Abstract
Numerous studies indicate that Sirtuin 1 (SIRT1), a mammalian nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylase (HDAC), plays a crucial role in p53-mediated stress responses by deacetylating p53. Nevertheless, the acetylation levels of p53 are dramatically increased upon DNA damage, and it is not well understood how the SIRT1–p53 interaction is regulated during the stress responses. Here, we identified Set7/9 as a unique regulator of SIRT1. SIRT1 interacts with Set7/9 both in vitro and in vivo. In response to DNA damage in human cells, the interaction between Set7/9 and SIRT1 is significantly enhanced and coincident with an increase in p53 acetylation levels. Importantly, the interaction of SIRT1 and p53 is strongly suppressed in the presence of Set7/9. Consequently, SIRT1-mediated deacetylation of p53 is abrogated by Set7/9, and p53-mediated transactivation is increased during the DNA damage response. Of note, whereas SIRT1 can be methylated at multiple sites within its N terminus by Set7/9, a methylation-defective mutant of SIRT1 still retains its ability to inhibit p53 activity. Taken together, our results reveal that Set7/9 is a critical regulator of the SIRT1-p53 interaction and suggest that Set7/9 can modulate p53 function indirectly in addition to acting through a methylation-dependent mechanism.
Keywords: p21waf1/cip1, posttranslational modifications, tumor suppression
The class III HDAC sirtuins, SIRT1 to SIRT7, are homologous to yeast silent information regulator 2 (Sir2) and play important biological roles during aging, metabolism, and autophagy (1, 2). Within the sirtuins, SIRT1 is the closest homolog of yeast Sir2. In contrast to class I and II HDACs, sirtuins require nicotinamide adenine dinucleotide (NAD+) as a coenzyme to act on their target proteins (3). Numerous studies have also suggested a role for SIRT1 in the development of tumors because SIRT1 is up-regulated in various cancerous tissues and cell lines, such as leukemia and prostate cancer (4, 5). In addition, SIRT1 is overexpressed in several p53-deficient tumor cell lines, and the transient knockdown of SIRT1 leads to increased apoptosis after DNA damage or oxidative stress (6). Moreover, tumor suppressors such as p53 (7, 8) and FoxO (9, 10) are deacetylated by SIRT1 and, thus, inactivated in response to DNA damage, which provides further evidence that SIRT1 is an oncogenic protein.
The tumor suppressor hypermethylated in cancer 1 (HIC1) has been shown to inhibit SIRT1 expression by forming a repressive complex with SIRT1 on its own promoter and sensitizing the p53 response to DNA damage (11). In addition, deleted in breast cancer 1 (DBC1) has also been reported to be a negative repressor of SIRT1 in various cell lines and leads to increased levels of p53 acetylation and up-regulation of p53 transcriptional activities (12, 13). However, active regulator of SIRT1 (AROS) interacts with SIRT1 and activates its deacetylase activity (14). In addition to protein interactions, posttranslational modifications of SIRT1 are also important for its activity. For instance, SIRT1 is phosphorylated by JNK2 at Ser27, and depletion of JNK2 reduces the half-life of the SIRT1 protein (15). SIRT1 is also sumoylated at K734, which in turn increases its activity (16). With these posttranslational modifications, SIRT1 deacetylase activity is substantially altered. Therefore, any newly identified regulator of SIRT1 activity would be helpful in understanding the regulation network of SIRT1 and its biological relevance in cancer development or tumor therapeutics.
Set7/9 was originally identified as a monomethyltransferase for histone H3K4 methylation and is thus involved in gene activation (17, 18). Recently, Set7/9-mediated lysine methylation has emerged as a key posttranslational modification that regulates the functions of nonhistone proteins such as p53 (19), TAF10 (20), and DNA methyltransferase 1 (DNMT1) (21, 22). Therefore, Set7/9 appears to function mainly through methylation of various target proteins, which in turn up- or down-regulates target protein activity.
In this study, we investigated a possible relationship between Set7/9 and SIRT1 to determine whether SIRT1 activity is regulated by Set7/9 in vivo and in vitro. We found that Set7/9 is able to both interact with and methylate SIRT1. However, the resulting methylation of SIRT1 is, by itself, dispensable for the deacetylase activity of SIRT1 on p53. Instead, the physical interaction of Set7/9 with SIRT1 disrupts SIRT1 binding to p53, such that p53 acetylation and transactivation are significantly enhanced by the dissociation of p53 from SIRT1.
Results
Set7/9 Regulates p53 Activity Through a p53 K372 Methylation-Independent Mechanism.
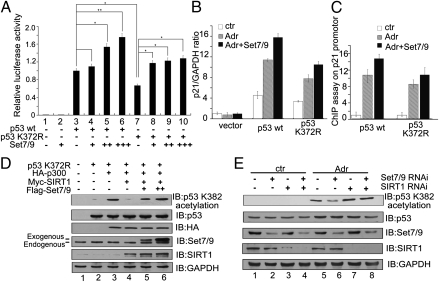
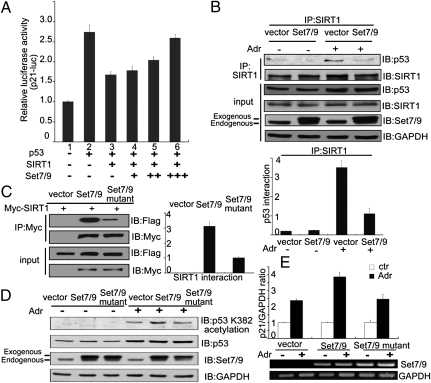
Set7/9 is a protein methyltransferase that has been reported to play a role in regulating cell cycle and DNA damage responses through the direct methylation of p53 at K372 (19). However, it has been an ongoing challenge to determine whether Set7/9 also modulates p53 activity independent of its methylation function. To address this question, we generated vectors that express either wild-type p53 (p53WT) or a mutant p53 (p53K372R) with a lysine-to-arginine mutation at amino acid residue 372. Both plasmids were then separately cotransfected, along with a luciferase-based expression plasmid containing 13 repeats of the p53-responsive element (pG13L) and a Flag-tagged Set7/9 expression plasmid, into HCT116 (p53−/−) cells, and relative luciferase activities were determined. Fig. 1A shows that the relative luciferase activity was significantly increased by p53WT expression (lane 3) and that coexpression of Set7/9 (lanes 4–6), which alone showed no effect (lane 2), significantly enhanced the p53WT-driven luciferase activity in a dose-dependent manner. p53K372R expression also increased pG13L activity (lane 7), but to a lower level than observed with p53WT, and this activity was further enhanced by Set7/9 (lanes 8–10). Because the magnitude of the stimulation by Set7/9 was comparable for p53WT and p53K372R, these results suggested that Set7/9 may regulate p53 activity through a mechanism other than methylation of p53 at K372. Next, HCT116 (p53−/−) cells were transfected with either the p53WT vector or the p53K372R vector, and expression of the endogenous p21Waf1/Cip1 gene (a direct target of p53) was measured by using real-time PCR in the presence or absence of the DNA-damaging agent adriamycin (Adr). Fig. 1B shows that p21Waf1/Cip1 expression was significantly increased in response to Adr treatment in cells expressing either p53WT (2.4-fold) or p53K372R (2.4-fold) compared with untreated sample. Moreover, Set7/9 further increased expression of p21Waf1/Cip1 in cells expressing either p53WT (3.5-fold) or p53K372R (3.2-fold) following Adr treatment compared with the untreated sample (Fig. 1B). A chromatin immunoprecipitation (ChIP) assay provided further evidence for the role of Set7/9 in regulating p53 activity in a K372 methylation independent manner. Thus, as shown in Fig. 1C, p53WT and p53K372R showed nearly equivalent levels of enrichment on the p21Waf1/Cip1 promoter after Adr treatment and, in both cases, coexpression of Set7/9 resulted in comparable enhancements of p53WT and p53K372R enrichments on this promoter.
Fig. 1.
p53 is activated by Set7/9 by a methylation-independent mechanism. (A) Expression plasmids for pG13L, p53WT, p53K372R, and Set7/9 were cotransfected into HCT116 (p53−/−) cells as indicated. Cells were harvested 24 h after transfection, and relative luciferase activity was measured. The luciferase activity was normalized to the amount of protein in the cell lysate. The cell sample transfected with p53WT alone served as the control and its value (relative luciferase activity) was set as 1. The relative luciferase activities of the other samples are normalized to this control. Data are means ± SD (n = 3). (B) An empty plasmid and an expression plasmid for p53WT or p53K372R were cotransfected with or without Set7/9 into HCT116 (p53−/−) cells and, after 24 h, cells were treated with 1 μM Adr for 6 h, and a quantitative PCR was performed to analyze p21Waf1/Cip1 expression. The signals were normalized to the expression of GAPDH. (C) An expression plasmid for p53WT or p53K372R was cotransfected with or without Set7/9 into HCT116 (p53−/−) cells treated under the same conditions as in B. Cells were harvested and subjected to a q-ChIP assay with anti-p53 to measure the DNA binding ability of p53 on the p21Waf1/Cip1 promoter. (D) Expression plasmids for p53K372R, HA-p300, Myc-SIRT1, and Flag-Set7/9 were cotransfected into HCT116 (p53−/−) cells and, after 24 h, derived cell lysates were subjected to Western blotting by using anti–acetyl-p53 (K382), anti-p53, anti-HA, anti-Set7/9, or anti-SIRT1. GAPDH served as the loading control. (E) HCT116 (p53+/+) cells were transfected with a nonspecific siRNA (lanes 1 and 5), an siRNA against Set7/9 (lanes 2 and 6), an siRNA against SIRT1 (lanes 3 and 7), or siRNAs against both Set7/9 and SIRT1 (lanes 4 and 8). Twenty-four hours after transfection, cells were treated with 1 μM Adr for 6 h, and the same lysates were subjected to Western blotting by using anti–acetyl-p53 (K382), anti-p53, anti-Set7/9, or anti-SIRT1. GAPDH served as the loading control.
The transcriptional activity of p53 depends on its acetylation status (23). To determine whether Set7/9 was still able not only to methylate p53 at K372, but also to regulate p53 acetylation, HCT116 (p53−/−) cells were transfected with plasmids expressing p53K372R and either HA-tagged p300 (a protein acetyltransferase) or Myc-tagged SIRT1. p53 acetylation was monitored in the presence or absence of exogenous Set7/9. As shown in Fig. 1D, p53 acetylation at K382 was markedly increased in p300-transfected cells (lane 3 vs. lane 2), and this p53 acetylation almost disappeared when the cells were cotransfected with SIRT1 (lane 4). However, the SIRT1-induced inhibition of p53 acetylation was remarkably alleviated when cells were cotransfected with Set7/9 (lanes 5 and 6). Furthermore, the critical role of Set7/9 in eliciting or maintaining endogenous p53 acetylation at K382 was demonstrated by Set7/9 RNAi treatment of HCT116 (p53+/+) cells. As shown in Fig. 1E, p53 acetylation at K382 was induced by treatment with Adr (lane 5 vs. lane 1), but this acetylation was largely blocked with concomitant knockdown of endogenous Set7/9 (lane 6). However, reduction in p53 acetylation by Set7/9-knockdown was significantly alleviated in the SIRT1-knockdown cells (lane 8 vs. lane 6). Together, these data suggest that Set7/9 is able to regulate p53 activity by inducing acetylation at K382 in addition to methylation at K372.
SIRT1 Interacts with Set7/9.
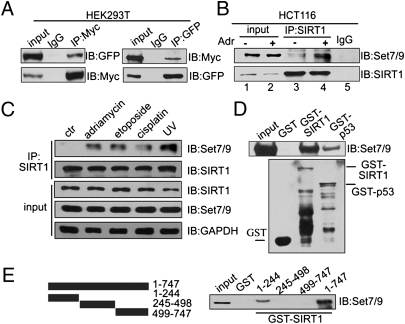
We next investigated the possibility that Set7/9 interacts with SIRT1 and, by decreasing its activity, enhances p53 acetylation and activity. To this end, plasmids expressing Myc-tagged SIRT1 and a GFP-tagged Set7/9 were cotransfected into HEK293T cells. Lysates of transfected cells were subjected to a co-immunoprecipitation (Co-IP) assay with either anti-Myc or anti-GFP followed by probing with anti-GFP or anti-Myc, respectively (Fig. 2A). The results clearly show an interaction between the exogenous SIRT1 and Set7/9 proteins, and both proteins were shown by confocal microscopy to be mainly colocalized in the nucleus (Fig. S1). Similarly, endogenous SIRT1 also showed an enhanced interaction with endogenous Set7/9 in HCT116 (p53+/+) cells in response to Adr treatment (Fig. 2B, lane 3 vs. lane 4). An enhanced interaction between Set7/9 and SIRT1 was also observed in HEK293T cells in response to Adr treatment (Fig. S2). In addition, the interaction between endogeneous SIRT1 and Set7/9 proteins was markedly increased after treatment with other DNA-damaging agents such as etoposide, cisplatin, or UV, which suggests that the interaction of Set7/9 and SIRT1 in response to DNA damage is a general phenomenon (Fig. 2C).
Fig. 2.
SIRT1 interacts with Set7/9 in vitro and in vivo. (A) Expression plasmids for GFP-Set7/9 and Myc-SIRT1 were cotransfected into HEK293T cells and, after 24 h, proteins were extracted and immunoprecipitated by using anti-Myc or anti-GFP, respectively. Western blotting was performed with antibodies as indicated. (B) HCT116 (p53+/+) cells were treated with 1 μM Adr for 6 h, and proteins were extracted for co-IP with anti-SIRT1 and probed with anti-Set7/9. The drug-treated sample was used for the IgG pull-down. (C) HCT116 (p53+/+) cells were treated with 1 μM Adr for 6 h, 20 μM etoposide for 12 h, 10 μM cisplatin for 6 h, or UV-C (60 J/m2). Proteins were extracted for co-IP with anti-SIRT1 and probed with anti-Set7/9. (D) His-Set7/9 protein (cloned into pET28 and expressed in bacteria) was purified in vitro and incubated with GST, GST-SIRT1, or GST-p53 fusion protein (purified from bacteria by using vector pGEX-4T3). Western blotting with anti-Set7/9 was performed to detect the interaction of SIRT1 and Set7/9 (Upper). GST, GST-SIRT1, or GST-p53 was detected by Western blotting with anti-GST (Lower). (E) Full-length SIRT1, an N-terminal fragment (aa 1–244), a middle fragment (aa 245–498), and a C-terminal fragment (aa 499–747) were cloned into pGEX (Left), expressed in and purified from bacteria, and incubated with HEK293T cell lysates. Western blotting was performed to detect the interaction of Set7/9 with SIRT1 (Right).
To investigate whether SIRT1 directly interacts with Set7/9, a His-tagged Set7/9 protein was expressed in bacteria, purified, and incubated with GST, GST-SIRT1, or GST-p53 (as a positive control). As shown in Fig. 2D, Set7/9 showed direct interactions with GST-SIRT1 and GST-p53, but not with GST alone. A reciprocal experiment using GST-Set7/9 to pull down His-tagged SIRT1 further confirmed the direct interaction of these proteins (Fig. S3). To further map the regions of SIRT1 responsible for its interaction with Set7/9, N-terminal (aa 1–244), middle (aa 245–498), and C-terminal (aa 499–747) fragments of SIRT1 were expressed, purified, and analyzed along with GST-SIRT1. The results indicate a selective interaction of Set7/9 with the N-terminal fragment (Fig. 2E). Using various deletion plasmids of SIRT1, the Set7/9-binding region of SIRT1 was further localized to a region between residues 121 and 295 (Fig. S4). These data raise the possibility that a direct interaction between SIRT1 and Set7/9 may play a role in the cellular DNA damage response.
SIRT1 Is Methylated at Multiple Lysines.
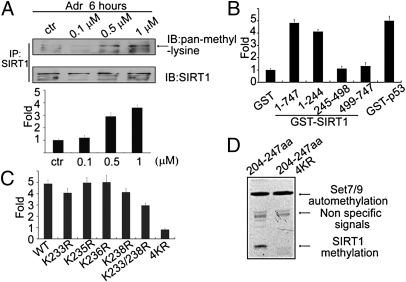
We subsequently asked whether SIRT1 is methylated by Set7/9. To this end, HCT116 (p53+/+) cells were treated with Adr at different doses and derived lysates were subjected to IP with anti-SIRT1 followed by probing with anti–pan-methyl-lysine antibody. The results indicate a significant Adr-dependent increase in SIRT1 methylation (Fig. 3A), indicating that SIRT1 methylation is enhanced after DNA damage. To determine the site of SIRT1 methylation, GST-tagged SIRT1 and SIRT1 fragments were incubated with recombinant Set7/9 in the presence of 3H-S-adenosylmethionine (3H-SAM) and relative levels of incorporated radioactivity were determined. As shown in Fig. 3B, the radiolabeling of either full-length SIRT1 or the N-terminal fragment of SIRT1 was fivefold higher than that of GST, and comparable to that observed with GST-p53, whereas the middle and C-terminal fragments of SIRT1 showed no significant radiolabeling above the control (GST) level. To identify the methylated lysines, three peptide fragments (GEPLRK35RPRR, RTILK203DLLPE, and PK233RK235K236RK238DIN) based on the sequence of the N terminus of SIRT1 were incubated with Set7/9 (plus S-adenosylmethionine) and then subjected to mass spectrometry. As shown in Fig. S5, methylation was detected only in the fragment containing K233/235/236/238. To determine which of these lysines is methylated by Set7/9, PK233RK235K236RK238DIN peptides with different lysine-to-arginine substitutions were incubated with 3H-SAM and Set7/9, and the relative radioactivity levels were determined. Fig. 3C shows that the relative radiolabeling of peptides with individually mutated lysines was not obviously decreased compared with that of full-length SIRT1, whereas radiolabeling was significantly reduced when lysines 233, 235, 236, and 238 were all changed to arginines (hereafter designated 4KR). In a further analysis, a SIRT1 fragment (aa 204–247) containing PK233RK235K236RK238DIN was subcloned into a pGEX plasmid, and an in vitro methylation assay using autoradiography was performed. A methylated band was clearly visible with this SIRT1 fragment in the presence of Set7/9, whereas no methylated band was observed when the corresponding 4KR fragment was incubated with Set7/9 (Fig. 3D). These data together suggest that Set7/9 methylates SIRT1 at lysines 233, 235, 236, and 238.
Fig. 3.
SIRT1 is methylated at multiple lysines in vitro and in vivo. (A) HCT 116 (p53+/+) cells were treated with increasing amounts of Adr (0.1, 0.5, 1 μM). Proteins were extracted for co-IP with anti-SIRT1 and probed with anti–pan-methyl-lysine (Upper). Methylation bands were scanned, and the relative band intensities were normalized to each SIRT1 band. The band intensity of the control was set as 1.0, and the numerical value of the intensity of each band was compared with the control (Lower). Data are means ± SD (n = 3). (B) GST-fusion fragments of SIRT1 (indicated in Fig. 2F) were purified from Escherichia coli, and an equal amounts were incubated with His-Set7/9 in HMT buffer at 30 °C for 3 h in the presence of 3H-SAM. The cpm value of each sample was measured by a scintillation counter. Data are means ± SD (n = 3). (C) Peptides including WT (PKRKKRKDIN), K233R (PRRKKRKDIN), K235R (PKRRKRKDIN), K236R (PKRKRRKDIN), K238R (PKRKKRRDIN), K233/238R (PRRKKRRDIN), and 4KR (PRRRRRRDIN) were incubated with His-Set7/9 in the presence of 3H-SAM and the signals were measured by using a scintillation counter. (D) A GST-fusion protein containing SIRT1 (aa 204–247) or mutated SIRT1 (aa 204–247; 4KR) was incubated with His-Set7/9 in the presence of 3H-SAM and exposed to film.
Methylation of SIRT1 Is Dispensable for Its Deacetylase Activity.
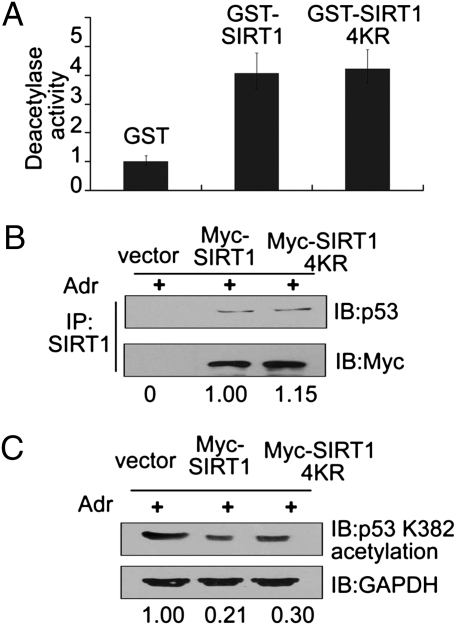
To determine whether the methylation of SIRT1 affects its activity, GST-tagged SIRT1 or GST-tagged SIRT1-4KR was incubated first with Set7/9 and then with acetylated p53 in vitro, and changes in p53 deacetylation were then measured. Contrary to expectations, levels of p53 deacetylation were comparable after treatment with SIRT1 or SIRT1-4KR (Fig. 4A). Next, to further assess the role of SIRT1 methylation in p53 deacetylation in vivo, a plasmid expressing Myc-tagged WT-SIRT1 or Myc-tagged SIRT1-4KR was transfected into HCT116 (p53+/+) cells that were then treated with Adr. As shown by a co-IP assay, both WT-SIRT1 and SIRT1-4KR bound p53 with nearly identical efficiencies (Fig. 4B). Similarly, the level of p53 deacetylation at K382 (approximately fivefold) was also comparable in cells transfected with either WT-SIRT1 or SIRT1-4KR (Fig. 4C). These data suggest that the methylation of SIRT1 does not affect its deacetylase activity toward p53, at least under the assay conditions analyzed here.
Fig. 4.
Methylation of SIRT1 is dispensable for its deacetylation activity. (A) GST-SIRT1-WT and GST-SIRT1-4KR mutant fusion proteins were generated and purified. The purified SIRT1 proteins were incubated first with Set7/9 for 2 h and then with acetylated p53 in the presence of NAD+. Deacetylase activity was then measured with an anti-acetylated p53 (K382). (B) A Myc WT-SIRT1 or a Myc SIRT1-4KR plasmid was transfected into HCT116 (p53+/+) cells and, after 24 h, cells were treated with 1 μM Adr for 6 h. Proteins were extracted for co-IP with anti-SIRT1 and probed with anti-p53 (Upper). The acetylated p53 bands were scanned and relative values are indicated below each band. The cell sample transfected with Myc-SIRT1 serves as the control and its value is set as 1 (Lower). (C) Under the same treatment conditions as in B, cells were harvested and subjected to Western blotting with anti-acetylated p53 (K382). GAPDH was the loading control. The acetylated p53 bands were scanned, and relative values are indicated below each band. The cell sample transfected with vector serves as the control, and its value is set as 1.
Interaction Between Set7/9 and SIRT1 Induces the Dissociation of SIRT1 from p53 and, in Turn, Increases p53 Activity.
To further explore the role of SIRT1 in Set7/9 regulated p53 activity, an assay for luciferase expression driven by the p21Waf1/Cip1 promoter was used to determine the effect of the interaction between SIRT1 and Set7/9 on p53 transactivation in HCT116 (p53−/−) cells. In this assay, exogenous p53 significantly increased the activity of the p21Waf1/Cip1 promoter, and exogenous SIRT1 effected a substantial reduction in transactivation by p53 (Fig. 5A, lanes 1–3). However, exogenous Set7/9 significantly blocked the inhibitory effect of SIRT1 on p53 transactivation in a dose-dependent manner (Fig. 5A, lanes 4–6). We next asked whether the interaction between Set7/9 and SIRT1 influences the binding of SIRT1 to p53. A Flag-tagged Set7/9 or an empty Flag plasmid was transfected into HCT116 (p53+/+) cells in the presence or absence of Adr. Derived cell lysates then were immunoprecipitated with anti-SIRT1, and SIRT1-associated proteins were probed with anti-p53. The data in Fig. 5B show an association of p53 with SIRT1 in response to Adr treatment, coincident with an increase in the p53 level, but this interaction was significantly decreased when exogenous Set7/9 was cotransfected. A statistical analysis showed that the association of SIRT1 with p53 decreased by ∼3.5-fold in cells transfected with the Set7/9 vector relative to cells transfected with the empty plasmid (Fig. 5B, Lower). These data suggest that although p53 acetylation and activity are not affected by Set7/9-catalyzed methylation of SIRT1, they are regulated by a direct interaction between SIRT1 and Set7/9.
Fig. 5.
Set7/9 interrupts the binding of SIRT1 to p53. (A) HCT116 (p53−/−) cells were cotransfected with a plasmid expressing p21-Luc, p53WT, Myc-SIRT1, or Flag-Set7/9 as indicated, and the luciferase assay was then performed. The luciferase activity was normalized to the amount of protein in the cell lysate. Data are means ± SD (n = 3). (B) A Flag-Set7/9 construct was transfected into HCT (p53+/+) cells, cells were treated with or without 1 μM Adr for 6 h, and proteins were extracted for co-IP with anti-SIRT1 and probed with anti-p53 or anti-SIRT1 (Upper). The bands for SIRT1-immunprecipitated p53 were scanned and normalized to each SIRT1 band (Lower). Data are means ± SD (n = 3). (C) HEK293T cells were cotransfected with a Myc-SIRT1 construct and either Flag-Set7/9 or Flag-Set7/9 H297A (Set7/9 mutant). Cell lysates were prepared for co-IP with anti-Myc and probed with anti-Flag (Left). The bands corresponding to Flag-Set7/9 and Flag-Set7/9 H297A were scanned and compared with the control (Right). Data are means ± SD (n = 3). (D) HCT116 (p53+/+) cells were transfected with an empty plasmid, Flag-Set7/9, or Flag-Set7/9 H297A 24 h after transfection, cells were treated with 1 μM Adr for 12 h, and cell lysates were prepared and subjected to Western blotting with anti-acetylated p53 (K382), anti-p53, or anti-Set7/9. GAPDH was the loading control. (E) Under the same conditions as in (Fig. 5D), cells were harvested and subjected to real-time PCR or RT-PCR to measure the expression of p21Waf1/Cip1 and Set7/9. GAPDH was the loading control.
To further demonstrate that the interaction between Set7/9 and SIRT1 is critical for the dissociation of p53 from SIRT1, we used a mutant Set7/9 (Set7/9 H297A) with a histidine 297-to-alanine replacement (19) that severely reduces binding to SIRT1 (Fig. 5C). A plasmid expressing either wild-type Set7/9 (WT-Set7/9) or mutant Set7/9 was separately cotransfected with a Myc-tagged SIRT1 construct into HCT116 (p53+/+) cells, and the cells were then treated with Adr. Derived cell lysates were subjected to co-IP to detect an interaction between p53 and SIRT1. As shown in Fig. 5D, exogenous WT-Set7/9 increased the level of p53 acetylation at K382, whereas mutant Set7/9 was unable to do so (Fig. 5D). Subsequently, p21Waf1/Cip1 expression was also determined in cells transfected with WT-Set7/9 or mutant Set7/9. Fig. 5E shows that the expression of endogeneous p21Waf1/Cip1 mRNA was increased in Adr-treated HCT116 (p53+/+) cells, and that this expression was further enhanced by exogenous WT-Set7/9 but not by mutant Set7/9. These data suggest that the interaction between Set7/9 and SIRT1 is required for the dissociation of p53 from SIRT1 as well as transactivation by p53.
Discussion
The data presented in this study provide evidence that the histone methyltransferase Set7/9 interacts with SIRT1 in vitro and in vivo. Although Set7/9 catalyses methylation of SIRT1 at several lysine sites, this methylation does not affect its deacetylase activity. However, the interaction of Set7/9 with SIRT1 does facilitate dissociation of SIRT1 from p53, with consequent enhancement of both p53 acetylation at K382 and p53-mediated transactivation in response to DNA damage.
Posttranslational modifications of SIRT1 influence SIRT1 activity (15, 16, 24–26). In view of our demonstration of a SIRT1–Set7/9 interaction, we also confirmed by autoradiography and mass spectrometry that Set7/9 catalyzes SIRT1 methylation. However, the Set7/9-catalyzed methylation of SIRT1 did not influence SIRT1 deacetylase activity. Functionally, the methylation of SIRT1 is different from the other posttranslational modifications of SIRT1. For example, SIRT1 is sumoylated at lysine 734, with a resultant increase in activity, in the human lung cancer cell line H1299, but desumoylated at this site by the nuclear desumoylase SENP1, with a resultant decrease in deactealyse activity, in response to UV or oxidative stresses (16). In addition, SIRT1 is phosphorylated at threonine 522 by the nuclear protein kinase DYRK1A upon treatment of cells with the DNA-damaging agent etoposide, which results in increased SIRT1 deacetylase activity (26). Similarly, phosphorylation of SIRT1 serine residues appears to enhance SIRT1 deacetylase activity. Thus, in response to ionizing radiation, SIRT1 is phosphorylated at several serines by casein kinase 2 (CK2) in HeLa cells; this modification results in increased SIRT1 deacetylase activity and subsequent induction of p53 deacetylation and protection of cells from apoptotic cell death (25). Although Set7/9 was found in the present study to catalyze the methylation of SIRT1 lysines 233, 235, 236, and 238 without any observed effect on deacetylase activity, we cannot exclude the possibility that the methylation of other lysines by another methyltransferase might influence SIRT1 activity or that the observed methylation events might regulate SIRT1 activity in another context.
Set7/9 has been reported to methylate p53, pRB, and DNMT1 (19, 21, 22, 27). However, Set7/9 methylation is not always related to functional changes of targeted molecules. For example, Set7/9 interacts with Pdx1, an insulin transactivator, and catalyzes Pdx1 methylation. Although the interaction of Pdx1 with Set7/9 is able to recruit Set7/9 to histone H3 and induce H3K4 methylation (28, 29), the methylation of Pdx1 is not directly associated with changes in Pdx1 function. In addition, although the methylation of DNMT1 leads to a decrease in the half-life of DNMT1, this modification is also not directly related to changes in DNMT1 methyltransferase activity (21, 30). Similarly, results in our study confirmed that methylation of SIRT1 did not change its function or its half-life (data not shown).
The interaction of DBC1 with SIRT1 decreases SIRT1 activity, whereas AROS has an opposite effect. DBC1 binds directly to the middle fragment of SIRT1 to block the catalytic domain of SIRT1, which in turn negatively regulates SIRT1 activity (12, 13). In contrast to DBC1, AROS binds to the noncatalytic domain of SIRT1 and subsequently increases SIRT1 activity (14). Although we showed here that the methylation of SIRT1 by Set7/9 did not affect its deacetylase activity, a direct interaction between these proteins was critical for the regulation of SIRT1 deacetylase activity on p53 because this interaction was also required for the dissociation of SIRT1 from p53 (Fig. 5B). It is unclear how the interaction between SIRT1 and Set7/9 influences the association of SIRT1 and p53. However, among other possibilities, this could reflect either a Set7/9-induced conformational change in SIRT1 that prevents SIRT1 from binding to p53 or a direct competition between Set7/9 and p53 for binding to SIRT1.
Our study reveals that exposure of cells to Adr results in a significantly decreased SIRT1 deacetylase activity on p53 concurrent with an increased SIRT1–Set7/9 interaction. The decreased SIRT1 activity on p53 results from the interaction of SIRT1 and Set7/9 because increased p53 acetylation was observed with WT-Set7/9 but not with a mutant Set7/9 that shows a significantly decreased ability to bind SIRT1. The p53 methylation at K372 by Set7/9 was partly associated with p53K382 acetylation and p53 activity because p53 acetylation or transactivation of the downstream target p21 promoter was decreased in cells expressing a mutant p53K372R relative to a p53WT. However, and importantly, overexpressed Set7/9 still effected a significant enhancement both of p53K382 acetylation and p53-dependent activation in p53K372R-transfected cells, which suggests that, in addition to methylating p53 at K372, Set7/9 may function on other p53-related proteins and, thus, induce changes in p53 transactivation. In this regard, the present results show that Set7/9 regulates p53 activity indirectly by negatively regulating SIRT1. Therefore, although the p53 methylation mediated by Set7/9 may contribute to the p53 transactivation potential, the interaction of SIRT1 and Set7/9 may have a more critical role in enhancing p53 activity. In addition, the interaction of SIRT1 and Set7/9 may explain why Set7/9 knockdown induces p53 deacetylation at K382 (31, 32), thus providing the mechanistic basis for linking p53 methylation at K372 to p53 acetylation at K382.
In conclusion, our data show that Set7/9 negatively regulates SIRT1, which results in an increased acetylation of p53 at K382. A more complete understanding how SIRT1 is regulated and how it in turn regulates its downstream targets will be valuable in designing new anticancer therapies.
Materials and Methods
Cell culture and treatments, plasmid construction, luciferase assays, RNA interference, Western blotting, co-IP, RT-PCR, and real-time PCR are explained in detail in SI Materials and Methods.
GST Pull-Down Assay.
GST or GST fusion proteins were expressed in bacteria induced with isopropyl-β-d-thio-galactoside and purified. Equal amounts of GST or GST fusion proteins were incubated with glutathione-Sepharose 4B beads (GE Healthcare) and then washed three times with TEN buffer (20-mM Tris at pH 7.4, 0.1 mM EDTA, and 100 mM NaCl). His-Set7/9 purified from bacteria was incubated with GST or GST fusion proteins. The beads were washed three times with TENT buffer (0.5% Nonidet P-40, 20 mM Tris at pH 7.4, 0.1 mM EDTA, and 300 mM NaCl) and analyzed by Western blotting with anti-His or anti-GST antibody.
In Vitro Methylation Assay.
Briefly, GST-fusion proteins were prepared as described above. Substrates (2 μg) were added into a 20-μL reaction containing 50 mM Tris at pH 9.0, 0.5 mM DTT, 1 mM PMSF, and 1 μg of His-Set7/9 enzyme in the presence of 0.5 μCi of 3H-SAM (PerkinElmer). Reactions were performed at 30 °C for 3 h. The methylation levels were determined either by scintillation counting or autoradiography.
In Vitro Deacetylation Assay.
GST-SIRT1 or GST-SIRT1-4KR was purified and incubated with Set7/9 in the presence of SAM at 30 °C for 3 h to induce SIRT1 methylation. The treated SIRT1 or SIRT1-4KR was then incubated with acetylated p53 (BIOMOL) in the presence of 50 μM NAD+ in deacetylase buffer (50 mM Tris·HCl at pH 9.0, 5% glycerol, 50 mM NaCl, 4 mM MgCl2, 0.5 mM DTT, and 0.1 mM PMSF) at 30 °C for 90 min. The reaction mixtures were subjected to immunoblotting by using anti–acetyl-p53 (K382) antibody.
Statistical Analysis.
Values are expressed as mean ± SEM. Significant differences between means were analyzed by two-tailed, unpaired Student's t test, and differences were considered significant at *P < 0.05 or **P < 0.01. Microsoft Excel was used to analyze all of the data.
Supplementary Material
Acknowledgments
Myc-tagged human SIRT1 was a gift from Wengong Wang. HA-p300 was a gift from Yanping Zhang. Deletion plasmids of SIRT1 were gifts from Zhenkun Lou. This study was supported by National Natural Science Foundation of China Grants 90919030, 31070691, 30921062, (to W.-G.Z), and 30628028 (to W.G); Ministry of Science and Technology of China Grants 2011CB504200 and 2011CB910103 (to W.-G.Z. and Y.Z); “111 project” from the Ministry of Education of China; grants from the Ministry of Science and Technology of China (to W.-G.Z.); and by National Institutes of Health Grant CA129325 (to R.G.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019619108/-/DCSupplemental.
References
- 1.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 3.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 4.Bradbury CA, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 5.Huffman DM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 6.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- 7.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 8.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 9.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 10.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 11.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 15.Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishioka K, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 19.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 20.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 21.Estève PO, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 24.Zschoernig B, Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem Biophys Res Commun. 2009;381:372–377. doi: 10.1016/j.bbrc.2009.02.085. [DOI] [PubMed] [Google Scholar]
- 25.Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J Biol Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munro S, Khaire N, Inche A, Carr S, La Thangue NB. Lysine methylation regulates the pRb tumour suppressor protein. Oncogene. 2010;29:2357–2367. doi: 10.1038/onc.2009.511. [DOI] [PubMed] [Google Scholar]
- 28.Francis J, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II elongation during activation of insulin transcription. J Biol Chem. 2005;280:36244–36253. doi: 10.1074/jbc.M505741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG. Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes. 2009;58:185–193. doi: 10.2337/db08-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pradhan S, Chin HG, Estève PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov GS, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurash JK, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell. 2008;29:392–400. doi: 10.1016/j.molcel.2007.12.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.