Abstract
It is well known that humans tend to associate with other humans who have similar characteristics, but it is unclear whether this tendency has consequences for the distribution of genotypes in a population. Although geneticists have shown that populations tend to stratify genetically, this process results from geographic sorting or assortative mating, and it is unknown whether genotypes may be correlated as a consequence of nonreproductive associations or other processes. Here, we study six available genotypes from the National Longitudinal Study of Adolescent Health to test for genetic similarity between friends. Maps of the friendship networks show clustering of genotypes and, after we apply strict controls for population stratification, the results show that one genotype is positively correlated (homophily) and one genotype is negatively correlated (heterophily). A replication study in an independent sample from the Framingham Heart Study verifies that DRD2 exhibits significant homophily and that CYP2A6 exhibits significant heterophily. These unique results show that homophily and heterophily obtain on a genetic (indeed, an allelic) level, which has implications for the study of population genetics and social behavior. In particular, the results suggest that association tests should include friends’ genes and that theories of evolution should take into account the fact that humans might, in some sense, be metagenomic with respect to the humans around them.
Keywords: social networks, gene–environment interaction, indirect genetic effects
Humans are unusual as a species in that virtually all individuals form stable, nonreproductive unions to one or more friends. Although people apparently choose their friends freely, genes are known to play a role in the formation (1), attributes (2–4), and network structures (5) of these unions. It remains unclear, however, whether variation in specific genes might be relevant to the process of friend selection or, distinctly, whether the process of choosing friends might result in correlated genotypes between friends. Both processes would have implications for our understanding of population genetics and gene–environment interactions.
Here, we examine whether or not phenotypic similarity between individuals connected in a social network is reflected in their genotypes. Associations between the genotypes of connected individuals are known to result from genetic correlations between mates or as a result of groupings established on the basis of kinship (e.g., matrilineal tribal groupings). Indeed, kin recognition has been shown in a variety of organisms, including plants (6), ants (7), and vertebrates (8), and is important for stabilizing cooperation and promoting inclusive fitness benefits in some species (9).
However, kinship may not be the only basis on which natural selection might possibly operate at the group level. For example, if genetic differences between social networks (or, conversely, genetic similarity within networks) were found at significant levels among humans, it would enhance the opportunity for natural selection to operate at the level of social groups established on a basis other than kinship. Such associations have long been postulated in the theoretical evolutionary genetics literature (10–13), but there is little extant evidence to support these theories.
Such a finding would also have relevance in the growing area of indirect genetic effects, wherein the phenotypic traits of a focal individual are influenced by genes in the genomes of its neighbors. This type of effect has recently been shown in a large SNP study in laying hens where it was found that an individual's feather condition is very strongly influenced by the genotypes of its neighbors (14). In fact, the number of such genes was an order of magnitude larger than the number of genes with a direct effect in the focal individual's own genome, and the average phenotypic effects of the “social” genes were more than two times greater than the genes with direct effect. Interestingly, some of the genes with indirect or associative effects in hens involve the serotonin pathway, which has also been shown to influence social behavior in humans (15, 16). Similar phenomena related to “social” fitness have also recently been observed in bacteria (17).
In humans, one of the most replicated findings in the social sciences is that people tend to associate with other people that they resemble, a process known as “homophily” (“birds of a feather flock together”) (18–20). Although phenotypic resemblance between friends might partly reflect the operation of social influence (21, 22), genotypes are not materially susceptible to change. Therefore, genotypic resemblance could result only from a process of selection. Such genotypic selection might in turn take several forms.
First, correlation in genes may be a trivial by-product of “population stratification,” resulting from the tendency for human groups with low mobility to reproduce with geographically proximate mates. Over time, the processes of genetic drift and local adaptation would cause groups that are geographically distant to develop distinct frequencies of genotypes (23). If people also tend to make friends with geographically proximate individuals, then their genotypes would appear to be more correlated within geographically separate groups than between them. Therefore, any attempt to discern whether some other selection process is involved in the correlation of genotypes must use very strict controls for population stratification.
Second, people may actively choose friends of a similar genotype. Although it is unlikely that people would observe the actual genotypes of others around them, they could observe their phenotypes, and these may be influenced by specific genotypes. For example, a person of normal weight may choose to associate only with others of normal weight, and this would cause people lacking the risk allele of the FTO gene [which has been associated with obesity (24)] to tend to befriend others with the same genotype. Similarly, people might choose to terminate relationships with people whose weight status differs from their own (25).
Third, people may actively choose environments they find convivial, environments where they are consequently likely to encounter people with similar phenotypes that are influenced by specific genotypes. If people tend to choose friends from within these environments (even at random), it would tend to generate correlated genotypes. For example, individuals interested in, and capable of, long-distance running may be drawn to clubs or locations where they have the opportunity to make similar friends.
Fourth, people may be chosen by others or otherwise selected into environments where they come into contact with similar people. For example, the admission process at a university or the hiring process in a workplace may select for people with specific phenotypes (e.g., cognitive skills) that are influenced by specific genotypes. Similarly, these institutions may get rid of individuals that do not exhibit certain phenotypes after they are admitted, and these phenotypes might also be influenced by genotypic variation.
In contrast to homophily, people might also exhibit “heterophily,” that is, they might actively choose to associate with people who are different with respect to some traits (“opposites attract”). It is noteworthy that if there is any substantial negative correlation between friends’ phenotypes or genotypes, this would be unlikely to result from population stratification or from people choosing, or being drawn to, the same environment (as in the case of homophily noted above). Instead, there are two other processes that might be at work. First, people may actively choose to befriend people of a different type. The classic example of this kind of negative correlation at the genetic level occurs during mate choice; human beings have a slight preference for mates with different HLA types, a process perhaps mediated by a chemical signaling mechanism (26). Second, certain environments may require specialization. For example, some workplaces may select people with different skills to work together, and if these traits are related to genotypes, then people may tend to be frequently exposed to dissimilar people with whom they may have a higher probability of becoming friends.
Results
As noted, genotypes tend to be correlated between people who live physically close to one another because of population stratification. However, no work has yet established that, net of such stratification, there are any genes that are correlated (either positively or negatively) between individuals in nonreproductive, friendship unions. To study whether such correlation exists, we analyzed two independent samples with information about respondents’ genes and about respondents’ friendship ties and social networks: the National Longitudinal Study of Adolescent Health (Add Health) and the Framingham Heart Study Social Network (FHS-Net). Add Health allowed respondents to name up to 10 friends at each of three waves over 6 y (27), and the FHS-Net captured up to two close friends at each of seven waves over 32 y (28).
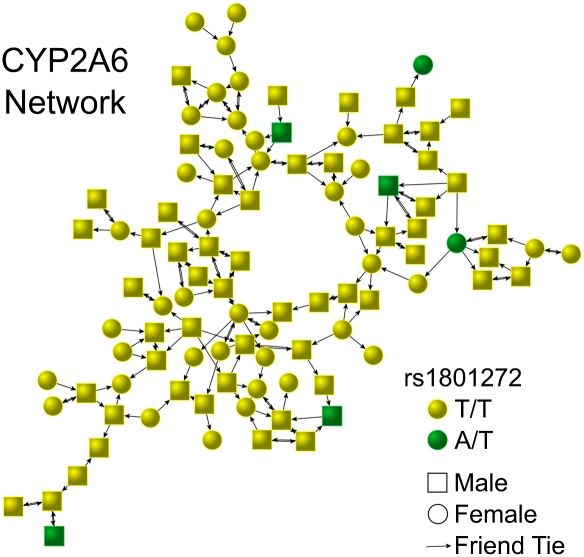
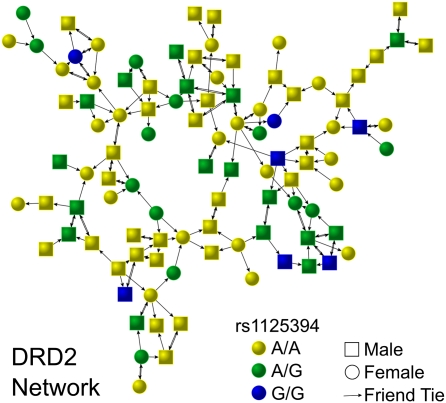
In Add Health, subjects were genotyped for one marker each in the DRD2, DRD4, CYP2A6, MAOA, SLC6A3, and SLC6A4 genes (Materials and Methods). Figs. 1 and 2 show how genotypes for two of these genes are distributed in the largest connected component of the friendship network. Notably, significant clusters of similar genotypes for DRD2 suggest the possibility of homophily, but the substantial absence of any connection between individuals with minor alleles of CYP2A6 suggests possible heterophily. In SI Appendix, Fig. S1, we show the results of simple Monte Carlo tests that suggest these clusters are unlikely to be the result of chance.
Fig. 1.
This social network figure shows genotypes for CYP2A6 in the largest connected component in the friendship network in the Add Health study. Each arrow indicates that the receiver was named as a friend by the sender; a double-headed arrow indicates both subjects named the other as a friend.
Fig. 2.
This social network figure shows genotypes for DRD2 in the largest connected component in the friendship network in the Add Health study. Each arrow indicates that the receiver was named as a friend by the sender; a double-headed arrow indicates both subjects named the other as a friend.
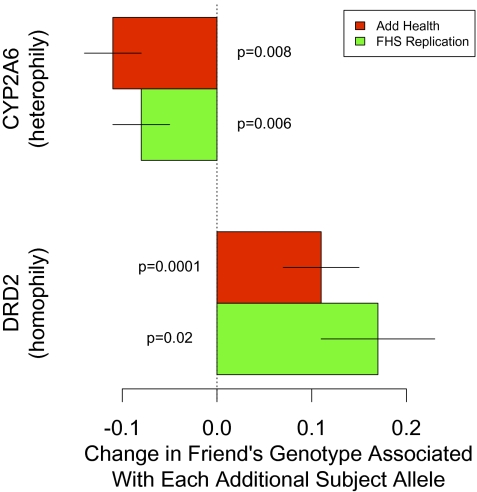
To evaluate statistically whether or not genotypes are correlated between friends, we regressed friend's genotype on subject's genotype (including controls for the age, sex, and race of both subject and friend) for each of the six available genotypes in Add Health (Materials and Methods). We restricted the sample to friends who were not related, and we conducted a sibling transmission disequilibrium test (sib-TDT), which includes the average sibling genotype in the model and has been shown to effectively control for population stratification (29–32). Fig. 3 shows the two genes that indicated significant correlation in both the permutation test and the sib-TDT after Bonferroni correction. The TaqI A repeat-fragment length polymorphism in the DRD2 gene (n = 1167, P = 0.008) showed significant positive correlation (homophily), and the SNP rs1801272 in the CYP2A6 gene (n = 1175, P = 0.0001) indicated significant negative correlation (heterophily).
Fig. 3.
Summary of the two associations in friends’ genotypes in Add Health that survive a Bonferroni-corrected 95% confidence threshold and replication results from the FHS. Horizontal lines indicate SEs. The genotype associations between friends for CYP2A6 and DRD2 replicate in the FHS.
We replicated the statistical procedure for these genotypes in the FHS-Net using a TDT that includes parental genotypes to control for population stratification (29–32). Imputed genotypes for DRD2 and CYP2A6 made possible direct comparison with the Add Health results (SI Appendix). Both genotypes, DRD2 (n = 3316, P = 0.006) and CYP2A6 (n = 1988, P = 0.02), show significant correlation in the same direction and at about the same magnitude in the FHS replication (Fig. 3).
Discussion
These results suggest that there is genotypic clustering in social networks that exceeds what might be expected solely from population stratification. People's friends may not only have similar traits, but actually resemble each other on a genotypic level, even at the level of specific alleles and nucleotides.
It may seem improbable that two of the six genotypes we test show signs of significant association, given that genome-wide studies often find only a handful of candidate genotypes. However, it is important to remember that the six genes chosen for genotyping by Add Health are those that were already known to have significant effects on human social behavior. We suspect that a genome-wide study would show that many genes in the serotonin and dopamine pathway exhibit similar correlation, and perhaps genes from other pathways relevant to social behavior or neuronal development as well.
Indeed, the fact that the genes investigated here have also been associated with certain behavioral and personality traits helps shed light on one possible mechanism for this observation (see SI Appendix for more detail regarding the known phenotypes of these genes). For example, DRD2 has been associated with alcoholism (33) and it is not hard to imagine that nondrinkers may actively avoid alcoholics, or that alcoholics may be drawn to environments that nondrinkers avoid. Less is known about CYP2A6. This gene has been associated with the trait of openness in one study (34), but it is not clear why people who score highly on this trait would tend to associate with those who do not. However, regardless of the causal mechanism, homophily and heterophily on a specific allele is especially noteworthy because the phenotypes with which these genes are related are likely polygenic (they are affected by many genes) and the genes are also likely pleiotropic (they affect many traits).
Whatever the specific phenotypes that might be involved, the significant negative correlation in CYP2A6 suggests that genetic structure in human social networks does not arise solely because of the tendency of people with similar genotypes to be drawn to similar environments. That is, the mechanism at play in the association between friends’ genotypes (whether homophily or heterophily) may reflect, at least in part, active friendship choice by individuals.
An important implication of these results is that genetic structure in human populations may result not only from the formation of reproductive unions, but also from the formation of friendship unions within a population. This finding implies that there may be a certain amount of omitted variable bias in association studies. For example, a person with a genotype that makes her susceptible to alcoholism may be directly influenced to drink. However, she may also be indirectly influenced to drink because she chooses friends with the same genotype (homophily) who are more likely to make alcohol available to her. If so, then an association test between alcoholism and genotype that does not include friends’ genotypes would overstate the effect of the genes. In contrast, if she chooses friends with a different genotype (heterophily), they may be less likely to give alcohol to her. If so, then the inhibitory effect of her friends’ genes could counteract the direct effect of her own genes and an association test that omitted friends’ genotypes would understate the effect of one's own genotype.
Thus, homophily and heterophily in friendships, expressed at the genetic level, may have notable implications for our understanding both of the way that our genes can shape our environmental exposures and the way that our social environment can influence our behavior (21, 22, 28, 35). A feedback process might emerge whereby a person's genes lead to the selection of friends with certain genotypes, which in turn facilitates or modifies the expression of a person's own genes. The idea of an evocative gene–environment interaction, proposed more than 30 y ago, suggests that a person's genes can lead him to seek out circumstances that are compatible with his genotype (2, 36). These circumstances could include not only the social behavior of a person's friends, but also their very genes. Such a process could also play out over longer time scales; the human evolutionary environment is not limited to the physical and biological environment, but also includes the social environment, which may itself be an evolutionary force (37, 38).
To the extent that genes play a role in the formation of nonreproductive unions, and to the extent that there is allelic similarity in connected but biologically unrelated individuals, this could constrain or facilitate a number of other biological and social processes, from the spread of germs to the spread of information. In some sense, humans might be “metagenomic” not just with respect to the microbes within them (39, 40), but also with respect to the humans around them. We could possibly view an individual's genetic landscape as a summation of the genes within the individual and those around him, just as in certain other organisms (14, 17).
There may even be genetic niches within social networks that promote or inhibit the evolution of certain kinds of social behavior. The people to whom we are connected provide important capabilities, from the ability to ward off infections, to the ability to exploit or transmit useful information, to the ability to reciprocate cooperative exchanges and thus enhance their payoffs (35). For example, some individuals might be “immune” to whatever pathogen is spreading in a population not because of their own constitution, but rather because they have come to surround themselves with others with particular genotypes. Personality traits associated with being a leader, based in part on certain genotypes, might best be matched with those in others associated with being a follower. Or cooperation might most easily arise and be sustained within social networks and friendship connections of a particular kind (41). Perhaps genetic properties of friendship groups might confer fitness advantages to individuals who choose them.
Materials and Methods
For our candidate sample, we use data from the National Longitudinal Study of Adolescent Health. Add Health is a large nationally-representative and publicly available study started in 1994 that explores the causes of health-related behavior of adolescents in grades 7 through 12 and their outcomes in young adulthood. In the first wave, information was gathered from subjects in 142 schools about their social networks. Student's were allowed to nominate up to five female and five male friends and were then asked more specific details about those friendships.
In Wave I of the Add Health study, researchers created a genetically informative sample of sibling pairs based on a screening of the in-school sample. These pairs include all adolescents who were identified as twin pairs, half siblings, or unrelated siblings raised together. Twins and half biological siblings were sampled with certainty. The Wave I sibling-pairs sample has been found to be similar in demographic composition to the full Add Health sample (42). Subjects sometimes name family members as “friends,” so we used this information to exclude from the friendship network all social ties to parents, siblings, half-siblings, adopted siblings, aunts, uncles, nephews, nieces, and cousins. Hence, remaining friends are unlikely to be genetically related.
Add Health produced genotype values from saliva samples collected in Wave III for six genes, CYP2A6 (cytochrome P450 2A6), DRD2 (dopamine D2 receptor), DRD4 (dopamine D4 receptor), MAOA (monoamine oxidase A-uVNTR), SLC6A3 (dopamine transporter), and SLC6A4 (serotonin transporter). We provide a detailed description of each of these markers and their associated phenotypes in the SI Appendix. DNA samples were genotyped at the University of Colorado, Boulder, Institute of Behavior Genetics. The Add Health study tested the validity of their genotyping protocols using preamplified DNA compared with genomic DNA for 315 individuals from non-Add Health samples, and found agreement in 98.7% of cases.
We identified all subject–friend pairs for which we had genetic information, and also race, sex, and age. To reduce the likelihood of false-positives caused by population stratification, we conduct a sibling family disequilibrium test, including siblings’ mean genotype as a control (30). For the purpose of this test, we only include as controls the genotypes of full siblings and nonidentical twins (half siblings, identical twins, and all other related pairs are excluded). To test for genetic homophily (positive association) or heterophily (negative association) in Add Health, we regressed friend's genotype on subject's genotype and included both the subject's and friend's age, sex, and race as controls. Genotype takes the value 0, 1, or 2 depending on the number of minor alleles. Because some subjects name multiple friends, we use generalized estimating equation regression (43) with clustering within family and an independent working covariance structure. Mean squared error fit statistics show the difference between predicted and observed values for the model and a null model with no covariates. Models with an exchangeable correlation structure yielded poorer fit.
Because we conducted tests on six genetic markers, a Bonferroni correction implies that the threshold for 95% confidence in an association is P = 0.05/6 = 0.008. SI Appendix, Table S1 A and B, show that three associations between subject and friend genotype are significant at this level, but only two are also significant in the permutation test shown in SI Appendix, Fig. S1.
For replication of these two candidate associations, we turned to the FHS. We had to make some modifications in our analytic approach, compared with the Add Health data, given the nature of the FHS data collection. In particular, we used a family-based TDT and we imputed SNPs to match the Add Health loci.
The FHS is a population-based, longitudinal, observational cohort study that was initiated in 1948 to prospectively investigate risk factors for cardiovascular disease. Since then, it has come to be composed of four separate but related cohort populations: (i) the “Original Cohort” enrolled in 1948 (n = 5,209); (ii) the “Offspring Cohort” (the children of the Original Cohort and spouses of the children), enrolled in 1971 (n = 5,124); (iii) the “Omni Cohort” enrolled in 1994 (n = 508); and (iv) the “Generation 3 Cohort” (the grandchildren of the Original Cohort), enrolled beginning in 2002 (n = 4,095). Published reports provide details about sample composition and study design for all these cohorts (44–46). As described elsewhere, we collected information identifying who was connected to whom via ties of friendship (and also ties of marriage, kinship, and so on) (21).
Out of the 14,428 members of the three main cohorts, a total of 9,237 individuals have been genotyped (4,986 women and 4,251 men). Genotyping was conducted using the Affymetrix 500 K array and an Affymetrix 50 K supplemental array. We had data on additional unobserved genotypes for 1,345 of the participants from an Affymetrix 100 K GeneChip array, so we used PLINK 1.06 (47) to merge the two datasets by subject. We also merged the data with European ancestry HapMap samples from Phase II and Phase III.
We applied a number of recommended quality control measures to the sample comprising all 9,237 individuals with genetic data (48, 49). First, we dropped individuals who were missing genotypic information for 5% or more of their available SNPs. High missingness indicates that a problem may have occurred in the genotyping procedure for the individual. Next, we deleted individual SNPs with a missing-data frequency greater than 2.5%. Then we eliminated SNPs with minor allele frequency less than 1% because low-frequency alleles are more likely to produce false high-magnitude estimates in association tests. Finally, we excluded SNPs that failed a test of Hardy-Weinberg equilibrium at the 10−6 level. The null hypothesis of this test is that the observed genotype frequencies are equal to their theoretical expectations under random mating. A large departure from Hardy-Weinberg equilibrium may be an indication of genotyping errors.
We used the SNPs that survived these quality checks to estimate the pairwise relatedness between every single pair in the sample. Friendship pairs that showed greater than zero percent of their genome identical by descent were removed from the data to ensure that association in friend genotypes is not, in fact, a result of people tending to make friends with family members.
None of the candidate genotypes from the Add Health study are available in the FHS, but there are nearby SNPs that may be in linkage disequilibrium with the Add Health markers. For CYP2A6 we used the proxy-impute command in PLINK to impute rs1801272. This procedure identified nine SNPs in high-linkage disequilibrium with the target SNP, rs7259265, rs2561543, rs2561528, rs2607414, rs41530251, rs7251418, rs8103444, rs10419393, rs8192719, and yielded imputation of rs1801272 for 6,802 individuals. The results should be treated with some caution because the INFO score indicating the goodness of fit for this imputation was 0.697 (scores above 0.8 are preferred). However, as we will show below, an analysis of rs7251418 (the unimputed, nearby SNP in highest linkage with rs1801272) also suggests significant heterophily for CYP2A6.
We also used PLINK to impute the rs1125394 for DRD2. This procedure identified two SNPs in high linkage disequilibrium with the target SNP, rs3897584 and rs2471854, and yielded imputation of rs1125394 for 1,841 individuals. The INFO score indicating the goodness of fit for this imputation was 0.997.
Although the individuals in the FHS are almost all of European ancestry, population stratification has been shown to be a concern even in samples of European Americans (50). Therefore, we used a family TDT (51) to partition between-family variance and within-family variance. This test includes the average number of alleles in the mother, father, or both (if known) as a control for the contribution of between-family variance (b). This value is subtracted from the genotype of the subject, and the coefficient on this variable indicates the extent to which within-family variance (w) is associated with the dependent variable. Results of these tests are shown in SI Appendix, Table S2.
Summary statistics for the variables in the regressions in SI Appendix, Table S1 A and B, are shown in SI Appendix, Table S3. Summary statistics for the variables in the regressions in SI Appendix, Table S2, are shown in SI Appendix, Table S4.
Finally, because the INFO score on the imputation for rs1801272 was somewhat low, in SI Appendix, Table S5, we show the results of a regression for rs7251418, the unimputed nearby SNP in highest linkage with rs1801272 (r = 0.30, distance = 12.9 K base pairs). These unimputed results also suggest the existence of significant heterophily (P = 0.04) in the region of CYP2A6.
Supplementary Material
Acknowledgments
We thank Ting Wu for helpful comments. This work was supported in part by Grants P-01 AG031093 and P30 AG034420 from the National Institute on Aging, Grant P-41 RR031228 from the National Institutes of Health, and SES-0719404 from the National Science Foundation. The Framingham Heart Study is supported by a contract from the National Heart, Lung, and Blood Institute (N01 HC25195).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011687108/-/DCSupplemental.
References
- 1.Burt A. A mechanistic explanation of popularity: Genes, rule breaking, and evocative gene-environment correlations. J Pers Soc Psychol. 2009;96:783–794. doi: 10.1037/a0013702. [DOI] [PubMed] [Google Scholar]
- 2.Kendler KS, Baker JH. Genetic influences on measures of the environment: A systematic review. Psychol Med. 2007;37:615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- 3.Iervolino AC, et al. Genetic and environmental influences in adolescent peer socialization: Evidence from two genetically sensitive designs. Child Dev. 2002;73:162–174. doi: 10.1111/1467-8624.00398. [DOI] [PubMed] [Google Scholar]
- 4.Guo G. Genetic similarity shared by best friends among adolescents. Twin Res Hum Genet. 2006;9:113–121. doi: 10.1375/183242706776402920. [DOI] [PubMed] [Google Scholar]
- 5.Fowler JH, Dawes CT, Christakis NA. Model of genetic variation in human social networks. Proc Natl Acad Sci USA. 2009;106:1720–1724. doi: 10.1073/pnas.0806746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudley SA, File AL. Kin recognition in an annual plant. Biol Lett. 2007;3:435–438. doi: 10.1098/rsbl.2007.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holldobler B, Wilson E. The Ants. Harvard, MA: Harvard University Press; 1990. [Google Scholar]
- 8.Griffin AS, West SA. Kin discrimination and the benefit of helping in cooperatively breeding vertebrates. Science. 2003;302:634–636. doi: 10.1126/science.1089402. [DOI] [PubMed] [Google Scholar]
- 9.Chaine AS, Schtickzelle N, Polard T, Huet M, Clobert J. Kin-based recognition and social aggregation in a ciliate. Evolution. 2010;64:1290–1300. doi: 10.1111/j.1558-5646.2009.00902.x. [DOI] [PubMed] [Google Scholar]
- 10.Wilson DS. The group selection controversy: History and current status. Annu Rev Ecol Syst. 1983;14:159–187. [Google Scholar]
- 11.Hamilton WD. Innate social aptitudes of man: An approach from evolutionary genetics. In: Fox R, editor. Biosocial Anthropology. London: Malaby Press; 1975. [Google Scholar]
- 12.Maynard Smith J. Group selection. Q Rev Biol. 1976;51:277–283. [Google Scholar]
- 13.Wilson DS. A theory of group selection. Proc Natl Acad Sci USA. 1975;72:143–146. doi: 10.1073/pnas.72.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biscarini F, et al. Across-line SNP association study for direct and associative effects on feather damage in laying hens. Behav Genet. 2010;40:715–727. doi: 10.1007/s10519-010-9370-0. [DOI] [PubMed] [Google Scholar]
- 15.Serretti A, et al. HTR2C and HTR1A gene variants in German and Italian suicide attempters and completers. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:291–299. doi: 10.1002/ajmg.b.30432. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Brookes K, Sun B, Ilott N, Asherson P. Investigation of the serotonin 2C receptor gene in attention deficit hyperactivity disorder in UK samples. BMC Res Notes. 2009;2:71. doi: 10.1186/1756-0500-2-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467:82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPherson M, Smith-Lovin L, Cook JM. Birds of a feather: Homophily in social networks. Annu Rev Sociol. 2001;27:415–444. [Google Scholar]
- 19.Christakis NA, Fowler JH, Imbens GW, Kalyanaraman K. An Empirical Model for Strategic Network Formation, National Bureau of Economic Research Working Paper No. 16039. Cambridge, MA: Natl. Bureau Econ. Res.; 2010. [Google Scholar]
- 20.Zeng Z, Xie YA. A preference-opportunity-choice framework with applications to intergroup friendship. AJS. 2008;114:615–648. doi: 10.1086/592863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christakis NA, Fowler JH. The spread of obesity in a large social network over 32 years. N Engl J Med. 2007;357:370–379. doi: 10.1056/NEJMsa066082. [DOI] [PubMed] [Google Scholar]
- 22.Fowler JH, Christakis NA. Dynamic spread of happiness in a large social network: Longitudinal analysis over 20 years in the Framingham Heart Study. BMJ. 2008;337:a2338. doi: 10.1136/bmj.a2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novembre J, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Malley AJ, Christakis NA. Health behaviors and traits and the formation and dissolution of friendship ties. Stat Med. 2011 doi: 10.1002/sim.4190. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ober C, et al. HLA and mate choice in humans. Am J Hum Genet. 1997;61:497–504. doi: 10.1086/515511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris KM, et al. The National Longitudinal Study of Adolescent Health: Research Design. URL: http://www.cpc.unc.edu/projects/Add Health/design, accessed July 28, 2010. [Google Scholar]
- 28.Christakis NA, Fowler JH. The collective dynamics of smoking in a large social network. N Engl J Med. 2008;358:2249–2258. doi: 10.1056/NEJMsa0706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spielman RS, Ewens WJ. A sibship test for linkage in the presence of association: The sib transmission/disequilibrium test. Am J Hum Genet. 1998;62:450–458. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldman ID, Robinson BF, Rowe DC. A logistic regression based extension of the TDT for continuous and categorical traits. Ann Hum Genet. 1999;63:329–340. doi: 10.1046/j.1469-1809.1999.6340329.x. [DOI] [PubMed] [Google Scholar]
- 32.Zou GY. Statistical methods for the analysis of genetic association studies. Ann Hum Genet. 2006;70:262–276. doi: 10.1111/j.1529-8817.2005.00213.x. [DOI] [PubMed] [Google Scholar]
- 33.Le Foll B, Gallo A, Le Strat Y, Lu L, Gorwood P. Genetics of dopamine receptors and drug addiction: A comprehensive review. Behav Pharmacol. 2009;20:1–17. doi: 10.1097/FBP.0b013e3283242f05. [DOI] [PubMed] [Google Scholar]
- 34.Waga C, Iwahashi K. CYP2A6 gene polymorphism and personality traits for NEO-FFI on the smoking behavior of youths. Drug Chem Toxicol. 2007;30:343–349. doi: 10.1080/01480540701522338. [DOI] [PubMed] [Google Scholar]
- 35.Fowler JH, Christakis NA. Cooperative behavior cascades in human social networks. Proc Natl Acad Sci USA. 2010;107:5334–5338. doi: 10.1073/pnas.0913149107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scarr S, McCartney K. How people make their own environments: A theory of genotype greater than environment effects. Child Dev. 1983;54:424–435. doi: 10.1111/j.1467-8624.1983.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 37.Distel MA, et al. Familial resemblance for loneliness. Behav Genet. 2010;40:480–494. doi: 10.1007/s10519-010-9341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrmann E, Call J, Hernàndez-Lloreda MV, Hare B, Tomasello M. Humans have evolved specialized skills of social cognition: The cultural intelligence hypothesis. Science. 2007;317:1360–1366. doi: 10.1126/science.1146282. [DOI] [PubMed] [Google Scholar]
- 39.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman E, Hauert C, Nowak MA. Evolutionary dynamics on graphs. Nature. 2005;433:312–316. doi: 10.1038/nature03204. [DOI] [PubMed] [Google Scholar]
- 42.Jacobson KC, Rowe DC. Genetic and shared environmental influences on adolescent BMI: Interactions with race and sex. Behav Genet. 1998;28:265–278. doi: 10.1023/a:1021619329904. [DOI] [PubMed] [Google Scholar]
- 43.Zorn C. Generalized estimating equation models for correlated data: A review with applications. Am J Pol Sci. 2001;45:470–490. [Google Scholar]
- 44.Cupples LA, D'Agnostino RB. Survival following initial cardiovascular events: 30 year follow-up. In: Kannel WB, Wolf PA, Garrison RJ, editors. The Framingham Study: An Epidemiological Investigation of Cardiovascular Disease. Bethesda, MD: National Heart, Lung and Blood Institute; 1988. publication #88-204029, section 35. [Google Scholar]
- 45.Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP. An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol. 1979;110:281–290. doi: 10.1093/oxfordjournals.aje.a112813. [DOI] [PubMed] [Google Scholar]
- 46.Quan SF, et al. The Sleep Heart Health Study: Design, rationale, and methods. Sleep. 1997;20:1077–1085. [PubMed] [Google Scholar]
- 47.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manolio TA, et al. GAIN Collaborative Research Group; Collaborative Association Study of Psoriasis; International Multi-Center ADHD Genetics Project; Molecular Genetics of Schizophrenia Collaboration; Bipolar Genome Study; Major Depression Stage 1 Genomewide Association in Population-Based Samples Study; Genetics of Kidneys in Diabetes (GoKinD) Study. New models of collaboration in genome-wide association studies: The Genetic Association Information Network. Nat Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan PF, Purcell S. Analyzing genome-wide association study data: A tutorial using PLINK. In: Neale BM, Ferreira MAR, Medland S, Posthuma D, editors. Statistical Genetics; Gene Mapping Through Linkage and Association. New York: Taylor & Francis Group; 2008. pp. 355–394. [Google Scholar]
- 50.Campbell CD, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 51.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.