Abstract
Cancer cells use different modes of migration, including integrin-dependent mesenchymal migration of elongated cells along elements of the 3D matrix as opposed to low-adhesion-, contraction-based amoeboid motility of rounded cells. We report that MDA-MB-231 human breast adenocarcinoma cells invade 3D Matrigel with a characteristic rounded morphology and with F-actin and myosin-IIa accumulating at the cell rear in a uropod-like structure. MDA-MB-231 cells display neither lamellipodia nor bleb extensions at the leading edge and do not require Arp2/3 complex activity for 3D invasion in Matrigel. Accumulation of phospho-MLC and blebbing activity were restricted to the uropod as reporters of actomyosin contractility, and velocimetric analysis of fluorescent beads embedded within the 3D matrix showed that pulling forces exerted to the matrix are restricted to the side and rear of cells. Inhibition of actomyosin contractility or β1 integrin function interferes with uropod formation, matrix deformation, and invasion through Matrigel. These findings support a model whereby actomyosin-based uropod contractility generates traction forces on the β1 integrin adhesion system to drive cell propulsion within the 3D matrix, with no contribution of lamellipodia extension or blebbing to movement.
Keywords: 3D migration, integrin, invasion
During metastasis, tumor cells encounter various extracellular matrix (ECM) environments with distinct composition and architecture, including basement membrane (BM) and interstitial collagen networks (1–3). Although basic mechanisms of cell motility on 2D substrata are generally well understood, much less is known regarding the mechanics of cell migration in 3D matrix environments (1, 4–7). Recent evidence supports the conclusions that 2D and 3D migration can differ considerably and that different types of 3D motility exist depending on the biophysical properties of the 3D environment (viscoelasticity, confinement, porosity) (8–15). Understanding the mechanics of cancer cell migration in these different 3D environments is therefore of paramount importance.
Schematically, two modes of 3D migration of invasive cells have been described; the models differ by the dependency on actomyosin contractility, requirement for integrin adhesion, and matrix remodeling. In the mesenchymal mode, which has some parallel with 2D migration of fibroblasts (1), invasive cells are elongated and require pericellular matrix proteolysis for extending Rac-dependent F-actin-based leading pseudopodia in an integrin-dependent manner (16–20). In contrast, some carcinoma cells invade with a low-adhesion amoeboid mode of migration, characterized by a round morphology, no stable intrinsic cell polarity, and a required high level of RhoA/ROCK-driven actomyosin contractility (17, 18). Amoeboid cell motility, which is typically associated with bleb formation, is independent of proteases, and it is generally thought that by forming blebs, tumor cells can squeeze themselves through preexisting voids in the matrix (5, 21). In vivo, bleb-based motility has been implicated in directional migration of primordial germ cells in zebrafish embryo (22). Depending on levels of RhoA, Rac, and Cdc42, matrix degradation capacity, and architecture of the ECM, carcinoma cells can perform different modes of motility in 3D and, hence, are thought to adapt to variable environmental clues (17, 18, 20, 23–25).
Several methods have been developed for visualizing and analyzing forces generated by cells during 2D migration. These studies revealed that propulsive forces that are produced by myosin II on the F-actin network, which is linked to adhesion sites underneath the protruding lamellipodium, support migration (26, 27). However, mechanics underlying 2D and 3D migration differ quite significantly (8–11, 13–15), and force repartition is generally unknown for cells migrating in 3D. Tracking of embedded beads within the matrix has been recently used to follow matrix deformations during mesenchymal migration of fibrosarcoma cells in a 3D type I collagen network (28).
In the present study, we address the mechanics of 3D migration of highly invasive MDA-MB-231 human breast cancer cells in Matrigel, a reconstituted BM matrix, by tracking micrometer-scale movements of fluorescent microbeads seeded in the gel. We found that MDA-MB-231 cells invade and migrate in 3D Matrigel with a striking round morphology, and they display neither lamellipodial extension nor blebbing at the leading edge, while cells form an F-actin/myosin II-rich uropod as a generator of contractile forces at the cell rear. Actomyosin-based contractility at the uropod, which is transmitted to and exerts traction forces on the ECM through β1 integrins, powers migration within the 3D matrix.
Results
Mechanics of MDA-MB-231 Rounded Cell 3D Migration.
Highly metastatic MDA-MB-231 cells stably expressing mCherry-Lifeact, which allows visualization of F-actin dynamics with minimal interference with actin assembly (29, 30), were seeded in 3D Matrigel matrix together with fluorescent microbeads (0.2-μm diameter) and followed by time-lapse fluorescence microscopy. MDA-MB-231 cells moved in random directions in 3D Matrigel with an average speed of 3.2 ± 0.14 μm/h during motile phase, similar to published data (Table S1; refs. 11 and 18). Motile cells exhibited a rounded morphology with no sign of membrane extension at the front and a striking accumulation of F-actin at the cell pole opposite to the direction of movement (Figs. 1A and 2A and Movies S1 and S2). By analogy with the rear of migrating leukocytes, we refer to the F-actin back of the cells as the uropod (31), although in contrast to leukocytes, MDA-MB-231 cells’ uropod does not protrude.
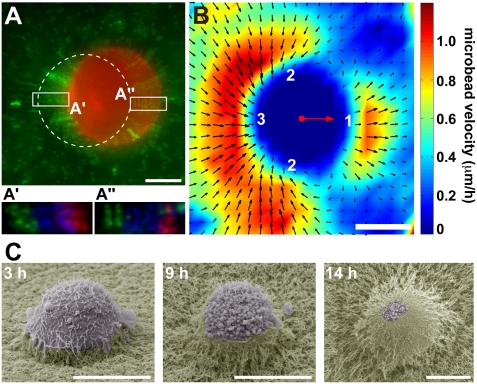
Fig. 1.
Matrix displacements during MDA-MB-231 rounded cell 3D migration in Matrigel. (A) MDA-MB-231 cells stably expressing mCherry-Lifeact seeded in Matrigel containing fluorescent microbeads (green) were imaged every 15 min (Movie S1). Shown is the last frame of mCh-Lifeact (red) superimposed on a 2-h time projection of the bead sequence (green). Dashed line indicates the position of the cell at the beginning of the time-lapse series. (A′ and A″) Bead positions at the back and front of the cell, respectively, with the first two frames (T0 and T15') color-coded in green, subsequent five frames (T30' to T90') in blue, and last two frames (T105' and T120') of the time series in red. (B) PIV analysis of matrix displacement around the cells. Individual velocity fields of eight cells were averaged. The mass center of cells is depicted with a red dot. Red arrow corresponds to the mean velocity vector of the cells. The amplitude of the bead velocity is color-coded, and direction of displacements is depicted with arrows. Matrix is pushed at the front (region 1), while it is pulled on the sides and at the back (regions 2 and 3, respectively). (C) Scanning electron micrographs of MDA-MB-231 cells invading a thick layer of Matrigel at indicated time after plating. (Scale bars, 10 μm.)
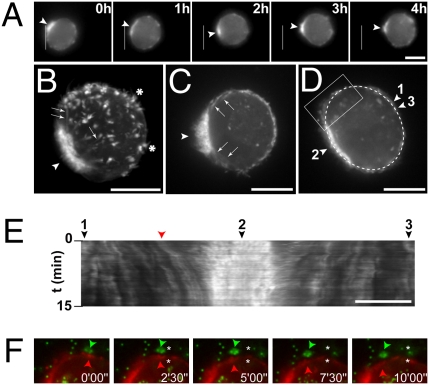
Fig. 2.
F-actin dynamics during MDA-MB-231 rounded cell 3D migration. MDA-MB-231 cells expressing mCh-Lifeact were seeded in Matrigel containing fluorescent microbeads (green in F) and imaged by wide-field (A) or confocal spinning disk (B–F) microscopy. (A) Selected frames from a time-lapse sequence with the white line showing the position of the rear of the cell at the beginning of the sequence. Arrowheads point to accumulation of F-actin at the cell rear in a uropod-like structure. (B) Z-projection of confocal sections (see Movie S3 for 3D reconstruction) showing the accumulation of F-actin at the uropod (arrowhead), in finger-like cortical extensions (asterisks), and in bundles radiating from the uropod toward the cortex (arrows). (C) Selected frame of a time-lapse series (Movie S4) showing intense blebbing activity restricted to the uropod region (arrowheads). Arrows point to radial F-actin bundles. (D and E) Selected frame (D) of a time-lapse series (Movie S6) showing the dashed line used for kymographic analysis of cortical movements in E (tags 1–3 are positioned on the x axis of the kymograph, as well as the cortical structure highlighted in F, shown by a red arrowhead). (F) Selected frames from the time series as in D and E (Movie S6). The region depicted is boxed in D. Images show F-actin labeled with mCh-Lifeact (red) and microbeads (green). Arrowheads point to parallel rearward movement of a cortical actin structure (red arrowhead) and microbeads (green arrowhead). Asterisks, position of the objects at time 0. (Scale bars, 10 μm.)
In contrast to the absence of microbead movement in cell-free regions of the matrix, indicating that beads are trapped in the meshwork, beads in the vicinity of motile cells underwent movement. Displacements of these beads reflect remodeling and deformation of the matrix induced by cell migration (Fig. 1 A and A″ and Movie S1). To provide a micrometer-scale quantitative description of matrix remodeling by invading cells, velocity fields were estimated from time-lapse image series of microbeads by using particle image velocimetry (PIV) analysis. Individual velocity fields of several cells were averaged (see Methods), revealing that Matrigel was pushed away in front of migrating cells with a ∼3.0 μm/h maximum velocity, similar to the migration speed of the cells (Fig. 1B, region 1, and Fig. S1B). In addition, the displacement field displayed two vortex-like structures originating from the poles perpendicular to the migration axis (Fig. 1B, region 2) and converging toward the cell rear (region 3). Axial vorticity field computed from average velocity field also revealed high shear motion in region 2, consistent with high tractions of the matrix exerted by migrating cells in 3D (Fig. S1A).
Cell movement was accompanied by the accumulation of beads at the cell rear (Fig. S2 A and B and Movie S2). Similarly, when MDA-MB-231 cells were plated atop a thick layer of Matrigel, they progressively invaded through the matrix with a round morphology with the matrix pulled on top of the cell and generating a track of modified ECM behind it (Fig. 1C; ref. 30). Of note, the matrix dome around invading cells was not caused by matrix retraction during scanning electron microscopy sample preparation, as it was also observed with lived unfixed MDA-MB-231 cells plated atop Matrigel and visualized with seeded fluorescent microbeads (Fig. S2C). In addition, we noticed the presence of intense membrane blebbing activity at the free rear of invading cells (Fig. 1C and below; ref. 30).
At the ultrastructural level, electron microscopy on thin sections of MDA-MB-231 cells embedded within 3D Matrigel confirmed extensive blebbing activity at one pole of the cell (Fig. S3). In addition, in comparison with normal porosity of Matrigel (Fig. S3A Inset 1, away from the cell), progressive reduction of the pore size and densification of the matrix was observed on the sides of the cell (Fig. S3A Inset 2), becoming maximal behind the cell (Fig. S3A Inset 3). Gel densification is in agreement with the observed accumulation of microbeads at the cell rear. In addition—possibly as a consequence of high shear forces (Fig. S1A) and gel cracking—voids in Matrigel were clearly observed in the immediate vicinity of the uropod (Fig. S3 A' and B), which could contribute to some extent to the settling of beads. Compaction of the gel was also visible at the front of the motile cell pushing on the matrix (Fig. S3A Inset 1).
Of note, inhibition of matrix metalloproteinase (MMP) activity had a modest inhibitory effect on the speed of migration of MDA-MB-231 cells in 3D Matrigel (∼15%; see Fig. 3E and Table S1), indicating no prominent contribution of MMP-based matrix degradation to this type of movement. Matrigel is a viscoelastic meshwork of matrix proteins of high elastic modulus (11). The observed voids in the gel, high shear forces, and progressive gel densification from the front to the rear rather support the view that cells move within this viscoelastic material by pulling on and pushing Matrigel aside.
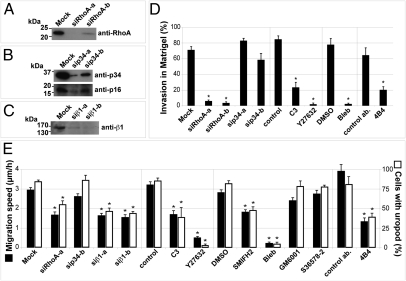
Fig. 3.
Inhibition of RhoA-ROCK-Myosin II and β1 integrin impairs uropod formation and invasion in Matrigel. (A–C) MDA-MB-231 cells were treated with the indicated siRNAs targeting RhoA (A), the p34-Arc (ARPC2) subunit of the Arp2/3 actin nucleating complex (B), or β1 integrin (C). Cell lysates were prepared after 72 h, and immunoblotting analysis was performed with indicated antibodies. Of note, knockdown of p34-Arc leads to decreased expression of p16-Arc (B). (D) Quantification of cell invasion by mCh-Lifeact MDA-MB-231 cells plated atop of a thick layer of Matrigel and treated with indicated siRNAs or drugs. Cell invasion was determined by analyzing the proportion of cells buried in Matrigel after 14 h from low magnification scanning electron micrographs. (E) Average migration speed (filled bars) and percentage of cells with uropod (open bars) in MDA-MB-231 cells seeded in Matrigel and treated with indicated siRNA and drugs. Asterisks indicate statistically significant differences compared with control cell populations (Tables S1–S3).
Rounded MDA-MB-231 Cells Migrating in 3D Matrigel Are Polarized.
High-resolution confocal spinning disk imaging of fixed cells in 3D Matrigel confirmed the accumulation of F-actin at the cell rear and in evenly distributed, finger-like cortical structures, as well as the absence of membrane extension and F-actin accumulation at the front (Fig. 2B and Movie S3). Filament bundles radiating from the uropod toward the cell front were visible (Fig. 2 B and C, arrows, and Movies S3 and S4). Although they migrated with a round morphology, MDA-MB-231 cells in 3D Matrigel were highly polarized with the nucleus positioned at the back and the Golgi apparatus localized at the cell front (Fig. S4 A and B).
The Arp2/3 complex is the main lamellipodial actin nucleating activity and is critical for 2D cell migration. Interestingly, silencing of the actin nucleating Arp2/3 complex by means of siRNA targeting of the p34-Arc subunit (causing also reduction of p16-Arc; see Fig. 3B) had no effect on vertical invasion of MDA-MB-231 cells when plated atop a thick layer of Matrigel (Fig. 3D; for statistics see Table S2) or on the migration speed and uropod formation in cells seeded in 3D Matrigel (Fig. 3E and Tables S1 and S3). In contrast—confirming our previous observation that another class of actin nucleators, the Diaphanous-related formins (DRFs), are required for MDA-MB-231 cell vertical invasion in Matrigel (30)—we found that a general inhibitor of FH2-domain containing formins (SMIFH2; ref. 32) led similarly to a ∼35% reduction of migration speed in 3D Matrigel and concomitant decrease of uropod formation (Fig. 3E). Thus, our data indicate that DRFs, but not the Arp2/3 complex, are implicated in 3D cell invasion and migration of MDA-MB-231 cells in Matrigel.
Live imaging also confirmed extensive blebbing activity at the cell rear (Fig. 2C, arrowhead, and Movies S4 and S5). In particular, large blebs were frequently observed at the edge of the uropod, which may correspond to a region of weakness of the cell cortex (Movie S5; ref. 5). Kymograph analysis of actin cortical structures revealed retrograde movement of the cortex toward the uropod during migration (Fig. 2 D and E and Movie S6). The retrograde flow of cortical F-actin was high close to the uropod region (Fig. 2E, arrowhead 2) and nearly zero at the opposite front side of the cell (arrowheads 1 and 3). Furthermore, rearward movement of the beads paralleled that of the cortex as an indication that the two were coupled (Fig. 2F), in agreement with observed high shear forces (Fig. S1A). Interestingly, the presence of an F-actin rich uropod-like structure was also observed when cells were plated on top of Matrigel (Fig. S4C and Movie S7), further indicating that vertical invasion of cells seeded atop or invasion of cells within Matrigel proceed through the same mechanism.
RhoA-ROCK-Myosin II–Mediated Contractility at the Uropod Is Required for 3D Migration.
The presence of blebs and convergent retrograde movement of the cell cortex were indicative of cell contraction. Immunolocalization analysis showed that myosin IIA heavy chain and the active (phospho-Ser19) form of myosin light chain (pS-MLC) were strongly accumulated at uropod together with F-actin (representative cells are shown in Fig. 4 A and B and Fig. S5). The functional contribution of actomyosin contractility for MDA-MB-231 cell invasion within Matrigel was assessed by inhibition of myosin II or its upstream activators. Pharmacological inhibition of myosin II with blebbistatin resulted in a strong decrease of vertical invasion capacity (Fig. 3D) and a concomitant and correlated reduction of migration speed and uropod formation in cells seeded in Matrigel (Fig. 3E and Tables S1–S3). Similarly, inhibition of RhoA by RNAi silencing (Fig. 3A) or treatment with Clostridium botulinum C3 exoenzyme, inhibition of ROCK kinase with Y27632 compound, or myosin II inhibition by blebbistatin led to correlated reduction of invasion capacity and uropod formation by MDA-MB-231 cells (Fig. 3 D and E and Tables S1–S3). In these cells, F-actin and pS-MLC accumulation, characteristic of uropod structure, was no longer visible (Fig. S5).
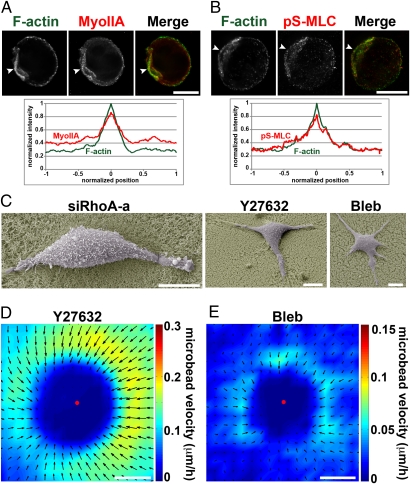
Fig. 4.
Actomyosin-based contractility is required for MDA-MB-231 cell invasion. (A and B) MDA-MB-231 cells seeded in Matrigel for 4 h, fixed, and stained for F-actin and myosin IIA (A) or pS-MLC (B). Arrowheads point to accumulation of myosin IIA and pS-MLC at F-actin positive uropod. Averaged normalized intensity profiles from 15 cells are shown. Position 0 corresponds to maximum normalized F-actin intensity (see SI Methods for details). For clarity, SEMs are not represented (< ±0.08 normalized intensity arbitrary unit). (C) Scanning electron micrographs of siRhoA-, Y27632-, or blebbistatin (bleb)-treated MDA-MB-231 cells seeded atop of Matrigel and fixed after 14 h. (D and E) Individual velocity fields of 13 Y27632-treated cells (D) or 4 blebbistatin-treated cells (E) were averaged as in Fig. 1B. Amplitude of the bead velocity is color-coded, and direction of displacements is depicted with arrows. (Scale bars, 10 μm.)
In addition, cells inhibited for RhoA, ROCK, or myosin II activity remained spread on the matrix when plated atop Matrigel, in contrast to the rounded invasive morphology of control MDA-MB-231 cells (compare Figs. 4C and 1C). PIV analysis of Y27632-treated samples showed isotropic low-force pulling of the matrix by the cells (Fig. 4D, Fig. S1B, and Movie S8), which did not support cell migration (Fig. 3E and Table S1). Finally, myosin II inhibition by blebbistatin almost completely abolished cell-induced matrix movements (Fig. 4E, Fig. S1B, and Movie S9). Together, these data demonstrate that RhoA, ROCK, and myosin II are required for formation of and contraction at uropod, and they suggest that uropod contractility is essential for MDA-MB-231 cell migration in 3D Matrigel.
β1 Integrin Is Required for Contraction at Uropod and Cell 3D Migration in Matrigel.
Contractile retrograde forces can support forward movement if coupled to the surrounding matrix through receptor (integrin)-mediated force transduction (4). We therefore assessed the involvement of β1 integrin in matrix remodeling and 3D migration of MDA-MB-231 cells in Matrigel. β1 integrin was present at the cell cortex and enriched at the uropod (Fig. 5A), although we cannot rule out the possibility that membrane accumulation at uropod resulted in apparent increased β1 staining. Addition of anti-human β1 integrin-blocking antibody 4B4 or silencing of β1 with two independent siRNAs (Fig. 3C) resulted in a significant 45–65% reduction of migration speed in 3D Matrigel correlated with impaired uropod formation (Fig. 3E and Tables S1–S3). In addition, 4B4-treated cells cultured atop the Matrigel layer were impaired in their vertical invasion capacity (Fig. 3D) and remained rounded and loosely attached to the surface of Matrigel, contrasting with the phenotype of control cells or MDA-MB-231 cells with impaired RhoA or myosin II activity (ref. 30; Fig. 5B; compare with Figs. 1C and 3C).
Fig. 5.
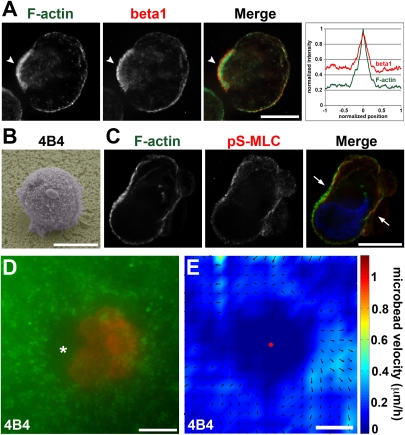
Role of β1 integrin in 3D migration of MDA-MB-231 cells in Matrigel. (A) MDA-MB-231 cells were incubated for 4 h in Matrigel, fixed, and stained for F-actin and β1 integrin. Arrowheads point to accumulation of β1 integrin at F-actin–rich uropod. Averaged normalized intensity profiles from 15 cells are shown as in Fig. 4 A and B. (B) Scanning electron micrographs of an MDA-MB-231 cell plated atop a thick layer of Matrigel in the presence of β1 integrin blocking antibody (4B4) for 14 h before fixation. (C) MDA-MB-231 cell seeded in Matrigel for 4 h in the presence of 4B4 antibody and stained for F-actin (green) and pS-MLC (red). Nucleus was labeled with DAPI (blue). Cells adopt irregular shapes with F-actin and pS-MLC accumulations and blebbing activity. (D) mCh-Lifeact MDA-MB-231 cells seeded in Matrigel in the presence of 4B4 mAb were imaged during 4 h by wide-field video microscopy. Shown is the superimposition of the 4-h time projection of the bead sequence and the last frame of mCh-Lifeact sequence. Asterisk points to region of cell detachment to the matrix. (E) Individual velocity fields of seven 4B4-treated cells were averaged as in Fig. 1B. Amplitude of the bead velocity is color-coded, and direction of displacements is depicted with arrows. (Scale bars, 10 μm.)
When seeded in microbead-containing Matrigel, 4B4-treated cells displayed saltatory movements, leading to matrix deformation and cortex/matrix detachment at the cell rear as visualized by dark regions surrounding the cells (Fig. 5D and Movies S10 and S11). These effects were correlated with strong cell constriction and blebbing activity localized in the middle plane of the cells where F-actin and pS-MLC accumulation was visible (Movie S11 and Fig. 5C, arrows). In contrast, inhibition of αV integrins with a nonpeptide RGD mimetic (compound S36578-2) had no effect on the migration of MDA-MB-231 cells in 3D Matrigel or on uropod formation (Fig. 3E). Together, these results indicate that β1 integrin binding to the ECM contributes to the mechanism of uropod formation, positioning of myosin II activation at uropod, and invasion of MDA-MB-231 metastatic breast cancer cells in Matrigel.
Discussion
We report on the mechanics of rounded-cell migration of MDA-MB-231 breast cancer cells invading through a reconstituted BM. Features of rounded-cell migration include the formation of an actomyosin contractile uropod as a generator of tensional forces to the matrix through β1 adhesion receptors and the absence of lamellipodial extensions and blebs as leading edge protrusions.
Model for MDA-MB-231 Rounded-Cell Invasion Through Matrigel.
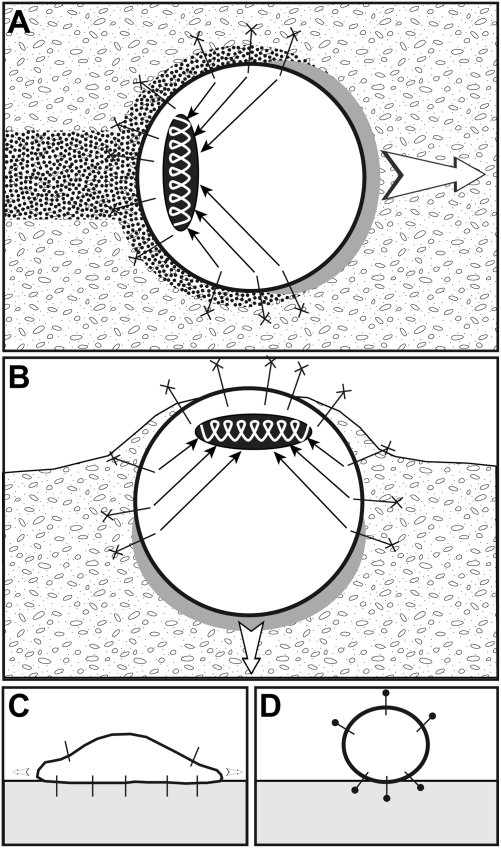
Using velocimetric analysis of seeded microbeads, we report a unique comprehensive analysis of matrix remodeling induced by the migration of rounded breast cancer cells within 3D Matrigel. In addition, two-color imaging of F-actin cortical structures and microbeads with high spatial and temporal resolution demonstrated tight coupling to rearward movement of the cell cortex. Based on these analyses, we propose the following model for MDA-MB-231 rounded-cell migration (depicted in Fig. 6A). An actomyosin cytoskeleton assembled at the cell rear in a uropod-like structure contracts (ellipsoid spring), and traction forces are transmitted to the surrounding matrix via β1 integrin transmembrane adhesion receptors connected to the cell cortex and to radially extending F-actin bundles (arrows). Ligands for β1 integrins in Matrigel include its major components laminin and type IV collagen and are represented by crosses at the end of integrins. Contractile convergent retrograde forces pull on the matrix in the rearward direction, generating forward movement of the cell that pushes the matrix at the front (gray area). Cell movement induces high shear motion in the matrix and matrix remodeling (dotted area).
Fig. 6.
Model of breast cancer round cell invasion and migration in 3D Matrigel. (A) MDA-MB-231 cells invade through Matrigel maintaining a spheroid shape. Actomyosin contractility is restricted to the cell rear (ellipsoid spring), generating tractional forces that are transmitted to the matrix through radial F-actin bundles (arrows) and β1 integrins bound to their matrix ligands (cross at the extremity of integrins). Gray and dotted areas represent regions of matrix compression and remodeling induced by cell movement, respectively. (B) Vertical invasion of cells atop a thick layer of Matrigel. (C) Inhibition of actomyosin contractility leads to cells spreading on the surface of Matrigel. (D) Inhibition of β1 integrin binding to its ligand(s) in the matrix (closed circle at the extremity of integrins) results in spheroid cells having minimal contact with the matrix and unable to invade. See text for details.
Invasion of MDA-MB-231 cells seeded atop the reconstituted BM proceeds through the same mechanism (Fig. 6B). Inhibition of actomyosin activity leads to spreading of the cells on the surface of the matrix through integrin engagement, while invasion is blocked (Fig. 6C). Cells blunted for β1 integrin function have minimal contact with the matrix and do not invade (Fig. 6D). Of note, the contraction- and adhesion-based 3D migration of rounded cells we describe here appears to be distinct from the adhesion-independent amoeboid migration of leukocytes in 3D (6).
Mechanics of Rounded-Cell Migration.
A striking observation of the present study is that there were neither lamellipodial extensions nor blebs as leading protrusions during rounded-cell movement. Blebbing is generally viewed as the driving mechanism for amoeboid motion by allowing cells to squeeze themselves through preexisting voids in the matrix (5, 21). Here, based on observation of hundreds of cells by optical and electron microscopy techniques, blebs were restricted to the rear of the cells where actomyosin contractility is highest. Beside the role of actomyosin in generating cortical tension underlying the mechanism of bleb formation (5, 33), it is also possible that bleb extension was facilitated by voids forming behind cells moving in the 3D matrix or at the free rear of cells invading atop Matrigel. Indeed, in cells with blunted β1 integrin function, large blebs formed in voids generated by cell-matrix detachment associated with sites of actomyosin-based cell constriction (Movies S10 and S11). In addition, in a control situation, matrix rigidity/porosity and pushing force of the moving cells on the matrix may oppose to bleb extension at the front. Therefore, our data support the conclusion that blebbing is simply an epiphenomenon caused by strong activation of cortical contractility at the rear and cannot contribute per se to rounded breast tumor cell movement that appears to depend solely on actomyosin contractility.
Our data indicate that lamellipodial extension and Arp2/3 complex, the main lamellipodial actin nucleating activity, are dispensable for MDA-MB-231 cell invasion and migration in Matrigel (Fig. 3). In contrast, formins of the DRF family contribute both to vertical invasion and migration of MDA-MB-231 cells in 3D Matrigel (ref. 30 and this study). Interestingly, actin filament bundles, which are connected to the substrate via focal adhesions, have been shown to assemble through DRF1-driven actin polymerization in cells plated on a 2D substratum (34). This finding leaves open the possibility that DRFs may also play a role in rounded-cell invasion at the level of tensional force transmission to the cortex by controlling the formation of radial F-actin bundles connecting the cortex to the uropod. Somewhat contrasting with the differential implication of the two classes of actin nucleators during rounded-cell invasion, both Arp2/3 complex and formin (DRF) activities are required for invadopodia formation and matrix proteolysis by invasive tumor cells on a rigid substrate (30, 35). Together, these data support the conclusion of at least partially distinct mechanisms underlying the processes of invadopodial degradation of a rigid matrix and invasion of reconstituted BM.
Nonmuscle myosin II and the actomyosin contractile apparatus comprise a generic force-generating system required for retraction of the trailing edge of the moving cell, for biomechanical coupling to actin assembly, lamellipodial protrusion, and adhesion at the cell front (36, 37). Myosin II similarly plays critical roles in tumor cell invasion in the elongated and amoeboid migration modes through upstream MRCK or ROCK regulatory pathways, respectively (18, 23, 24, 38, 39). Here, we characterize the mechanism of breast cancer cell invasion in Matrigel, which is based on activation of the RhoA/ROCK/Myosin II cascade at the cell rear and transmission of traction forces to the cortex, whereas blebbing is only a contractility reporter with no contribution to migration per se.
Methods
See SI Methods for details on cell culture, inhibitors, siRNA treatments, antibodies, and indirect immunofluorescence analysis.
Analysis of Matrix Displacements by PIV.
See SI Methods for details.
Quantification of Cell Migration and Uropod Formation.
Cells were tracked manually with ImageJ software to measure migration speeds. Dividing, aggregated, and out-of-focus cells were excluded from quantifications. Uropod was defined as polarized accumulation of actin filaments decorated with mCh-Lifeact that persisted for at least 2 h over a 4-h sequence.
Scanning Electron and Transmission Electron Microscopy.
See SI Methods for details.
Statistical Analyses.
Statistical analyses were performed with SPSS (Version 12.0). See the legends of Tables S1–S3 for details.
Supplementary Material
Acknowledgments
We thank the PICT-IBiSA imaging facility of the Institut Curie for help with image acquisition and processing; Drs. G. Raposo (Curie Institute) and M.-C. Prévost (Ultrastructural Microscopy Platform of Pasteur Institute) for help with transmission and scanning electron microscopy, respectively; Drs. E. Dérivery, M. Heuzé, S. Miserey-Lenkei, M. H. Verlhac, G. C. Tucker, C. Albiges-Rizo, and M. Bornens for providing reagents; Drs. P. Silberzan and A. Buguin for helpful discussion during this study; and Dr. C. Sykes for critical reading of the manuscript. This work was supported by grants from Ligue Nationale contre le Cancer “Equipe Labellisée 2008” and Agence Nationale de la Recherche (ANR-08-BLAN-0111) and core funding support from Institut Curie and Centre National de la Recherche Scientifique (to P.C.). R.P. was supported by grants from Association pour la Recherche contre le Cancer (ARC) and Institut National du Cancer (INCa). F.L. and O.C. were supported by postdoctoral fellowship from Fondation pour la Recherche Médicale (FRM) and Institut Curie.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010396108/-/DCSupplemental.
References
- 1.Friedl P, Wolf K. Tumour-cell invasion and migration: Diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 2.Rowe RG, Weiss SJ. Breaching the basement membrane: Who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Poincloux R, Lizárraga F, Chavrier P. Matrix invasion by tumour cells: A focus on MT1-MMP trafficking to invadopodia. J Cell Sci. 2009;122:3015–3024. doi: 10.1242/jcs.034561. [DOI] [PubMed] [Google Scholar]
- 4.Lauffenburger DA, Horwitz AF. Cell migration: A physically integrated molecular process. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 5.Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–736. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 6.Lämmermann T, Sixt M. Mechanical modes of ‘amoeboid’ cell migration. Curr Opin Cell Biol. 2009;21:636–644. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 9.Beningo KA, Dembo M, Wang YL. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci USA. 2004;101:18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Even-Ram S, Yamada KM. Cell migration in 3D matrix. Curr Opin Cell Biol. 2005;17:524–532. doi: 10.1016/j.ceb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 11.Zaman MH, et al. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc Natl Acad Sci USA. 2006;103:10889–10894. doi: 10.1073/pnas.0604460103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faure-André G, et al. Regulation of dendritic cell migration by CD74, the MHC class II-associated invariant chain. Science. 2008;322:1705–1710. doi: 10.1126/science.1159894. [DOI] [PubMed] [Google Scholar]
- 13.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Packard BZ, Artym VV, Komoriya A, Yamada KM. Direct visualization of protease activity on cells migrating in three-dimensions. Matrix Biol. 2009;28:3–10. doi: 10.1016/j.matbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: Three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabeh F, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf K, et al. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 19.Wolf K, et al. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 20.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 21.Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaser H, et al. Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow. Dev Cell. 2006;11:613–627. doi: 10.1016/j.devcel.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson S, Paterson HF, Marshall CJ. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat Cell Biol. 2005;7:255–261. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- 24.Wyckoff JB, Pinner SE, Gschmeissner S, Condeelis JS, Sahai E. ROCK- and myosin-dependent matrix deformation enables protease-independent tumor-cell invasion in vivo. Curr Biol. 2006;16:1515–1523. doi: 10.1016/j.cub.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 25.Gadea G, Sanz-Moreno V, Self A, Godi A, Marshall CJ. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr Biol. 2008;18:1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 26.Munevar S, Wang YL, Dembo M. Distinct roles of frontal and rear cell-substrate adhesions in fibroblast migration. Mol Biol Cell. 2001;12:3947–3954. doi: 10.1091/mbc.12.12.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 28.Bloom RJ, George JP, Celedon A, Sun SX, Wirtz D. Mapping local matrix remodeling induced by a migrating tumor cell using three-dimensional multiple-particle tracking. Biophys J. 2008;95:4077–4088. doi: 10.1529/biophysj.108.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lizárraga F, et al. Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 2009;69:2792–2800. doi: 10.1158/0008-5472.CAN-08-3709. [DOI] [PubMed] [Google Scholar]
- 31.Sánchez-Madrid F, Serrador JM. Bringing up the rear: Defining the roles of the uropod. Nat Rev Mol Cell Biol. 2009;10:353–359. doi: 10.1038/nrm2680. [DOI] [PubMed] [Google Scholar]
- 32.Rizvi SA, et al. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tinevez JY, et al. Role of cortical tension in bleb growth. Proc Natl Acad Sci USA. 2009;106:18581–18586. doi: 10.1073/pnas.0903353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi H, et al. Molecular mechanisms of invadopodium formation: The role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 2005;168:441–452. doi: 10.1083/jcb.200407076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giannone G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itoh K, et al. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat Med. 1999;5:221–225. doi: 10.1038/5587. [DOI] [PubMed] [Google Scholar]
- 39.Dulyaninova NG, House RP, Betapudi V, Bresnick AR. Myosin-IIA heavy-chain phosphorylation regulates the motility of MDA-MB-231 carcinoma cells. Mol Biol Cell. 2007;18:3144–3155. doi: 10.1091/mbc.E06-11-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.