Abstract
Imprinted genes are expressed primarily or exclusively from either the maternal or paternal allele, a phenomenon that occurs in flowering plants and mammals. Flowering plant imprinted gene expression has been described primarily in endosperm, a terminal nutritive tissue consumed by the embryo during seed development or after germination. Imprinted expression in Arabidopsis thaliana endosperm is orchestrated by differences in cytosine DNA methylation between the paternal and maternal genomes as well as by Polycomb group proteins. Currently, only 11 imprinted A. thaliana genes are known. Here, we use extensive sequencing of cDNA libraries to identify 9 paternally expressed and 34 maternally expressed imprinted genes in A. thaliana endosperm that are regulated by the DNA-demethylating glycosylase DEMETER, the DNA methyltransferase MET1, and/or the core Polycomb group protein FIE. These genes encode transcription factors, proteins involved in hormone signaling, components of the ubiquitin protein degradation pathway, regulators of histone and DNA methylation, and small RNA pathway proteins. We also identify maternally expressed genes that may be regulated by unknown mechanisms or deposited from maternal tissues. We did not detect any imprinted genes in the embryo. Our results show that imprinted gene expression is an extensive mechanistically complex phenomenon that likely affects multiple aspects of seed development.
Keywords: epigenetics, gene imprinting, angiosperm reproduction, DNA demethylation
Genomic imprinting, the differential expression of alleles of the same gene depending on parent of origin, independently evolved in mammals and flowering plants (1). Imprinted expression is a clear example of inheritance of epigenetic states, because genetically identical sequences are differentially transcribed depending on the sex of the parent from which the gene originates. A widely accepted evolutionary explanation of genomic imprinting is the parental conflict theory (1–3), which argues that, when females mate with multiple males and allocate resources directly to the developing embryo, males will favor expression of genes that maximize resource extraction for their offspring, whereas females will favor genes that equalize resource allocation to all offspring.
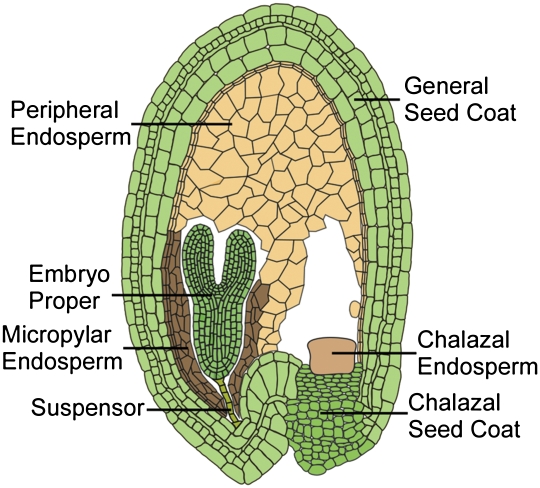
Double fertilization is unique to flowering plants and underlies the distinctive cellular programming of plant gene imprinting (4). In the ovule, a haploid megaspore undergoes three mitoses to form the female gametophyte with egg, central, synergid, and antipodal cells. The central cell initially has two haploid nuclei that fuse to create the diploid central cell. In stamens, haploid microspores undergo cell divisions to produce the male gametophyte with two sperm cells and a vegetative cell, which transport the sperm to the egg and central cell. The diploid embryo and triploid endosperm develop from the fertilized egg and central cell, respectively. In flowering plants, all known cases of imprinted expression, with one exception, occur in endosperm (5, 6). Endosperm is a nutrient tissue, acquiring and storing resources from the maternal chalazal seed coat and underlying vasculature to nourish the embryo (7, 8) (Fig. 1), and thus, is the tissue where conflict over resource allocation would be expected to unfold.
Fig. 1.
Drawing of an A. thaliana seed with a linear cotyledon stage embryo showing the major seed compartments.
Imprinted expression of all known plant genes depends on differential DNA methylation, activity of polycomb repressive complex 2 (PRC2), or both. Maternally inherited mutations in Arabidopsis thaliana genes that encode PRC2 proteins FERTILIZATION INDEPENDENT ENDOSPERM (FIE; WD40 protein), MULTICOPY SUPRESSOR OF IRA 1 (MSI1; WD40 protein), FERTILIZATION INDEPENDENT SEED 2 (FIS2; zinc finger protein), and MEDEA (MEA; SET domain protein that methylates H3K27) cause endosperm overproliferation, embryo abortion, and seed lethality (9). The MEA gene is self-imprinted, with maternal MEA protein activity required to silence the paternal allele after fertilization (10). Maternal PRC2 proteins also silence the paternal allele of the actin regulator, ARABIDOPSIS FORMIN HOMOLOG 5 (FH5) (11).
Active DNA demethylation is catalyzed by the DNA glycosylase DEMETER (DME) that excises 5-methylcytosine in the A. thaliana central cell (10). The maternal alleles of FIS2, FLOWERING WAGENINGEN/HOMEODOMAIN GLABROUS 6 (FWA/HDG6) homeodomain leucine zipper transcription factor gene, and MATERNALLY EXPRESSED PAB C-TERMINAL (MPC) polyA binding protein gene are activated by this demethylation. These genes are biallelically expressed in the endosperm if the male (sperm) genome is demethylated by mutation of the MET1 DNA methyltransferase (12–14). Passive DNA demethylation caused by inhibited expression of MET1 during female gametophyte cell proliferation might also contribute to imprinted expression (15). Because FIS2 activation is mediated by DME-dependent DNA demethylation, proper imprinting of genes regulated by PRC2 may also require DME.
Three paternally expressed imprinted transcription factor genes, HDG3, MYB THREE REPEAT 2 (MYB3R2), and PHERES 1 (PHE1), have been identified (16, 17). Silencing of the maternal PHE1 allele depends on a functional PRC2 complex, and maternally inherited mutations in PRC2 cause biallelic expression of PHE1 (18, 19). In addition, silencing of the maternal PHE1 allele is thought to require maternal demethylation at the PHE1 gene (17, 20).
Hundreds of mammalian imprinted genes have been described that are thought to regulate nutrient transfer capacity of fetal placenta, embryonic growth, childhood development, and adult brain function (21, 22). Imprinting disorders affect fetal growth, hormone systems after birth, and behavior. By contrast, only 11 imprinted genes are known in A. thaliana, some of which control growth and likely influence nutrient transfer capacity of the endosperm (5). Here, we report identification of imprinted A. thaliana genes by deep sequencing of cDNA libraries from polymorphic F1 seeds. We discovered 43 genes regulated by the DNA-demethylating glycosylase DME, the DNA methyltransferase MET1, or the core Polycomb group (PcG) protein FIE that are preferentially expressed from either the paternal or maternal allele in endosperm, including transcription factors, proteins involved in auxin and ethylene signaling, components of the ubiquitin-26S proteosome pathway, regulators of histone and DNA methylation, and small RNA pathway proteins. We also identified maternally expressed genes for which allele-specific expression was not obviously altered by mutations affecting DNA methylation or PcG function, suggesting that paternal silencing of these genes might be caused by an unknown pathway or that the mRNA is deposited in endosperm from maternal tissues. In contrast to endosperm, we did not identify any imprinted genes in embryo. Our study has significantly expanded the known set of imprinted genes in plants, showing that imprinting is a major epigenetic process affecting endosperm gene expression.
Results
Identification of Genes Imprinted in Endosperm.
To identify imprinted genes, we prepared cDNA libraries from endosperm derived from two pairs of reciprocal crosses between the Col and Ler accessions (two independent library pairs). cDNA libraries were sequenced using the Illumina GA2 platform and aligned to both Col and Ler genomic scaffolds (Dataset S1 and SI Methods). Reads that preferentially aligned to either scaffold were assigned to that accession. Each gene received Col and Ler expression scores equal to the number of reads assigned to each ecotype.
To gauge the performance of our method, we examined all 11 genes previously shown to be imprinted in endosperm (Table S1). Two of these genes (MEA and PHE1) lack SNPs between Col and Ler, and another (MPC) lacked reads. With the partial exception of FH5, other genes behaved consistently with published results, although most lacked sufficient reads for statistical significance (Dataset S2). For example, HDG3 is paternally expressed in both crosses, HDG9 and MYB3R2 are maternally expressed in both crosses, and HDG8 is maternally expressed when Col is female (CxL) and biallelic when Ler is female (LxC) (Table S1), as previously described (16). FH5 was reported as reciprocally maternally expressed in a cross between the Ler and C24 ecotypes (11), whereas we find that it is maternally expressed when Col is female but biallelic when Ler is female, a discrepancy that may be explained by the different ecotypes used.
At a P value cutoff of 0.05, we identified 1,081 genes preferentially or exclusively expressed from the female genome and 25 genes preferentially or exclusively expressed from the male genome in endosperm (Dataset S2), with 739 maternally expressed and nine paternally expressed genes at a stricter P value cutoff of 0.001 (Fisher's exact test) (Dataset S2, and Fig. S1). A potential complication in identifying genes expressed from the female genome is that contamination with RNA from maternal tissues such as the seed coat will mimic imprinting. Indeed, several genes in our P < 0.001 maternal dataset, such as TRANSPARENT TESTA 10 (At5g48100), are highly expressed in the seed coat (Dataset S2). To address this issue, we sequenced cDNA derived from CxL F1 endosperm tissue obtained through laser capture microdissection (LCM), a technique that allows much greater precision than manual dissection. We considered genes to be imprinted if their expression levels in both of our manually dissected endosperm library pairs were no more than fourfold greater than those from the LCM dataset. We also included genes close to the above cutoff if their imprinted status was significantly altered by mutations that would not affect expression in maternal tissues (met1, fie, and dme). These filtering steps reduced the number of maternally expressed imprinted genes to 114 (P < 0.001) (Dataset S2), which includes two previously reported genes, MYB3R2 and HDG9 (Table S1). We focused further analyses on the LCM-filtered maternal P < 0.001 set and the paternal P < 0.001 set, because we believe that these best represent genes with imprinted endosperm expression (Tables 1 and 2, Table S1 and S2, and Fig. S1).
Table 1.
Selected maternal genes
| Number | Annotation | CxL M/P | LxC M/P | met1 M/P | fie M/P | dme M/P | Endo exp* | Emb exp* | met1 exp | fie exp | dme exp |
| AT1G59930 m/d | PHE-related | 495/1 | 465/1 | 202/189 | 1,418/5 | 32/1 | 418/1,123 | 22/0 | 1,854 | 3,287 | 93 |
| AT2G17690 m/d | SDC | 124/1 | 119/2 | 33/87 | 111/3 | 10/0 | 59/96 | 4/2 | 186 | 66 | 8 |
| AT2G24740 # | SUVH8 | 48/0 | 50/19 | 3/1 | 248/1 | 77/0 | 31/24 | 1/0 | 19 | 178 | 40 |
| AT2G28380 m/d/f | DRB2 | 806/198 | 737/171 | 94/60 | 37/5 | 69/11 | 936/343 | 789/490 | 562 | 156 | 272 |
| AT2G34880 #/d | JMJ15 | 3/0 | 12/0 | 0/0 | 14/0 | 0/0 | 8/3 | 1/2 | 3 | 24 | 2 |
| AT4G00220 m/d/f | JLO | 666/11 | 836/15 | 116/45 | 22/1 | 36/1 | 478/186 | 33/1 | 469 | 33 | 37 |
| AT4G18650 m/d/f | TF-related | 807/89 | 92/14 | 25/72 | 2/1 | 1/0 | 434/32 | 73/3 | 416 | 6 | 4 |
| AT4G31060 m/d/f | ERF/AP2 TF | 719/9 | 229/7 | 45/62 | 0/0 | 17/0 | 498/150 | 41/25 | 472 | 2 | 13 |
| AT5G03280 m/d/f | EIN2 | 1,346/207 | 1,725/169 | 155/80 | 305/67 | 269/65 | 730/610 | 212/84 | 393 | 200 | 142 |
| AT5G35490 m/d/f | MRU1 | 43/0 | 30/0 | 4/6 | 3/2 | 0/0 | 32/16 | 0/0 | 23 | 3 | 1 |
A partial list of previously undescribed maternally expressed imprinted genes at a P < 0.001 (except #). Total maternal (M) and paternal (P) reads are shown for the indicated genotypes as well as transcriptional scores (number of reads per kilobase of sequence per 10 million aligned reads) for endosperm (Endo exp), embryo (Emb exp), and the indicated mutant genotypes. #, identified by Sanger sequencing; m, biallelic in met1 endosperm; d, down-regulated in dme endosperm; f, down-regulated in fie endosperm; TF, transcription factor.
*Transcriptional scores derived from manually dissected and LCM tissue are shown before and after the slash, respectively.
Table 2.
Paternally expressed imprinted genes
| Number | Annotation | CxL M/P | LxC M/P | met1 M/P | fie M/P | dme M/P | Endo exp* | Emb exp* | met1 exp | fie exp | dme exp |
| AT1G17770 #/F/D/M | SUVH7 | 3/5 | 5/13 | 0/0 | 368/213 | 295/254 | 3/6 | 0/0 | 0 | 159 | 121 |
| AT1G31640 F/M | AGL92 | 1/6 | 1/14 | 0/0 | 150/131 | 8/49 | 2/16 | 1/0 | 0 | 115 | 14 |
| AT1G48910 F/D/M | YUC10 | 36/70 | 30/279 | 6/5 | 124/131 | 703/372 | 78/2 | 0/0 | 15 | 194 | 651 |
| AT1G57800 F/D | VIM5 | 249/2,818 | 343/3,513 | 10/246 | 3,734/3,966 | 1,066/694 | 458/208 | 22/2 | 157 | 1,965 | 324 |
| AT1G60410 F/M | F-box | 4/33 | 6/32 | 0/0 | 552/301 | 17/160 | 8/10 | 3/0 | 0 | 351 | 56 |
| AT2G21930 F/M | F-box | 2/15 | 0/18 | 0/0 | 16/24 | 0/7 | 17/4 | 1/3 | 0 | 109 | 17 |
| AT2G36560 F/D/M | 0/55 | 3/42 | 0/0 | 826/227 | 84/334 | 8/17 | 1/0 | 0 | 263 | 78 | |
| AT4G11940 F/D | 1/6 | 1/15 | 0/0 | 124/141 | 67/36 | 3/9 | 0/0 | 1 | 183 | 54 | |
| AT5G63740 F/D | 16/34 | 16/59 | 0/0 | 11,420/12,497 | 1,110/1,391 | 24/9 | 7/0 | 0 | 15,141 | 1,159 |
A list of previously undescribed paternally expressed imprinted genes at a P < 0.001 (except #). Total maternal (M) and paternal (P) reads are shown for the indicated genotypes as well as transcriptional scores (number of reads per kilobase of sequence per 10 million aligned reads) for endosperm (Endo exp), embryo (Emb exp), and the indicated mutant genotypes. #, identified by Sanger sequencing; F, biallelic in fie endosperm; D, biallelic in dme endosperm; M, paternal allele down-regulated in met1 endosperm.
*Transcriptional scores derived from manually dissected and LCM tissue are shown before and after the slash, respectively.
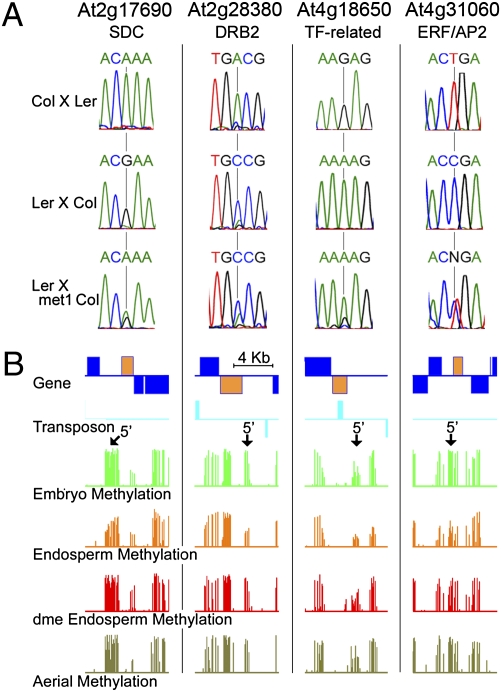
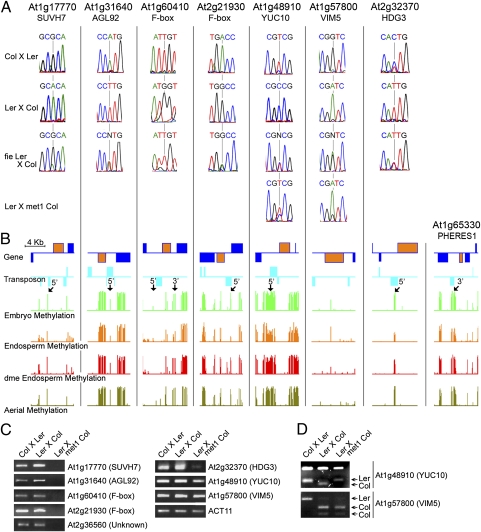
We examined allele-specific expression of 52 genes by RT-PCR followed by conventional DNA sequencing, and the results agreed closely with those obtained by sequencing cDNA libraries using the Illumina GA2 platform (Figs. 2A and 3A, Fig. S2A, and Datasets S2 and S3). In both datasets, 43 genes showed clear monoallelic expression in reciprocal crosses, and 9 genes showed clear monoallelic expression in one cross, with a greater tendency to biallelic expression in the reciprocal cross. Similar effects of ecotypes on parent of origin expression have been reported for imprinted genes in A. thaliana and mammals (22, 23). We also identified three imprinted genes with a P value that was outside our statistical selection criteria, maternally expressed SUVH8 (At2g24740) and JUMONJI C DOMAIN 15 (JMJ15/PKDM7C; At2g34880) (Table 1 and Fig. S2) and paternally expressed SUVH7 (At1g17770) (Fig. 3 and Table 2). In total, we identified 116 maternally expressed genes (Table 1 and Dataset S2), including 2 that were previously described (HDG9 and MYB3R2) (Table S1), and 10 paternally expressed genes, including previously described HDG3 (Table 2, Table S1, and Fig. S1).
Fig. 2.
Maternally expressed imprinted genes. (A) RT-PCR sequencing chromatographs at selected SNP regions measuring allele-specific expression in reciprocal crosses between Ler and Col ecotypes and in female Ler crossed to male met1-6 Col-gl. (B) CG methylation profiles in WT embryo, endosperm, aerial tissues, and dme endosperm for genes shown in A are displayed. Genes and transposable elements oriented 5′ to 3′ and 3′ to 5′ are shown above and below the line, respectively. Gene models indicated in yellow represent the imprinted genes as shown in A. Arrows indicate the 5′ end of imprinted genes where CG demethylation is seen in WT endosperm.
Fig. 3.
Paternally expressed imprinted genes. (A) RT-PCR sequencing chromatographs at selected SNP regions measuring allele-specific expression in reciprocal crosses between Ler and Col ecotypes, in female fie Ler crossed to male Col for all genes, and in female Ler crossed to male met1-6 Col-gl. (B) CG methylation profiles of genes shown in A and PHE1 are displayed. Genes and transposable elements oriented 5′ to 3′ and 3′ to 5′ are shown above and below the line, respectively. Gene models indicated in yellow color represent the imprinted genes shown in A. Arrows indicate 5′ and 3′ ends of imprinted genes where CG demethylation is detected in WT endosperm. (C) Expression analysis by semiquantitative RT-PCR in WT reciprocal crosses between Ler and Col ecotypes and in female WT Ler crossed to male met1-6 Col-gl. (D) Allele-specific expression of At1g48910 (YUC10) and At1g57800 (VIM5). RT-PCR analysis using F1 endosperm RNA isolated from Col females crossed to Ler males, Ler females crossed to Col males, and Ler females crossed to Col-gl met1-6 males. For YUC10 RT-PCR products, HpaII enzyme cuts the Ler allele into a 212- and 77-bp band, whereas the Col allele is cut into 212-, 53-, and 24-bp bands. For VIM5 RT-PCR products, enzyme BsmI cuts the Col allele but not the Ler allele.
Imprinted Expression Not Detected in Embryo.
We prepared embryo cDNA libraries from one pair of reciprocal crosses between the Col and Ler accessions, sequenced using the Illumina GA2 platform, and processed the sequencing data as we did for endosperm libraries. We identified one paternally expressed gene, VARIATION IN METHYLATION 5 (VIM5) (24), and 37 maternally expressed genes at a P value cutoff of 0.001 (Fisher's exact test) (Dataset S2). Erroneous identification of imprinted genes in embryo can be caused by contamination with endosperm or seed coat tissue, a concern highlighted by the fact that VIM5 is by far the most highly paternally expressed imprinted gene in endosperm (Table 2), and 30 of the maternally expressed genes were identified as imprinted in endosperm at a P value below 0.001 before LCM filtering (Dataset S2). To address this issue, we sequenced cDNA derived from CxL F1 LCM-isolated embryos and filtered our embryo dataset as we did for endosperm data. Only two of the putatively imprinted embryo genes (At1g70830 and At5g47150) survived this treatment, and both were discarded, because they were scored as imprinted in endosperm but filtered out as likely maternal tissue contaminants (Dataset S2). Our results indicate that imprinted gene expression is either very rare or does not occur in A. thaliana embryos, which is consistent with imprinted genes not being detected in vegetative rice tissues (25) but in contrast with extensive imprinted expression of genes in the mammalian fetus and adult (21, 22). Alternatively, imprinted genes in the embryo may be difficult to detect because of low RNA levels or expression that is restricted to specific cell types or tissues.
Effects of Paternal met1 and Maternal dme and fie Mutations on Maternally Expressed Genes.
To better understand the mechanisms underlying imprinted gene expression, we sequenced cDNA libraries from endosperm generated from crosses between Ler and Columbia glabrous (Col-gl) where the male was homozygous for the met1-6 mutation, Ler and Col where the female was heterozygous for fie-1, and Col-gl and Ler where the female was heterozygous for dme-2. Endosperm with a maternal mutant dme or fie allele was identified by its abnormal development. RNA was isolated, and libraries were constructed, sequenced, and analyzed as described for WT endosperm above.
Previous studies showed that DNA methylation silences the paternal alleles of the maternally expressed imprinted genes FWA, FIS2, and MPC, and these genes exhibit activation of the paternal allele in endosperm fertilized with met1 pollen (12–14). Consistent with this model, we identified nine maternally expressed genes that displayed biallelic expression caused by a paternally inherited met1 mutation (Fig. 2A, Table 1, Figs. S1 and S2A, and Table S1). Genes affected by met1 include transcription factors MYB3R2 and ERF/AP2 (At4g31060), At1g59930, which encodes a truncated PHE1-related MADS box transcription factor gene, and three genes known to be regulated by DNA methylation: SDC (At2g17690) and MRU1 (At5g35490), which are overexpressed in lines lacking non-CG methylation (26, 27), and At4g18650, a transcription factor gene down-regulated by mutation of the DME homolog REPRESSOR OF TRANSCRIPTIONAL GENE SILENCING 1 (ROS1) in seedlings (28). SDC encodes an F-box gene that is predicted to confer specificity to the E3 ligase complex that ubiquitylates proteins targeted for degradation by the 26S proteasome (29). Among met1-affected genes are two regulators of hormone signaling: JAGGED LATERAL ORGANS (JLO; AT4G00220), a transcription factor that affects transport of the plant hormone auxin by regulating the expression of PINFORMED auxin-efflux carrier genes (30), and ETHYLENE INSENSITIVE 2 (EIN2; At5g03280), a membrane protein crucial for perception of the gaseous hormone ethylene that is also required for proper auxin, abscisic acid, jasmonic acid, salicylic acid, and cytokinin signaling (31) (Table 1). DOUBLE-STRANDED RNA BINDING 2 (DRB2; At2g28380) is a predicted component of the small RNA pathway (32). Available microarray data (http://seedgenenetwork.net; GEO accession no. GSE12404) show that the met1-affected genes are expressed primarily in endosperm (Fig. S3A). Although small in number, many maternally expressed imprinted genes affected by met1 are likely endosperm-specific key regulators that activate or repress other genes.
Active DNA demethylation catalyzed by DME in the central cell (10) has been implicated in the activation of maternal FWA, FIS2, and MPC alleles, because their maternal allele expression is absent or reduced when a dme mutation is maternally inherited (12–14). All nine above-described genes that are biallelically expressed in met1 exhibited down-regulation of the maternal allele in dme endosperm (Table 1 and Table S1). Examination of our previously published DNA methylation data (33) revealed that these genes show DNA methylation that either overlaps or is just upstream of their transcriptional start sites (TSS) and that is reduced in endosperm in a DME-dependent manner (Fig. 2B and Fig. S2B), which is consistent with the model that DME-mediated DNA demethylation activates maternal allele expression of these genes.
JMJ15 (At2g34880) is closely related to JMJ14 (PKDM7B), which is thought to demethylate trimethylated lysine 4 of histone H3 (H3K4me3) and is involved in DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2)-mediated maintenance of DNA methylation (34). Maternal JMJ15 expression was not detected in a dme mutant, suggesting that DME is required to demethylate and activate maternal allele expression (Fig. S2C). However, the expected activation of paternal allele expression in a met1 mutant was not detected (Fig. S2A). It is possible that other MET1 homologs expressed in the endosperm silence the JMJ15 paternal allele by maintaining CG methylation in the region of its transcriptional start site (Fig. S2B).
Another important regulator that maintains the silent state of paternal alleles of imprinted genes is the maternal PRC2 complex. Previous analysis of imprinted genes that do not exhibit activation of the paternal allele in endosperm fertilized with met1 pollen (FH5 and MEA) revealed that the maternally expressed PRC2 complex silences the paternal allele (10, 11). In accordance with this idea, we identified 20 genes that exhibited activation of the paternal allele caused by a maternally inherited fie mutation (Fig. S1 and Table S2). Other than the SKP2B F-box protein (At1g77000) and two zinc-finger proteins (At1g08050 and At5g22920), most of these genes function in intermediary metabolism or signaling. Metabolism genes encode the ADS2 lipid desaturase (At2g31360), an acylphosphatase (At5g03370) that might function in glycolysis, the TPK5 potassium channel protein (At4g01840), and the FPS1 farnesyl diphosphate synthase (At5g47770) that is in the isoprenoid biosynthesis pathway. Signaling genes encode the PP2C-related protein phosphatase (At3g17250), which may negatively regulate protein kinase pathways, a phosphoinositide binding protein (At3g22810) potentially involved in lipid signaling, and the ACX1 acyl-CoA oxidase (At4g16760) that is in the jasmonate hormone biosynthesis pathway (35). Most of these genes are expressed primarily in the endosperm (Fig. S3A).
Two of twenty imprinted genes affected by fie, At1g69900 and At5g47770 (FPS1), display biallelic expression caused by a maternally inherited dme mutation (Fig. S1 and Table S2), consistent with the role of DME in activating maternal expression of the core PRC2 components FIS2 and MEA. One possible explanation for a more limited effect of dme compared with fie might be that FIE is a single copy gene required for all PRC2 complex formation (9), whereas MEA and FIS2 are members of gene families. PcG proteins related to MEA (SWINGER and CURLY LEAF) and FIS2 (VERNALIZATION 2 and EMBRYONIC FLOWER 2) are expressed in endosperm (http://seedgenenetwork.net). These proteins can interchangeably form PRC2 complexes (36) and might provide redundant PRC2 functionality in a dme mutant background.
We found that the PRC2 complex is required for proper expression levels but not imprinting of some genes (Table 1, Table S1, and Dataset S2). A good example of this is the maternal FWA allele, which is activated by DME-dependent DNA demethylation (12). In fie mutant endosperm, the maternal allele of FWA is massively overexpressed, whereas paternal expression is unaffected (Table S1). These results reveal a hierarchical control of epigenetic marks. For genes like FWA, DNA demethylation is required for activation, whereas PRC2 regulates the expression level of the activated allele.
Effects of Paternal met1 and Maternal dme and fie Mutations on Paternally Expressed Genes.
We identified nine paternally expressed genes (Fig. S1 and Table 2) that are expressed primarily in the endosperm within the seed (Fig. S3B). Many of these genes encode potential regulatory proteins (Table 2), including the transcription factor AGAMOUS LIKE 92 (AGL92), YUCCA10 (YUC10), a homolog of monooxygenase enzymes that synthesize auxin (37), and two F-box genes (At1g60410 and At2g21930). SUVH7 (At1g17770) is a SET domain protein related to SUVH4 (KRYPTONITE), SUVH5, and SUVH6 histone H3 lysine 9 (H3K9) methyltransferases required for CHROMOMETHYLASE 3 (CMT3)-mediated non-CG DNA methylation (38), and it is the closest homolog of maternally expressed SUVH8 (39). VARIATION IN METHYLATION 5 (VIM5) belongs to a protein family required for maintenance of CG methylation.
PHE1 is a paternally expressed imprinted gene that is biallelically expressed in endosperm with maternally inherited mutations in PRC2 (18, 19). Maternal demethylation of tandem repeats downstream of PHE1 is also thought to be required for maternal PHE1 silencing. This idea is supported by the observation that loss of methylation in the paternal genome because of a met1 mutation reduced expression of the paternal PHE1 allele (17, 20). Indeed, we detected DME-dependent endosperm hypomethylation of these tandem repeats (33) (Fig. 3B). Thus, the current model explaining regulation of PHE1 imprinting proposes that maternal DNA demethylation near the gene exposes a PRC2 binding site, thereby allowing PcG-mediated silencing of the maternal allele (17). Supporting this model, Weinhofer et al. (40) recently reported that DNA hypomethylation allows targeting by PcG proteins in endosperm. This model predicts that demethylation of the paternal genome by met1 should silence similarly regulated genes by exposing the paternal allele to PRC2-mediated repression, whereas a maternal fie mutation should cause biallelic expression by disabling PRC2.
The silenced maternal alleles of all nine paternally expressed imprinted genes that we identified and HDG3 are activated by a maternal fie mutation, in some cases (e.g., At5g63740) accompanied by massive overexpression of both alleles, indicating that maternal alleles of these genes are silenced by the PRC2 complex in WT endosperm (Table 2, Fig. S1, and Table S1). Paternal allele expression of seven genes (SUVH7, AGL92, At1g60410 F-box, At2g21930 F-box, YUC10, At2g36560, and HDG3) is reduced in endosperm fertilized with met1 pollen, indicating that DNA methylation is required for WT paternal allele expression and likely prevents the establishment of repressive PRC2 complexes on the paternal allele (Fig. 3C). For YUC10, the met1 mutation activates expression of the maternal allele (Fig. 3 A and D), which was also reported for PHE1 (20). These results are consistent with the model proposed for PHE1 regulation. However, the imprinted status of VIM5 is unaffected by met1 (Fig. 3 A and D and Table 2), and little DNA methylation is present at or near the VIM5 gene (Fig. 3B), suggesting that VIM5 maternal allele repression may be mediated by PRC2 independent of DNA demethylation.
As described above, a dme mutation can theoretically lead to activation of the maternal allele, because retention of DNA methylation prevents binding of the repressive PRC2 complex or PRC2 activity is compromised. Among the seven genes that show reduced paternal allele expression in a met1 mutant, we find that the dme mutation causes biallelic expression of four genes (SUVH7, YUC10, At2g36560, and HDG3) (Table 2, Table S1, and Fig. S1). For two genes (AGL92 and At1g60410 F-box), maternal allele expression is activated; however, paternal allele expression still predominates (Table 2), which may reflect the complex interactions between DNA methylation and PRC2 function that were reported for the regulation of PHE1 (20).
Maintenance of CG Methylation in Endosperm Is Carried Out by Distinct MET and VIM Genes.
The A. thaliana VIM family is comprised of five genes, of which only VIM1, VIM2, and VIM3 are expressed in leaves and flowers (24). These three genes are mostly redundant, and the corresponding triple mutant plants lack CG DNA methylation—the same phenotype as animal cells without the VIM ortholog Uhrf1 (24, 41). Uhrf1 contains an SRA (SET and RING-associated) domain that specifically binds to DNA hemimethylated at CG sites and thus, is postulated to provide the specificity that allows the MET1/Dnnmt1 methyltransferases to maintain CG DNA methylation after DNA replication (42). Although VIM5 expression is low in leaves, flowers (24), and embryos, VIM5 is the predominant VIM gene in endosperm (Table 3). Thus, the DNA methyltransferases of the MET1 family primarily depend on VIM5 expressed from the paternal allele to maintain CG DNA methylation after DNA replication in endosperm.
Table 3.
VIM and MET genes
| Number | Annotation | CxL M/P | LxC M/P | met1 M/P | fie M/P | dme M/P | Endo exp* | Emb exp* | met1 exp | fie exp | dme exp |
| AT1G57820 | VIM1 | 233/144 | 168/442 | 23/51 | 215/178 | 289/72 | 114/47 | 717/632 | 55 | 136 | 95 |
| AT1G66050 | VIM2 | 22/7 | 28/24 | 1/0 | 206/126 | 81/85 | 69/73 | 74/67 | 11 | 856 | 436 |
| AT5G39550 | VIM3 | 2/17 | 30/13 | 1/0 | 151/20 | 14/58 | 35/38 | 138/141 | 19 | 329 | 111 |
| AT1G66040 | VIM4 | NA | NA | NA | NA | NA | 50/62 | 56/53 | 8 | 678 | 380 |
| AT1G57800 | VIM5 | 249/2,818 | 343/3,513 | 10/246 | 3,734/3,966 | 1,066/694 | 458/208 | 22/2 | 157 | 1,965 | 324 |
| AT5G49160 | MET1 | 1/0 | 2/1 | 2/0 | 7/0 | 6/6 | 87/87 | 327/375 | 34 | 264 | 287 |
| AT4G08990 | MET2 | 93/13 | 60/55 | 1/1 | 141/81 | 326/81 | 36/78 | 6/4 | 3 | 136 | 130 |
| AT4G13610 | MET3 | 12/17 | 59/11 | 2/0 | 1,204/461 | 115/479 | 13/19 | 1/1 | 1 | 434 | 121 |
| AT4G14140 | MET4 | 176/72 | 199/117 | 11/14 | 619/344 | 681/222 | 66/205 | 7/5 | 29 | 261 | 172 |
A list of all A. thaliana VIM and MET genes. Total maternal (M) and paternal (P) reads are shown for the indicated genotypes, as well as transcriptional scores (number of reads per kb of sequence per 10 million aligned reads) for endosperm (Endo exp), embryo (Emb exp), and the indicated mutant genotypes.
*Transcriptional scores derived from manually-dissected and LCM tissue are shown before and after the slash (/), respectively.
In addition to VIM5 imprinting, the expression of MET1 (At5g49160) and its three A. thaliana homologs (At4g08990, At4g13610, and At4g14140) is altered in endosperm. In embryo, MET1 is by far the predominantly expressed maintenance methyltransferase gene, with expression levels about 50-fold higher than those of its homologs (Table 3). This pattern is also seen in vegetative and floral tissues (43). By comparison, although none of the MET genes seem to be imprinted in endosperm, MET1 is down-regulated, whereas the other three MET genes are up-regulated relative to their expression in embryo (Table 3). Thus, maintenance of CG methylation in endosperm is carried out by distinct MET and VIM gene family members.
Maternally Expressed Genes That Are Not Affected by Paternal met1 and Maternal dme and fie Mutations.
Among the 116 maternally expressed genes that we analyzed, we only scored 35 as affected by mutations in met1, dme, or fie (Fig. S1, Dataset S2, and SI Methods). Many of the remaining genes show significant expression in seed coat (Fig. S3C), raising the possibility that some of these are false positives. However, even in cDNA from CxL LCM-derived endosperm, some of these genes show clear maternal expression, including ARGONAUTE 9 (AGO9; At5g21150), a protein kinase (At1g29730), and HDG9 (Fig. S4), indicating that these genes are either imprinted or deposited from maternal tissues.
That a gene did not meet our statistical significance criteria does not necessarily prove that its imprinting is not affected by met1, dme, or fie. For this reason, we used a more stringent cutoff—a gene had to be expressed at least 16-fold higher from the maternal than paternal genome in met1, dme, and fie—to identify 19 genes that were still clearly maternally expressed in all three mutant lines (Fig. S1). These genes include AGO3 (At1g31290), a MYB transcription factor gene (At3g10590), ARABIDOPSIS SKP1-LIKE E3-ligase component genes ASK8 (At3g21830) and ASK10 (At3g21860), and cytidine deaminase genes (At4g29570 and At4g29640) (Table 4 and Dataset S2). Seed microarray data (GEO accession no. GSE12404) are available for 16 of these genes, of which seven are expressed only in chalazal seed coat and chalazal endosperm (Table 4 and Figs. S1 and S5A). Only 26 of 22,533 genes on the array, including the seven genes mentioned above, have this pattern of expression (P = 3.6 × 10−16; Fisher's exact test) (Dataset S2). This group includes ASK7 (At3g21840) and three more cytidine deaminase genes (At4g29580, At4g29600, and At4g29620), which show a clear bias to maternal allele expression in our sequenced cDNA libraries (Dataset S2). The remaining genes all either show a maternal bias, have no SNPs between Col and Ler, or had no reads detected because of the low abundance of their transcripts in endosperm (Dataset S2). Chalaza is an active site of nutrient transfer from maternal seed coat to the developing endosperm (8), suggesting that the mRNA for these genes might be synthesized in the maternal chalazal tissues and transported into the chalazal endosperm. As a control, we identified 48 genes from the microarray dataset with expression only in the chalazal seed coat (Dataset S2 and Fig. S5B), none of which are on our list (LCM-filtered; P < 0.001) of maternally expressed imprinted genes (P = 0.0004; Fisher's exact test) (Dataset S2), indicating that seed coat contamination is unlikely to account for our results. To further rule out contamination, we analyzed expression of four genes (AGO3, MYB transcription factor, ASK8, and ASK10) by RT-PCR in CxL LCM-derived endosperm, and all four are clearly maternally expressed (Fig. S4). A mechanism of gene imprinting that does not require MET1, FIE, or DME is also consistent with our data, but the characteristic mRNA profile makes transport much more likely.
Table 4.
Selected maternally expressed genes unaffected by met1, fie, or dme
| Number | Annotation | CxL M/P | LxC M/P | met1 M/P | fie M/P | dme M/P | Endo exp* | Emb exp* | met1 exp | fie exp | dme exp |
| AT1G31290 | AGO3 | 324/33 | 280/22 | 177/3 | 199/9 | 562/7 | 160/169 | 7/0 | 294 | 181 | 508 |
| AT1G61090 | 244/0 | 419/2 | 373/2 | 187/1 | 629/0 | 125/127 | 0/0 | 639 | 161 | 375 | |
| AT3G10590 | MYB TF | 47/3 | 69/3 | 59/0 | 59/1 | 115/0 | 62/57 | 0/0 | 279 | 118 | 178 |
| AT3G21830 | ASK8 | 252/1 | 318/1 | 220/1 | 228/3 | 723/1 | 149/96 | 0/0 | 507 | 264 | 651 |
| AT3G21860 | ASK10 | 34/0 | 99/0 | 49/0 | 77/0 | 258/0 | 34/48 | 1/0 | 69 | 50 | 353 |
| AT4G29570 | CDA | 34/4 | 82/5 | 60/1 | 98/1 | 150/0 | 49/75 | 0/0 | 221 | 119 | 178 |
| AT4G29640 | CDA | 32/8 | 127/14 | 109/2 | 49/0 | 44/0 | 87/73 | 2/0 | 368 | 146 | 155 |
A list of maternally expressed genes, the imprinted status of which is not affected by met1, fie, or dme, that are expressed only in chalazal endosperm and seed coat. Total maternal (M) and paternal (P) reads are shown for the indicated genotypes as well as transcriptional scores (number of reads per kilobase of sequence per 10 million aligned reads) for endosperm (Endo exp), embryo (Emb exp), and the indicated mutant genotypes; TF, transcription factor; CDA, cytidine deaminase.
*Transcriptional scores derived from manually dissected and LCM tissue are shown before and after the slash, respectively.
Discussion
Our study has significantly expanded the number of known genes with parent of origin-specific expression in A. thaliana endosperm. We estimate (based on SNP availability and sequencing depth) that our dataset is sufficiently deep to ascertain the imprinted status of 10,755 endosperm genes or roughly one-half of the endosperm transcriptome, assuming about two-thirds of the 28,244 A. thaliana genes that we examined are expressed in endosperm (Fig. S1). However, taking into account our rather stringent statistical cutoff (P < 0.001) and filtering using LCM data, this estimate should be revised closer to 20–30% of the endosperm transcriptome. Consistent with this fraction, only 3 of 10 previously described imprinted genes (not counting FH5) passed all of our filters (Table S1). Thus, there may be 30–50 paternally expressed endosperm genes, about 200 maternally expressed genes regulated by DNA methylation or PcG activity, and potentially over 500 maternally biased genes if genes regulated by unknown mechanisms or deposited from maternal tissues are considered. Allele-specific gene expression is clearly a major phenomenon in plant endosperm that is comparable with the extensive imprinting recently reported in mouse brain (22).
Parental Conflict May Occur at Many Regulatory Levels.
The parental conflict theory (1) proposes that nutrient allocation is the driving force for the evolution of gene imprinting in mammals and plants. Although the effect on nutrient allocation of the imprinted genes described here is not yet known, the potential lines of conflict between maternal and paternal parents have significantly expanded. At the chromatin level, in addition to the previously discovered maternally expressed PRC2, paternally expressed proteins potentially silence target genes by promoting maintenance DNA methylation (VIM5) and H3K9 methylation (SUVH7), and maternally expressed genes potentially silence targets by regulating the small RNA pathway (DRB2), H3K9 methylation (SUVH8), H3K4 demethylation, and DRM2-mediated DNA methylation (JMJ15). Parental conflict may occur at the posttranslational level, mediated by degradation of specific proteins through the ubiquitin-26S proteasome system, which rivals transcription as a dominant regulatory mechanism in A. thaliana (29). Parental conflict may also take place through protein–protein interactions. At1g59930, a maternally expressed imprinted gene, encodes a truncated MADS box transcription factor that lacks the MADS box domain. Although it is unlikely to bind DNA, this protein may inhibit other MADS box transcription factors through dimerization (44), including the activity of a close full-length relative, the paternally expressed PHE1. Imprinting of hormone synthesis (YUC10 and ACX1) and response (JLO and EIN2) genes suggests that hormone action may also be involved in parental conflict.
Imprinted Genes Regulated by DNA Methylation Often Encode Regulatory Proteins.
Imprinted expression of genes with regulatory potential is frequently regulated by DNA methylation, whereas the PRC2 complex regulates imprinting of genes that participate in cellular metabolism and signaling (Tables 1 and 2, Dataset S2, and Table S2). Polycomb group proteins function in maintaining rather than establishing the silent state (45). It is possible that DNA demethylation in the central cell initially imprints genes encoding regulatory proteins that, in turn, activate or repress other genes, the transcriptional states of which are cemented by PRC2 activity. If so, mutations in imprinted genes directly regulated by DNA methylation would be predicted to affect the transcriptional status of other imprinted genes, particularly those dependent on PRC2.
Genes Not Regulated by DME, FIE, or MET1.
A subset of maternally expressed genes (ASK, cytidine deaminases, and AGO3) do not seem to be regulated by DME, FIE, or MET1. It is possible that their imprinted expression is regulated by an unknown mechanism for paternal allele silencing. Alternatively, because their mRNA is detected only in chalazal seed coat and chalazal endosperm (Fig. S5A), the mRNA for these genes might be synthesized in the chalazal seed coat and transported into the chalazal endosperm. If this is the case, these genes would represent an additional mechanism by which the maternal parent genetically controls seed development. There is evidence for intercellular movement of plant RNAs through plasmodesmata (46), although it remains to be experimentally tested whether RNAs can navigate the apoplastic pathways that connect the chalazal seed coat and endosperm (47). The three ASK genes and five cytidine deaminase genes are organized in tight clusters and likely represent recent duplication events, which might serve to coordinate this expression pattern. Such clustering was not detected among the other A. thaliana imprinted genes in this study, confirming that plant genes, in contrast to mammalian imprinted genes, are singletons (1). Transport of AGO9 RNA (itself maternally biased in endosperm) (Dataset S2) from somatic companion cells mediates small RNA-dependent transposon silencing necessary for specification of gametophyte precursor cells (48), and maternally deposited AGO3 may likewise influence transposon silencing in the endosperm. Although the significance of converting cytidine to uridine by cytoplasmic cytidine deaminase proteins in the chalazal endosperm is not clear, picomolar concentrations of uridine enhance cell division responses to auxin and cytokinin in root cortical cells (49).
Control of Genome Hypomethylation in the Endosperm by DME, VIM, and MET Genes.
We previously showed that virtually the entire A. thaliana endosperm genome is demethylated in the CG context and that this demethylation is largely dependent on DME (33). Here, we show that the VIM5 gene is primarily expressed from the paternal genome, MET1 is down-regulated in endosperm (as has been previously shown in the female gametophyte), and VIM and MET genes are up-regulated in dme-deficient and fie-deficient endosperm (Table 3). These results suggest that CG hypomethylation in the central cell and endosperm might be orchestrated by regulation of VIM and MET genes in addition to direct DME activity. To affect CG methylation in the central cell, VIM and MET genes would have to be down-regulated before central cell differentiation to allow for passive demethylation by DNA replication. As DME is expressed specifically in the central cell, the initial down-regulation would not require DME. However, maintenance of VIM and MET repression, which might be necessary for genome-wide hypomethylation, would require a functional PRC2 complex (Table 3), which, in turn, requires DME. Thus, one possible model would be that DME directly demethylates a number of discrete loci, whereas global demethylation is caused, at least in part, by DME-dependent VIM and MET repression. This hypothesis predicts that WT global demethylation should be dependent on PRC2 activity. This prediction is consistent with our observation that some maternally expressed genes apparently activated by removal of DNA methylation (i.e., paternal allele up-regulated by met1 and maternal allele down-regulated by dme) are down-regulated in fie endosperm (Table 1).
Methods
Illumina cDNA libraries were constructed with the Ovation RNA-seq System (NuGen Technologies) using total RNA isolated from manually or LCM-dissected seeds 7–8 d after pollination. Each gene received Col and Ler scores, each equal to the number of reads aligned to that gene that were assigned to the respective ecotype. Each gene also received a transcriptional score, equal to the number of reads aligned to the cDNA model (irrespective of ecotype) per 1 kb of sequence per 10 million aligned reads. We calculated the probability that a gene's expression deviates from expectation or that a gene's imprinted status is altered by mutation using Fisher's two-tailed exact test. SI Methods has details.
Supplementary Material
Acknowledgments
We thank Leath Tonkin for performing Illumina sequencing. This work was funded by National Science Foundation Grants DBI-0501720 (to J.J.H.) and MCB 0918821 (to D.Z. and R.L.F.) and National Institutes of Health Grant R01-GM069415 (to R.L.F.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE24644).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019273108/-/DCSupplemental.
References
- 1.Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Haig D, Westoby M. Parent-specific gene expression and the triploid endosperm. American Naturalist. 1989;134:147–155. [Google Scholar]
- 3.Moore T, Haig D. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- 4.Huh JH, Bauer MJ, Hsieh T-F, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–744. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Berger F, Chaudhury A. Parental memories shape seeds. Trends Plant Sci. 2009;14:550–556. doi: 10.1016/j.tplants.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol. 2009;19:1677–1681. doi: 10.1016/j.cub.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen H, Brown RC, Lemmon BE. The specialized chalazal endosperm in Arabidopsis thaliana and Lepidium virginicum (Brassicaceae) Protoplasma. 2000;212:99–110. [Google Scholar]
- 8.Ingram GC. Family life at close quarters: Communication and constraint in angiosperm seed development. Protoplasma. 2010;247:195–214. doi: 10.1007/s00709-010-0184-y. [DOI] [PubMed] [Google Scholar]
- 9.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: Same rules, different players? Trends Genet. 2009;25:414–423. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitz Gerald JN, Hui PS, Berger F. Polycomb group-dependent imprinting of the actin regulator AtFH5 regulates morphogenesis in Arabidopsis thaliana. Development. 2009;136:3399–3404. doi: 10.1242/dev.036921. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303:521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 13.Jullien PE, Kinoshita T, Ohad N, Berger F. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell. 2006;18:1360–1372. doi: 10.1105/tpc.106.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tiwari S, et al. MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell. 2008;20:2387–2398. doi: 10.1105/tpc.108.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jullien PE, et al. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villar CBR, Erilova A, Makarevich G, Trösch R, Köhler C. Control of PHERES1 imprinting in Arabidopsis by direct tandem repeats. Mol Plant. 2009;2:654–660. doi: 10.1093/mp/ssp014. [DOI] [PubMed] [Google Scholar]
- 18.Köhler C, et al. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 2003;17:1540–1553. doi: 10.1101/gad.257403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler C, Page DR, Gagliardini V, Grossniklaus U. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet. 2005;37:28–30. doi: 10.1038/ng1495. [DOI] [PubMed] [Google Scholar]
- 20.Makarevich G, Villar CBR, Erilova A, Köhler C. Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci. 2008;121:906–912. doi: 10.1242/jcs.023077. [DOI] [PubMed] [Google Scholar]
- 21.Constância M, Kelsey G, Reik W. Resourceful imprinting. Nature. 2004;432:53–57. doi: 10.1038/432053a. [DOI] [PubMed] [Google Scholar]
- 22.Gregg C, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinoshita T, Yadegari R, Harada JJ, Goldberg RB, Fischer RL. Imprinting of the MEDEA polycomb gene in the Arabidopsis endosperm. Plant Cell. 1999;11:1945–1952. doi: 10.1105/tpc.11.10.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo HR, Dittmer TA, Richards EJ. Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 2008;4:e1000156. doi: 10.1371/journal.pgen.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson IR, Jacobsen SE. Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading. Genes Dev. 2008;22:1597–1606. doi: 10.1101/gad.1667808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurihara Y, et al. Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem Biophys Res Commun. 2008;376:553–557. doi: 10.1016/j.bbrc.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 29.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 30.Bureau M, Rast MI, Illmer J, Simon R. JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol Biol. 2010;74:479–491. doi: 10.1007/s11103-010-9688-2. [DOI] [PubMed] [Google Scholar]
- 31.Stepanova AN, Alonso JM. Ethylene signaling and response: Where different regulatory modules meet. Curr Opin Plant Biol. 2009;12:548–555. doi: 10.1016/j.pbi.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleris A, et al. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep. 2010;11:950–955. doi: 10.1038/embor.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schilmiller AL, Koo AJK, Howe GA. Functional diversification of acyl-coenzyme A oxidases in jasmonic acid biosynthesis and action. Plant Physiol. 2007;143:812–824. doi: 10.1104/pp.106.092916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:13244–13249. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, et al. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 38.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baumbusch LO, et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinhofer I, Hehenberger E, Roszak P, Hennig L, Köhler C. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 2010;6:e1001152. doi: 10.1371/journal.pgen.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraft E, Bostick M, Jacobsen SE, Callis J. ORTH/VIM proteins that regulate DNA methylation are functional ubiquitin E3 ligases. Plant J. 2008;56:704–715. doi: 10.1111/j.1365-313X.2008.03631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genger RK, Kovac KA, Dennis ES, Peacock WJ, Finnegan EJ. Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol Biol. 1999;41:269–278. doi: 10.1023/a:1006347010369. [DOI] [PubMed] [Google Scholar]
- 44.de Folter S, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Kehr J, Buhtz A. Long distance transport and movement of RNA through the phloem. J Exp Bot. 2008;59:85–92. doi: 10.1093/jxb/erm176. [DOI] [PubMed] [Google Scholar]
- 47.Stadler R, Lauterbach C, Sauer N. Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 2005;139:701–712. doi: 10.1104/pp.105.065607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olmedo-Monfil V, et al. Control of female gametophyte formation by a small RNA pathway in Arabidopsis. Nature. 2009;464:628–632. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smit G, et al. Uridine, a cell division factor in pea roots. Plant Mol Biol. 1995;29:869–873. doi: 10.1007/BF00041176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.