Abstract
Internal ribosome entry site (IRES) RNAs are elements of viral or cellular mRNAs that bypass steps of canonical eukaryotic cap-dependent translation initiation. Understanding of the structural basis of IRES mechanisms is limited, partially due to a lack of high-resolution structures of IRES RNAs bound to their cellular targets. Prompted by the universal phylogenetic conservation of the ribosomal P site, we solved the crystal structures of proposed P site binding domains from two intergenic region IRES RNAs bound to bacterial 70S ribosomes. The structures show that these IRES domains nearly perfectly mimic a tRNA•mRNA interaction. However, there are clear differences in the global shape and position of this IRES domain in the intersubunit space compared to those of tRNA, supporting a mechanism for IRES action that invokes hybrid state mimicry to drive a noncanonical mode of translocation. These structures suggest how relatively small structured RNAs can manipulate complex biological machines.
Keywords: ribosome structure, RNA structure, tRNA mimicry
The translation machinery is remarkably conserved among all organisms, but initiation of protein synthesis differs dramatically between eukarya and bacteria. Bacteria use only three initiation factor proteins, and recruitment of the ribosome to the mRNA is achieved largely through base pairing to the rRNA (1). In contrast, there are two known mechanisms by which translation is initiated in eukaryotes. The first is the canonical cap-dependent mechanism that is used by the vast majority of eukaryotic mRNAs, which requires an m7G cap at the 5′ end of the mRNA, initiator Met-tRNAmet, more than a dozen initiation factor proteins, directional scanning, and GTP hydrolysis to place a translationally competent ribosome at the start codon (Fig. 1A) (2). Hence, canonical eukaryotic translation initiation is essentially a protein-driven process of considerable complexity. The second mechanism is cap-independent initiation that is used by some mRNAs as well as many eukaryote-infecting viruses. This mechanism bypasses the need for the cap and often many of the protein factors, using cis-acting RNA elements called internal ribosome entry sites (IRESs) to recruit the ribosome and initiate protein synthesis (3). For some IRESs, the number of required protein factors is small and initiation from these IRESs is essentially RNA-driven. Many viruses of medical and economic importance use an IRES, including poliovirus, hepatitis A virus, hepatitis C virus, foot-and-mouth-disease virus, human immunodeficiency virus-1, and many others. There is great diversity among viral IRES RNAs in terms of their sequences, proposed secondary structures, and functional requirements for protein factors, but all drive a mode of translation initiation that depends on specific RNA sequences and likely specific RNA structures in the IRES (4). IRES RNA structures and their mechanisms are potential targets for new antiviral therapeutics, but this goal requires more insight into the detailed structure-based mechanisms of IRES function than currently exists.
Fig. 1.
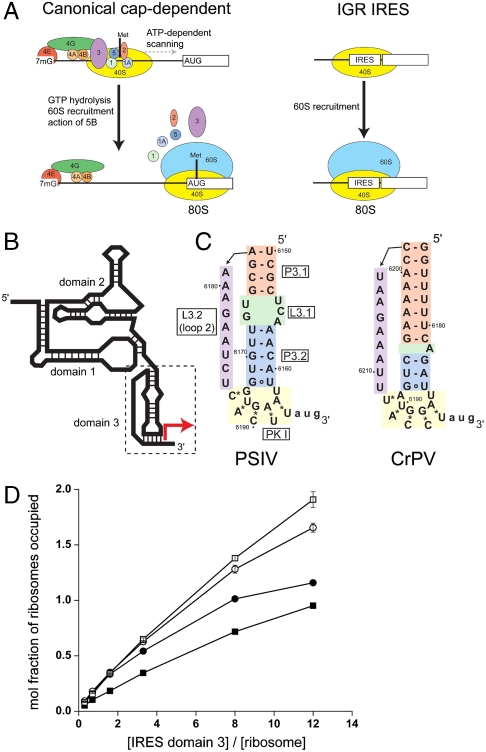
Translation initiation by the IGR IRESs and binding of domain 3 to the ribosome. (A) Comparison of canonical eukaryotic cap-dependent and IGR IRES-driven 80S ribosome recruitment. The IGR IRES does not use any protein factors or GTP hydrolysis. Protein initiation factors and the ribosomal subunits are depicted as colored ovals and labeled, and the initiator Met-tRNAmet is shown as a black vertical line. For clarity, an exhaustive list of factors involved in canonical initiation is not shown. (B) Secondary structure cartoon of the IGR IRES RNAs. The locations of the three domains that comprise the structure are shown, domain 3 is boxed, and the location of the start of protein synthesis is shown with a red arrow. (C) Secondary structures of the PSIV and CrPV IGR IRES domain 3 RNAs used in binding and crystallization studies. Elements of the structures are colored and labeled. The 3′-most nucleotides (lowercase) were mutated. (D) Binding of the PSIV (•,◯) and CrPV (▪,□) domain 3 RNAs shown in panel C to (•,▪) 70S or (◯,□) 80S ribosomes. See also Figs. S1 and S2.
The diversity of IRES structures and potential mechanisms of action demands that model systems be used to understand some basic tenets of this type of translation initiation. Useful models are the Dicistroviridae intergenic region (IGR) IRESs, which use the most streamlined IRES mechanism known (5). The IGR IRESs recruit both the large and small ribosome subunits and assemble ribosomes without tRNA, initiation factors, or GTP hydrolysis (Fig. 1A) (5, 6). IGR IRES RNAs bind directly to host cell ribosomes, initiate translation of the downstream message using a non-AUG codon, and induce translocation before a peptide bond is made (7–17). Thus, the IGR IRESs are direct manipulators of the translation machinery, and they show how an RNA can drive its own translation using direct ribosome recruitment without protein intermediaries, reminiscent of an RNA world. The IGR IRESs are therefore a powerful model system to show how structured RNAs can manipulate cellular machines, to understand mechanisms of viral IRESs, and to observe how the normally complicated and multistep process of recruiting and activating a eukaryotic ribosome can be reduced to a few steps, revealing core features of ribosome function.
The IGR IRES mechanism depends on the three-dimensional folded structure of the IRES RNA. The IGR IRES RNAs fold before encountering the ribosome into a conformation with three structural domains: Domains 1 and 2 fold together and are important for initial recruitment of the ribosome (14, 17, 18), whereas domain 3 is implicated in correct positioning of the IRES-containing viral RNA with respect to the ribosomal reading frame (Fig. 1B) (14). Despite some sequence differences and secondary structure variations, the members of the IGR IRES family fold into similar three-dimensional architectures (19). The crystal structures of the unbound domains 1 and 2 of the Plautia stali intestine virus (PSIV) IRES (20) and domain 3 of the cricket paralysis virus (CrPV) IRES (21) have been solved by X-ray crystallography (Fig. S1). These structures, when combined with biochemical and functional data, lend insight into the structural basis of IRES function.
An important part of IGR IRES function is placement of the coding portion of the viral RNA in the ribosome’s decoding groove and establishment of the reading frame. It has been proposed that this is accomplished by occupation of the P site of the small subunit by pseudoknot I (PK I) of IGR IRES domain 3, based on toeprinting experiments (9, 13). Functional studies have suggested that PK I of domain 3 has some tRNA-like characteristics (22). Indeed, the crystal structure of unbound domain 3 from CrPV supports the proposal that PK I mimics an initiator tRNA anticodon stem-loop (ASL) base-paired to its cognate start codon in the small subunit P site (21), and that docking of PK I into the P site places the next (non-AUG) codon into the A site. However, it has not been possible to test this proposal crystallographically, largely because of the unavailability of suitable crystals of eukaryotic ribosomes. A 7.3-Å cryo-EM reconstruction provided a low-resolution electron density map and model of the CrPV IRES bound to an 80S ribosome (23), but could not reveal details of IRES-ribosome interactions.
We developed a strategy to explore crystallographically the detailed interactions of IGR IRES domain 3 with the ribosome. We reasoned that the IGR IRES domain 3 might interact with bacterial 70S ribosomes in the same way that it interacts with eukaryotic 80S ribosomes, because features that comprise the main structural determinants of the ribosomal P site are universally phylogenetically conserved (24–26). Thus, an IGR IRES domain 3•70S ribosome complex could be used to study atomic-resolution details of IRES-ribosome interactions. Using this strategy, we solved the structure of domain 3 RNAs from both the PSIV and CrPV IGR IRESs bound to Thermus thermophilus 70S ribosomes by X-ray crystallography to resolutions of 3.5 and 3.4 Å, respectively. These structures of an IRES RNA domain bound to a ribosome reveal details of ribosome-IRES interactions and suggest how the IRES may enable noncanonical translocation during initiation.
Results
To test whether isolated IGR IRES domain 3 RNA is able to bind to 70S ribosomes, we used a filter-binding assay with in vitro transcribed and purified RNA. The RNA constructs contained just domain 3 from the PSIV and CrPV IGR IRES, with three nucleotides following PK I (Fig. 1C). Domain 3 RNAs from both PSIV and CrPV bind to T. thermophilus 70S ribosomes with Kd = ∼ 0.7 μM, ∼2-fold weaker than their affinity for 80S ribosomes (Fig. 1D). Previous reports have implicated domains 1 and 2 as the parts of the IGR IRES primarily responsible for ribosome recruitment (14, 17, 18); our assays indicate that domain 3 by itself can bind to ribosomes, but at a much lower affinity than does the full-length IRES (∼2–10 nM) (18). Binding of the domain 3 RNA to 70S ribosomes was reduced by competitor tRNA, suggesting that the IRES domain occupies a tRNA binding site (Fig. S2).
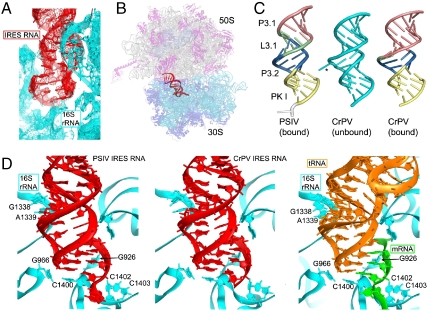
Based on the results of the binding assays, we cocrystallized domain 3 from the PSIV and CrPV IRES RNAs (Fig. 1C) with T. thermophilus 70S ribosomes and solved their structures to 3.5- and 3.4-Å resolution, respectively, by X-ray crystallography. Phases were obtained using molecular replacement with the vacant 70S ribosome (i.e., a ribosome containing no bound mRNA or tRNA) as a search model. In both structures, domain 3 electron density occupies the 30S subunit P site (Fig. 2A and Fig. S3). The domain 3 RNA structures were built into the density and the structures of the 70S complexes were refined to R/Rfree values of 0.23/0.26 and 0.23/0.27, respectively (Fig. 2B and Table S1). The structures of the IRES domains from CrPV and PSIV are very similar and make identical contacts to the ribosome. Domain 3 elements helix P3.1, loop L3.1, helix P3.2, and PK I (Fig. 1C) all are represented by strong electron density, but the nine nucleotides of loop L3.2 are disordered in both structures. P3.1 of the IRES RNA stacks coaxially on the distal end of P3.2, forming an extended A-form RNA helix (Fig. 2C). Electron density for the three nucleotides that follow PK I, which are expected to correspond to the A site codon, is weak. This codon is likely disordered because of the absence of an A site tRNA, as is also observed in ribosome structures bound with conventional mRNAs (27, 30, 31).
Fig. 2.
Structures of the PSIV and CrPV IGR IRES domain 3 RNAs bound to the T. thermophilus 70S ribosome. (A) σA-weighted 3Fo-2Fc electron density (contoured at 1.5σ of the PSIV domain 3 in the P site of the small subunit). (B) Placement of the PSIV domain 3 (red) within the ribosome structure. The ribosome is shown semitransparent to allow visualization of the IRES domain between the two subunits, which are labeled. (C) Comparison of the bound PSIV and CrPV domain 3 structures (Left and Right, colored and labeled to match Fig. 1C) with the previously determined unbound CrPV domain 3 structure (Middle, cyan) (21). (D, Left and Middle) Interactions made between the PSIV and CrPV IRES domains (red) and the 16S rRNA (cyan). At right are the interactions between the 16S rRNA (cyan) and a P site-bound tRNA (orange, Right) paired to its cognate codon within a mRNA (green) (PDB ID code 2J00) (27–29). Specific rRNA nucleotides listed in Table 1 are labeled. See also Figs. S3 and S4.
To determine the degree to which docking into the ribosomal P site alters the structure of domain 3, we compared the structure of unbound domain 3 to the bound structure solved here. The structures of the unbound CrPV domain 3 (21) and the ribosome-bound CrPV or PSIV domain 3 RNA are similar but not identical (Fig. 2C). In both structures, PK I mimics a tRNA ASL-mRNA codon structure, and in both, helices P3.2 and P3.1 are stacked coaxially. In the unbound structure, ordering of L3.2 was induced by crystal contacts (21), whereas in the bound structures this loop is disordered with no visible density, revealing that it is not directly involved in interactions with the ribosome and is thus unlikely to play a critical role in this step of initiation. The lack of stable structure in L3.2 contrasts with structural studies of other RNA pseudoknots, where the bridging loop interacts with the minor groove of the stacked helices (32). The fact that L3.2 is unstructured in the bound forms of both CrPV and PSIV domain 3 suggests that this disorder is conserved and might have a functional role. Another difference is that A6182 of the CrPV IRES bulges out from the helix in the unbound structure (forming a crystal contact) but in the bound structure is stacked between P3.1 and P3.2 (Fig. 2C and Fig. S4); likewise, all of the nucleotides in loop 3.1 in the bound PSIV domain 3 are stacked into the extended helix. The overall similarity between the structures of the bound and unbound domain 3 RNAs and across viral species shows that domain 3 prefolds prior to binding to the ribosome.
The crystal structures of the ribosome-bound domains from CrPV and PSIV allow us to examine the molecular details of the interactions between an IRES RNA and the ribosome. In both RNAs, PK I is a near-perfect mimic of a tRNA bound to its cognate mRNA codon in the 30S subunit P site, forming contacts with the 30S subunit that are virtually identical to those in a canonical tRNA-mRNA-containing preinitiation complex. The IRES domain contacts 16S rRNA bases G926, G966, G1338, A1339, C1400, C1402, and C1403, all of the bases previously found to position tRNA and mRNA in the 30S subunit P site (Fig. 2D and Table 1) (27, 28, 30). This suggests that the requirement for an initiator tRNA in the P site can be overcome by an IRES domain acting as a precise and accurate structural mimic (9, 16, 22). Although our structures represent a heterologous interaction between a bacterial ribosome and a eukaryotic IRES domain, the mode of binding to the small subunit of the 70S ribosome is likely identical to that of the eukaryotic ribosome, for two reasons. First, structural elements of the small subunit P site are composed almost exclusively of 16S or 18S rRNA and are virtually identical between 30S and 40S subunits (24–26, 33). Six of the seven 16S rRNA bases that contact the tRNA or domain 3 are identical in 18S rRNA (Table 1); the single base that differs stacks against the ribose of the wobble nucleotide 34 of tRNA and thus is not base-specific. Second, eukaryotic tRNAs function well in bacterial translation systems, demonstrating that their corresponding binding sites share close structural and functional similarities (35–38). Thus, the interactions observed in these domain 3•70S ribosome structures are likely to be identical to those formed in complexes between the IRES RNA and 80S ribosomes.
Table 1.
Interactions of IRES RNA, tRNA, and mRNA with 16S rRNA bases in the 30S subunit P site
| 16S rRNA | 18S rRNA* | PSIV IRES | mRNA† | tRNA† | Contact |
| G926 | G1207 | C6190 | A1 | Phosphate | |
| G966 | U1249 | A6163 | C34 | Ribose | |
| G1338 | G1639 | U6171 | C41 | A minor | |
| A1339 | A1640 | G6170 | G30 | A minor | |
| C1400 | C1701 | A6163 | C34 | Stacking | |
| C1402 | C1703 | U6192 | G3 | Phosphate | |
| C1403 | C1704 | U6192 | G3 | Phosphate |
*Homologous nucleotides in corresponding positions of eukaryotic 18S rRNA (34). Bases that are identical between the bacterial and human rRNAs are shown in boldface. The numbering of eukaryotic rRNA is according to the human 18S rRNA sequence.
†Corresponding contacts between mRNA, P site tRNA, and 16S rRNA are taken from ref. 27.
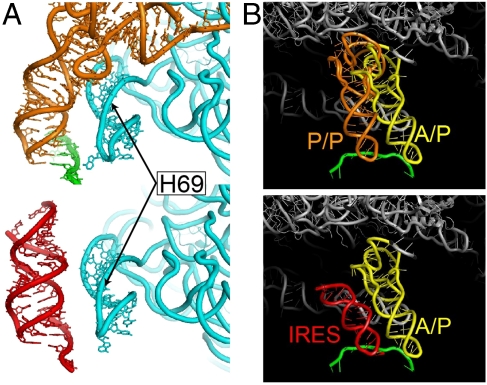
Although the contacts between the domain 3 RNA and the ribosome are identical to those of a tRNA ASL, the structure of the IRES RNA diverges from that of tRNA outside the small subunit P site, with potential mechanistic significance. Specifically, the coaxially stacked P3.1/P3.2 stem resembles an extended A-form helix and thus diverges from the trajectory of the anticodon arm of a P site-bound tRNA, which is kinked by about 25° at the anticodon stem-D stem junction. As a result, whereas the D stem of P site tRNA contacts helix 69 of 23S rRNA (27, 28, 39), domain 3 makes no such interaction (Fig. 3A). The closest approach of domain 3 to any element of the large subunit is between phosphate 6155 of the IRES domain (PSIV numbering) and phosphate 1923 of helix 69 of 23S rRNA (∼4 Å). The orientation of domain 3 directs the connecting elements of the IRES toward the E site of the large subunit, but avoids any contacts with the 50S subunit P site (Fig. 3A).
Fig. 3.
Different trajectories of the IRES domain and tRNA. (A) Comparison of the interactions formed between a P site tRNA (Top, orange) and rRNA helix H69 (labeled), and the lack of interactions formed between the bound PSIV IRES domain 3 (red, Bottom) and H69. See also Fig. S4. (B, Top) Structure of a P/P state tRNA (orange), and mRNA (green) in the decoding groove of the 70S ribosome (gray) (28), showing a steric clash with a hybrid A/P state tRNA modeled into position (yellow). (Bottom) Our structure of the PSIV IRES domain 3 (red) in the P site, with a mRNA (green) and a modeled hybrid A/P state tRNA (yellow). The IRES domain would not sterically hinder movement of the A/A tRNA to an A/P hybrid state.
The position of domain 3 in the intersubunit space is consistent with the possibility that the IGR IRES promotes formation of the hybrid state, allowing translocation before a peptide bond is made. During translocation, movement of the tRNAs into the A/P and P/E hybrid states is accompanied by intersubunit rotational movement of the ribosome (40, 41). To avoid steric clash, the A site tRNA can enter the A/P hybrid state only if the P site tRNA moves into the P/E hybrid state. In our complex, the ribosome is in the nonrotated (classical) state and so the observed orientation of the IRES toward the E site might serve to avoid potential steric clash that would result from movement of the aminoacyl-tRNA into the A/P hybrid state prior to intersubunit rotation (Fig. 3B). Thus, within an IGR IRES-ribosome complex, the A site tRNA could move into the A/P hybrid state prior to peptide bond formation, promoting noncanonical translocation (42).
Discussion
The IGR IRESs bind directly to ribosomes and initiate translation without using any initiation factor proteins or initiator tRNA (5, 10). In so doing, they eliminate the need for over a dozen specific protein factors in a process that is dependent on specific RNA structures in the IRES. A remarkable aspect of the IGR IRES mechanism is its ability to induce translocation of an aminoacylated elongator tRNA from the A site into the P site before the first peptide bond is made; this event lies at the core of IGR IRES function and thus understanding how it occurs is critical for understanding how the IRES works and how they manipulate ribosomes.
Translocation by the IGR IRES must have different requirements compared to conventional translocation. Although the minimal requirements for translocation in 80S ribosomes have not been described, on 70S ribosomes there is a strict requirement for a deacylated full-length tRNA in the P site and at least an ASL bound to the A site (44), and there is evidence that a backbone contact between the acceptor stem of the deacylated hybrid P/E state tRNA and helix 68 of 23S rRNA is crucial (45). During IGR IRES-driven initiation, there is no tRNA of any type in the P site and thus critical contacts between the ribosome and P site or P/E hybrid state tRNA are missing. Clearly, specific but noncanonical interactions between IRES domains and the ribosome must overcome this.
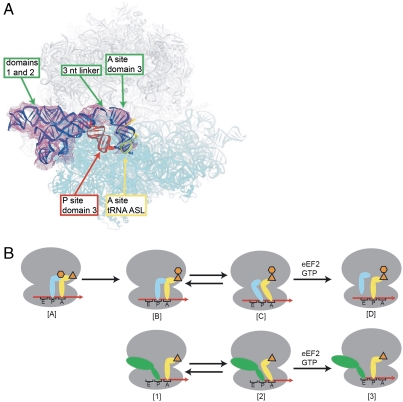
It has been proposed previously that the IGR IRES could induce noncanonical translocation of a tRNA from the A to the P site (and thus IRES domain from the P to the E site) by mimicking a P/E hybrid state tRNA (Fig. 4B) (21, 42). In this model, IGR IRES domain 3 lies in the small subunit’s decoding groove, precisely mimicking the tRNA ASL•mRNA codon interaction in the P site and satisfying all the contacts normally associated with tRNA interactions with the small subunit in this site. However, as mentioned above, there are normally contacts to the large subunit made by a deacylated tRNA in the P/E hybrid state; thus, some other IRES elements must provide these critical tRNA-like contacts. In this model, delivery of an aminoacyl-tRNA to the A site is followed by sampling of the A/P hybrid state by this tRNA. The ribosomal complex is then in a pretranslocation state that is recognized by eEF2, which catalyzes translocation in a GTP-dependent manner. This translocation event moves the tRNA fully into the P site (i.e., the P/P state), allowing entry of the next aminoacyl-tRNA into the A site so that protein synthesis can proceed.
Fig. 4.
Proposed mechanism for the action of IGR IRES domain 3. (A) Results of superposition of the domain 3 RNA-70S ribosome structure presented here into the 7.3 Å cryo-EM density of a CrPV IGR IRES bound to an 80S ribosome (23), followed by docking of the PSIV IGR IRES domains 1 and 2 crystal structure (20) into the density (green). Only placement of domain 3 into the A site of the cryo-EM map matches the density (green) and is the proper distance from domains 1 and 2 to allow fitting of the 3-nucleotide linker (green). The location of an A site tRNA ASL (28) is shown (yellow). Placement of domain 3 into the P site (red) creates steric clash. See also Fig. S5. (B) Proposed mechanism for IGR IRES translocation (Bottom) compared to canonical translation initiation and translocation (Top). (Top) [A] The aminoacylated (orange) initiator P (cyan) and A site tRNAs (yellow) are bound to the ribosome (gray). [B] Peptidyl transfer results in a deacylated tRNA in the P site and a peptidyl tRNA in the A site. [C] The tRNAs sample the hybrid states. [D] The hybrid state is a substrate for eEF2-catalyzed translocation. (Bottom) [1] The IRES (green) assembles on an 80S ribosome. In the state shown, domain 3 is in the P site. [2] The orientation of domain 3 allows movement of the A site tRNA into an A/P hybrid state and this, combined with domains 1 and 2 of the IRES, promotes movement of the ribosome into a hybrid state. [3] An eEF2-catalyzed translocation event moves the IRES into the E site and the tRNA into the P site, completing the initiation process. Translation now can proceed through the normal elongation cycle.
The crystal structure presented here strongly supports the above described mechanism, as do several other lines of evidence. First, our structure shows that PK I is a precise and accurate mimic of the ASL•codon interaction and satisfies all of the required interactions in the P site (Fig. 2D), as predicted by the model. Second, the position of helix 3.1 deviates from that of the tRNA D stem (Fig. 3A), orienting the distal end of domain 3 toward the E site, where domains 1 and 2 are observed in cryo-EM reconstructions of complexes containing complete IGR IRES RNAs bound to 80S ribosomes (23, 43). This orientation, similar to that of a P/E hybrid state tRNA (46), would potentially allow an A site tRNA to enter the A/P hybrid state without steric clash (Fig. 3B). Thus, domains 1 and 2 are positioned to make critical contacts normally made by a deacylated tRNA in the P/E hybrid state. Third, the presence of an IGR IRES bound to 80S ribosomes stimulates the GTPase activity of elongation factor eEF2, consistent with the proposal that the bound IRES moves the ribosome into a pretranslocation state (42). Thus, although additional experiments are needed to test this mechanistic model thoroughly, it is consistent with a wealth of published functional and structural studies.
Mutational analysis of IGR IRES RNAs has shown that the primary determinants for binding to the ribosome reside in domains 1 and 2; deletion of domain 3 has only a minor effect on ribosome binding affinity (17, 42). In contrast, certain localized changes in domain 3 have profound effects on gene expression. Disruption of base pairing in stems P3.1 or P3.2 of domain 3 (Fig. 1C) abolished translation initiation from the IRES, which was restored by introduction of compensating base changes (16). This result highlights the importance of the ASL-like stem structure for proper positioning of domain 3 in the tRNA binding sites of the small ribosomal subunit. Similarly, mutations that create mismatches between the codon- and anticodon-like triplets of PKI abolishes initiation (16); again, these effects are rescued by introducing compensating mutations that restore pairing. Our structure shows that proper binding of domain 3 to the small subunit P site is mediated by numerous stacking and H-bonding interactions involving bases and backbone elements of the IRES RNA, in both the stem and codon-anticodon-like regions (Fig. 2 and Table 1) that would be disrupted by structural distortions introduced by the mutations. Toeprinting studies showed that mutations that disrupt base pairing in PKI caused loss of correct positioning of the viral mRNA in the ribosome, indicating the role of PKI in establishing the translational reading frame (14). This result is in good agreement with the structure of the ribosomal complex, which shows that binding of domain 3 in the small subunit P site positions the mRNA to place the first codon of the open reading frame in the A site.
Finally, the fact that PK I so closely mimics a tRNA ASL-mRNA codon structure is compatible with the possibility that domain 3 can also bind to the ribosomal A site, and possibly the E site, of the small subunit. Indeed, the structures of tRNA ASLs bound to their codons in the A and P sites (28) are virtually identical (rmsd < 1.0 Å), suggesting that domain 3 should be capable of binding to either the A or P sites on the small subunit. Structural evidence that domain 3 can occupy the A site comes from the 7.3-Å cryo-EM map of the CrPV IRES-80S ribosome complex (Fig. 4A and Fig. S5) (23). In this reconstruction, the position of domain 3 was interpreted as overlapping the A and P sites but close to features known to contact the ASL of an A site-bound tRNA. To reexamine these data in light of our structure, we aligned the 80S cryo-EM map with the all-atom structure of the 70S ribosome. The crystal structures of domains 1 and 2 of the PSIV IRES (20) could be fitted readily into the EM map, in close agreement with their previously modeled positions (23). However, domain 3 could only be fit to density unambiguously present in the A site as there was no density in the P site (Fig. 4A and Fig. S5). In our fitting, domain 3 is rotated about 180° from its previously modeled position. This placement results in a 20-Å spacing between domains 2 and 3 that matches the length of the three-nucleotide linker connecting the two domains (Fig. 4A and Fig. S1), whereas placement of domain 3 in the P site leads to substantial steric clash. Thus, within the 7.3-Å cryo-EM reconstruction of the CrPV IGR IRES-80S ribosome complex, domain 3 occupies the 40S subunit A site (Fig. 4A), also in agreement with earlier low-resolution cryo-EM observations (43). This observation suggests that initiation begins with domain 3 bound to the small subunit A site, followed by translocation to the P site, followed by the steps discussed above.
Different combinations of structural strategies are undoubtedly used by different classes of viral IRES RNAs to recruit, position, and activate host cell ribosomes. Our structures, combined with previous functional evidence, show that the IGR IRES uses both direct structural mimicry and precise interactions with the ribosome to drive non-canonical translocation and initiation. In addition, this study shows how folded RNA structures can manipulate the ribosome, a strategy that might extend beyond viral IRESs to other noncoding RNAs.
Experimental Procedures
IRES RNA—Ribosome Binding Assays.
A nitrocellulose filter-binding assay was performed to determine the binding of PSIV or CrPV IRES domain 3 RNA to 70S or 80S ribosomes. [32P]-labeled IRES domain 3 was mixed with 6.6 mM unlabeled IRES domain 3 in buffer F (25 mM Tris-Cl, pH 7.0, 100 mM KCl, 10 mM MgCl2, 2 mM spermine) and heated at 70 °C for 3 min followed by gradual cooling to room temperature over 20 min. A series of 10 mL PSIV or CrPV domain 3 solutions at 6.6, 4.9, 3.3, 1.6, 0.8, and 0.4 mM in buffer F was made and 5 pmol of 70S or 80S ribosomes in 30 mL buffer F were added. The reaction mixtures were incubated at 37 °C for 30 min and then split into two 20-mL samples to spot on two 0.45-μm HA nitrocellulose filters (Millipore) for duplicate results. The filters were washed three times with 5 mL of buffer G (25 mM Tris-Cl, pH 7.6, 100 mM KCl, 25 mM MgCl2) at room temperature. Filters were dried and the radioactivity retained on the filters was measured by liquid scintillation counting.
Cocrystallization of IRES RNA with 70S Ribosomes.
Three hundred micromoles of PSIV or CrPV domain 3 were annealed by heating at 65 °C for 5 min in buffer H (10 mM K+-Hepes, pH 7.5, 2.5 mM MgCl2, 2 mM spermine) and slowly cooled to room temperature over 20 min. To form the complex, reassociated T. thermophilus 70S ribosomes were incubated with an 8- to 10-fold excess of refolded IRES domain 3 at 37 °C for 40 min. The final concentration of the complex was 10 mg/mL (4 μM) 70S ribosomes with 32–40 mM PSIV or CrPV domain 3 in buffer F supplemented with 2.8 mM deoxy-Bigchap (Hampton Research). The formed ribosome complex was then clarified by centrifugation at 16,000 × g for 5 min at room temperature before being subjected to crystallization.
Initial crystallization screening was performed around conditions previously reported (31, 48) by dispensing 0.2 + 0.2 mL sitting drops with a Phoenix robotic liquid handling system (Art Robbins) on 96-well plates. Once optimal crystallization conditions were determined, crystals were grown by the sitting-drop vapor-diffusion method using drops dispensed by the Phoenix with 1- to 2-mL ribosome complexes mixed with 1–2 mL of reservoir solution [100 mM Tris-OAc, pH 7.0, 200 mM potassium thiocyanate (KSCN), 3.6–5% PEG 20,000, 6–14% 2-methyl-2,4-pentanediol (MPD)] at 22.5 °C. Crystals emerged after 5–7 d and matured between 2–3 wk. Crystals were then subjected to cryoprotection by gradually replacing the mother liquor with cryoprotection buffer I (100 mM Tris-OAc, pH 7.0, 200 mM KSCN, 5% PEG 20,000, 25% MPD, 14% PEG 200, and 10 mM Mg(OAc)2). The crystals then were flash-frozen by plunging into liquid nitrogen.
X-Ray Data Collection and Structure Determination.
Crystals were screened at beamlines 7.1, 9.1, 9.2, 11.1, and 12.2 at the Stanford Synchrotron Radiation Laboratory. X-ray diffraction data for the 70S-PSIV IRES complex were recorded at beamline 23 ID-D at the Advanced Photon Source at Argonne National Laboratory using an X-ray wavelength of 1.0332 Å and an oscillation angle of 0.2°. Data from four datasets obtained from different positions of the same crystal were integrated and merged using the XDS package (48), scaled in SCALA (49), and truncated in TRUNCATE (50). X-ray data for the 70S-CrPV IRES complex were recorded at beamline 12.2 at the Stanford Synchrotron Radiation Laboratory using the PILATUS 6M detector; data reduction was carried out similarly to that for the 70S-PSIV IRES complex. In both datasets, 1% of reflections were marked as test-set (Rfree set) reflections to monitor the progress of refinement.
Structure determination started with rigid-body refinement (51) of the previously determined structure of the RF2 termination complex, which was obtained from the same crystal form (52); the release factor and tRNAs were omitted from the structure. Electron density corresponding to domain 3 of the IRES was clearly visible at the P site of the small subunit in the Fourier difference map calculated from the rigid-body refined model. Noncrystallographic symmetry (NCS)-averaged simulated-annealing maps were used to build domain 3 of the PSIV or CrPV IRES RNAs using the structure of the isolated domain 3 of the CrPV IRES (21) as a guide. Simulated-annealing and grouped B-factor refinements were performed in CNS (51). NCS restraints as well as RNA and protein secondary structure restraints were used throughout the refinement as described (47). PyMOL (53), O (54), and local real-space refinement (55) were employed for structure visualization and model building. Figures were rendered using PYMOL (53).
Supplementary Material
Acknowledgments.
We thank the members of the Noller and Kieft labs for helpful discussions, the staffs at Stanford Synchrotron Radiation Lightsource and Advanced Proton Source for their expert support during screening and data collection, and T. Pestova, P. Sarnow, and C. Spahn for comments on the manuscript. This work was supported by Grants GM-17129 and GM-59140 from the National Institutes of Health (NIH) and MCB-723300 from the National Science Foundation (to H.F.N.) and Grants GM-72560 and GM-81346 from the NIH (to J.S.K.). J.S.K. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
The authors declare no conflict of interest.
Data deposition: Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PSIV complex: PDB ID codes 3PYN, 3PYO, 3PYQ, and 3PYR; CrPV complex: PDB ID codes 3PYS, 3PYT, 3PYU, and 3PYV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018582108/-/DCSupplemental.
References
- 1.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestova T, Lorsch JR, Hellen CU. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 87–128. [Google Scholar]
- 3.Doudna JA, Sarnow P. In: Translational Control in Biology and Medicine. Mathews MB, Sonenberg N, Hershey JWB, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. pp. 129–153. [Google Scholar]
- 4.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009;139:137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Jan E, et al. Initiator Met-tRNA-independent translation mediated by an internal ribosome entry site element in cricket paralysis virus-like insect viruses. Cold Spring Harb Sym. 2001;66:285–292. doi: 10.1101/sqb.2001.66.285. [DOI] [PubMed] [Google Scholar]
- 8.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 10.Deniz N, Lenarcic EM, Landry DM, Thompson SR. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA. 2009;15:932–946. doi: 10.1261/rna.1315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cevallos RC, Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki J, Nakashima N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci USA. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishiyama T, et al. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J Mol Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfingsten JS, Costantino DA, Kieft JS. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science. 2006;314:1450–1454. doi: 10.1126/science.1133281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat Struct Mol Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan E, Kinzy TG, Sarnow P. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci USA. 2003;100:15410–15415. doi: 10.1073/pnas.2535183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 24.Moazed D, Noller HF. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- 25.Moazed D, Noller HF. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16 S rRNA. J Mol Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- 26.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Re. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 27.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 29.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 30.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korostelev A, et al. Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome. Proc Natl Acad Sci USA. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilbers CW, Michiels PJ, Heus HA. New developments in structure determination of pseudoknots. Biopolymers. 1998;48:137–153. doi: 10.1002/(SICI)1097-0282(1998)48:2<137::AID-BIP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 33.Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 34.Graifer D, et al. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004;32:3282–3293. doi: 10.1093/nar/gkh657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caskey CT, Redfield B, Weissbach H. Formylation of guinea pig liver methionyl-sRNA. Arch Biochem Biophys. 1967;120:119–123. doi: 10.1016/0003-9861(67)90605-4. [DOI] [PubMed] [Google Scholar]
- 36.Berthelot F, Bogdanovsky D, Schapira G, Gros F. Interchangeability of factors and tRNA’s in bacterial and eukaryotic translation initiation systems. Mol Cell Biochem. 1973;1:63–72. doi: 10.1007/BF01659939. [DOI] [PubMed] [Google Scholar]
- 37.Laycock DG, Hunt JA. Synthesis of rabbit globin by a bacterial cell free system. Nature. 1969;221:1118–1122. doi: 10.1038/2211118a0. [DOI] [PubMed] [Google Scholar]
- 38.Von Ehrenstein G, Lipmann F. Experiments on hemoglobin biosynthesis. Proc Natl Acad Sci USA. 1961;47:941–950. doi: 10.1073/pnas.47.7.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 40.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 41.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J Biol Chem. 2007;282:7770–7776. doi: 10.1074/jbc.M610887200. [DOI] [PubMed] [Google Scholar]
- 43.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Joseph S, Noller HF. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feinberg JS, Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc Natl Acad Sci USA. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 47.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 48.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 49.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 50.The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 51.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Korostelev A, et al. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA. 2008;105:19684–19689. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 54.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 55.Korostelev A, Bertram R, Chapman MS. Simulated-annealing real-space refinement as a tool in model building. Acta Crystallogr D. 2002;58:761–767. doi: 10.1107/s0907444902003402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.