Abstract
During plant development, because no cell movement takes place, control of the timing and extent of cell division and coordination of the direction and extent of cell expansion are particularly important for growth and development. The plant hormone gibberellins (GAs) play key roles in the control of these developmental processes. However, little is known about the molecular components that integrate the generic GA signaling into a specific cell/tissue to coordinate cell division and cell expansion. Here we report that SCARECROW-LIKE 3 (SCL3), a GRAS protein, acts as a positive regulator to integrate and maintain a functional GA pathway by attenuating the DELLA repressors in the root endodermis. The tissue-specific maintenance of GA signaling in the root endodermis plays distinct roles along the longitudinal root axis. While in the elongation/differentiation zone (EDZ), the endodermis-confined GA pathway by SCL3 controls primarily coordination of root cell elongation; in the meristem zone (MZ) SCL3 in conjunction with the SHORT-ROOT/SCARECROW (SHR/SCR) pathway controls GA-modulated ground tissue maturation. Our findings highlight the regulatory network of the GRAS transcription regulators (SCL3, DELLAs, and SHR/SCR) in the root endodermis, shedding light on how GA homeostasis is achieved and how the maintenance of GA signaling controls developmental processes in roots.
Keywords: formative division, hormonal regulation, middle cortex formation, root development
Because no cell movement takes place during plant development, control of cell division (formative and proliferative) and coordination of cell expansion are crucial in growth and development. The plant hormone gibberellins (GAs) play key roles in the control of these developmental processes (1–6). With molecular, genetic, and biochemical approaches, molecular components of GA signaling have been well characterized (7, 8). The soluble GA receptors GIBBERELLIN INSENSITIVE DWARF 1 (GID1) interact with DELLA proteins (DELLAs), major negative regulators of GA signaling, in a GA-dependent manner (9–13). Bioactive GAs promote the GID1–DELLA interaction and, in turn, lead to rapid degradation of the DELLA repressors via the ubiquitin/26S proteasome pathway (11–14). DELLAs accumulate when bioactive GA levels are low, whereas degradation of DELLAs is accelerated when bioactive GA levels are elevated. Thus, bioactive GAs act as “inhibitors of inhibitors” (7), promoting degradation of the DELLA repressors and thus GA-mediated growth responses (14). DELLAs belong to the GRAS transcription regulator family (9, 10, 15, 16), and in Arabidopsis thaliana there are five DELLAs (GAI, RGA, RGL1, RGL2, and RGL3), which have overlapping but distinct roles in plant growth and development (7–10, 17). Although the GA signaling pathway has become increasingly well characterized, still little is known about its integration into a specific cell/tissue to regulate developmental processes in the plant life cycle.
The SCARECROW (SCR) and SHORT-ROOT (SHR) transcription regulators, which also belong to the GRAS family, control specification of stem cell niche (18) and ground tissue formation in the Arabidopsis root (19, 20). Mutations in SCR and SHR cause defects in the formative periclinal (parallel to the growth axis) division that generates cortex and endodermis (19, 20). Recent work demonstrated that the SHR/SCR pathway, in conjunction with the GA pathway, controls the timing and extent of additional formative periclinal division for endodermis and additional cortex (termed middle cortex; MC) at later stages (2, 4, 21). These findings imply the involvement of additional regulatory components to integrate the GA signaling pathway in the root endodermis, because neither SHR nor SCR is subject to regulation by bioactive GAs or GA signaling per se. Recently, it was also shown that GA signaling controls cell proliferation in the root meristem (5, 6) and that the endodermis-specific disruption of GA signaling results in uncoordinated cell expansion in the root (3). However, the molecular components that integrate the generic GA signaling into the root endodermis to coordinate cell division and cell expansion are largely unknown.
In this study, we show that the GRAS transcription regulator SCARECROW-LIKE 3 (SCL3) serves as a tissue-specific integrator of the GA pathway in the Arabidopsis root endodermis. Our genetic and physiological results indicate that SCL3, acting downstream of RGA, is likely a positive regulator in GA signaling. Furthermore, our findings reveal that the spatial integration of the GA pathway by SCL3 plays distinct roles along the longitudinal root axis. In the elongation/differentiation zone (EDZ), SCL3, acting as an attenuator of GAI and RGA, controls coordination of root cell elongation. In the meristem zone (MZ), the maintenance of a functional GA signaling by SCL3–DELLA interaction, in conjunction with the SHR/SCR pathway, modulates the timing and extent of the formative division for ground tissue maturation.
Results and Discussion
SCL3 Acts as an Integrator of the GA/DELLA and SHR/SCR Pathways.
Recent microarray analysis revealed that among GA/DELLA-regulated genes, RGA was associated with the promoter of SCL3, which also belongs to the GRAS family, like DELLAs (22). In addition, the SHR and SCR transcription regulators, key in root radial patterning (19, 20), form a heterodimer that regulates SCL3 transcription being associated with its promoter (23, 24). Thus, we started with the premise that SCL3 acts downstream of both the GA and SHR/SCR pathways, serving as a convergent point.
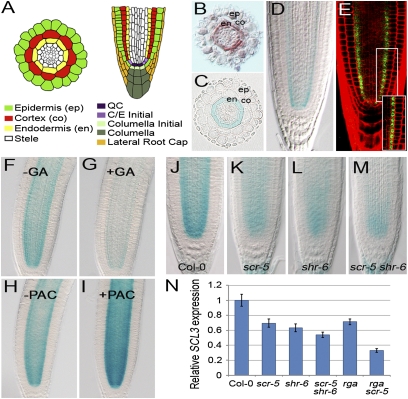
In the Arabidopsis root (Fig. 1A), mRNA in situ hybridization analysis showed that SCL3 transcripts localized primarily to the endodermis (Fig. 1B) (16). Additionally, a transcriptional fusion between the SCL3 promoter and β-glucuronidase (GUS) (pSCL3::GUS) basically recapitulated the mRNA in situ hybridization pattern (Fig. 1 C and D). Furthermore, a translational fusion to green fluorescent protein (GFP) (pSCL3::SCL3-GFP) conferred nuclear localization in the cells of the quiescent center (QC), cortex/endodermis initial (CEI), and endodermis (Fig. 1 A and E), suggesting that SCL3 acts as a transcription regulator.
Fig. 1.
Transcriptional control of SCL3 in the root by the GA/DELLA and SHR/SCR pathways. (A) Schematic of the Arabidopsis root. (B–E) Localization of SCL3 mRNA and its protein. Expression of SCL3 is detected primarily in the endodermis by in situ hybridization (B) and a transcriptional fusion (pSCL3::GUS) (C and D). Blue staining is observed in a manner similar to the SCL3 in situ hybridization pattern. (E) Translational fusion (pSCR::SCL3-GFP) in the root. GFP signals are observed in the nuclei of the QC and the endodermis lineage. (F and G) pSCL3::GUS in WT in the absence (F) or presence (G) of 10 μM of exogenous GA3 for 6 h. (H and I) pSCL3::GUS in WT in the absence (H) or presence (I) of 10 μM of PAC for 6 h. Expression of SCL3 is modulated by bioactive GA contents. (J–M) Expression of pSCL3::GUS in Col-0 (J), scr-5 (K), shr-6 (L), and scr-5 shr-6 (M). (N) qRT-PCR of SCL3 transcripts in Col-0, scr, shr, scr shr, rga, and rga scr roots. The SCL3 transcript level is substantially reduced in rga scr double mutants. The SCL3 mRNA level in Col-0 is arbitrarily set to 1. Error bars indicate SD from three biological replicates.
Next, we analyzed expression of SCL3 with the pSCL3::GUS transcriptional fusion and reverse transcription-based quantitative PCR (qRT-PCR). In the presence of exogenous bioactive GA (GA3), SCL3 expression was reduced substantially (Fig. 1 F and G and Fig. S1). By contrast, its expression was up-regulated by the GA biosynthesis inhibitor paclobutrazol (PAC) (Fig. 1 H and I and Fig. S1). Consequently, in the GA-deficient ga1-3 mutant (25), SCL3 expression level was higher than that in wild type (WT), and it was modulated by bioactive GA levels: decreased by GA3 and increased by PAC (Fig. S1). Our data are in agreement with the previous findings that SCL3 expression is modulated by bioactive GA levels (22). We further investigated the SCL3 transcript levels in loss-of-function mutants of DELLAs: gai (SALK_082622), rga (SALK_089146), and gai rga and della (gai rga rgl1 rgl2 rgl3). As reported previously (22), SCL3 expression is regulated, at least in part, through accumulation of DELLAs that is modulated by bioactive GA contents.
It is noteworthy that mRNA and protein localization of SCL3 in the QC, CEI, and endodermis exactly overlaps with the spatial domains of SCR mRNA and protein as well as SHR protein, implying a molecular interplay between SCL3 and the SHR/SCR module at the cellular levels. We therefore examined the expression pattern of SCL3 in scr-5, shr-6, and scr-5 shr-6 mutants. In scr-5, the expression level of SCL3 was reduced to ∼70%, and in shr-6 its expression was about 60%, compared with that in WT roots (Fig. 1 J–L and N). We also found a reduction of the SCL3 expression level in scr-5 shr-6 (∼50% of the WT level) (Fig. 1 M and N), further verifying that SCL3 transcription in the root is controlled by the SHR/SCR pathway.
Intriguingly, we observed only modest reduction of SCL3 expression even in the presence of exogenous GA application or with mutations in both SCR and SHR. Thus, we reasoned that SCL3 transcription is modulated by both the GA/DELLA and SHR/SCR pathways. To test this, we generated rga scr double mutants that harbor loss-of-function mutations in the two direct upstream genes and investigated the SCL3 expression. Indeed, the level of SCL3 mRNA was markedly reduced to only 30% of WT roots (Fig. 1N). Our findings indicate that SCL3 likely plays as a cell/tissue-specific integrator, acting downstream of both the GA/DELLA and SHR/SCR pathways.
SCL3 Acts as a Positive Regulator in the GA Response Pathway.
Because SCL3 expression is actively modulated by bioactive GAs through DELLAs, we initially postulated that SCL3 likely acts as another negative regulator in GA signaling. To test this, we first used a genetic approach by introducing either loss or gain of SCL3 function in backgrounds with defective GA biosynthesis and GA response. Interestingly, the scl3 ga1-3 double mutant exhibited a shorter root phenotype compared with ga1-3 in our root growth assay (Fig. S2 A and C). In contrast, root growth of the SCL3 overexpressors (35S::SCL3) in ga1-3 was indistinguishable from that of ga1-3 (Fig. S2 A and C). In the presence of exogenous GA3, the phenotype of scl3 ga1-3 was restored, even though both ga1-3 and scl3 ga1-3 roots were slightly shorter than WT roots (Fig. S2 B and C). Furthermore, root growth of scl3 was more sensitive to PAC treatment, whereas 35S::SCL3 seedlings conferred resistance to PAC (Fig. S2 D and E). Additionally, we observed that both rga and scl3 rga were indistinguishable in root growth in the presence of PAC (Fig. S2 D and E), indicating that RGA is epistatic to SCL3. Taken together, our observations indicate that SCL3 is likely a positive regulator in GA signaling. In accordance with our findings, Zhang et al. (26) in this issue of PNAS describe the molecular details on the role of SCL3 as an activator in the GA response pathway.
To maintain GA homeostasis, transcription of GA metabolism genes is subject to feedback regulation in which DELLA repressors play key roles (27, 28). Like DELLAs, SCL3 also belongs to the GRAS transcription regulator family (15, 16, 29). Thus, to investigate the involvement of SCL3 in the maintenance of GA homeostasis, we analyzed expression profiles of the GA metabolism genes with qRT-PCR in the absence or presence of exogenous GA or PAC, as well as in ga1-3 and scl3 ga1-3. Of the GA metabolism genes examined, transcript levels of GA20ox1, GA20ox2, and GA20ox3 were notably up-regulated in scl3 under GA-deficient conditions (in the presence of PAC or ga1-3) (Fig. S2 F and G). Our findings are in accordance with the results by Zhang et al. (26) in this issue of PNAS that the transcription of GA20ox1, GA20ox2, and particularly GA20ox3 is subject to regulation by the SCL3 transcription regulator to maintain GA homeostasis.
Endodermis-Confined GA Signaling by SCL3 Coordinates Root Cell Elongation in the EDZ.
Recent reports demonstrated that GA controls root meristem size by promoting proliferative cell divisions (5, 6) and that GA response in the root endodermis also coordinates cell elongation (3). Endodermis-specific SCL3 expression and root growth inhibition of scl3 in GA-deficiency raised the question as to whether SCL3 plays a role in promoting cell division, cell elongation, or both. To address this question, we analyzed GA-mediated root growth of WT and scl3 seedlings in the absence or presence of ga1-3 or PAC. As described previously (5, 6), we measured the number and length of ground cells from the QC as parameters for root meristem size (Fig. S3A). Under standard conditions, root meristem sizes of both WT and scl3 seedlings were indistinguishable (Fig. S3 B and C). We also monitored the CYCB1::GUS mitotic marker that was shown to correlate with division potential in the root meristem (5, 6). In the presence of PAC, the number of dividing cells was reduced in scl3 to a level similar to that of WT roots (Fig. S3 D–F). In addition, the root meristem size of scl3 ga1-3 double mutants was indistinguishable from that of ga1-3 (Fig. S3 G and H), suggesting that GA-mediated cell proliferation in the root meristem is likely independent of SCL3 function. Thus, we investigated the role of SCL3 in cell elongation, because rapid elongation of cells that exit from the meristem drives postembryonic root growth (30). In the presence of PAC or ga1-3, root cell elongation in scl3 appeared to be reduced when compared with WT, whereas 35S::SCL3 roots were PAC resistant (Fig. S4 A and B). We also found that scl3 rga exhibited a PAC-resistant phenotype in cell elongation indistinguishable from rga (Fig. S4A), further corroborating that SCL3 acts downstream of RGA. Notably, root cell elongation of scl3, 35S::SCL3, rga, and scl3 rga correlated well with the EDZ length of the individuals (Fig. S4 C and D), suggesting that the sum of each cell's elongation contributes primarily to the EDZ length, and consequently to the whole root length.
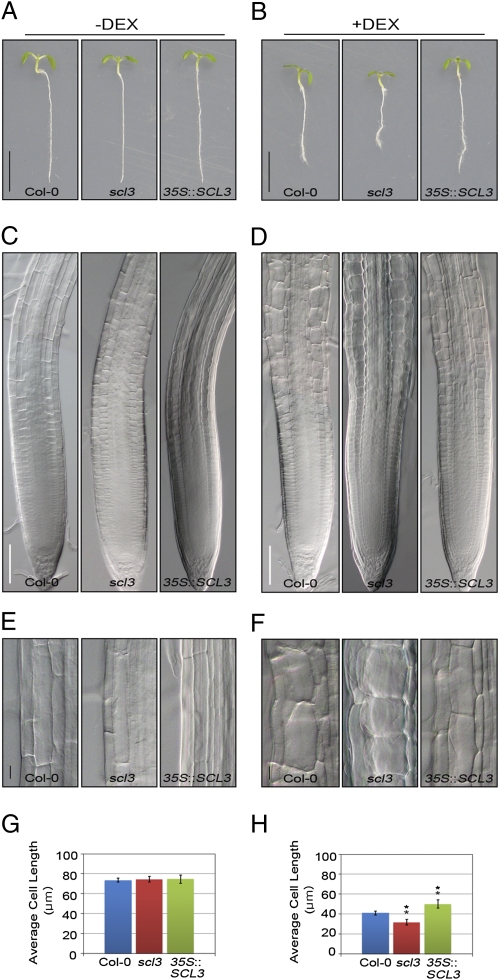
Recently, the endodermis-specific disruption of a functional GA response by expressing the nondegradable gai under the SCR promoter (pSCR::gai-GR-YFP) was shown to affect root cell elongation and cell morphology in the EDZ (3). Thus, we introgressed the pSCR::gai-GR-YFP construct into WT, scl3, and 35S::SCL3 plants, respectively. In the absence of dexamethasone (−DEX), we found no difference in root cell elongation in WT, scl3, and 35S::SCL3 seedlings (Fig. 2 A, C, E, and G). When induced with DEX (+DEX), root growth of pSCR::gai-GR-YFP seedlings in the WT background was reduced (Fig. 2B), and cells in the EDZ showed inhibition of elongation (Fig. 2 D and F). Notably, cell elongation of pSCR::gai-GR-YFP seedlings in scl3 was more severely inhibited (Fig. 2 D and F). Under prolonged DEX treatment (+DEX for 3 d), the surface of pSCR::gai-GR-YFP roots in scl3 appeared bulged because the direction of cortex cell elongation was shifted perpendicularly compared with the untreated roots (Fig. S5 A and B). As a result, the average length of individual cells in scl3 was more significantly reduced than in the WT background (Fig. 2H and Fig. S5 B and C). We also observed similar results in WT and scl3 roots harboring the pSCR::rga-GR-YFP construct (Fig. S5 D–F). Interestingly, 35S::SCL3 seedlings with pSCR::gai-GR-YFP suppressed the inhibition of cell elongation when induced with DEX (Fig. 2 B, D, F, and H). These observations indicate that SCL3 attenuates the effects of nondegradable gai and rga in the root endodermis. The results from ref. 26 in this issue of PNAS lend support for the notion that SCL3 acts as an attenuator of DELLAs. Thus, the maintenance of GA signaling by SCL3–DELLA interaction in the endodermis controls coordination of cell elongation for root growth.
Fig. 2.
Regulation of GA-mediated root cell elongation by SCL3–DELLA interaction in the EDZ. The pSCR::gai-GR-YFP seedlings in Col-0, scl3, and 35S::SCL3 were grown in MS agar plates for 4 d, transferred to MS agar plates supplemented with or without 10 μM of DEX, and incubated for another 3 d (A and B) or for 12 h (C–H). In the absence of DEX (−DEX), neither inhibition of root growth nor cell elongation was observed (A, C, E, and G). In the presence of 10 μM of DEX (+DEX) (B, D, F, and H), pSCR::gai-GR-YFP seedlings in Col-0 showed inhibition of root growth and cell elongation. Root growth and cell elongation of pSCR::gai-GR-YFP seedlings in scl3 were more severely inhibited. Notably, overexpression of SCL3 (pSCR::gai-GR-YFP in 35::SCL3) suppressed the inhibition of root growth and cell elongation. As a result, the average cell length is reduced significantly in scl3, whereas increased in 35S::SCL3 (n > 20) (H). (Scale bars, 5 mm in A and B; 30 μm in C and D; and 10 μm in E and F, respectively.) Statistical significance of differences was determined by Student's t test (error bars: SE, **P < 0.01).
SCL3 and Bioactive GA Levels Modulate the Timing and Extent of the Formative Division for Ground Tissue Maturation.
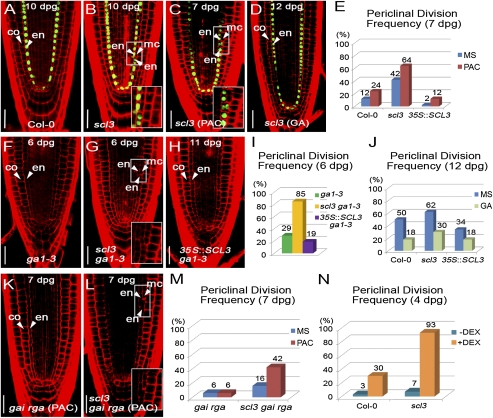
The asymmetric cell division that generates cortex and endodermis is established during embryogenesis and perpetuated in postembryonic development (31). At later stages, Arabidopsis seedlings undergo the additional formative division giving rise to endodermis and another cortex (MC) for ground tissue maturation (21). Recent work demonstrated that the GA pathway, in conjunction with the SHR/SCR module, controls the timing and extent of MC formation (2, 4). Our results that SCL3 serves as an integrator of the GA/DELLA and SHR/SCR pathways in the endodermis raised the possibility that SCL3 plays a role in the regulation of MC formation. To test this possibility, we focused on the timing and extent of the formative division in WT, scl3, and 35S::SCL3 roots. In WT roots, occurrence of the formative division was first observed at 6 d postgermination (dpg), and half of the seedlings in a given population showed MC formation at 12 dpg under our experimental conditions (Fig. S6A). As roots became mature, scl3 seedlings had already undergone the periclinal division (Fig. 3 A and B). For instance, only 12% of WT roots exhibited the formative ground tissue division, whereas 42% of scl3 seedlings had already formed MC at 7 dpg (Fig. 3E and Fig. S6A). By contrast, 35S::SCL3 roots at 7 dpg exhibited a delay in occurrence of the formative division compared with WT roots (2 vs. 12%) (Fig. 3E and Fig. S6A). Interestingly, a decreased frequency of MC formation conferred by 35S::SCL3 had diminished from 9 dpg onward, implying that SCL3 is likely subject to its own regulation, as demonstrated by Zhang et al. (26) in this issue of PNAS. Our data indicate that SCL3 modulates the timing and frequency of the formative ground tissue division for MC formation.
Fig. 3.
Control of the formative ground tissue division in the MZ by the interaction of SCL3 and GA/DELLA pathways. MC formation in Col-0 (A) and scl3 roots (B) at 10 dpg. The scl3 root exhibited the additional formative periclinal division giving rise to endodermis (en) and middle cortex (mc). Arrowheads indicate endodermis (en), middle cortex (mc), and cortex (co), respectively. The nascent MC layer loses the endodermis identity marked by pSCR::GFP-SCR (Insets in B and C). Occurrence of the formative division in scl3 in the presence of 1 μM of PAC (C) and 10 μM of GA3 (D). The treatment with PAC increased, whereas exogenous GA3 decreased the MC formation. (E) Quantitative evaluation of MC formation confirms that GA deficiency causes an increased frequency of MC formation in Col-0, scl3, and 35::SCL3. In addition, loss and gain of SCL3 function also have opposing effects on the MC formation: increase in scl3 and decrease in 35::SCL3. (F–I) Occurrence of MC formation in ga1-3, scl3 ga1-3, and 35::SCL3 in ga1-3. Similarly, the frequency of the formative division was also modulated by loss and gain of SCL3 function. (J) Quantitative evaluation of MC formation confirms that exogenous bioactive GA, conversely, decreases the frequency in Col-0, scl3, and 35::SCL3. (K–M) MC formation in gai rga and scl3 gai rga in the absence or presence of 1 μM of PAC. As predicted, gai rga roots were extremely resistant to PAC, whereas scl3 gai rga triple mutants showed far more increased occurrence of the formative division in the presence of PAC. (N) In the absence or presence of DEX, the pSCR::gai-GR-YFP seedlings in Col-0 and scl3 were grown in MS agar plates for 4 d, transferred to MS agar plates supplemented with or without 10 μM of DEX, and incubated for 12 h. In the presence of DEX (+DEX), pSCR::gai-GR-YFP seedlings in scl3 showed a dramatically increased frequency of MC formation compared with pSCR::gai-GR-YFP seedlings in Col-0. (Scale bar, 30 μm.)
Previously, it was shown that GA deficiency (either by PAC or ga1-3) caused premature MC formation, and conversely exogenous bioactive GA application delayed MC formation, indicating that bioactive GA levels regulate ground tissue maturation (2, 4). Thus, we next investigated the mutual relation between SCL3 and bioactive GA levels in the control of the timing and extent of the formative division. In the presence of PAC, WT roots showed precocious MC formation compared with the untreated WT roots at 7 dpg (24 vs. 12%) (Fig. 3E and Fig. S6 B and C). In the same GA-deficient condition, scl3 roots also showed an increase in frequency of MC formation compared with the untreated scl3 roots (64 vs. 42%) (Fig. 3 B, C, and E). Similarly, we observed an increased frequency of MC formation in scl3 ga1-3 than in ga1-3 roots (85 vs. 29% at 6 dpg) (Fig. 3 F, G, and I). These observations indicate that scl3 and GA deficiency additively or synergistically cause an increased frequency of precocious MC formation. In contrast, 35S::SCL3 roots in GA-deficient conditions (by PAC or ga1-3) showed a resistant phenotype in the MC formation compared with WT and scl3 (Fig. 3 H and I). In the presence of exogenous GA3, only 18% of WT roots exhibited MC formation at 12 dpg compared with the untreated WT (50%) (Fig. 3J and Fig. S6 D and E). Similarly, when applied with exogenous GA3, scl3 roots showed a decreased frequency of MC formation at 12 dpg compared with the untreated scl3 (30 vs. 62%) (Fig. 3 D and J). In addition, application of exogenous GA3 further reduced the frequency of MC formation in 35S::SCL3 roots compared with the untreated 35S::SCL3 (18 vs. 34%) (Fig. 3J). Taken together, our results indicate that SCL3 and bioactive GA levels additively or synergistically regulate the timing and extent of the formative division for ground tissue maturation.
Maintenance of a Functional GA Signaling by SCL3–DELLA Interaction Regulates the MC Formation.
Next, we interrogated genetically the mutual relation between SCL3 and GA response in MC formation. To this end, we generated gai rga double mutants (loss-of-function mutations in two major DELLAs) and scl3 gai rga triple mutants. Under standard conditions, gai rga roots showed a decreased frequency of MC formation compared with scl3 gai rga roots (6 vs. 16%) (Fig. 3M). In the presence of PAC, gai rga roots, as predicted, were extremely PAC resistant. Surprisingly, the difference in the frequency of MC formation between gai rga and scl3 gai rga became clearly evident at 7 dpg (6 vs. 42%) in the presence of PAC (Fig. 3 K–M). Our data indicate that a PAC-sensitive phenotype of scl3 gai rga in MC formation is likely conferred by loss of SCL3 function. The results from ref. 26 in this issue of PNAS also support the notion that SCL3 regulates its own expression to maintain a functional GA signaling. Next, we examined the MC formation in roots with defects in GA response by targeting the nondegradable rga in the endodermis under the SCR promoter (pSCR::rga-GR-YFP). In the presence of DEX, pSCR::rga-GR-YFP in WT showed an ∼10-fold increase in the occurrence of the MC formation compared with the untreated seedlings (Fig. 3N). Notably, pSCR::rga-GR-YFP in scl3 seedlings dramatically increased the frequency of the MC formation in the given population (Fig. 3N), further corroborating that SCL3 attenuates the endodermis-specific disruption of GA signaling conferred by the GA-insensitive rga function.
SCL3 and the SHR/SCR Module Regulate the MC Formation.
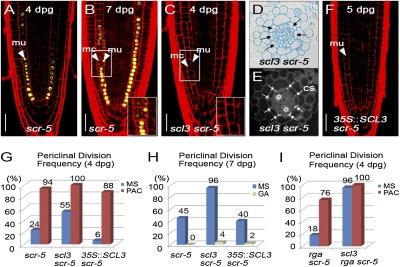
At later stages, the radial patterning mutant scr also occasionally undergoes the additional formative division, resulting in the sporadic production of two ground tissue layers: an inner layer with fused characteristics of endodermis and cortex (termed mutant layer; mu) and an outer layer with cortex characteristics (Fig. 4 A and B) (2, 4). Thus, we genetically interrogated the mutual relation between SCL3 and the SHR/SCR module in the control of the formative division. We generated the double mutant combinations of scl3 shr and scl3 scr, respectively, and examined the MC formation. In scl3 shr double mutants, we observed no additional formative division regardless of PAC or GA3, resulting in the single ground tissue layer (Fig. S7 A–H). These observations are in accordance with the previous results that SHR is required for the formative ground tissue division (2). Interestingly, scl3 scr roots showed much earlier occurrence of the formative division than in scr; at 4 dpg, 55% of scl3 scr had already generated two ground tissue layers (Fig. 4 C–E and Fig. S7I). By contrast, 35S::SCL3 in scr delayed the MC formation compared with scr roots (8 vs. 32% at 5 dpg) (Fig. 4F and Fig. S7I). In addition, we found no effects of 35S::SCL3 on the delay in the MC formation from 6 dpg onward (Fig. S7I), further corroborating that SCL3 regulates its own expression. Our findings indicate that scl3 and scr additively or synergistically cause precocious MC formation.
Fig. 4.
Control of MC formation by the interplay of the SCL3, SHR/SCR, and GA/DELLA pathways. (A and B) MC formation in scl3 roots marked by pCO2::H2B-YFP. At 7 dpg, scl3 roots showed the additional formative periclinal division giving rise to two ground tissue layers. Arrowheads indicate mutant layer (mu) and middle cortex (mc), respectively. (C–E) Occurrence of the formative division in scl3 scr. The scl3 scr double mutants exhibited uncoordinated formative divisions, which generate an inner layer with the Casparian stripe (CS) and an additional cortex layer. Black arrows indicate the additional formative division (D), and white arrows indicate the Casparian stripe (CS) (E). (F) 35::SCL3 in scr reduced the occurrence of the formative division (at 5 dpg). Quantitative evaluation of MC formation in scr, scl3 scr, and 35::SCL3 in scr in the absence or presence of PAC (G) or GA3 (H). Bioactive GA levels additively or synergistically have influence on MC formation with opposing effects: increase by PAC and decrease by GA3. (I) Occurrence of MC formation in rga scr and scl3 rga scr in the absence or presence of 1 μM of PAC. The scl3 rga scr roots showed a far more increased frequency of the formative division regardless of PAC. (Scale bar, 30 μm.)
Interplay of the SCL3, SHR/SCR, and GA/DELLA Modules Regulates the MC Formation.
The results of our genetic experiments between scl3 and mutants defective in the GA pathway and in the SHR/SCR module indicate that the two pathways converge on SCL3 to control the formative ground tissue division. To interrogate the interplay of the GA/DELLA and SHR/SCR pathways, we examined the MC formation in scr, scl3 scr, and 35S::SCL3 in scr in the presence of PAC or GA3. Interestingly, all of the seedling combinations including scr single mutants were sensitive to PAC (10 nM), resulting in premature MC formation (Fig. 4G). In contrast, scr, scl3 scr, and 35S::SCL3 in scr showed inhibition of MC formation in the presence of GA3 (Fig. 4H). These findings suggest that bioactive GA levels in combination with scl3 and scr have a more profound effect on the MC formation. We further genetically analyzed the MC formation in rga scr (loss-of-function mutations in the two direct upstream genes of SCL3) in the absence or presence of SCL3 function. In the presence of PAC, the frequency of MC formation in both rga scr and scl3 rga scr increased substantially (Fig. 4I), indicating that a PAC-resistant phenotype of rga disappeared in the scr background. Notably, scl3 rga scr triple mutants exhibited a dramatically increased MC formation even in the absence of PAC compared with rga scr (96 vs. 18%), resulting in a lack of control in the MC formation (Fig. 4I). Our findings suggest that SCL3–DELLA interaction, in conjunction with the SHR/SCR module, plays an important role in the GA-mediated coordination of MC formation for ground tissue maturation.
Conclusions
Bioactive GAs promote growth by regulating cell division and cell elongation (1–8). Although the molecular components of GA signaling have become available and increasingly well characterized, still little is known about the spatial integration of the GA pathway to control these developmental processes in the plant life cycle. Here, we provide compelling evidence that SCL3 is a tissue-specific integrator of the GA pathway. Acting as a positive regulator by attenuating the DELLA repressors, SCL3 maintains GA homeostasis. Along the longitudinal root axis, the maintenance of GA signaling by SCL3–DELLA interaction in the endodermis plays distinct roles: coordination of cell elongation in the EDZ (Fig. S8A) and of MC formation in the MZ (Fig. S8B). In the EDZ, the loss of SCL3 function aggravates uncoordinated cell expansion caused by endodermis-specific disruption of GA signaling, whereas the gain of SCL3 function restores cell elongation. Intriguingly, the SCL3–DELLA interaction for the maintenance of the GA pathway also employs the SHR/SCR module in the MZ to control MC formation. In the MZ, scl3 roots show an increase of MC formation, whereas roots of 35S::SCL3 delay the timing and extent of the ground tissue division. Moreover, bioactive GA levels in combination with the SHR/SCR module additively or synergistically act in the control of MC formation: promotion by GA deficiency and inhibition by exogenous bioactive GA. Notably, scl3 rga scr triple mutants exhibited a lack of control in the formative division even under normal conditions, resulting in premature MC formation. Thus, our results reveal the network of the GRAS transcription regulators (SCL3, DELLAs, and SHR/SCR) in the endodermis of the MZ for the coordination of MC formation. This study and Zhang et al. (26) in this issue of PNAS shed light on how GA homeostasis is achieved and how the maintenance of GA signaling controls developmental processes in roots. Together with recent work in the shoot, in which the brassinosteroid signaling pathway in the epidermis modulates shoot growth (32), our study provides a molecular framework for the cell/tissue-specific integration of hormone signaling pathways in plant growth and development.
Materials and Methods
Plant Material and Growth Conditions.
All plants were of A. thaliana Columbia (Col-0) background except for della quintuple mutant in Landsberg erecta (Ler). Origins of mutant and transgenic lines are described in SI Materials and Methods. Seeds were sterilized and incubated as previously described (29, 33). Primers used for genotyping are listed in Table S1.
Treatment and Root Assay.
Root growth analysis was performed as described previously (6). For detailed description on the analysis of root meristem size and periclinal division, see SI Materials and Methods.
Molecular Cloning and Transgenic Plants.
To generate pSCL3::GUS, pSCL3::SCL3-GFP, and 35S::SCL3, the Gateway recombination cloning technology (Invitrogen) was used as described previously (29, 33). Primers used for plasmid construction are listed in Table S2. For detailed description on plasmid construction and transgenic plant production, see SI Materials and Methods.
Expression Analysis.
Total RNA extraction and reverse transcription-associated quantitative RT-PCR (qRT-PCR) were performed as described previously (29, 33). Primers used for qRT-PCR are listed in Table S3. mRNA in situ hybridization was performed as described previously (19, 20). (See SI Materials and Methods for detailed procedures.)
Histology and Microscopy.
GUS histochemical staining was performed and visualized as described previously (29, 33). Confocal laser scanning microscopy was performed as described previously (29). (See SI Materials and Methods for detailed description.)
Supplementary Material
Acknowledgments
We thank Philip Benfey, Malcom Bennett, John Celenza, Renze Heidstra, Ben Scheres, Tai-ping Sun, Susana Ubeda-Tomás, the Arabidopsis Biological Resource Center, and the Nottigham Arabidopsis Stock Center for materials. We are grateful to Tai-ping Sun for sharing unpublished results and critical reading of the manuscript. We also thank Philip Benfey for critical reading and suggestions. This work was supported by National Research Foundation Grant 2009-0079304/R0602494 (to M.M.L.) and Grant NRF2009-0077753 (to J.L.) and by the Konkuk University (J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012215108/-/DCSupplemental.
References
- 1.Davies PJ, editor. Plant Hormones: Physiology, Biochemistry and Molecular Biology. The Netherlands: Kluwer, Dordrecht; 2004. [Google Scholar]
- 2.Paquette AJ, Benfey PN. Maturation of the ground tissue of the root is regulated by gibberellin and SCARECROW and requires SHORT-ROOT. Plant Physiol. 2005;138:636–640. doi: 10.1104/pp.104.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ubeda-Tomás S, et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10:625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]
- 4.Cui H, Benfey PN. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009;58:1016–1027. doi: 10.1111/j.1365-313X.2009.03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achard P, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19:1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- 6.Ubeda-Tomás S, et al. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Harberd NP, Belfield E, Yasumura Y. The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell. 2009;21:1328–1339. doi: 10.1105/tpc.109.066969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun T-p. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2008. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng J, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silverstone AL, Ciampaglio CN, Sun T-p. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell. 1998;10:155–169. doi: 10.1105/tpc.10.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murase K, Hirano Y, Sun T-p, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 12.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 13.Willige BC, et al. The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell. 2007;19:1209–1220. doi: 10.1105/tpc.107.051441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang C, Fu X. GA action: Turning on de-DELLA repressing signaling. Curr Opin Plant Biol. 2007;10:461–465. doi: 10.1016/j.pbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–692. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 16.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 17.Tyler L, et al. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135:1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Laurenzio L, et al. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- 20.Helariutta Y, et al. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell. 2000;101:555–567. doi: 10.1016/s0092-8674(00)80865-x. [DOI] [PubMed] [Google Scholar]
- 21.Baum SF, Dubrovsky JG, Rost TL. Apical organization and maturation of the cortex and vascular cylinder in Arabidopsis thaliana (Brassicaceae) roots. Am J Bot. 2002;89:908–920. doi: 10.3732/ajb.89.6.908. [DOI] [PubMed] [Google Scholar]
- 22.Zentella R, et al. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levesque MP, et al. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui H, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 25.Sun T-p, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z-L, et al. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:2160–2165. doi: 10.1073/pnas.1012232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedden P, Phillips AL. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 28.Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M-H, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- 30.Beemster GT, Baskin TI. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 1998;116:1515–1526. doi: 10.1104/pp.116.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 32.Savaldi-Goldstein S, Peto C, Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199–202. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- 33.Yu N-I, et al. Characterization of SHORT-ROOT function in the Arabidopsis root vascular system. Mol Cells. 2010;30:113–119. doi: 10.1007/s10059-010-0095-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.