 10–11 wk compared with that in either untreated NOD mice or mice treated with 2D2-Scrm cells. Moreover, only 30% of 2D2-shKIR cell-treated NOD mice eventually developed T1D, compared with 75–80% of untreated or 2D2-Scrm cell-treated NOD mice. Although invasive insulitis has already developed in 10-wk-old NOD mice, 2D2-shKIR cell treatment was still able to inhibit the progression of T1D significantly in these mice. We then performed studies to determine whether transfer of 2D2-shKIR Tregs to NOD mice at a younger age (6 wk) could prevent the initiation of T1D. Fig. 5B shows that none of the 6-wk-old NOD mice transferred with 2D2-shKIR cells developed diabetes at 30 wk of age (24 wk after cell transfer). In comparison, mice transferred with 2D2-Scrm cells developed diabetes at a rate comparable to that found in untreated NOD mice. Therefore, in young mice, 2D2-shKIR Tregs are very effective in preventing the development of T1D.
Fig. 5.
Down-regulation of KIR3DL1 significantly increases the ability of 2D2-shKIR cells to inhibit spontaneous T1D in both early-preonset and late-preonset models of NOD mice. NOD mice were i.v. injected with cells (1 × 107 cells per mouse) at either the late-preonset age (A; 10 wk) or the early-preonset age (B; 6 wk). The mice were monitored up to 30 wk of age. (C) Histological analyses of islets from NOD recipient mice. H&E staining of frozen pancreas sections obtained from 21-wk-old NOD mice treated with 2D2-Scrm cells or 2D2-shKIR cells. Images are representative of sections from at least three mice per group. (D) Cumulative percentage of different types of insulitis detected in NOD mice treated with either 2D2-Scrm or 2D2-shKIR cells. At least 30 sections were examined.
Because invasive insulitis develops before T1D development, we histologically examined the insulitis status of islets using pancreas sections obtained from 2D2-shKIR–treated or 2D2-Scrm–treated animals. Fig. 5 C and D shows that islets in 2D2-shKIR–treated mice remained intact or not invasively infiltrated with leukocytes, whereas the islets of 2D2-Scrm–treated NOD mice were heavily infiltrated with leukocytes. Altogether, our data support the model that 2D2-shKIR Tregs are able to inhibit spontaneous T1D, at least partly, through the suppression of invasive insulitis development.
Discussion
It is known that the regulatory function of autoantigen-specific CD4+ Tregs can be dependent on cytokine production and, depending on the Treg line, may or may not require contact with target cells (1–4, 9–12, 32, 34). Beyond this, the molecular mechanisms that control their regulatory function are still largely elusive. We report here the identification of a gene that may play a key role in modulating the regulatory function of autoantigen-specific Tregs. We found that KIR3DL1 is expressed at a high level in two GAD peptide-specific Tregs but not in either pathogenic BDC cells or neutral nonpathogenic N79 cells. We then found that down-regulation of KIR3DL1 in the 2D2 Tregs, which did not affect their proliferation and survival in response to stimulation, dramatically enhanced their suppressive effect on the proliferation of target T cells in vitro and their ability to prevent spontaneous T1D in NOD mice. The enhanced Treg function is partly attributable to increased IL-10 production, because blockade of IL-10 partially abrogated the suppressive function of Tregs. Altogether, our results support the hypothesis that KIR3DL1 is a negative regulator of Treg function both in vitro and in vivo.
KIRs play a critical role in regulating NK cell function (13–19). T cells normally express very few or no KIRs, although KIR expression can be detected on a small subset of T cells in patients with autoimmune diseases, such as lupus and rheumatoid arthritis (22, 26–31). It is suspected that KIR expression on the subset of T cells in these conditions may play a role in regulating aberrant T-cell responses that are associated with the initiation and/or progression of the diseases (13, 15, 22, 29, 30). Beyond this, little is known about the role of KIRs in T cells, and expression of KIRs in Tregs has not been studied. Previous studies, mostly in humans, have suggested that expression of different sets of inhibitory or activating KIRs could contribute to the function of NK cells by modulating immune responses to target cells, including tumor cells and autoreactive pathogenic cells (20, 21, 35–38). Inhibitory KIRs have a long cytoplasmic domain containing immune receptor tyrosine-based inhibitory motifs (ITIMs) (13–19). Both mouse and human KIR3DL1 contains ITIMs and may function as inhibitory KIRs (13–19, 39). Therefore, a hypothesis consistent with our results is that expression of KIR3DL1 in Tregs serves as a functional brake to fine-tune their suppressive effect on immune responses. Without such a molecular brake, Tregs may overly inhibit immune responses. According to this model, expression of KIRs, such as KIR3DL1, in Tregs may play a key regulatory role in orchestrating immunity and autoimmunity.
Down-regulation of KIR3DL1 in 2D2 Tregs significantly increased both IL-10 and IL-4 production. Moreover, blockade of IL-10 but not IL-4 partially reversed the effect of KIR3DL1 down-regulation, suggesting that KIR3DL1 exerts its negative effect, in part, by controlling production of suppressive cytokines, such as IL-10. Previous studies on human KIRs have demonstrated that several KIRs are capable of mediating their function by recognizing HLA class I molecules as their ligands. For example, human KIR3DL1 recognizes HLA-B Bw4 (13–18). Based on the amino acid sequence homology and functional similarity between mouse and human KIR, it is likely but not yet experimentally shown that mouse KIR3DL1 also recognizes MHC class I molecules. One likely outcome following recognition of self-MHC molecules by KIRs is inactivation of NK cells so that they will not attack self but will only attack foreign cells not presenting the correct MHC or malignant host cells that have lost self-MHC expression. If the mouse KIR3DL1 also recognizes self-MHC class I molecules, it could inactivate or down-regulate Treg function. One possible biological consequence is self-control of the functional strength of Treg during the induction of immune tolerance to prevent autoimmune disease outcomes while ensuring effective immunity against infection or malignant cells. Considering the important role of KIR3DL1 in modulating Treg function and the potential of using it as a target to enhance Treg function in inducing more effective immune tolerance, it would be important to identify the ligands for mouse KIR3DL1 and whether MHC class I molecules can bind to KIR3DL1. Further studies would be necessary to examine how ligand-activated KIR3DL1 in 2D2 cells may modulate their function at both biochemical and molecular levels.
We found that adoptive transfer of a Treg with down-regulated KIR3DL1 into NOD mice dramatically decreases the incidence of T1D. Even after invasive insulitis had already developed in 10-wk-old NOD mice, 2D2-shKIR cell treatment was still able to prevent T1D. Importantly, the protective effect against the disease was further improved when the mice were treated at an early-preonset stage at 6 wk of age. In addition, our results from histological analyses of insulitis development in NOD mice showed that the 2D2-shKIR cells were able to inhibit T1D, perhaps, at least partly, through the suppression of spontaneous invasive insulitis development in treated NOD mice. Based on these results, it is likely that down-regulation of KIR3DL1 improves the ability of 2D2 Tregs to inhibit Tpath trafficking and infiltration into the islets.
In summary, we report that KIR3DL1 expression in Tregs can have a negative effect on their in vitro and in vivo function. Down-regulation of KIR3DL1 was able to enhance the regulatory function of Tregs and their ability to inhibit insulitis and T1D. Our studies provide evidence that expression of a KIR, previously thought to function in NK cells, may play an important role in CD4+ Tregs by regulating their function in modulating immune responses and inducing immune tolerance. This suggests that KIRs may be good molecular targets for improvement of Treg functional potency. A requirement, and challenge, for successful Treg-based cellular immunotherapy is to produce a sufficient number of Tregs for clinical use. By down-regulating KIR3DL1 expression in Tregs, significantly fewer cells may be needed for treatment of T1D or other autoimmune diseases and in preventing graft rejection and graft-vs.-host diseases.
Materials and Methods
Mice and Cells.
NOD mice were purchased from the Jackson Laboratory. BDC2.5 T-cell receptor (TCR) transgenic NOD (BDC) mice were a gift from D. Mathis and C. Benoist (Joslin Diabetes Center/Harvard Medical School, Boston, MA) (40). All animals were housed in a specific pathogen-free animal facility at The Beckman Research Institute of the City of Hope. About 75% of female NOD mice develop diabetes by the age of 6 mo. 2D2 cells are a Treg clone derived from the previously described N206 Treg line (9). C1i cells are a Treg clone derived from the NR206 Treg line (11). The previously described N79 cells, recognizing a mimotope p79 peptide, function neither as Tregs nor as Tpaths and are used as functionally neutral control cells (41). CD4+ BDC cells, isolated from the BDC mice, were cultured and activated three times by the p79 peptide in vitro and were used as the Tpaths in the gene array assays. These BDC cells did not express Foxp3 and were able to induce an aggressive form of T1D when transferred to NOD/SCID mice.
Microarray Analysis.
For microarray analyses, total RNA was extracted from each cell line using an RNeasy Miniprep kit (Qiagen). The Affymetrix GeneChip Mouse Gene 1.0-ST array was used to determine gene expression profiles. Synthesis and labeling of cDNA targets as well as hybridization and scanning of GeneChips were carried out by the Microarray Core Facility at the City of Hope. Raw intensity measurements of all probe sets were background-corrected, normalized, and converted into expression measurements using the Affymetrix Expression Console v1.1.1. The Bioconductor “ArrayTools” package was then used to identify the genes differentially expressed between the 2D2, C1i, BDC, and N79 cells. The corresponding P values were adjusted using the false discovery rate method. Significant genes were selected with a cutoff of adjusted P < 0.05 and log2 ratio of 1.7.
Lentivirus Production and T-Cell Transduction.
Lentivirus-based vectors containing shRNA against the KIR3DL1 (target sequence CCTTTCCTCTTGATTCTACAA) or KIR3DL2 (target sequence CAATAAACAGACAGCTTTCTA) were purchased from Sigma–Aldrich. A scrambled shRNA was used as a control. VSV-G (G protein of vesicular stomatitis virus) pseudotyped lentiviral vectors were produced using a four-plasmid transfection system as described (42). Supernatant containing recombinant lentivirus was collected 3 d after transfection, filtered, and concentrated by ultracentrifugation. Viral pellets were resuspended in culture medium and frozen at −80 °C until use. After sequential concentration, lentiviral titers were determined using HT1080 cells and reached 1.5–3 × 108 TU/mL. For KIR knockdown, 2D2 cells were infected overnight with recombinant lentivirus at multiplicity of infection of 10 in the presence of 8 μg/mL polybrene and selected with puromycin (18 μg/mL) for 28 d. Cells showing effective suppression of KIR3DL1 or KIR3DL2 expression were identified using real-time PCR. Separate transduction experiments were done to obtain at least two independent cell lines for each construct.
Real-Time RT-PCR.
For KIR3DL1 gene expression analyses, total RNA was prepared using the RNeasy Miniprep kit (Qiagen) and converted to cDNA using SuperScript III First-strand Synthesis System (Invitrogen) according to the manufacturer’s protocols. Real-time PCR was performed by SYBR Green Gene Expression Assays (Applied Biosystems). All reactions were performed in triplicate for 40 cycles. The PCR primers were as follows: KIR3DL1, 5′-ACACAGAAGCAGATACCAAAACAA-3′ and 5′-GTCTTTCAAAGTCCTGTGTG-3′, and β-actin, 5′-GTATGGAATCCTGTGGCATCCATG-3′ and 5′-GGACTCATCGTACTCCTGCTTGCT-3′. Equal amounts of RNA from cell samples were used as PCR templates to obtain the threshold cycle (Ct), and the Ct was normalized using the known Ct from β-actin RNAs. For RT-PCR analysis, KIR3DL1 transcripts were amplified using the following primers with an annealing temperature of 56 °C: 5′-CACTCCCGGGTCATTTCATAATTTCCATGTACAC-3′ and 5′-TATACTCGAGAGCCATGCTGCTCTGGTTCCTC-3′.
Cell Proliferation Assays.
The 2D2, 2D2-Scrm, or 2D2-shKIR cells (5 × 104 per well) were stimulated with soluble anti-mouse CD3/CD28 (Invitrogen) or GAD p206 peptide in the presence of antigen-presenting cells (APCs; irradiated NOD mouse splenocytes) in 96-well plates for 72 h. A total of 0.5 μCi of 3H-thymidine (Amersham Biosciences) was added per well during the last 24 h of culture. The cells were harvested onto glass fiber filter mats, and the incorporated radioactivity was measured.
Analyses of Cell Death.
Cells (1 × 105 per well) were activated in 96-well plates coated with the anti-TCR Ab H57 (25 μg/mL) plus soluble anti-CD28 (5 μg/mL) for 24 h. The percentage of apoptotic cells was measured by Annexin V (BD PharMingen) staining. Specific cell death was calculated as follows: specific cell death = (% of activated cell death − % of nonactivated cell death)/(100 − % of nonactivated cell death) × 100.
Analyses of Cytokine Production.
Cell culture supernatant was harvested after activating cells with anti-CD3/CD28 or GAD p206 and irradiated APCs for 24 h. Mouse IFN-γ, IL-4, and IL-10 OptEIA ELISA kits (BD PharMingen) were used to measure the amount of cytokines according to the manufacturer's instruction.
In Vitro Inhibition Assays.
These assays have been described (9–12). CD4+ T cells from BDC mouse splenocytes were purified by negative selection using magnetic beads (Miltenyi Biotec). The purified CD4+ BDC cells (the target cells) cultured in the absence of Tregs with or without stimulation with p79 peptide (0.1 μg/mL) were used as controls. To measure inhibition, the CD4+ T cells were cultured with p79 (0.1 μg/mL) plus irradiated (2,000 rad) 2D2, 2D2-Scrm, or 2D2-shKIR cells. Irradiated CD4+ T cell-depleted splenocytes were used as APCs. The cells were cultured in RPMI medium 1640 containing 5% FCS (vol/vol) for 4 d in 96-well plates and pulsed with 0.5 μCi of 3H-thymidine during the last 24 h of culture.
To determine the effect of different cytokines on target cell proliferation, anticytokine antibodies were added to the cell cultures of in vitro inhibition assays. In these cytokine blockade assays, a saturating amount of antibodies against IL-10, IL-4, or IFN-γ (24 μg/mL) was added to the culture and the cells were incubated for 4 d before being harvested for analyses.
Adoptive Transfer and Histology.
For adoptive transfer experiments, either 6- or 10-wk-old female NOD mice received a single i.v. injection of 1 × 107 cells of 2D2-Scrm or 2D2-shKIR Tregs. Recipient mice were monitored for up to 30 wk of age and were considered diabetic after 2 consecutive weeks of glycosuria ≥2% and a blood glucose level ≥250 mg/dL. At 21 wk of age, pancreases were removed from treated mice for histological analyses. Pancreatic tissues were fixed in 10% formalin/PBS (vol/vol), followed by 70–100% ethanol and xylene, and were then paraffin-embedded and sectioned. Sections were stained with H&E to detect islet-infiltrating leukocytes.
Statistical Analysis.
Kaplan–Meier survival analysis was used to compare cumulative diabetes incidence. The Student's t test was used for data analyses of all other studies. Significance was set at P ≤ 0.05.
Supplementary Material
Acknowledgments
This manuscript is dedicated to the memory of Dr. Zunde Wang, who did not survive to see completion of the work he started. We thank Megan Kiyohara for technical assistance. This work was supported, in part, by funds from the Juvenile Diabetes Research Foundation, the American Heart Association, and the American Diabetes Association, by the National Institutes of Health, and by the H. L. Snyder Medical Foundation.
Footnotes
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE26456).
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019082108/-/DCSupplemental.
References
- 1.Sakaguchi S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Shevach EM. Regulatory/suppressor T cells in health and disease. Arthritis Rheum. 2004;50:2721–2724. doi: 10.1002/art.20500. [DOI] [PubMed] [Google Scholar]
- 3.Ziegler SF. FOXP3: Of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 4.O'Garra A, Vieira P. Regulatory T cells and mechanisms of immune system control. Nat Med. 2004;10:801–805. doi: 10.1038/nm0804-801. [DOI] [PubMed] [Google Scholar]
- 5.Tarbell KV, Yamazaki S, Olson K, Toy P, Steinman RM. CD25+ CD4+ T cells, expanded with dendritic cells presenting a single autoantigenic peptide, suppress autoimmune diabetes. J Exp Med. 2004;199:1467–1477. doi: 10.1084/jem.20040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Q, et al. In vitro-expanded antigen-specific regulatory T cells suppress autoimmune diabetes. J Exp Med. 2004;199:1455–1465. doi: 10.1084/jem.20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masteller EL, Tang Q, Bluestone JA. Antigen-specific regulatory T cells—Ex vivo expansion and therapeutic potential. Semin Immunol. 2006;18:103–110. doi: 10.1016/j.smim.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Hori S, Haury M, Coutinho A, Demengeot J. Specificity requirements for selection and effector functions of CD25+4+ regulatory T cells in anti-myelin basic protein T cell receptor transgenic mice. Proc Natl Acad Sci USA. 2002;99:8213–8218. doi: 10.1073/pnas.122224799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Lee WH, Yun P, Snow P, Liu CP. Induction of autoantigen-specific Th2 and Tr1 regulatory T cells and modulation of autoimmune diabetes. J Immunol. 2003;171:733–744. doi: 10.4049/jimmunol.171.2.733. [DOI] [PubMed] [Google Scholar]
- 10.You S, et al. Presence of diabetes-inhibiting, glutamic acid decarboxylase-specific, IL-10-dependent, regulatory T cells in naive nonobese diabetic mice. J Immunol. 2004;173:6777–6785. doi: 10.4049/jimmunol.173.11.6777. [DOI] [PubMed] [Google Scholar]
- 11.Chen C, Lee WH, Zhong L, Liu CP. Regulatory T cells can mediate their function through the stimulation of APCs to produce immunosuppressive nitric oxide. J Immunol. 2006;176:3449–3460. doi: 10.4049/jimmunol.176.6.3449. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Liu CP. Regulatory function of a novel population of mouse autoantigen-specific Foxp3 regulatory T cells depends on IFN-gamma, NO, and contact with target cells. PLoS ONE. 2009;4:e7863. doi: 10.1371/journal.pone.0007863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: Multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 14.Moretta L, Moretta A. Unravelling natural killer cell function: Triggering and inhibitory human NK receptors. EMBO J. 2004;23:255–259. doi: 10.1038/sj.emboj.7600019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moretta L, Moretta A. Killer immunoglobulin-like receptors. Curr Opin Immunol. 2004;16:626–633. doi: 10.1016/j.coi.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Long EO, et al. Inhibition of natural killer cell activation signals by killer cell immunoglobulin-like receptors (CD158) Immunol Rev. 2001;181:223–233. doi: 10.1034/j.1600-065x.2001.1810119.x. [DOI] [PubMed] [Google Scholar]
- 17.Vilches C, Parham P. KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 18.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 19.Joncker NT, Raulet DH. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakker AB, Phillips JH, Figdor CG, Lanier LL. Killer cell inhibitory receptors for MHC class I molecules regulate lysis of melanoma cells mediated by NK cells, gamma delta T cells, and antigen-specific CTL. J Immunol. 1998;160:5239–5245. [PubMed] [Google Scholar]
- 21.Beelen DW, et al. Genotypic inhibitory killer immunoglobulin-like receptor ligand incompatibility enhances the long-term antileukemic effect of unmodified allogeneic hematopoietic stem cell transplantation in patients with myeloid leukemias. Blood. 2005;105:2594–2600. doi: 10.1182/blood-2004-04-1441. [DOI] [PubMed] [Google Scholar]
- 22.Ugolini S, et al. Involvement of inhibitory NKRs in the survival of a subset of memory-phenotype CD8+ T cells. Nat Immunol. 2001;2:430–435. doi: 10.1038/87740. [DOI] [PubMed] [Google Scholar]
- 23.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 24.Alter G, Altfeld M. NK cell function in HIV-1 infection. Curr Mol Med. 2006;6:621–629. doi: 10.2174/156652406778195035. [DOI] [PubMed] [Google Scholar]
- 25.Wilson EB, Parachoniak CA, Carpenito C, Mager DL, Takei F. Expression of murine killer immunoglobulin-like receptor KIRL1 on CD1d-independent NK1.1(+) T cells. Immunogenetics. 2007;59:641–651. doi: 10.1007/s00251-007-0216-8. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Yu M, Weyand CM, Goronzy JJ. Epigenetic regulation of killer immunoglobulin-like receptor expression in T cells. Blood. 2009;114:3422–3430. doi: 10.1182/blood-2009-01-200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhrberg M, et al. The repertoire of killer cell Ig-like receptor and CD94:NKG2A receptors in T cells: Clones sharing identical alpha beta TCR rearrangement express highly diverse killer cell Ig-like receptor patterns. J Immunol. 2001;166:3923–3932. doi: 10.4049/jimmunol.166.6.3923. [DOI] [PubMed] [Google Scholar]
- 28.Goronzy JJ, Weyand CM. T-cell co-stimulatory pathways in autoimmunity. Arthritis Res Ther. 2008;10(Suppl 1):S3–S4. doi: 10.1186/ar2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. J Exp Med. 2005;201:1025–1029. doi: 10.1084/jem.20050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu D, et al. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J Immunol. 2009;183:3481–3487. doi: 10.4049/jimmunol.0900034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Chen Y, Richardson B. Decreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4(+)CD28(−) T cells. Clin Immunol. 2009;132:257–265. doi: 10.1016/j.clim.2009.03.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 33.Levings MK, Roncarolo MG. T-regulatory 1 cells: A novel subset of CD4 T cells with immunoregulatory properties. J Allergy Clin Immunol. 2000;106:S109–S112. doi: 10.1067/mai.2000.106635. [DOI] [PubMed] [Google Scholar]
- 34.von Boehmer H. Mechanisms of suppression by suppressor T cells. Nat Immunol. 2005;6:338–344. doi: 10.1038/ni1180. [DOI] [PubMed] [Google Scholar]
- 35.Gumperz JE, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 36.Gati A, et al. CD158 receptor controls cytotoxic T-lymphocyte susceptibility to tumor-mediated activation-induced cell death by interfering with Fas signaling. Cancer Res. 2003;63:7475–7482. [PubMed] [Google Scholar]
- 37.Passweg JR, Huard B, Tiercy JM, Roosnek E. HLA and KIR polymorphisms affect NK-cell anti-tumor activity. Trends Immunol. 2007;28:437–441. doi: 10.1016/j.it.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Moretta A, Locatelli F, Moretta L. Human NK cells: From HLA class I-specific killer Ig-like receptors to the therapy of acute leukemias. Immunol Rev. 2008;224:58–69. doi: 10.1111/j.1600-065X.2008.00651.x. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Kuick R, Hanash S, Richardson B. DNA methylation inhibition increases T cell KIR expression through effects on both promoter methylation and transcription factors. Clin Immunol. 2009;130:213–224. doi: 10.1016/j.clim.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- 41.You S, et al. Detection and characterization of T cells specific for BDC2.5 T cell-stimulating peptides. J Immunol. 2003;170:4011–4020. doi: 10.4049/jimmunol.170.8.4011. [DOI] [PubMed] [Google Scholar]
- 42.Kowolik CM, Yam P, Yu Y, Yee JK. HIV vector production mediated by Rev protein transduction. Mol Ther. 2003;8:324–331. doi: 10.1016/s1525-0016(03)00166-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.