Abstract
We identify a gene, ORGAN BOUNDARY1 (OBO1), by its unique pattern of enhancer- driven GFP expression at the boundaries between the apical meristems and lateral organs in Arabidopsis embryos, seedlings, and mature plants. OBO1 also is expressed at the root apical meristem and in distinct cell files surrounding this area. OBO1 is one of a 10-member plant-specific gene family encoding a single small domain (133 amino acids) with unknown function. One member of this gene family, OBO2, is identical to a previously studied gene, LIGHT-SENSITIVE HYPOCOTYL1. Overexpression of OBO1 causes an abnormal number and size of petals and petal–stamen fusions. The patterns of OBO1 gene expression are distinct but overlap with other genes involved in boundary formation in the Arabidopsis shoot apical meristem, including CUP-SHAPED COTYLEDON, LATERAL ORGAN BOUNDARIES, BLADE-ON-PETIOLE, ASYMMETRIC LEAVES, and LATERAL ORGAN FUSION. Nuclear localization of OBO1 suggests that it might act with one or more of the transcription factors encoded by the foregoing genes. Ablation of the specific cells expressing OBO1 leads to loss of the shoot apical meristem and lateral organs. Thus, the cells expressing OBO1 are important for meristem maintenance and organogenesis in Arabidopsis.
Keywords: enhancer trap, meristem boundary, plant development, domain of unknown function 640
The continuous growth potential of plants derives mainly from the regenerative activity of plant meristems. Typically plants have two classes of meristems, the shoot apical meristem (SAM) and the root apical meristem (RAM) below the ground. The SAM contains a central organizing center that maintains upper layers of stem cells capable of continuous cell division (1). After cell division, some cells at the center remain as stem cells, whereas others shift to the periphery and differentiate into primordia, groups of specialized cells that become lateral organs, such as leaves, branches, and flowers (2). As development progresses, a shoot organ primordium is distinguished from its meristem by the creation of a groove containing narrow files of nondividing cells that define the boundaries between the meristem and organs or between adjacent organs (3–5). Establishment of such boundaries is temporally coordinated with changes in morphology and gene expression patterns in the meristem (5, 6).
Following apical–basal axis formation in Arabidopsis, the quiescent center of the RAM inhibits differentiation of a single layer of surrounding stem cells (7, 8) that generate all root tissues, including endodermis, cortex, and epidermis, in a radial pattern (8, 9). Cell position, not lineage, determines the fate of cells according to signals originating from overlying mature cells (10). Secondary root meristems that give rise to lateral roots are generated de novo from differentiated cells in inner layers of the root, in contrast to secondary shoot meristems, which arise directly from the SAM (11).
Here we identify and characterize ORGAN BOUNDARY 1 (OBO1), which displays a unique pattern of expression at both the SAM and RAM. We became interested in OBO1 because of its meristem-specific pattern of expression in the J2341 enhancer trap line (12) (http://www.plantsci.cam.ac.uk/Haseloff/). Haseloff (13) designed a T-DNA vector carrying a minimal promoter adjacent to GAL4 coding sequences that are linked to a GAL4- activated promoter driving endoplasmic reticulum-tethered GFP (ER-GFP) expression. GAL4 expression is dependent on enhancer elements at the site of T-DNA insertion. The GAL4 reporter reveals the temporal and spatial pattern of expression specific to the enhancer element and its cognate gene.
We discuss OBO1 gene expression compared with other genes essential for SAM and organ boundary formation, including SHOOT MERISTEMLESS (STM), CUP-SHAPED COTYLEDON (CUC1, CUC2, and CUC3), LATERAL ORGAN BOUNDARIES (LOB), BLADE-ON-PETIOLE (BOP1 and BOP2), ASYMMETRIC LEAVES (AS1 and AS2), and LATERAL ORGAN FUSION (LOF) (14–23). OBO1 has overlapping expression patterns with many of these genes, but also exhibits unique expression patterns.
Results
Enhancer-Driven ER-GFP Expression in J2341 Embryos.
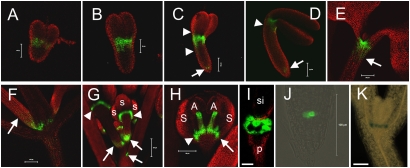
ER-GFP expression in the J2341 enhancer line was first detected at the shoot apex in mid-heart stage embryos (Fig. 1A). In early- and mid-torpedo embryos, this expression broadened to a “bowtie”-like pattern around the SAM, with lower GFP expression in the center of the SAM and vertically increasing expression in the regions away from the center (Fig. 1 B and C). In late-torpedo embryos, ER-GFP expression narrowed to a small region apparently encircling the SAM (Fig. 1D). The J2341 enhancer also drove ER-GFP expression near the root meristem starting at the mid-torpedo stage (Fig. 1 C and D, arrows). These patterns of expression have been confirmed and extended by more sensitive in situ mRNA studies.
Fig. 1.
ER-GFP expression in enhancer line J2341. (A–D) Mid-heart (A), early-torpedo (B), mid-torpedo (C), and late-torpedo (D) embryos. In C and D, arrows indicate ER-GFP fluorescence in the root apex. In C, arrowheads indicate a gradient of fluorescence surrounding the SAM; in D, arrowheads indicate diminished expression of ER-GFP around the SAM. (E and F) 5-d-old (E) and 15-d-old (F) seedlings. Arrows denote a gradient of fluorescence around the SAM. (G) ER-GFP fluorescence in the inflorescence axis. (H) A single flower showing fluorescence in anther filaments. In G and H, arrows indicate adaxial junctions between a main stem and floral petioles, and arrowheads indicate fluorescence in the floral meristem at the base of lateral floral organs. (I) Fluorescence between the petiole and the base of the silique. (J) Fluorescence in the root apex of 5-d-old seedlings. (K) GUS signal at the shoot apex in a 5-d-old seedling. Red indicates chlorophyll autofluorescence; green, ER-GFP fluorescence. S, sepal; A, anther; si, silique; p, petiole. (Scale bars: 50 μm in A and B; 100 μm in C, D, and H–K; 200 μm in E–G.).
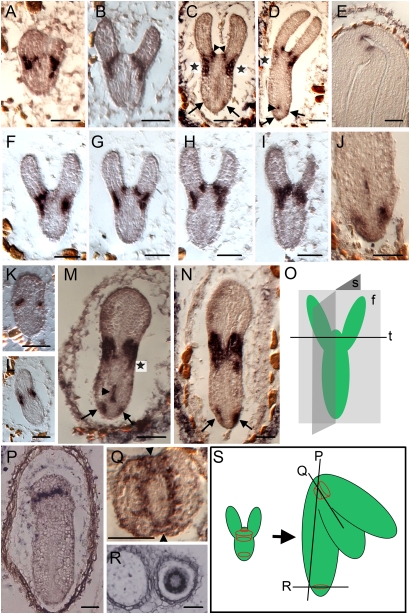
J2341 enhancer-driven expression of ER-GFP and its mRNA were first detected in cells around the shoot apex in mid- and early-heart stage embryos (Figs. 1A and 2A). A sagittal section at the early heart stage exhibited ER-GFP mRNA expression at the junction of two cotyledon margins (Fig. 2K); this section corresponds to a longitudinal section through the points of the two arrows in a transverse section (Fig. 2Q). Frontal and lateral sagittal sections of late-heart stage embryos showed ER-GFP mRNA expression confined to a continuous single cell layer surrounding the SAM and absent from the center (Fig. 2 B and L). In contrast, frontal sections of early-torpedo embryos showed two separate regions of ER-GFP mRNA, an inner region demarcating the adaxial side of the boundaries between cotyledons and the SAM (Fig. 2C, arrowheads) and an outer region demarcating the abaxial boundaries of the cotyledons (Fig. 2C, asterisks). Serial sections of an embryo in transition from late-heart to early-torpedo stage exhibited two separate regions of ER-GFP mRNA expression creating two concentric rings at the boundaries of cotyledon margins around the SAM (Fig. 2 F–I, diagrammed in Fig. 2S). Sagittal sections of the central plane of early-torpedo embryos showed that the ER-GFP mRNA signal covered the entire region surrounding the SAM and expanded outward and downward toward the epidermis (Fig. 2M, asterisk); this expression pattern continued at the mid-torpedo stage (Fig. 2N). Just as GFP fluorescence diminished upward toward the cotyledons and downward to the hypocotyl (Fig. 1C, arrowheads), ER-GFP mRNA expression also decreased in a frontal plane section (Fig. 2D, asterisk). In late-torpedo embryos, ER-GFP expression was maintained only as a narrow band around the SAM (Figs. 1D, arrowhead and 2E). A lateral sagittal section of the hypocotyl including the SAM demonstrated mRNA expression in a line of cells (Fig. 2P). Transverse sections of the SAM (Fig. 2Q) in late-torpedo embryos showed mRNA expression forming a ring that included the boundaries of both the cotyledon margins (arrowheads) and the SAM.
Fig. 2.
J2341-specific ER-GFP mRNA expression during embryo development. Early-heart (A and K), late-heart (B and L), early-torpedo (C and M), mid-torpedo (D and N), and late-torpedo (E and P) embryos showing ER-GFP mRNA expression in frontal (A–E) and sagittal (K–N and P) sections. In C, arrowheads indicate adaxial sides of junctions between the SAM and cotyledons, and asterisks indicate abaxial expression. In D, the asterisk indicates diminished expression of ER-GFP mRNA in the abaxial region. In M, the asterisk represents a gradient of ER-GFP mRNA expression. In D and M, arrowheads denote the signal above the root apex. In C, D, M, and N, arrows indicate mRNA signals in the root apex. (F–I) Serial sections of an early-torpedo embryo. (J) Root of a late-torpedo embryo. (O) Frontal (f), sagittal (s), and transverse (t) sections of a hypocotyl. (P) Lateral sagittal section of a hypocotyl. (Q and R) Transverse sections of the shoot (Q) and root (R) apices. (S) Diagram of ER-GFP mRNA expression observed in P–R along with their corresponding planes of section. (Scale bars: 50 μm.)
ER-GFP mRNA was first detected on either side of the root apex in early-torpedo embryos (Fig. 2 C and M, arrows) and as a single column of cells above the RAM (Fig. 2M, arrowhead). Two small dots of GFP signal were first detected in the root apex of mid-torpedo embryos (Fig. 1C, arrow). The pattern of expression at the root apex in early-torpedo embryos was maintained through the mid-torpedo stage (Fig. 2 C, D, M, and N); ER-GFP mRNA was observed just above the root apex (Fig. 2D, small arrowhead) and around the root apex (Fig. 2 D and N, large arrows). In late-torpedo embryos, mRNA expression was detected in two limited regions around the RAM and in a column of cells above the RAM, as in early- and mid-torpedo embryos (Fig. 2J). In the root apex from late-torpedo embryos, transverse sections revealed mRNA expression in a ring (Fig. 2R), as seen in the shoot apex as well (Fig. 2Q).
ER-GFP Expression During Seedling Development.
After 5 d, J2341 seedlings demonstrated an intense ER-GFP signal at the shoot apex with decreasing expression downward from the SAM into the upper region of the hypocotyls, similar to that seen in embryos (compare Fig. 1E, arrow and Fig. 1C, lower arrowhead). After 15 d, seedlings exhibited an intense GFP signal around the SAM and a weak GFP signal along the basal petioles of true leaves (Fig. 1F, arrow). J2341 was crossed to a transgenic plant carrying GUS coding sequences under control of GAL4 upstream activating sequence (UAS). At 5 d, F1 seedlings exhibited a GUS signal in four spots (seen on the zoomed image in Fig. 1K) around the SAM. Each spot was a single cell that marked the junction between the basal cotyledons or leaf petioles and the SAM; two cells expressed GUS at the base of each cotyledon, and two cells expressed GUS at the base of each leaf primordium (Fig. 1K). The GUS signal did not detect expression in the root tip; however, J2341-specific ER-GFP fluorescence was detected in a few cells at the quiescent center of the RAM in 5-d-old seedlings (Fig. 1J). This indicates that ER-GFP expression was maintained at the boundaries of the SAM and RAM, as observed in torpedo stage embryos.
Identification of the Gene at the J2341 Insertion Site.
To clone the genomic DNA flanking the T-DNA in J2341, we performed thermal asymmetric interlaced (TAIL)-PCR using degenerate primers and right or left T-DNA border primers. We obtained a 450-bp product from the reaction using left T-DNA border primers. This product mapped to BAC clone T16B12 in an intergenic region 3,405 bp downstream of the 3′ end of At2g31150 and 936 bp upstream of the translational start of At2g31160. The gene interrupted by the T-DNA insert in J2341 should have the same expression pattern as seen for its enhancer driving ER-GFP. We tested the mRNA expression pattern of At2g31160, closest to the T-DNA insertion site. At2g31160 mRNA expression was not detected by in situ hybridization in WT embryos, suggesting that this gene is not controlled by the J2341 enhancer, or that expression level of the At2g31160 is below the limit of detection. Given that there are five copies of the GAL4 UAS binding site upstream of ER-GFP, expression of ER-GFP is expected to be significantly higher than that of the native mRNA at the J2341 insertion site. Thus, we tested At2g31160 expression at other developmental times.
J2341 ER-GFP expression was low in seedlings but readily detected during reproductive development at the junctions between flowers and the inflorescence axis (Fig. 1G, arrows), at the boundaries between the floral meristem and base of floral organs (Fig. 1G and H, arrowheads), and at the adaxial sides of sepals (Fig. 1H, arrow). Boundary- specific expression was maintained in mature siliques, revealing two concentric rings of ER-GFP expression at the base (Fig. 1I). Among floral organs, anther filaments in young flowers showed an ER-GFP signal (Fig. 1H), with expression disappearing as flowers mature.
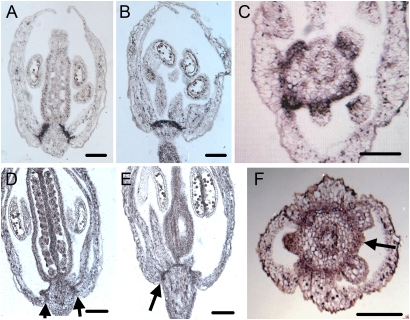
We compared ER-GFP mRNA and At2g31160 mRNA expression in flowers by in situ hybridization. ER-GFP mRNA expression reiterated the patterns observed for ER-GFP fluorescence and was detected at the bases of the second (petal) and third (stamen) whorls of floral organs (Fig. 3 A and B), most clearly at the bottom of anthers (Fig. 3C), and at the boundary between the sepals and the second and third whorls, as demonstrated by adaxial expression at the base of the sepals (Fig. 3A). Similar, albeit weaker, expression was seen in WT flowers probed with antisense DNA to At2g31160 mRNA (Fig. 3 D–F). Both ER-GFP and At2g31160 mRNAs were expressed at the base of floral organs, except for the gynoecium in J2341 and WT (Fig. 3 A and D, arrows). Lateral sagittal sections that pass obliquely through the base of a flower demonstrated mRNA signals encircling the floral base (Fig. 3 B and E, arrow). The expression pattern of At2g31160 in a cross-section (Fig. 3F) was identical to that of ER-GFP mRNA shown in Fig. 3C, with the signal clearly absent in the center of the floral meristem. The similar expression patterns with antisense probes to ER-GFP mRNA in J2341 and At2g31160 mRNA in WT suggest that At2g31160 is the gene at the T-DNA insertion site in J2341. WT tissues probed with control sense At2g31160 mRNA exhibited no signal.
Fig. 3.
Expression of ER-GFP in J2341 and At2g31160 in WT flowers. (A–C) ER-GFP mRNA expression in J2341. (D–F) At2g31160 mRNA in WT. Longitudinal (A and D), oblique, (B and E), and transverse (C and F) sections of stage-12 flowers before opening are shown. Arrows in D–F indicate At2g31160 mRNA expression at the base of floral organs. (Scale bars: 100 μm.)
We named At2g31160 ORGAN BOUNDARY1 (OBO1) based on its specific expression pattern at the boundaries of vegetative and reproductive organs. OBO1 is a member of a plant-specific single-domain gene family in Arabidopsis consisting of 10 genes predicted (24) to encode small proteins ranging from 164 to 219 amino acids (Fig. S1). These 10 Arabidopsis genes, including OBO1, consist of a single exon encoding a protein with a conserved 133-aa domain of unknown function in the center and flanking unique sequences at both ends. To underscore the high conservation of the OBO domain, we also examined the amino acid sequence of a homolog, G1L1 (25), from the fern Selaginella moellendorffi. G1L1 is phylogenetically most related to OBO5 (Fig. S1 and Fig. 4), containing 82% identical amino acids. The 11 proteins are overall 51% identical, with an additional 23% similarity. The C-termini of OBO1 and six other homologs contain predicted nuclear localizing sequences (NLS). GFP fusions to OBO1 were transiently expressed in Nicotiana leaves. GFP-OBO1 was nuclear-localized; in contrast, OBO1-GFP was not nuclear-localized, likely due to masking of the C-terminal NLS by GFP.
Fig. 4.
The DUF640 single-domain family in Arabidopsis. Phylogenetic tree of OBO/LSH proteins. Numbers on the right indicate branch lengths proportional to the amount of inferred evolutionary change.
OBO2 has been previously identified as LIGHT-DEPENDENT SHORT HYPOCOTYL 1 (LSH1) (26). LSH1 was identified by activation tagging; LSH1-D exhibits a dominant short hypocotyl phenotype in response to red, blue, and far-red light. Ten LSH homologs have been described (26); OBO1 corresponds to LSH3 (Figs. S1 and S2). The conserved domain in the OBO/LSH family is termed DUF640 (domain of unknown function 640) on the Pfam Web site (http://www.sanger.ac.uk/cgi-bin/Pfam). The rice g1 mutation causes sterile lemma formation in the rice spikelet, and its WT gene is known as ALOG (Arabidopsis, LSH1, and Oryza G1) due to its homology to LSH1 (25). To avoid confusion, Figs. S1 and S2 use both LSH and OBO in the names of genes and gene products; new nomenclatures will undoubtedly arise as more functions are assigned to this family.
ER-GFP mRNA in J2341 and OBO1 mRNA in WT plants have the same expression pattern as detected in the flower, suggesting that enhancer and promoter regions are likely intact in J2341. The J2341 insertion occurs 936 bp before the start of OBO1 translation; this region may contain both the enhancer and promoter of OBO1, or the OBO1 enhancer may be upstream of the insertion site. There are no developmental defects in J2341 or in enhancer trap lines displaying root tissue-specific ER-GFP expression (27). Alternatively, normal development in J2341 may be due to redundant gene activity in the OBO/LSH gene family. In support of this, we found no phenotype after gene silencing with siRNA specific only to OBO1.
Mediation of Cell Ablation by the J2341 Enhancer.
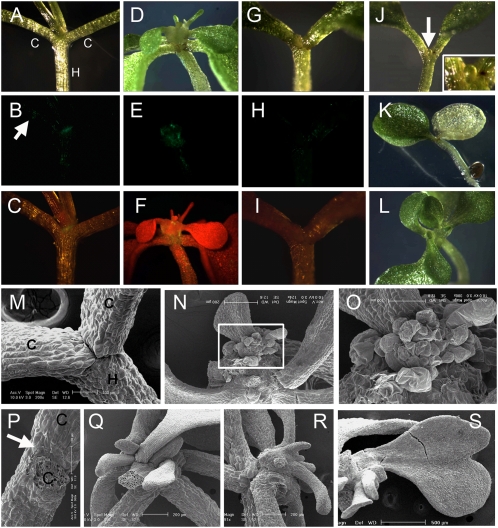
In genetic ablation experiments performed to study OBO1 function, we crossed homozygous J2341 to a transgenic plant homozygous for the diphtheria toxin chain A (DTA) gene under the control of the GAL4 upstream activating sequence (UAS) (27). Crossing the UAS-DTA line to J2341 that expresses GAL4 leads to DTA expression in zygotic tissues. DTA inhibits translation cell autonomously by catalyzing ADP ribosylation of eukaryotic elongation factor 2, leading to its inactivation (28). At 16 d after germination, 21 F1 plants were observed by epifluorescence microscopy and scanning electron microscopy (SEM) (Table 1 and Fig. 5). Three F1 plants exhibited normal shoot and root development, with between two and six true leaves as in WT plants, and GFP expression around the SAM, petioles, and the hypocotyl, as in J2341 (Figs. 1E and 5 A–C). Six F1 plants had a disordered phyllotaxy, often with multiple shoot apexes (Fig. 5 D–F, L, N, Q, and R), and one plant had a heart-shaped leaf (Fig. 5S). As evidence of ablation of cells expressing OBO1, we detected little ER-GFP signal at the SAM (Fig. 5 B, C, H, and I). Ablation may provoke other cells to attempt to express OBO1, resulting in abnormal morphology and expression at the basal boundaries of leaves and the upper part of the hypocotyl (Fig. 5 E and F). A cluster of amorphous cells at the SAM was seen in all six plants (Fig. 5 N and O). Three F1 plants had normal cotyledons but retarded true leaves. Eight F1 plants had no true leaves, and four of these plants had pale-yellow cotyledons (Fig. 5K). Two of the four plants with green cotyledons had small bulbous structures at the presumptive SAM (Fig. 5J, arrow and Inset). The remaining two plants did not have either SAM or leaf primordia in the space between the cotyledons (Fig. 5 M and P, arrow), and had a barely visible GFP signal (Fig. 5 H and I). Growth of one of the 21 plants, with small pale-yellow cotyledons, was completely inhibited after germination (Table 1). Root elongation was severely inhibited in plants exhibiting phenotypes resulting from OBO1-driven DTA expression. The variable and severe phenotypes detected indicate that the cells expressing OBO1 are critical for embryo and consequent seedling development.
Table 1.
Distribution of phenotypes from cell ablation in J2341
| No. of plants | No. of true leaves | Plant morphology | Root length, mm |
| 3 | 2–6 | WT | 18.0 ± 1.9 |
| 6 | 4–7 | Disturbed phyllotaxy | 16.4 ± 2.3 |
| Multiple shoot apexes | |||
| Abnormal leaf morphology | |||
| 3 | 2 | Retarded true leaves | 13.1 ± 6.7 |
| 8 | 0 | One pale-yellow cotyledon (3*) | 6.2 ± 3.9 |
| All pale-yellow cotyledons (1*) | |||
| WT green cotyledons (4*) | |||
| 1 | 0 | All pale-yellow cotyledons and no growth after germination | 1 |
*Number of plants with the phenotype.
Fig. 5.
OBO1 enhancer-driven cell ablation. (A–C) A phenotypically WT plant with normal cotyledons (c) and a hypocotyl (h). In B, the arrow indicates very weak GFP fluorescence in petioles. (D–F and L) Plants with disordered phyllotaxy and multiple shoot apexes. (G–K) Plants without true leaves. In J, the arrow indicates a bulbous structure at the presumptive SAM, (enlarged in the Inset). Images were obtained by light microscopy (A, D, G, and J–L) or epifluorescence microscopy to detect GFP only (B, E, and H) or GFP with chlorophyll autofluorescence (C, F, and I). (M–S) SEM of the cell-ablated plants. (M and P) Plants without a SAM. In P, the arrow indicates the presumptive location of the SAM. (N, Q, and R) Plants with disordered phyllotaxy. (O) Enlargement of the Inset in N. (S) Plant with disordered phyllotaxy and abnormal leaves.
Disruption of Floral Organ Number and Identity from Overexpression of OBO1.
Transgenic plants expressing OBO1 under the 35S promoter exhibited abnormal numbers and morphology of petals, including flowers with five petals (Fig. 6A) and elongated (Fig. 6B, arrow) or shortened (Fig. 6B, arrowhead) petals. Floral organ identity was disrupted as well, with stamenoid petals with a pollen sac fused to the boundary region between a petal base and a petal lobe along the margin (Fig. 6 C–E, arrows). The area of petal tissue varied among stamenoid petals displaying a fused pollen sac. Otherwise, the 35S::OBO1 plants were normal.
Fig. 6.
Floral phenotype of 35S::OBO1 transgenic plants. (A and B) Flowers of transgenic plants overexpressing OBO1 have an abnormal number (A) and/or shape (B) of petals. (C–E) Stamenoid petals of 35S::OBO1 transgenic flowers show different degrees of fusion between a petal and a stamen. Arrows indicate a pollen sac fused to the marginal lobe of a petal.
Discussion
Here we characterize the gene expression patterns in enhancer trap line J2341 and identify the gene interrupted by the enhancer trap T-DNA insert. The J2341 GAL4 enhancer drives the ER-GFP expression first detected in embryos in a pattern that surrounds the SAM at the early-heart stage. This ER-GFP expression marks a boundary between the meristem and the cotyeldons. In seedlings, this expression pattern continues and marks the region between the SAM and leaf primordia. During floral development, ER-GFP expression retains its circular pattern around the floral meristem at the bases of petals and stamen, in anther filaments, and at the adaxial junctions between the inflorescence stem and floral petioles. In the root, J2341expression first appears at the mid-torpedo stage within a specific subset of cells encircling the RAM and marks a boundary between the RAM and its surrounding cells. These latter cells resemble endodermis/cortex initial cells in pattern and location.
The gene interrupted by the enhancer trap T-DNA insert is At2g31160, a member of a single-domain family encoding a plant-specific conserved region of 133 amino acids and flanking short sequences of unknown function. Given the gene's pattern of expression at the boundaries of lateral organs, we named At2g31160 OBO1. OBO1 mRNA expression is low compared with ER-GFP, because there are five copies of the GAL4 UAS binding site upstream of ER-GFP in J2341. OBO1 mRNA is first detected during floral development in a pattern identical to that of ER-GFP mRNA expression in flowers of J2341.
The generation of organ primordia from the meristem is orchestrated by the activities of various genes in Arabidopsis (2, 29). The STM gene encodes a homeodomain-type transcription factor expressed throughout embryogenesis in the exact center of the SAM between the cotyledons (14). Expression of STM begins at the late globular stage, when the embryo initiates radial patterning. As development proceeds, STM expression is excluded from organ primordia and remains at the center of the SAM, maintaining stem cell fate (14).
Arabidopsis CUC1, CUC2, and CUC3 encode NAM-ATAF-CUC (NAC)-type transcription factors required for STM expression (16, 23). STM and CUC are critical for initiation of the SAM and establishment of its boundary with the cotyledons (15, 23). CUC3 expression begins in the apical center of globular stage embryos and occurs between the meristem and lateral organs later during development (15). OBO1 expression is distinct from STM and CUC3 expression, never occurring in the SAM center.
STM represses expression of AS1 and AS2. AS1 and AS2 are ectopically expressed in stm mutants, leading to repression of KNAT1 expression and meristem arrest (30, 31). as2 mutants cause ectopic expression of KNOTTED-LIKE homeobox genes (31–33) and are epistatic to stm mutants (18). AS2 is a member of the LOB domain gene family, a family of transcription factors encoded by 42 Arabidopsis genes (17, 18, 34). LOB expression (17) resembles OBO1 expression when it occurs in a band of cells at the adaxial bases of lateral organs; however, LOB expression also occurs at the base of lateral roots. Ectopic expression of LOB leads to dwarf plants that are sterile (18). This phenotype is distinct from the limited alterations in petal number and morphology observed with OBO1 overexpression.
BOP1, a gene required for leaf morphogenesis in Arabidopsis, regulates transcription of AS1 and AS2 (35). BOP1 carries two types of domains, a BTB/POZ domain for protein dimerization and four copies of an ankyrin repeat to mediate protein–protein interaction (22). BOP1 expression begins at the torpedo stage (22). Like OBO1, BOP1 expression occurs at the junction between the meristem and lateral organs during embryo and seedling development, and its expression occurs at the bases of sepals and petals during floral development (20, 22). Their overlapping expression in petals suggests that BOP1 and OBO1 may act together. However, overexpression of BOP1 leads to dramatic increases in leaf and floral number (20), a significantly different phenotype than that seen in petals for OBO1 overexpression.
LOF1 and LOF2 encode MYB domain-type transcription factors expressed at the boundaries of lateral organs, similar to OBO1, CUC, LOB, and BOP (19). lof1 mutants display a phenotype of organ fusion and reduced axillary meristems, consistent with a role in organ boundary formation. lof1 mutants also increase the meristem defects of cuc3 and stm-10, suggesting that LOF1 acts either upstream of or in concert with CUC3 and STM.
OBO1 expression at the boundary of the SAM and lateral organs resembles similar patterns of expression of CUC, LOB, BOP, and LOF. The expression of OBO1 differs significantly from that of these genes, however. During embryogenesis, the OBO1 enhancer in J2341 drives ER-GFP expression not only in the cells immediately surrounding the SAM, but also in cells extending outward to the edges of the epidermis at the junction between the cotyledons and hypocotyl, forming two concentric rings around the SAM. Potentially, CUC, LOB, and BOP enhancers might drive similar patterns when expressed at fivefold higher levels, as in the J2341 enhancer trap line.
An independent study recently identified LSH3 and LSH4 as direct targets of the CUC1 transcription factor (36). LSH3 is identical to OBO1, and LSH4 is identical to OBO4; OBO4 is most homologous to OBO1 among the 10 members of the OBO family (Fig. 4 and Figs. S1 and S2). These data place OBO1 downstream of CUC1 activity. GFP-OBO1 localization to the nucleus suggests that OBO1 might interact with one or more transcription factors (LOB, BOP, and LOF) or with chromatin-remodeling factors that regulate CUC3 expression (37) to control the development of lateral organs. The fact that both STM and OBO1 are targets of CUC suggests that they act in the same pathway(s) to control embryo and seedling development.
In support of the critical role of the boundary between the SAM and lateral organs, when cells expressing OBO1 were destroyed in J2341 by cell ablation, 86% of seedlings displayed abnormal development and 50% did not produce true leaves. Elongation of roots was severely inhibited as well. Seedlings with only minor defects displayed ER-GFP fluorescence at the shoot apex, whereas seedlings with more severe defects had barely detectible ER-GFP. The moderate and severe phenotypes observed in seedlings likely reflect the result of DTA-induced cell ablation in boundary cells expressing OBO1, causing abnormal patterning during embryo development. That embryos form at all to produce the seedlings observed might possibly reflect the plasticity of plant development. Cells expressing OBO1 that are killed might be replaced by others, allowing development to occur. Variability in the timing and severity of phenotypes resulting from DTA-induced expression in embryos has been reported and suggested to arise from stochastic variability in gene expression, especially in transgenes (38).
Stamenoid tissues occur on petal margins in 35S::OBO1 flowers, likely reflecting OBO1-specific expression at the bases of petal and stamen primordia and along anther filaments. Defects in floral organ identity may result from a failure to establish floral organ boundaries or identities when OBO1 is overexpressed. Stamenoid petals also arise in weak mutants of APETALA2 (39). Mutations of UNUSUAL FLORAL ORGANS (UFO) result in reduced number of petals and petal–stamen fusions. UFO expression occurs at the base of developing petal primordial, similar to the region of OBO1 expression (40), and during embryogenesis, UFO expression occurs in a cup-shaped boundary at the basal boundary of the SAM (14).
Abnormal floral organs were observed in the rice mutant long sterile lemma (g1) (25). G1 encodes a small protein of 276 amino acids containing the OBO/LSH conserved domain. Thus, disrupted function of the conserved plant-specific DUF640 domain results in defects in floral development in both rice and Arabidopsis.
OBO1 (LSH3) is a player in the formation of the quintessential boundary for organ initiation off the flanks of the SAM in Arabidopsis. It will be interesting to address the requirement for OBO1 function during embryo, seedling, and floral development by testing for interactions between OBO1 and other organ boundary-specific genes and floral identity genes, especially class B genes, with directed yeast two-hybrid studies, double-mutant analyses, and gene-silencing strategies.
Materials and Methods
Microscopy.
For SEM, seedlings were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) and postfixed with 1% osmium tetroxide in the same buffer (pH 7.2). Dehydration and critical point drying are described at http://em-lab.berkeley.edu/EML/protocols/psem.php.
In Situ Hybridization.
Tissue fixation and in situ hybridization were performed as described previously (http://www.imbv.uio.no/gen/groups/narc/insitu.html). A 729-bp EcoRI-SacI fragment of ER-GFP–specific coding sequence was used to probe ER-GFP mRNA by hybridization for 16 h at 51 °C. OBO1 cDNA was obtained from the Arabidopsis Biological Resource Center (stock no. U14267). An OBO1-specific probe was designed by comparing the nucleotide sequences of OBO homologs (Fig. S2). Unique 5′ (256 bp) and 3′ (287 bp) fragments of OBO, excluding the conserved DUF640 domain, were linked by overlap PCR (http://www.bio.net/bionet/mm/methods/2004-April/098106.html), amplified, and inserted into pBluescript II SK(-) (Stratagene). Hybridization was performed for 17 h at 51 °C.
TAIL-PCR.
Genomic DNA was prepared from homozygous J2341 by the “simple DNA prep” method using shorty buffer as described previously (http://www.hos.ufl.edu/meteng/HansonWebpagecontents/NucleicAcidIsolation.html). Three left border-specific primers (PB4: 5′-CCGATTTCGGAACCACCATC-3′; PB5: 5′-TGAAGGGCAATCAGCTGTTG-3′; and PB6: 5′-GTCCGCAATGTGTTATTAA G-3′) and three degenerate primers [AD1: 5′-NTCGA(G/C)T(A/T)T(G/C)G(A/T)GT T-3′; AD2: 5′-NGTCGA(G/C)(A/T)GANA(A/T)GAA-3′; AD3: 5′-(A/T)GTGNAG(A/T)A NCANAG A-3′] were a generous gift from Jim Haseloff (University of Cambridge, Cambridge, UK). TAIL-PCR was performed, and the resulting products were cloned into pCR2.1-TOPO (Invitrogen) and sequenced.
Overexpression of OBO1.
GUS coding sequences downstream of the 35S promoter in pCAMBIA3301 (http://www.cambia.org/daisy/cambia/2071/version/1/part/4/data/pCAMBIA3301.pdf?branch=main&anguage=default) were removed by NcoI/PmlI digestion, replaced by OBO1 coding sequences, transformed into Agrobacterium, and transformed into WT C24 Arabidopsis.
Supplementary Material
Acknowledgments
We thank Steve Ruzin and Denise Schichnes of the College of Natural Resources Biological Imaging Facility for excellent advice and Jim Haseloff for PCR primers and UAS-DTA seeds. This research was supported by National Institutes of Health Grant GM45244 (to P.C.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018542108/-/DCSupplemental.
References
- 1.Mayer KFX, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 2.Clark SE. Cell signalling at the shoot meristem. Nat Rev Mol Cell Biol. 2001;2:276–284. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- 3.Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- 4.Breuil-Broyer S, et al. High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J. 2004;38:182–192. doi: 10.1111/j.1365-313X.2004.02026.x. [DOI] [PubMed] [Google Scholar]
- 5.Aida M, Tasaka M. Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol Biol. 2006;60:915–928. doi: 10.1007/s11103-005-2760-7. [DOI] [PubMed] [Google Scholar]
- 6.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 7.Benfey PN, Scheres B. Root development. Curr Biol. 2000;10:R813–R815. doi: 10.1016/s0960-9822(00)00814-9. [DOI] [PubMed] [Google Scholar]
- 8.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 9.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 10.van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- 11.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124:33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 12.Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:2227–2231. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haseloff J. GFP variants for multispectral imaging of living cells. Methods Cell Biol. 1999;58:139–151. doi: 10.1016/s0091-679x(08)61953-6. [DOI] [PubMed] [Google Scholar]
- 14.Long JA, Barton MK. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- 15.Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;15:1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takada S, Hibara K, Ishida T, Tasaka M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development. 2001;128:1127–1135. doi: 10.1242/dev.128.7.1127. [DOI] [PubMed] [Google Scholar]
- 17.Shuai B, Reynaga-Peña CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee DK, Geisler M, Springer PS. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development. 2009;136:2423–2432. doi: 10.1242/dev.031971. [DOI] [PubMed] [Google Scholar]
- 20.Hepworth SR, Zhang YL, McKim S, Li X, Haughn GW. BLADE-ON-PETIOLE–dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell. 2005;17:1434–1448. doi: 10.1105/tpc.104.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha CM, et al. The BLADE-ON-PETIOLE 1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development. 2003;130:161–172. doi: 10.1242/dev.00196. [DOI] [PubMed] [Google Scholar]
- 22.Ha CM, Jun JH, Nam HG, Fletcher JC. BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1361–1370. doi: 10.1093/pcp/pch201. [DOI] [PubMed] [Google Scholar]
- 23.Aida M, Ishida T, Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- 24.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida A, Suzaki T, Tanaka W, Hirano HY. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci USA. 2009;106:20103–20108. doi: 10.1073/pnas.0907896106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L, et al. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. 2004;37:694–706. doi: 10.1111/j.1365-313x.2003.01993.x. [DOI] [PubMed] [Google Scholar]
- 27.Laplaze L, et al. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot. 2005;56:2433–2442. doi: 10.1093/jxb/eri236. [DOI] [PubMed] [Google Scholar]
- 28.Czakó M, Jang JC, Herr JM, Jr., Márton L. Differential manifestation of seed mortality induced by seed-specific expression of the gene for diphtheria toxin A chain in Arabidopsis and tobacco. Mol Gen Genet. 1992;235:33–40. doi: 10.1007/BF00286178. [DOI] [PubMed] [Google Scholar]
- 29.Laux T, Würschum T, Breuninger H. Genetic regulation of embryonic pattern formation. Plant Cell. 2004;16(Suppl):S190–S202. doi: 10.1105/tpc.016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrne ME, et al. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- 31.Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- 32.Semiarti E, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 33.Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- 34.Husbands A, Bell EM, Shuai B, Smith HMS, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35:6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ha CM, Jun JH, Nam HG, Fletcher JC. BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell. 2007;19:1809–1825. doi: 10.1105/tpc.107.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda S, et al. 21st International Conference on Arabidopsis Research. Stanford, CA: The Arabidopsis Information Resource; 2010. LSH4 and LSH3, two members of the ALOG gene family in Arabidopsis thaliana, are activated in shoot organ boundary cells by the transcription factor CUP-SHAPED COTYLEDON1; p. 169. [Google Scholar]
- 37.Kwon CS, et al. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development. 2006;133:3223–3230. doi: 10.1242/dev.02508. [DOI] [PubMed] [Google Scholar]
- 38.Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R. Diphtheria toxin–mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 2003;133:1882–1892. doi: 10.1104/pp.103.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durfee T, et al. The F-box–containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc Natl Acad Sci USA. 2003;100:8571–8576. doi: 10.1073/pnas.1033043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.