Abstract
The transcription factor p73 plays critical roles during development and tumorigenesis. It exhibits sequence identity and structural homology with p53, and can engage p53-like tumor-suppressive programs. However, different pathways regulate p53 and p73, and p73 is not mutated in human tumors. Therefore, p73 represents a therapeutic target, and there is a critical need to understand genes and noncoding RNAs regulated by p73 and how they change during treatment regimens. Here, we define the p73 genomic binding profile and demonstrate its modulation by rapamycin, an inhibitor of mammalian target of rapamycin (mTOR) and inducer of p73. Rapamycin selectively increased p73 occupancy at a subset of its binding sites. In addition, multiple determinants of p73 binding, activity, and function were evident, and were modulated by mTOR. We generated an mTOR-p73 signature that is enriched for p73 target genes and miRNAs that are involved in mesenchymal differentiation and tumorigenesis, can classify rhabdomyosarcomas by clinical subtype, and can predict patient outcome.
Keywords: p63, ChIP-on-Chip, stem cell, muscle, consensus binding site
No single target gene can explain more than a fraction of the tumor-suppressive activity of p53. Instead, p53 regulates the transcription of an extensive network of genes involved in diverse functions, such as cell cycle arrest, apoptosis, senescence, and autophagy (1). Genome-wide technologies, such as ChIP coupled with sequencing (ChIP-Seq or ChIP-PET) or microarray (ChIP-on-Chip) confirm that p53 regulates complex gene networks (2, 3). In addition, mammals have two homologs of p53: p63 and p73, each with multiple isoforms (4). All three p53 family members share a core structural architecture and significant sequence identity and, although their precise roles remain unknown, p63 and p73 can also act as tumor suppressors (4).
It is clear that p63 and p73 can regulate many of the same functions as p53. However, comprehensive whole-genome analyses are needed to identify shared or unique target genes that underlie these functions, as well as biochemical determinants, such as sequence-specificity, use of cofactors, and epigenetic context. Analysis of p63 binding sites in cervical cancer cells has revealed both similarities and differences in the way that p53 family members regulate gene expression (5). Such insights into p53 and p63 highlight the need to investigate p73 transcriptional networks. Unlike p53 and p63, p73 is neither mutated nor amplified in human tumors (4). It is unclear how upstream factors regulate the binding distribution of p73 in cancer cells and whether this can be manipulated to affect tumor growth.
In the present study we report a comprehensive, whole-genome analysis of p73 binding in human cells. We also used a cellular stressor to assess p73 binding under nonsteady state conditions. Mammalian target of rapamycin (mTOR) is a serine/threonine kinase that regulates cell growth, proliferation, and survival. It is a known inhibitor of p73 in cancer cells (6). In our study rapamycin, an mTOR inhibitor, selectively increased p73 occupancy at ∼9% of genomic binding sites. In addition, we identified determinants of p73 binding, many of which were modulated by mTOR. We created an mTOR-p73 gene signature that is enriched for genes and noncoding RNAs (ncRNAs) involved in mesenchymal differentiation and rhabdomyosarcoma tumorigenesis, and can classify tumors by clinical subtype and predict patient outcome.
Results
Mammalian TOR Modulates the p73 Cistrome.
To perform a comprehensive analysis of p73 binding sites in the genome (the p73 cistrome), we sought a cell line that expresses abundant p73 with detectable binding to chromatin. We chose the rhabdomyosarcoma line, Rh30, because p73 exhibited robust binding to representative target genes in these cells (Fig. S1A). Although Rh30 cells express mutant p53, rapamycin does not alter p53 levels in these cells and there is no detectable interaction between p73 and mutant p53 (6). It is possible for p73 to be expressed as one of multiple isoforms that either contain (TA) or lack (ΔN) an N-terminal transactivation domain (4). For genome-wide ChIP analysis, we used an antibody that only recognizes p73 proteins with a transactivation domain. These TAp73 isoforms are considered “active” as they are capable of inducing expression of target genes. In addition, p73 exhibits C-terminal diversity, and TAp73 can be expressed as any of nine variants (4). We detect two isoforms in Rh30 cells at the protein level, TAp73α and TAp73β. The levels of both are increased in rapamycin-treated Rh30 cells, and both isoforms bind DNA (Fig. 1 A and B).
Fig. 1.
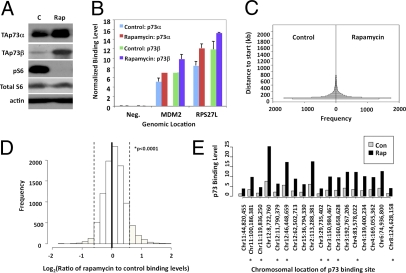
Rapamycin increases p73 binding to specific regions of the genome. (A) Protein levels of p73 increase in Rh30 cells treated with 40 nM rapamycin for 24 h, as analyzed by Western blot. “C” is vehicle control; pS6 is a marker of mTOR activity. (B) ChIP-qPCR was performed to assess p73 occupancy at MDM2 and RPS27L promoters using two antibodies that preferentially recognize either p73α or p73β. Error bars indicate SD from triplicate analyses. (C) Histogram analysis was performed to assess the distribution of p73 binding sites relative to transcriptional start sites. The control sample (Left) and rapamycin-treated sample (Right) were compared using the two-sample Kolmogorov-Smirnov test. The P value was 0.6, indicating no significant difference. (D) Histogram analysis was performed to assess the impact of rapamycin on overall p73 binding level. The ratio of binding levels in rapamycin-treated vs. control samples was calculated and log-transformed. A skew to the right is evident in this plot, indicating higher binding in rapamycin-treated samples. The degree of skew is 0.34, with P < 0.0001. (E) Binding levels of p73 were plotted for 18 loci that exhibited the greatest increase in binding after rapamycin treatment. Chromosomal locations within 10 kb of known genes are indicated by an asterisk. From left to right, these genes are: FLJ32810, ARHGEF12, ETV6, RAPGEF3/FLJ20489, RBMS3, C3orf16, SCHIP1, HNRPDL/ENOPH1, and FBXO32.
ChIP-on-Chip was performed on both control and rapamycin-treated Rh30 cells, based on previously published data showing a robust increase in p73 protein levels at 24 h after 40 nM rapamycin treatment (6). DNA recovered from ChIP was hybridized to oligonucleotide tiling arrays that cover the entire nonrepetitive human genome at 35-bp resolution, and p73 binding was measured as fold-enrichment after ChIP compared with unenriched input. A total of 7,678 sites in control cells and 8,165 sites in rapamycin-treated cells showed greater than 2.5-fold enrichment after p73 ChIP. The false-discovery rate at this cutoff is ∼0.86%, giving us high confidence that these sites were indeed bound by p73.
The accuracy of ChIP is highly reliant on antibody specificity. We validated our ChIP-on-Chip results on 30 randomly chosen p73-binding sites by performing ChIP-qPCR compared with a nonspecific antibody, and with two additional C-terminal p73 antibodies (Fig. S1B). To further validate our ChIP-on-Chip results, we used phastCons as a measure of evolutionary conservation at every nucleotide in the genome (7). Sequence conservation is an established property of functional genomic elements. p73 binding sites were conserved relative to nonbound, distant regions (Fig. S1 C and D). Thus, ChIP-on-Chip identified high-confidence p73 binding sites of likely functional significance.
To determine if inhibition of mTOR resulted in alteration of the p73 genomic binding profile, we measured the effect of rapamycin on p73 binding relative to two different parameters. First, we measured the p73 cistrome by distance to nearest transcriptional start site. As shown in Fig. 1C, rapamycin treatment did not alter p73 binding distribution. As a second measure of p73 occupancy, ChIP enrichment levels were compared before and after rapamycin treatment. There was a significant increase in p73 binding level after rapamycin treatment (Fig. 1D) (P < 0.0001, skew test). The p73 binding did not increase uniformly at all sites in the genome after rapamycin treatment. At 8.7% of p73 binding sites, we observed a greater than 50% increase in binding level after treatment. Eighteen sites to which p73 exhibited greater than 2.5-fold increased binding are shown in Fig. 1E. Collectively, these data suggest that mTOR inhibition increases p73 protein occupancy only at select loci. Therefore, we explored determinants of p73 binding, such as sequence-specificity and association with cofactors, which may contribute to selective changes in p73 binding or activity.
Annotation and Analysis of the p73 Cistrome Reveals Multiple Determinants of p73 binding.
We annotated the 7,678 p73 binding sites from the untreated Rh30 sample by their location relative to genes. A total of 4,083 genes in the genome were bound by p73, with some genes containing multiple p73 binding sites (Fig. S2A). Rapamycin treatment resulted in an ∼5% increase in this number (Fig. S2B). In both treated and untreated cells, p73 bound predominantly to introns, proximal promoters, and distant sites that may be enhancers (Fig. S2C). When p73-bound genes were assigned to functional and biochemical pathways, diverse signaling pathways and cancer types were identified. Many of these associations were found to have increased significance in rapamycin-treated samples (Fig. S2D).
Transcription factors use a specific DNA sequence or motif to aid in selective binding. We used a motif-finding algorithm to identify a sequence enriched in p73-bound loci. Treatment with rapamycin did not significantly alter this motif (Fig. S2E), which is nearly identical to the p53 and p63 response elements (2, 8). However, as indicated by the arrows in Fig. S2E, p73 shares certain degenerate motif elements with p63 compared with p53. These degenerate elements are thought to decrease selectivity of binding, leading to a larger pool of potential binding sites (8). Consistent with these data, p73 binds ∼4,000 genes, compared with ∼3,000 for p63 and ∼1,500 for p53 (3, 5).
In Fig. S3, despite differences in cell lines, methodology, and treatments, when comparing our data set with those previously published for p53 (2) and p63 (5) we were able to identify a significant set of shared targets (SI Appendix, Dataset S1). The p73 cistrome was determined for TAp73 (Fig. S3D) and assessed in the absence of p63 (Fig. S3E), which may explain the identification of unique as well as shared p73 targets in the present study.
Based on our analysis of the p73 binding motif, p73 binds only a fraction of its potential binding sites. Thus, other factors must determine whether or not p73 binds a particular motif both at baseline and during mTOR inhibition. One such determinant may be the association of p73 with other transcription factors and cofactors. To obtain a list of DNA-binding factors associated with p73-bound loci, we used the TRANSFAC and JASPAR databases of eukaryotic cis-acting regulatory DNA elements that contain a total of ∼800 nucleotide distribution matrices (7). By searching for enriched motifs in p73-bound regions, 58 DNA-binding factors were found to have significant association (binomial test, P < 0.00001) with the p73 cistrome (SI Appendix, Dataset S2).
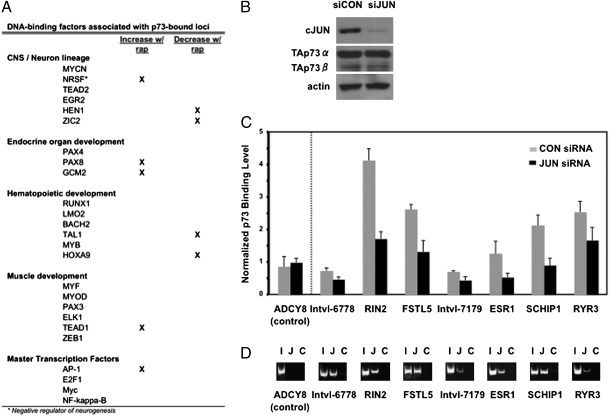
Many of these cofactors are involved in tissue-specific functions, such as neuron lineage specification, endocrine organ development, hematopoietic development, and muscle development (Fig. 2A). Interestingly, p73-null mice suffer from a number of tissue-specific defects, such as neuron cell death and immune cell infiltration (9). We validated the set of cofactors through additional analysis of c-Jun (AP-1), a master transcription factor that is also a regulator of myogenic differentiation (10). Knock-down of c-Jun did not alter p73 protein levels in Rh30 cells (Fig. 2B); however, depletion of c-Jun did result in decreased p73 binding at genomic loci in the p73 cistrome (Fig. 2C). We performed ChIP analysis of c-Jun to confirm that c-Jun was binding within 500 bp of each p73-bound locus (Fig. 2D). These data confirm that c-Jun can bind near p73 and promote binding of p73 to genomic loci, validating this approach for identification of cofactors.
Fig. 2.
Tissue-specific factors associate with p73-bound loci. (A) DNA-binding factors with motifs overrepresented in the p73 cistrome are organized by tissue-specific function (binomial test, P < 0.0001). Marked factors have ≥ 20% increased enrichment in rapamycin-treated or control samples. (B) Small interfering RNA-mediated depletion of c-Jun does not alter p73 protein levels in Rh30 cells, as assessed by Western blot. (C) ChIP-qPCR is shown assessing p73 binding to select genomic loci (indicated by interval number or by nearest target gene, where possible) in Rh30 cells treated with siRNA targeting c-Jun or control siRNA. Error bars indicate SD from triplicate analyses. (D) ChIP of c-Jun binding to sites within 500 bp of p73 binding sites is shown. Primers flank c-Jun motifs as assessed by sequence analysis for the same genomic loci as in C. PCR product was resolved by acrylamide gel electrophoresis for reactions using the following DNA samples: genomic input (“I”), DNA purified after ChIP with a c-Jun-specific antibody (“J”), and DNA purified after ChIP with a control, nonspecific, isotype-matched antibody (“C”). ADCY8 in C and D is a negative control locus that does not have a c-Jun consensus binding site within 500 bp of the p73 interval.
Of note, the presence of several cofactor DNA binding motifs is differential at p73-bound loci in rapamycin-treated or untreated cells (Fig. 2A). Cofactor binding is one mechanism by which increased p73 protein may be recruited to specific sites after rapamycin treatment.
Mammalian TOR Regulates the Expression of Genes, miRNAs, and ncRNAs Through p73.
Microarray analyses were performed to determine which p73-binding sites are associated with transcriptional regulation by the mTOR-p73 axis. Rh30 cells were treated with vehicle or rapamycin, as above. In addition, two different RNAi targeting sequences were used to deplete p73 in independent samples to assess p73-dependence. Knockdown was confirmed by Western blot, as has been reported previously for these constructs (6).
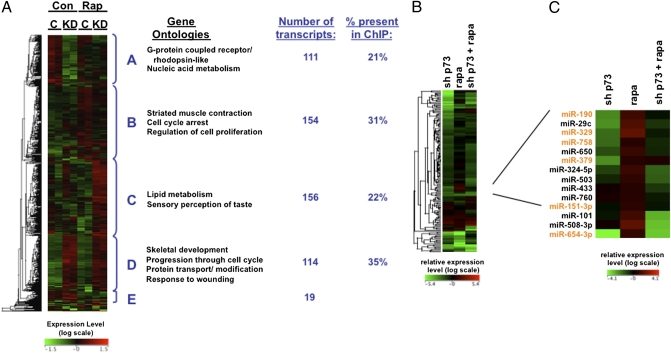
Gene-expression patterns using the two p73 RNAi constructs (pan-p73 and TA-specific) showed a high level of correlation (Pearson correlation coefficient r2 = 0.996 in rapamycin-treated samples), and were treated as duplicates to calculate fold-change differences in expression. Clustering analysis revealed that 554 transcripts, which comprise an mTOR-p73 gene signature, segregated into discrete groups (Fig. 3A, Clusters A–E). Cluster A contains 111 transcripts that decreased after knockdown of p73, including known targets of p73, such as mdm2 (4). We also found 154 transcripts that were increased in a p73-dependent manner after mTOR inhibition (Fig. 3A, Cluster B). However, these genes were only a subset of those regulated by mTOR and p73. Intriguingly, more than half of the genes in the signature increased rather than decreased after knockdown of p73, and thus were targets of p73-mediated repression (Fig. 3A, Clusters C and D).
Fig. 3.
Genes and miRNAs regulated by mTOR and p73 display distinct patterns of regulation. (A) Microarray analysis was performed in duplicate using Rh30 cells transduced with shRNA targeting p73 (KD) or GFP (C) for 3 d, cells were treated with vehicle (con) or 40 nM rapamycin (rap) for 24 h. Depicted by heat map are genes that changed >50% with rapamycin treatment or p73 depletion. Hierarchical clustering (average linkage) reveals distinct groups of transcripts: A, increased by p73; B, increased by p73 in an mTOR-dependent manner; C, decreased by p73 in an mTOR-dependent manner; D, decreased by p73; E, regulated by mTOR alone. Major gene ontologies (hypergeometric test, P < 0.05), total transcript numbers, and the percentage of genes present in the ChIP-on-Chip dataset are indicated for each cluster. (B) Hierarchical clustering was used to segregate 142 miRNAs whose expression levels were regulated by mTOR or p73 into discrete groups. Rh30 cells were treated with p73 RNAi (shp73) or 40 nM rapamycin (rapa), and the ΔΔCT method was used to calculate expression relative to control samples. (C) A total of 14 miRNAs whose expression levels increase after rapamycin treatment in a p73-dependent manner are shown. Those miRNAs that are located within 10 kb of a p73 binding site are highlighted in orange text.
There were too few transcripts regulated by mTOR independent of p73 for additional statistical analysis (Fig. 3A, Cluster E). Gene ontologies were used to functionally categorize the genes in the other four clusters (hypergeometric test, P < 0.05) (Fig. 3A). Comparison of these p73-regulated genes to our ChIP-on-Chip database showed that 29% of p73-regulated genes were direct targets of p73. A comprehensive list of genes and ncRNAs directly regulated by p73 can be found in SI Appendix, Dataset S3, and contains both known and novel p73 targets.
Finally, we addressed one class of transcript that is not covered by whole-genome oligonucleotide arrays: short miRNAs. Through direct modulation of miRNAs, p73 may regulate genes indirectly. Indeed, analysis of p73 ChIP data revealed 116 miRNAs that are within 10 kb of p73 binding sites (SI Appendix, Dataset S4). Less is known about the regulatory regions of miRNAs than of genes. However, miRNA promoters were recently mapped across the human genome (11). We found that p73 bound within the promoters of eight of these miRNAs (Table S1).
Interrogation of 667 miRNA expression levels after rapamycin treatment and p73 depletion in Rh30 cells was performed using TaqMan low-density arrays and two different RNAi constructs. A total of 142 miRNAs were identified whose expression levels changed more than 50% with either p73 knockdown or rapamycin treatment. Unlike p73-regulated genes, most of these miRNAs were transcriptionally activated by p73 (Fig. 3B). We overlapped the 142 miRNAs that were modulated after p73 knockdown with the 116 p73-bound miRNA loci. A total of 41 p73-regulated miRNAs were in the vicinity of p73 binding sites, and thus are likely directly regulated by p73 (SI Appendix, Dataset S4), although in some cases this may be because of the regulation of large polycistrons of miRNAs by p73. In addition, 14 miRNAs were regulated by mTOR in a p73-dependent manner (Fig. 3C). This finding suggests extensive regulation of genes and ncRNAs by mTOR through regulation of p73 binding and transcriptional activity.
Genes Regulated by mTOR and p73 Classify Rhabdomyosarcoma Subtypes and Patient Outcomes.
Our genomic analyses gave us an unprecedented opportunity to explore p73 function using the mTOR-p73 gene signature (Fig. 3A, Clusters A–D). We were intrigued by the pathways enriched in p73-bound loci that are associated with muscle function and development (Table S2). It has been reported that p53, p63, and p73 play at least partially redundant roles in myogenic differentiation and rhabdomyosarcoma formation (12). Alteration of the p53 pathway for example, through a dominant-negative isoform of p73, can block skeletal muscle cell differentiation and promote transformation in vitro (12), and mTOR activity, perhaps by acting upstream of p73, is negatively associated with pediatric rhabdomyosarcoma outcome (13).
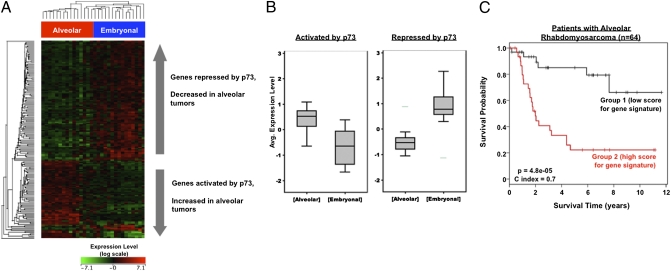
Wachtel et al. analyzed the gene expression patterns of 29 primary rhabdomyosarcomas (14). We filtered these tumor data against the mTOR-p73 signature, and used hierarchical clustering to segregate tumors according to the status of the mTOR-p73 signature (Fig. 4A). Interestingly, rhabdomyosarcoma patients segregated into two distinct clusters that were equivalent to the major rhabdomyosarcoma subtypes: alveolar and embryonal.
Fig. 4.
The p73-regulated genes are differentially expressed in rhabdomyosarcoma subtypes. The mTOR-p73 gene signature was assessed using a publicly available dataset in which 29 rhabdomyosarcoma tumors were profiled by microarray (14). (A) All genes that changed >20% with rapamycin or p73 RNAi were included in this initial analysis to increase overlap across microarray platforms. Hierarchical clustering demonstrates that the mTOR-p73 gene signature segregates tumors to two classes, corresponding to the clinical alveolar and embryonal subtypes. (B) Genes regulated > 50% by p73 were analyzed in ARMS and ERMS. Box plots demonstrate opposing gene expression trends in these two tumor subtypes. (C) The expression levels of 17 direct p73 target genes were used to segregate an independent cohort of 64 ARMS patients to two groups, using Cox proportional hazards modeling and Kaplan-Meier analysis. This 17-gene p73 signature segregates patients with ARMS but not ERMS by clinical survival time.
These two subtypes of rhabdomyosarcoma are associated with different outcomes, molecular alterations, age groups, and histologic appearances (15). Alveolar rhabdomyosarcoma (ARMS) is more common in older children and exhibits small round cells with primitive myoblast differentiation, embryonal rhabdomyosarcoma (ERMS) typically presents at an earlier age and is histopathologically analogous to embryonic skeletal muscle (15). We found that genes positively regulated by p73 were increased in ARMS (Fig. 4 A and B). Conversely, genes transcriptionally repressed by p73 were decreased in ARMS (Fig. 4 A and B). These data suggest that p73, downstream of mTOR, is actively engaged in transcriptional regulation in the alveolar subtype that is associated with a worse prognosis. A similar effect was observed in an independent cohort containing 139 rhabdomyosarcoma patients, previously reported by Davicioni et al. (16) (Fig. S4 A and B).
Clinical outcome data were available for 134 rhabdomyosarcoma patients in the Davicioni et al. cohort (16). In this cohort we could identify genes in the p73 signature that correlated with 5-y survival after treatment for ARMS (Fig. S4C). The tumor expression levels of 17 direct p73 target genes, selected by t test and shown in Fig. S4C (orange text), were used to create a compound score model (described in SI Materials and Methods) that could segregate rhabdomyosarcoma patients into subgroups. Alveolar tumors with a low compound score for p73 target genes were associated with a 5-y survival rate of ∼65%, whereas tumors with a high compound score were associated with a 5-y survival rate of ∼20% (Fig. 4C). This effect was independent of tumor size, patient age, or tumor stage. In contrast to ARMS, there was no significant difference in survival from ERMS based on these genes (Table S3). Thus, genes in the p73 signature are associated with clinical outcome, suggesting that the mTOR-p73 axis plays a role in ARMS pathogenesis.
We hypothesized that this role could be related to functions of p73 during myogenic differentiation (12). Although the precise cell of origin remains unknown, studies of molecular translocations provide evidence for mesenchymal stem cells (MSCs) as possible cells of origin for ARMS (15). We analyzed the fate of p73-regulated genes during MSC differentiation. We first confirmed that p73 does bind known target genes in MSCs (Fig. 5A). MSCs can be induced to differentiate along the myogenic lineage by treating them with 5-azacytidine (5AZA) (17). We identified a group of p73 target genes, both direct and indirect, whose expression levels changed during MSC differentiation (corrected P value < 0.05) (Fig. 5B). Furthermore, we confirmed that 5AZA treatment decreased phospho-S6 levels in MSCs (Fig. 5C). The direct p73 target genes with altered expression during differentiation that were sufficient to segregate alveolar and embryonal tumors are shown in Fig. 5D. Through RNAi targeting of p73 in MSC cultures, we discovered that several of these genes were regulated by 5AZA in a p73-dependent manner (Fig. 5E).
Fig. 5.
Genes and miRNAs regulated by p73 and mTOR are associated with MSC differentiation. (A) Analysis of p73 binding level to select target genes by ChIP-qPCR, performed in human MSCs. Error bars indicate SD from triplicate analysis. (B) p73/mTOR-regulated genes, p73-bound genes (ChIP-on-Chip), and genes significantly altered during 5AZA-induced myogenic differentiation of MSCs (multiple testing, corrected P value < 0.05) were compared. MSC microarray data were publicly available (17). The Venn diagram shows the number of Affymetrix probes in each category. (C) Treatment of MSCs with 5AZA results in decreased mTOR activity as shown by Western blot analysis of pS6, total S6, and actin levels. (D) This core set of nine direct p73 target genes from those regulated by 5AZA was sufficient to segregate ARMS and ERMS. Fold-changes are indicated in two bar charts for: 5AZA-treated MSCs relative to control (Left), and ARMS relative to ERMS (Right). (E) Quantitative RT-PCR was performed in MSCs treated with 5AZA, after knock-down of p73 (shp73) or control (shGFP) for the indicated genes, normalized to GAPDH levels and control. (F) Quantitative RT-PCR analysis of mature miR-133b levels was performed in Rh30 cells treated with control RNAi (shGFP), or three different RNAi constructs that target p73 (shp73-1, -2, and -3), normalized to RNU19 levels. Error bars represent SDs from three independent experiments.
Interestingly, many genes indirectly regulated by p73 were also associated with mesenchymal differentiation (120 genes) (Fig. 5B). The expression of these genes might be regulated by p73 through direct regulation of miRNAs. Three miRNAs with established roles in myogenic differentiation, miR-133b, miR-133a-2, and miR-1-1 (18), mapped to p73-bound loci. These three miRNAs are up-regulated in certain human sarcomas, including ARMS (19). MicroRNA-133b was the closest of the three to a p73 binding site (∼5 kb upstream of miR-133b transcriptional start site). We used real-time PCR and three different p73 RNAi constructs to confirm that p73 regulates miR-133b expression levels in Rh30 cells (Fig. 5F). Taken together, these data suggest that p73 regulates transcriptional programs common to both MSC differentiation and rhabdomyosarcoma tumorigenesis. These data may explain the ability of p73 gene signatures to subclassify rhabdomyosarcoma and predict patient outcome.
Discussion
Herein we demonstrate that, like p53 and p63, p73 exhibits complex patterns of binding and activity across the genome. In addition, we show that mTOR modulates p73 occupancy in a selective, locus-specific manner. Although p73 has affinity for a particular 20-bp DNA motif, this only loosely defines the boundaries of the p73 cistrome. The motifs of numerous cofactors are found at p73-bound loci, and likely serve as additional determinants of transcriptional specificity. Cofactor interactions provide the potential for exquisite control of p73 binding, and are regulated in part by mTOR. In addition, mTOR may regulate p73 function by altering the TAp73α:TAp73β ratio, as both isoforms bind DNA in these cells but TAp73β appears more robustly regulated by mTOR. Consistent with this finding, the majority of p73-regulated genes were transcriptionally repressed, and the TAp73α isoform can serve as a transcriptional inhibitor (20). The end product of this intricate regulation is the expression of genes and noncoding RNAs, for which we demonstrate functional significance through analysis of rhabdomyosarcoma tumorigenesis.
The definition of the p73 cistrome allows comparison with the p53 and p63 cistromes (2, 5). Although it has previously been shown that p63 and p73 can co-occupy DNA binding sites and have similar genome binding profiles (21), our analysis of p73 in the absence of other family members demonstrates that it has unique target genes, is regulated by unique upstream signaling pathways, and interacts with diverse cofactors representing widespread tissue expression. In contrast to p63, which is a marker of squamous differentiation and is overexpressed in many squamous-type tumors, p73 is expressed in many normal tissues types in which p63 expression is absent, as well as in tumors of the brain, breast, lung, ovary, skin, prostate, colon, thymus, and muscle (4). The activity of p73 in both the presence and absence of p63 warrants extensive investigation, and may yield insight to potential targets in a wide range of cancer types.
The p73 cistrome can be used to expand on previous studies of p53 family members in myogenic differentiation. For example, in one study an inhibitory isoform of p73 was overexpressed in a mouse myogenic cell line undergoing differentiation (12). By microarray, this p73 isoform modulated the expression of a large number of genes; however, because of lack of information on the p73 cistrome the authors were unable to identify which of these genes were direct targets (12). Through overlay of the mouse genes identified in the previous gene-expression analysis to human orthologs, and comparison with the p73 cistrome, we were able to identify 76 target genes that are directly regulated by p73 in the murine system (SI Appendix, Dataset S5).
Recent studies show that p73 can be engaged to activate p53 apoptotic pathways in tumor cells that have mutated p53 (22). There has been substantial interest both in drugs that target p73 and in potential prognostic markers of p73 activity. We have shown that mTOR inhibition induces p73 and selectively alters the p73 cistrome. Even more striking, the majority of transcriptional changes that followed rapamycin treatment were dependent on p73. Mammalian TOR inhibitors are currently in clinical trials for a number of human cancers, and it will be interesting to see if these drugs activate p73 in vivo or synergize with other drugs that can activate p73.
Because p73 is not mutated in tumors, the key to a p73-based treatment approach will lie in the ability to predict response based on an assessment of p73 status. Perhaps to a greater extent than its family members, the activity of p73 frequently does not correlate well with its RNA expression level. Indeed, we found that p73 RNA levels varied by only 2 to 4% across alveolar and embryonal subsets of rhabdomyosarcomas in the Davicioni et al. dataset (16). Because the latter tumor specimens were not available to assess the p73 protein levels or mTOR activity, we obtained other rhabdomyosarcoma specimens and found that 60% of the tumors analyzed by immunohistochemistry had robust p73 and phospho-S6 staining (a surrogate marker of mTOR activity) (SI Material and Methods). These results are consistent with prior studies describing mTOR (13) and p73 (12) in rhabdomyosarcomas. Changes in p73 activity are likely a combination of the isoforms of p73 that are expressed, the interacting proteins that regulate its activity, and the other p53 family members that are interacting with it or a target gene in any given tumor. Using gene signatures as a biomarker of p73 activity would be a composite of all of the above factors. Such signatures might find utility in the molecular profiling of sarcomas and other tumor types for therapeutic decision-making. The advantage of using a gene signature as a readout of a complex cellular change lies not only in the ability to connect pathways, but also in the detection of signaling modules that can stratify tumors to clinically relevant subtypes. Transcription factors are no longer intractable therapeutic targets, but instead could be used for their high degree of “connectivity.”
Materials and Methods
ChIP-on-Chip, ChIP-qPCR, and Statistical Analyses.
Formaldehyde cross-linking and chromatin preparation and immunoprecipitation were carried out as described previously (6). For ChIP-on-Chip and semiquantitative ChIP experiments, cells were cross-linked and submitted to GenPathway, Inc., according to their FactorPath protocol. See SI Materials and Methods for additional details on this protocol and statistical analyses.
Dataset Accession Numbers.
Microarray and ChIP-on-Chip data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession numbers GSE-15703 and GSE-15704.
Protocols for cell transfection/infection and shRNA, protein lysate preparation and Western analysis, and miRNA isolation and expression analysis can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank C. Coffin for tumor samples, C. Backendorf and G. Melino for p73 expression constructs, K. Johnson and C. Pendleton for isolation of MSCs, and E. Burgess for modifying the pSicoR vectors. This work was supported by National Institutes of Health Grants CA70856 and CA105436 (to J.A.P.), ES00267 and CA68485 (core services), US Army Medical Research and Materiel Command Grant W81XWH-04-1-0304 (to J.M.R.), and GM07347 (Medical Scientist Training Program).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE15719).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011936108/-/DCSupplemental.
References
- 1.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 3.Smeenk L, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: An orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 5.Yang A, et al. Relationships between p63 binding, DNA sequence, transcription activity, and biological function in human cells. Mol Cell. 2006;24:593–602. doi: 10.1016/j.molcel.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;28:5951–5964. doi: 10.1128/MCB.00305-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34(Web Server issue):W551–W554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez CA, Ott J, Mays DJ, Pietenpol JA. p63 consensus DNA-binding site: Identification, analysis and application into a p63MH algorithm. Oncogene. 2007;26:7363–7370. doi: 10.1038/sj.onc.1210561. [DOI] [PubMed] [Google Scholar]
- 9.Yang A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 10.Ostrovsky O, Bengal E, Aronheim A. Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J Biol Chem. 2002;277:40043–40054. doi: 10.1074/jbc.M205494200. [DOI] [PubMed] [Google Scholar]
- 11.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cam H, et al. p53 family members in myogenic differentiation and rhabdomyosarcoma development. Cancer Cell. 2006;10:281–293. doi: 10.1016/j.ccr.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 13.Petricoin EF, 3rd, et al. Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Res. 2007;67:3431–3440. doi: 10.1158/0008-5472.CAN-06-1344. [DOI] [PubMed] [Google Scholar]
- 14.Wachtel M, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64:5539–5545. doi: 10.1158/0008-5472.CAN-04-0844. [DOI] [PubMed] [Google Scholar]
- 15.Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: Is the cell of origin a mesenchymal stem cell? Cancer Lett. 2009;279:126–136. doi: 10.1016/j.canlet.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 16.Davicioni E, et al. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66:6936–6946. doi: 10.1158/0008-5472.CAN-05-4578. [DOI] [PubMed] [Google Scholar]
- 17.Mishra PJ, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–4339. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian S, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–2026. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 20.Vilgelm A, El-Rifai W, Zaika A. Therapeutic prospects for p73 and p63: Rising from the shadow of p53. Drug Resist Updat. 2008;11:152–163. doi: 10.1016/j.drup.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang A, et al. Genome-wide mapping indicates that p73 and p63 co-occupy target sites and have similar dna-binding profiles in vivo. PLoS ONE. 2010;5:e11572. doi: 10.1371/journal.pone.0011572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell HS, Ryan KM. Targeting the p53 family for cancer therapy: ‘Big brother’ joins the fight. Cell Cycle. 2007;6:1995–2000. doi: 10.4161/cc.6.16.4614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.