Abstract
The diterpenoid phytohormone gibberellin (GA) controls diverse developmental processes throughout the plant life cycle. DELLA proteins are master growth repressors that function immediately downstream of the GA receptor to inhibit GA signaling. By doing so, DELLAs also play pivotal roles as integrators of internal developmental signals from multiple hormone pathways and external cues. DELLAs are likely nuclear transcriptional regulators, which interact with other transcription factors to modulate expression of GA-responsive genes. DELLAs are also involved in maintaining GA homeostasis through feedback up-regulating expression of GA biosynthesis and receptor genes. However, the molecular mechanisms by which DELLAs restrict growth and development are largely unknown. This study reveals an important step of the mechanism. Previous microarray studies identified SCARECROW-LIKE 3 (SCL3) as a direct target gene of DELLA in Arabidopsis seedlings. SCL3 expression is induced by DELLA and repressed by GA. Unexpectedly, a scl3 null mutant displays reduced GA responses and elevated expression of GA biosynthesis genes during seed germination and seedling growth, indicating that SCL3 functions as a positive regulator of GA signaling. SCL3 seems to act as an attenuator of DELLA proteins. Transient expression, ChIP, and co-IP studies show that SCL3 autoregulates its own transcription by directly interacting with DELLA. Our data further show that SCL3 and DELLA antagonize each other in controlling both downstream GA responses and upstream GA biosynthetic genes. This work is beginning to shed light on how this complex regulatory network achieves GA homeostasis and controls GA-mediated growth and development in the plant.
Keywords: gibberellin-regulated development, hormone homeostasis, SCARECROW-LIKE 3–DELLA interaction, DELLA attenuator
Bioactive gibberellins (GAs) are a class of phytohormones that plays critical roles in modulating plant growth and development in response to internal developmental programs and environmental cues (1–4). DELLA proteins are likely nuclear transcriptional regulators that function as master growth repressors by inhibiting all aspects of GA responses (1, 5, 6). Binding of GA to its receptor GA INSENSITIVE DWARF1 (GID1) enhances the GID1–DELLA interaction, which, in turn, leads to the rapid proteolysis of DELLA through the ubiquitin-proteasome pathway and allows transcriptional reprogramming of GA-responsive genes (7–11). A specific ubiquitin E3 ligase SCFSLY1/GID2 (Skp1-Cullin-F-box protein complex) is responsible for recruiting DELLA for polyubiquitination (12–15). DELLA proteins belong to a subfamily of the plant-specific GRAS family [for GA INSENSITIVE (GAI), REPRESSOR OF gal-3 (RGA), and SCARECROW (SCR)] of regulatory proteins (5, 16). In addition to the C-terminal GRAS domain that is common in all GRAS family members, DELLA protein also contains a unique DELLA domain in its N terminus that is required for GID1 binding and GA-induced degradation (7, 17–20). Arabidopsis contains five DELLAs [RGA, GAI, RGA-LIKE1 (RGL1), RGL2, and RGL3], which display overlapping but also some distinct functions in repressing GA responses (21–24). RGA and GAI are the major repressors of vegetative growth and floral induction (23, 24). Without a canonical DNA binding domain, DELLA seems to modulate gene expression by interacting with other transcription factors (25). Recent findings indicate that interaction between DELLA and a subset of bHLH transcription factors, PHYTOCHROME INTERACTING FACTORs (PIFs), blocks transcription of the target genes of PIF and hence, inhibits PIF-induced hypocotyl elongation in Arabidopsis (26, 27).
In an effort to elucidate how DELLA proteins regulate plant growth and development, several DELLA target genes were identified in our previous microarray studies (25). Among them, SCARECROW-LIKE 3 (SCL3; AT1G50420) was found to be a GA-repressed and DELLA-induced gene, suggesting that SCL3 may function as a downstream negative regulator of GA signaling. Like DELLA, SCL3 is also a GRAS protein, but it does not contain the GA-responsive DELLA domain. Interestingly, in the primary root, SCL3 mRNA is mainly expressed in the endodermis (16), which has been shown to be the primary site of GA-induced DELLA degradation (28). Expression of a GA-resistant (gain of function) DELLA mutant protein in the endodermis (but not other cell types) inhibits root elongation (28). In addition, the SCL3 promoter is directly induced by the SCR and SHORT-ROOT (SHR) heterodimer, two GRAS proteins that are essential in root endodermis specification and stem cell maintenance (29, 30). Taken together, these observations suggest that SCL3 may play an important role in regulating root elongation. In the current report, we show that SCL3 participates in root and above-ground organ development. Surprisingly, SCL3 is a positive regulator of GA signaling, which is contrary to what was originally proposed. Importantly, SCL3 antagonizes DELLA function in modulating downstream GA responses as well as GA homeostasis by direct protein–protein interaction.
Results
Higher SCL3 mRNA Levels in Germinating Seeds, Roots, and Seedlings.
Previous microarray analysis showed that SCL3 is a GA-repressed and DELLA-induced gene (25). The elevated SCL3 transcript levels by DELLA correlate with increased accumulation of the SCL3 protein (Fig. S1A), suggesting that SCL3 may function as a negative regulator of GA signaling acting downstream of DELLA. To understand the role and action site of SCL3 in the GA response pathway, we first investigated its developmental expression profile by real-time quantitative RT-PCR (qRT-PCR). SCL3 mRNA is expressed throughout all developmental stages tested (Fig. S1B). The highest amounts of the SCL3 mRNA were detected in germinating seeds, whole seedlings, and seedling roots, suggesting that SCL3 plays a role in germination, root, and seedling development.
Increased Sensitivity of the scl3 Mutant to GA Biosynthesis Inhibitor.
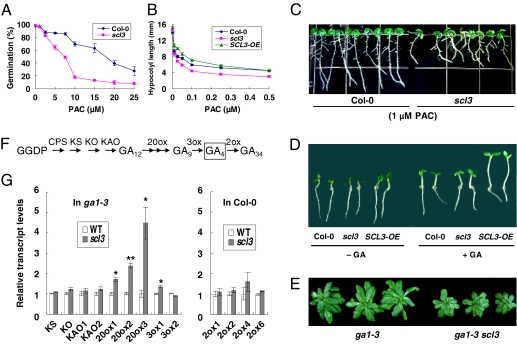
To study the physiological function of SCL3, we characterized a scl3 transfer DNA (T-DNA) mutant (SALK_002516, also named scl3-1) (31), in which the T-DNA is inserted into the second exon (326 bp downstream of the ATG start codon) of this gene. RT-PCR analysis confirmed that this mutant is a null scl3 allele, because no WT transcript was detected. Under regular growth conditions, scl3-1 did not show any phenotype compared with WT Col-0 (Fig. 1A–B and Fig. S1D). This could be because of functional redundancy with other GRAS protein(s). Therefore, we examined scl3-1 phenotypes in the presence of GA biosynthesis inhibitor paclobutrazol (PAC). Interestingly, the germination and root-length assays showed that scl3 was more sensitive to PAC treatment than WT (Fig. 1 A and C). The increased sensitivity of scl3 to GA biosynthesis inhibitor suggests that this mutant either contains lower levels of active GAs and/or is partially defective in GA responses. Therefore, SCL3 should function as a positive regulator of GA production or GA responses. We confirmed that this increased sensitivity to PAC phenotype is caused by the scl3 mutation, because expression of SCL3 promoter:SCL3 genomic DNA in scl3-1 rescued its root growth defect in the presence of GA biosynthesis inhibitor (Fig. S1C). Moreover, overexpression of SCL3 in transgenic Col-0 containing Cauliflower Mosaic Virus (CaMV) 35S promoter:SCL3 cDNA (SCL3-OE) conferred a longer root phenotype than WT in the presence of PAC, indicating that SCL3-OE lines are resistant to PAC (Fig. S1D).
Fig. 1.
Phenotypes of the scl3 mutant and SCL3 overexpression line. (A) Germination assay. Seed coat rupture after 8 d was scored as germination. (B) Hypocotyl elongation assay of etiolated seedlings in response to PAC at day 6. (C) Root elongation assay at day 10. (D) Hypocotyl elongation assay of light-grown seedlings in response to 1 μM GA4 at day 4. (E) Rosettes of ga1-3 and the ga1-3 scl3 double homozygous mutant at 45 d old on soil. (F) The major GA biosynthesis and catabolism pathways in Arabidopsis. GGDP, geranylgeranyl diphosphate. Enzyme for each step is listed above each arrow. CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; KAO, ent-kaurenoic acid oxidase; 20ox, GA20-oxidase; 3ox, GA3-oxidase; 2ox, GA2-oxidase. GA4 is the major active GA in Arabidopsis. (G) Relative transcript levels in 8-d-old seedlings. (Left) ga1-3 and ga1-3 scl3; (Right) Col-0 and scl3. Data represent the average of three qRT-PCR measurements ± SE. The housekeeping gene GAPC (glyceraldehyde-3-phosphate dehydrogenase C subunit), whose expression is not responsive to GA (12), was used to normalize different samples. The mRNA level of each corresponding gene in ga1-3 (for GA biosynthetic genes) or Col-0 (for GA catabolic genes) was arbitrarily set to 1. *P < 0.05; **P < 0.01.
Similarly, etiolated scl3 and SCL3-OE seedlings had shorter and longer hypocotyls, respectively, than that of WT in the presence of PAC (Fig. 1B). Interestingly, in the constant light conditions, the SCL3-OE seedlings had slightly longer hypocotyls than WT, even without PAC treatment (Fig. 1D). Moreover, in response to 1 μM GA4 treatment, the hypocotyl of the SCL3-OE line was dramatically longer than that of WT, although there was no difference between WT and scl3 (Fig. 1D and Fig. S1E). We also found that scl3 enhanced the dwarf phenotype of the GA-deficient mutant ga1-3 (Fig. 1E), further supporting the positive role of SCL3 in the GA pathway.
Up-Regulated Expression of GA Biosynthetic Genes in scl3.
To determine whether SCL3 functions to promote GA accumulation or GA signaling, we analyzed the effect of scl3 mutation on the expression of GA biosynthetic genes and GA catabolic genes that are known to be expressed in seedlings (Fig. 1 F and G) (32–36). If SCL3 directly promotes bioactive GA accumulation, scl3 mutation may decrease the expression of GA biosynthetic genes and/or increase the expression of GA catabolic genes. Because transcript levels of some of the GA20ox and GA3ox genes are more readily detected under GA-deficient background, we performed qRT-PCR assays for all GA biosynthetic genes in the ga1-3 background and compared mRNA levels in ga1-3 vs. ga1-3 scl3. Conversely, transcripts of the GA catabolic genes (GA2ox) are known to be higher in the GA-producing WT background. Therefore, we analyzed GA2ox gene expression in WT Col-0 vs. scl3 single mutant. The expression levels of the early GA biosynthetic genes [ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase 1 (KAO1), and KAO2] and GA catabolic genes (GA2ox) were not altered by scl3 (Fig. 1G). However, expression of several GA biosynthetic genes, including GA20ox1, GA20ox2, GA20ox3, and GA3ox1, was up-regulated significantly by scl3. These results indicated that PAC-sensitive phenotypes of scl3 are unlikely because of reduced GA levels. Instead, scl3 causes reduced GA signaling activities, which, in turn, feedback up-regulates expression of GA biosynthetic genes. Therefore, SCL3 is likely an activator of the GA signaling pathway.
rga and spindly Mutations Are Epistatic to scl3.
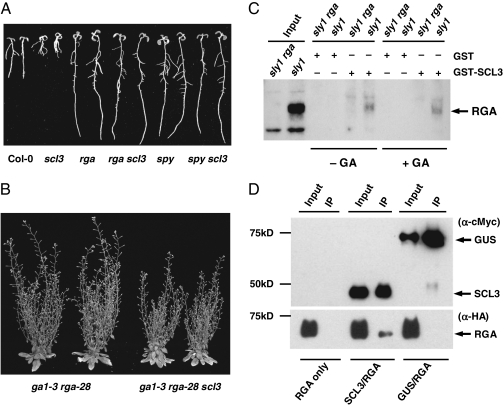
To place SCL3 in the GA signaling pathway, genetic interactions between SCL3 and two known GA signaling repressors, RGA (an Arabidopsis DELLA) (37) and SPINDLY (SPY) (38), were studied by epistasis analysis using the root-length assays. SPY is an O-linked N-acetylglucosamine (O-GlcNAc) transferase, which was proposed to activate DELLA by O-GlcNAc modification (39, 40). As predicted, rga-28 (a null allele) and spy-3 single mutants were resistant to PAC and displayed longer root phenotypes than WT (Fig. 2A and Fig. S2A). The root lengths of the rga scl3 and spy scl3 double mutants were similar to rga and spy single mutants, respectively (Fig. 2A and Fig. S2A), suggesting that both rga and spy are epistatic to scl3 in the GA pathway. However, at later developmental stages, rga was only partially epistatic to scl3 in controlling rosette diameter, flowering time, and plant height (Fig. 2B and Fig. S2B).
Fig. 2.
Interactions between SCL3, RGA, and SPY. (A) rga and spy are epistatic to scl3 in the root-length assays in response to 1 μM PAC (10-d-old seedlings). (B) rga is partially epistatic to scl3 in the ga1-3 background (65-d-old plants) (Fig. S2B). (C) In vitro pull-down of RGA with recombinant GST-SCL3. Protein extracts from sly1-10 and sly1-10 rga-24 double mutant were incubated with GST or GST-SCL3 from Escherichia coli separately. − GA, in the absence of GA; + GA, in the presence of 100 μM GA4. Input and pull-down samples were analyzed by immunoblotting using affinity-purified RGA antibody. (D) co-IP of SCL3 and RGA in planta. HA-RGA was transiently expressed alone (RGA only) or coexpressed with cMyc-SCL3 (SCL3/RGA) or cMyc-GUS (GUS/RGA) in N. benthamiana. The nuclear protein extracts were immunoprecipitated with cMyc antibody-conjugated agarose beads, and the input and IP samples were analyzed by immunoblotting using antibodies for cMyc and HA, separately.
Direct Interaction Between DELLA and SCL3 Proteins.
Our previous microarray, qRT-PCR, and ChIP-qPCR data indicated that SCL3 mRNA levels are directly induced by DELLA. Surprisingly, the current study showed that SCL3 is a positive regulator of GA signaling pathway, and rga is partially epistatic to scl3. We, therefore, hypothesized that DELLA and SCL3 may interfere with each other's function by direct protein–protein interaction, and the up-regulation of SCL3 mRNA levels by DELLA may be a part of the feedback mechanism to maintain GA homeostasis. Like DELLA proteins (5), the SCL3-GFP fusion protein was detected in the nuclei of root cells of a transgenic line carrying 35S:SCL3-GFP (Fig. S2C). In support of the idea of direct interaction between SCL3 and DELLA, we also detected weak interactions between SCL3 and three DELLA proteins (RGA, GAI, and RGL1) in yeast two-hybrid assays (Fig. S2D). SCL31, a GRAS protein (encoded by At1g07520) that is divergent from both the DELLA subfamily and SCL3 (41), did not show any interaction with SCL3, suggesting that SCL3–DELLA interactions are specific (Fig. S2D).
In vitro pull-down assays further showed that purified recombinant GST-SCL3 protein bound to endogenous RGA proteins in plant extracts (Fig. 2C). The F-box mutant sleepy1-10 (sly1-10) background was used in these assays, because it accumulates high levels of RGA (13). The sly1-10 rga-24 double mutant, which lacks the endogenous RGA protein (12), was used as a negative input control. RGA–SCL3 interaction seems to be independent of GA, because RGA was pulled down by GST-SCL3 in the presence or absence of GA (Fig. 2C).
To confirm SCL3–RGA interaction in planta, we performed coimmunoprecipitation (co-IP) assays by transiently coexpressing 35S:cMyc-SCL3 and 35S:HA-RGA constructs in leaves of Nicotiana benthamiana through Agrobacterium-mediated transformation. Tissues infiltrated with 35S:HA-RGA alone or coinfiltrated with 35S:cMyc-GUS-NLS (cMyc epitope tagged β-glucuronidase fused with SV40 nuclear localization signal) were included as negative controls. From cross-linked leaf tissues, nuclear proteins were extracted and immunoprecipitated using anti-cMyc antibody-conjugated agarose beads. Fig. 2D shows that HA-RGA was coimmunoprecipitated only when it was coexpressed with cMyc-SCL3 but not when it was expressed alone or coexpressed with cMyc-GUS-NLS. Therefore, SCL3 and RGA physically interact when overexpressed in N. benthamiana leaves.
Antagonistic Effects of SCL3 and RGA on the Expression of SCL3.
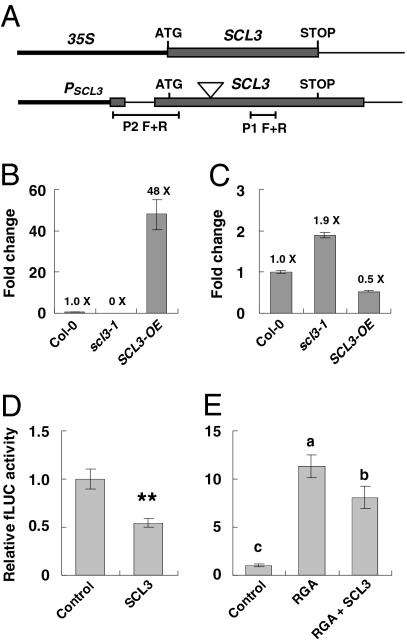
Our previous microarray study had identified several early DELLA-induced and GA-repressed genes, including SCL3, and two GA biosynthetic genes, GA20ox2 and GA3ox1 (25). These genes are likely direct targets of DELLA, because their expression decrease in response to GA correlates with the disappearance of DELLA—after 10 min of 2 μM GA4 treatment, 90% of DELLA is degraded; mRNA levels of these DELLA target genes are reduced three- to fivefold at 30 min (25). Moreover, by ChIP-qPCR assays, we found that the RGA fusion protein with a tandem affinity purification tag is associated with the SCL3 promoter in vivo, although no interaction was detected with the promoters of GA20ox2 or GA3ox1 (25). In this study, we found that SCL3 down-regulates GA20ox2 and GA3ox1 transcript levels (Fig. 1G), opposite to the effect of DELLA. Because SCL3 itself is also a DELLA-induced gene, we tested whether SCL3 down-regulates its own expression. Indeed, overexpression of SCL3 (in the SCL3-OE line) dropped the amounts of endogenous SCL3 transcript by one-half (Fig. 3 A–C). Moreover, the scl3-1 mutant produces truncated SCL3 transcripts upstream of the T-DNA insertion site, and the amounts of the truncated transcript were about 1.9-fold higher than the SCL3 transcripts in Col-0 (Fig. 3C). These results support that SCL3 down-regulates its own promoter expression.
Fig. 3.
SCL3 interferes with RGA to regulate the expression of the SCL3 promoter. (A) Schematics of 35S:SCL3 in the SCL3-OE line and the endogenous SCL3 locus. The triangle symbol indicates the T-DNA insertion site in scl3. Primers P2 F+R only amplify the endogenous SCL3 transcripts, whereas primers P1 F+R amplify SCL3 transcripts produced by both the transgene and endogenous SCL3 locus. (B) Total SCL3 transcript levels in Col-0, scl3, and SCL3-OE lines. qRT-PCR analysis was performed using primers P1 F+R. (C) WT or truncated endogenous SCL3 transcripts in Col-0, scl3, and SCL3-OE lines. qRT-PCR analysis was performed using primers P2 F+R. In B and C, data represent the average of three measurements ± SE. A GA nonresponsive gene (At4g33380) (7) was used to normalize different samples. The amount of SCL3 mRNA in Col-0 was set to 1. (D) SCL3 repressed its own promoter expression. (E) SCL3 antagonized RGA-induced SCL3 promoter expression in the transient coexpression assays. In D and E, the reporter construct (PSCL3:fLUC) contains 2 kb SCL3 promoter plus the 35S minimal promoter (−45- to 1-bp region that includes the TATA box) (49) fused to fLUC. 35S:Renilla LUC (rLUC) served as an internal control for normalization of transformation efficiency. Effector constructs were 35S:RGA and/or 35S:SCL3, and the empty vector was used as a negative control. PSCL3:fLUC and 35S:rLUC constructs were cobombarded into 11-d-old triple-mutant ga1-3 rga-28 scl3 seedlings with various combinations of effector constructs using the same molar ratio. The relative fLUC activity of the empty effector control was set to 1. Data represent average value ± SE of 14 replicates from three independent experiments. Pair-wise t tests were performed. (D) **P < 0.01. (E) When two samples show different letters (a–c) above the bars, the difference between them is significant (a–c and b–c, P < 0.01; a–b, P < 0.05). Another reporter construct containing a 1-kb SCL3 promoter with its native TATA box fused to fLUC rendered similar results (Fig. S3).
Given the evidence that SCL3 and RGA interact in vitro and in vivo, we tested whether transiently coexpressed SCL3 and DELLA in Arabidopsis compete to modulate transcription of SCL3, GA20ox2, and GA3ox1 using the dual luciferase (LUC) reporter assay (42). The reporter constructs contain promoter sequences of SCL3, GA20ox2, and GA3ox1 genes, respectively, which were fused to the firefly LUC gene (fLUC) (Fig. S3A). The 35S:Renilla LUC (rLUC) was used as an internal standard. The effector constructs contain 35S:RGA or 35S:SCL3. We used the ga1 rga scl3 triple mutant in these assays to enhance the effects of overexpression of SCL3 and/or RGA. Overexpression of SCL3 alone caused repression of PSCL3:fLUC by about twofold compared with the empty effector control (Fig. 3D), whereas overexpression of RGA alone up-regulated PSCL3:fLUC by 11.4-fold (Fig. 3E). In contrast, when RGA and SCL3 were coexpressed, PSCL3:fLUC expression was induced less dramatically (8.1-fold) than when induced by RGA alone (Fig. 3E). These results support the notion that RGA and SCL3 play opposing roles in regulating the SCL3 promoter.
Transient coexpression experiments using a PGA3ox1:fLUC or a PGA20ox2:fLUC reporter construct did not detect any significant effects caused by RGA or SCL3 effectors (1- to 0.9-fold compared to the empty effector control, P > 0.5).
Association of SCL3 with Its Own Promoter in Vivo.
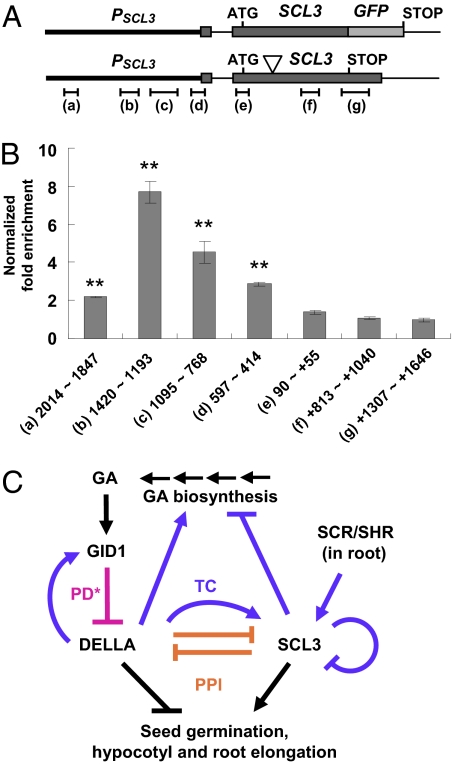
To verify whether the SCL3 protein interacts with its own promoter to down-regulate its expression in planta, we performed ChIP-qPCR assays using a transgenic Arabidopsis line containing PSCL3:SCL3-GFP. The SCL3-GFP fusion protein is functional in planta to rescue the PAC-sensitive root phenotype of scl3 (43). The cross-linked chromatin from the control scl3 or the scl3 PSCL3:SCL3-GFP transgenic line, separately, was incubated with anti-GFP antibody followed by pull-down with protein A-coated agarose beads. Real-time qPCR analysis was carried out to quantify the fold enrichment of different regions of the SCL3 promoter (Fig. 4 A and B). A 2.2- to 7.7-fold enrichment of the SCL3 promoter sequences was observed, with the peak of enrichment at 1,420 to 1,193 bp upstream of the ATG start site (Fig. 4B). In contrast, there was no enrichment for the coding sequence and the 3′-UTR of the SCL3 gene. These results convincingly support that SCL3 protein associates with its own promoter to self-regulate its expression. Consistent with the data showing protein–protein interaction between SCL3 and RGA, their strongest binding sites in the SCL3 promoter overlap in the 1,420- to 768-bp region (25) (Fig. 4B). We did not observe enrichment of the promoter sequences of GA biosynthetic genes, including GA3ox1, GA20ox1, GA20ox2, and GA20ox3 (Fig. S4), although the expression of these genes was up-regulated in scl3 (Fig. 1G).
Fig. 4.
Interaction of SCL3-GFP with the SCL3 promoter in vivo and a model for SCL3–DELLA interaction. (A) Schematics of the transgene PSCL3 (2.5 kb) :SCL3-GFP and the endogenous SCL3 locus. The T-DNA insertion site in scl3 is indicated by the triangle symbol. The regions tested in the ChIP-qPCR assay are indicated underneath the SCL3 genomic DNA structure. (B) SCL3 promoter scanning by ChIP-qPCR. Chromatin isolated from scl3 or scl3 PSCL3:SCL3-GFP seedlings was immunoprecipitated using anti-GFP antibody and followed by qPCR. The 18S rRNA gene was used to normalize the qPCR results in each ChIP sample. Fold enrichment of each region in the scl3 PSCL3:SCL3-GFP line was calculated by comparing with the control scl3 and then, was normalized to the copy numbers of each corresponding region (one copy for region +1,307 to +1,646 and three copies for the rest of the regions) (Fig. S4A). The normalized values of fold enrichment are the average ± SE of three qPCR reactions from one ChIP experiment. Similar results were obtained in an independent ChIP experiment. **P < 0.01 (t tests). The numbers underneath each bar indicate base pairs upstream of the ATG of the SCL3 gene. The plus symbol indicates base pairs downstream of the ATG. (C) A model for antagonistic interaction between SCL3 and DELLA in regulating upstream GA accumulation and downstream GA responses. In the root, SCL3–DELLA interaction coordinates the GA signaling activity with the developmental program controlled by the SCR/SHR pathway. Activation or inhibition could be through different modes of action. PD, protein degradation (magenta line); PPI, protein–protein interaction (orange lines); TC, transcriptional regulation (purple lines). The asterisk indicates that, in addition to PD, proteolysis-independent inactivation of DELLA by GID1 binding (PPI) has been shown to occur in the GID1 overexpression Arabidopsis line (50).
Discussion
The nuclear DELLA proteins are highly conserved growth repressors in angiosperms. GA activates its signaling pathway by enhancing the GID1–DELLA interaction, which then induces rapid degradation of DELLA. This GA-GID1-DELLA signaling module plays a pivotal role in controlling plant growth in response to endogenous developmental programs and external cues (1, 3, 4). Here, we reveal that another GRAS protein, SCL3, acts as a positive regulator of GA signaling in modulating seed germination as well as hypocotyl and root elongation in Arabidopsis (Fig. 4C). This is unexpected, because SCL3 was initially identified as a direct target gene of DELLA, and DELLA induces SCL3 transcription (25). The scl3 mutant phenotype was only apparent under GA-deficient conditions. This could be because of functional redundancy of other GRAS (SCL) protein(s), because a large number of other SCL genes (over 10) are also expressed in the root endodermis (44). Our results show a complex regulatory circuit of the GA signaling pathway modulating plant growth and development (Fig. 4C). The newly characterized SCL3 seems to act as an attenuator of DELLA proteins. By direct protein–protein interaction, DELLA and SCL3 play opposing roles in regulating downstream GA responses. SCL3 also down-regulates its own expression by interfering with DELLA. This conclusion is supported by our transient expression results and ChIP-qPCR analyses showing that the peak of SCL3 association with its own promoter overlaps with the peak of DELLA binding site. Our expression studies further indicate that DELLA and SCL3 antagonize each other in maintaining GA homeostasis by feedback regulating upstream GA biosynthetic genes. GA20ox2 and GA3ox1 are likely direct targets of DELLA, because similar to SCL3, transcript levels of these two genes decrease within 30 min after GA treatment (25). This timing tightly follows the disappearance of DELLA. However, we were unable to detect in vivo association of DELLA or SCL3 with the promoter sequences of GA20ox2 and GA3ox1 by ChIP-qPCR. Similarly, our transient expression assays did not reveal any significant effects of DELLA or SCL3 on expression of these GA biosynthesis genes. DELLA and SCL3 are likely to associate with target DNA indirectly by binding to other transcription factors, because these proteins do not contain any known DNA binding domain. Indirect association of DELLA and SCL3 at the promoters of the GA biosynthesis genes may be too weak to be detected by ChIP-qPCR.
Recently, the root endodermis was shown to be the rate-limiting cell type for coordinating elongation of the entire root (28). Our report and an accompanying paper by Heo et al. (43) show that the endodermis-expressed SCL3 mediates GA-promoted cell elongation in the root. Heo et al. (43) further show that SCL3 also plays a role in determining the timing of the root ground tissue divisions, acting downstream of SCR and SHR. Taken together, the findings in our paper and the accompanying paper by Heo et al. (43) show that SCL3–DELLA interaction coordinates the GA signaling activity with the developmental program controlled by the SCR/SHR pathway during root development.
Materials and Methods
Plant Materials and Mutant Characterization.
All of the Arabidopsis mutants and transgenic lines were derived from ecotype Col-0 unless otherwise noted. The homozygous scl3-1 T-DNA mutant line (Salk_002516) was identified by PCR (http://signal.salk.edu/tdnaprimers.2.html) (Table S1) and backcrossed one time to Col-0. Homozygous double and triple mutants (ga1-3 scl3, rga-28 scl3, spy-3 scl3, and ga1-3 rga-28 scl3) were generated by crossing scl3-1 to ga1-3 (backcrossed six times to Col-0), ga1-3 rga-28 (22), and spy-3 (45), respectively. Transgenic Arabidopsis lines were generated by the floral dip method (46). Detailed information on mutant characterization is described in SI Materials and Methods.
Real-Time qRT-PCR Analyses and Plasmid Construction.
Total RNA was isolated as previously described (25), and cDNA was synthesized with a first-strand cDNA synthesis kit (Roche Diagnostics). Real-time qPCR using SYBR Green and the LightCycler (Roche Diagnostics) was performed as previously described (22). A Student t test was performed using the statistical package SPSS version 17.0. Detailed information on how constructs were generated is in SI Materials and Methods. Sequences of primers used in this study are listed in Table S1.
In Vitro Pull-Down and ChIP-qPCR Assays.
These assays were performed as described previously (25, 29, 47) with some modifications (SI Materials and Methods).
Transient Expression in N. benthamiana by Agro-Infiltration and Co-IP of SCL3 and RGA.
Transient coexpression of HA-RGA/cMyc-SCL3 and HA-RGA/cMyc-GUS-NLS in N. benthamiana by Argobacterium-mediated transformation was performed as described (48) with slight modifications. Nuclear proteins were extracted from cross-linked tissues as described (25), and co-IP was performed using anti-cMyc agarose-conjugated beads (A7470; Sigma) following the manufacturer's protocol with slight modifications. SI Materials and Methods has detailed information on Agro-infiltration and co-IP experiments.
Transient Expression Assays by Particle Bombardment of Arabidopsis Seedlings.
Particle bombardment was carried out using the PDS-1000/He particle gun delivery system (Bio-Rad Laboratories) as described previously (42), except that, instead of detached leaves, whole seedlings were used. A dual-luciferase reporter assay (DLRA) system (Promega) was used to quantify fLUC and rLUC activities. SI Materials and Methods has a detailed protocol.
Supplementary Material
Acknowledgments
We thank Philip Benfey for providing the SCL3 genomic clone. This work was supported by National Science Foundation Grants NRF2009-0077753 (to J.L.), IOS-0641548 (to T.-p.S.), and MCB-0923723 (to T.-p.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012232108/-/DCSupplemental.
References
- 1.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–198. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- 2.Fleet CM, Sun TP. A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr Opin Plant Biol. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Achard P, Genschik P. Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J Exp Bot. 2009;60:1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- 5.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004;55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753. [DOI] [PubMed] [Google Scholar]
- 6.Thomas SG, Sun TP. Update on gibberellin signaling. A tale of the tall and the short. Plant Physiol. 2004;135:668–676. doi: 10.1104/pp.104.040279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths J, et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willige BC, et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;19:1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueguchi-Tanaka M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima M, et al. Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 2006;46:880–889. doi: 10.1111/j.1365-313X.2006.02748.x. [DOI] [PubMed] [Google Scholar]
- 11.Silverstone AL, et al. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dill A, Thomas SG, Hu J, Steber CM, Sun T-p. The Arabidopsis F-box protein SLEEPY1 targets GA signaling repressors for GA-induced degradation. Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGinnis KM, et al. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki A, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- 15.Fu X, et al. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell. 2004;16:1406–1418. doi: 10.1105/tpc.021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 17.Dill A, Jung H-S, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc Natl Acad Sci USA. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell. 2002;14:57–70. doi: 10.1105/tpc.010319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 20.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 21.Lee S, et al. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2002;16:646–658. doi: 10.1101/gad.969002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyler L, et al. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dill A, Sun T. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King KE, Moritz T, Harberd NP. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zentella R, et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 28.Ubeda-Tomás S, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10:625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- 29.Cui HC, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 30.Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MH, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- 32.Sun T-p. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2008. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 34.Mitchum MG, et al. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- 35.Rieu I, et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 36.Rieu I, et al. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20:2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverstone AL, et al. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 2007;143:987–1000. doi: 10.1104/pp.106.091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimada A, et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006;48:390–402. doi: 10.1111/j.1365-313X.2006.02875.x. [DOI] [PubMed] [Google Scholar]
- 41.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 42.Yamakawa S, et al. Systematic transient assays of promoter activities for leaf-specific genes identified by gene-expression profiling with cDNA microarrays in Arabidopsis thaliana. J Biosci Bioeng. 2004;98:140–143. doi: 10.1016/S1389-1723(04)70257-1. [DOI] [PubMed] [Google Scholar]
- 43.Heo J-O, et al. Funneling of gibberellin signaling by the GRAS transcription regulator SCARECROW-LIKE 3 in the Arabidopsis root. Proc Natl Acad Sci USA. 2011;108:2166–2171. doi: 10.1073/pnas.1012215108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 45.Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 47.Bowler C, et al. Chromatin techniques for plant cells. Plant J. 2004;39:776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- 48.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 49.Fang RX, Nagy F, Sivasubramaniam S, Chua NH. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell. 1989;1:141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ariizumi T, Murase K, Sun TP, Steber CM. Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell. 2008;20:2447–2459. doi: 10.1105/tpc.108.058487. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.