Abstract
Cell populations able to generate a large repertoire of genetic variants have increased potential to generate tumor cells that survive through the multiple selection steps involved in tumor progression. A mechanism for the generation of aneuploid cancer cells involves passage through a tetraploid stage. Supernumerary centrosomes, however, can lead to multipolar mitosis and cell death. Using tissue culture and transgenic mouse models of breast cancer, we report that Cut homeobox 1 (CUX1) causes chromosomal instability by activating a transcriptional program that prevents multipolar divisions and enables the survival of tetraploid cells that evolve to become genetically unstable and tumorigenic. Transcriptional targets of CUX1 involved in DNA replication and bipolar mitosis defined a gene expression signature that, across 12 breast cancer gene expression datasets, was associated with poor clinical outcome. The signature not only was higher in breast tumor subtypes of worse prognosis, like the basal-like and HER2+ subtypes, but also identified poor outcome among estrogen receptor-positive/node-negative tumors, a subgroup considered to be at lower risk. The CUX1 signature therefore represents a unique criterion to stratify patients and provides insight into the molecular determinants of poor clinical outcome.
Keywords: tetraploidy, transcription factor, spindle assembly checkpoint
Aneuploidy is one of the most common aberrations in cancer (1). One route to aneuploidy involves the generation of tetraploid cells that readily become aneuploid owing to frequent chromosome missegregation and rearrangements (2). The presence of multiple centrosomes can lead to multipolar mitosis from which nonviable daughter cells are generated (3), but centrosome clustering to two poles enables cancer cells to undergo bipolar mitosis (3–5). Supernumerary centrosomes, however, increase the possibility of chromosome missegregation, which results mostly from merotelic kinetochore–microtubule attachments. This is manifested at anaphase by the presence of lagging chromosomes and chromosome bridges in a fraction of cells in the population (3, 6).
Elevated Cut homeobox 1 (CUX1) expression was reported in various human tumors and was associated with poor prognosis (reviewed in ref. 7). Transgenic mice expressing various CUX1 isoforms develop malignancies in the hematopoietic system and the mammary gland (8, 9). CUX1 plays a role in at least two distinct processes with relevance to cancer: cell cycle progression and cell motility (10, 11). Identification of p110 CUX1 transcriptional targets revealed a striking enrichment for genes that play a role during two phases of the cell cycle: S phase and mitosis (12). Constitutive expression of p110 CUX1 was shown to accelerate entry into S phase (11). Here we identified a role for p110 CUX1 in mitotic regulation. We show that p110 CUX1 contributes to the establishment of a transcriptional program that enables tetraploid cells to undergo bipolar mitosis in the presence of multiple centrosomes. The survival of tetraploid cells was associated with chromosomal instability and the generation of an aneuploid population that evolved to become tumorigenic. A gene expression signature made of CUX1 targets involved in S phase and mitosis is able to predict patients with poor clinical outcome in breast cancers.
Results
Long-Term Expression of p110 CUX1 Causes Tetraploidization in HEK293 and NMuMG Cells.
We initially noticed that HEK293 cells stably expressing p110 CUX1 eventually formed a population composed exclusively of polyploid cells (Fig. S1A). Progressive polyploidization was repeatedly observed after p110 CUX1 expression both in NMuMG mouse mammary epithelial cells and HEK293 cells (Fig. 1A and Fig. S1B). Moreover, polyploidization was accelerated with a mutant, p110S1237,1270A CUX1, that cannot be phosphorylated by cyclin A/Cdk1 and remains active in G2 (Fig. 1B and Fig. S1 C and D) (13). Tetraploidization was not observed after 35 passages in five other cell lines, suggesting that p110 CUX1 does not itself induce tetraploidization but could facilitate the survival of polyploid cells in cell lines where cytokinesis failure occurs at a higher frequency. Consistent with this hypothesis, the spontaneous rate of binucleation was significantly higher in NMuMG cells (47 of 3,029) than in Rat1 (15 of 3,208; P < 0.0001) and U2OS (22 of 3,541; P < 0.0004) cells.
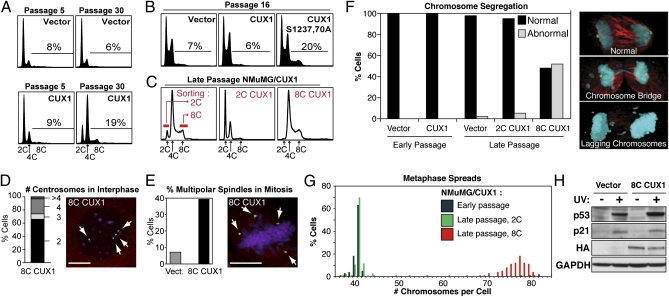
Fig. 1.
Tetraploidy and chromosomal instability in p110 CUX1-expressing NMuMG cells. (A) NMuMG cells stably expressing p110 CUX1 (amino acids 747–1505) were generated by retroviral infection. More than 500 colonies were pooled after selection, which was considered passage 1. Cell cycle distribution was determined by FACS. (B) FACS analysis was performed on independent populations of NMuMG cells expressing either p110 CUX1 or p110S1237,1270A CUX1. (C) Late passage NMuMG/CUX1 cells were FACS-sorted according to DNA content after Hoechst 33342 staining and put back in culture. 2C represent G1 diploid cells and 8C are G2/M tetraploids. (D and E) Number of centrosomes during interphase (D, n = 304) or spindle poles during early mitosis (E, n = 250 for each cell line) was determined by indirect immunofluorescence according to γ-tubulin staining (green), whereas microtubules and DNA were revealed by α-tubulin (red) and DAPI (purple) staining, respectively. (Scale bars, 10 μm.) (F) Chromosome segregation during anaphase was studied by confocal microscopy for late-passage NMuMG/vector (n = 31), early-passage NMuMG/CUX1 (n = 50), and late-passage 2C-sorted (n = 57) 8C-sorted (n = 23) NMuMG/CUX1 cells. Cells were stained for γ-tubulin (green), α-tubulin (red), and DNA (DAPI, blue). Images are a composite of z-sections encompassing entire cells. (G) Chromosome counts were determined on metaphase spreads prepared from early-passage NMuMG/p110 CUX1 or late-passage 2C- or 8C-sorted NMuMG/p110 CUX1 cells (n ≥ 100 each). (H) NMuMG/vector and 8C NMuMG/CUX1 cells were left untreated or exposed to 20 mJ/cm2 UV. Cell lysates were prepared 24 h later and analyzed by SDS/PAGE and Western blotting using the indicated antibodies.
Chromosomal Instability and Aneuploidy in Late-Passage NMuMG/CUX1 Cells.
Subsequent experiments focused on the nontumorigenic mammary epithelial NMuMG cells because these cells initially displayed a near-diploid karyotype (14). Diploid (2C) and tetraploid (8C) subpopulations were FACS-sorted from late-passage NMuMG/CUX1 cells (Fig. 1C). 8C NMuMG/CUX1 cells maintained a 4C-8C DNA content and displayed two or four centrosomes per cell during interphase according to γ-tubulin staining (Fig. 1 C and D). During early mitosis, 38% of 8C NMuMG/CUX1 cells harbored multiple spindle poles, vs. 8% for late-passage NMuMG/vector cells (Fig. 1E; P < 0.001). In time-lapse microscopy, we did not observe multipolar divisions in 8C NMuMG/CUX1 cells, indicating that extra centrosomes were efficiently nucleated into two poles before anaphase (n = 698; Table S1). However, the duration of mitosis was extended by 10 min in these cells (48 min vs. 38.5 min; P < 0.0001; Table S1), in agreement with the notion that a longer mitosis may be an intrinsic characteristic of viable tetraploid cells (15). Although 8C NMuMG/CUX1 cells underwent bipolar mitoses like the 2C cells, a much higher proportion of 8C cells exhibited chromosome segregation defects during anaphase (Fig. 1F; 51% 8C vs. 4% 2C; P < 0.001), such that almost all 8C cells displayed a subtetraploid chromosome count, ranging from 70 to 80 chromosomes per cell (Fig. 1G). In contrast, chromosomal instability was a rare event in 2C NMuMG/CUX1 cells (Fig. 1 F and G). Of note, sorted 8C NMuMG/CUX1 cells maintained a functional p53 pathway, as judged from the stabilization of p53 and the up-regulation of p21 after UV irradiation (Fig. 1H). These results indicate that 8C NMuMG/CUX1 cells are prone to chromosomal instability and evolve to become a heterogeneous population of aneuploid cells.
p110 CUX1 Expression Favors Bipolar Mitoses in Newly Formed Tetraploid Cells.
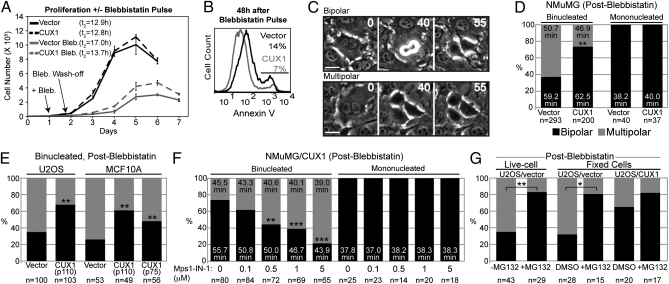
To verify the effect of p110 CUX1 on newly arising tetraploid cells, we took advantage of the reversible myosin II inhibitor blebbistatin to induce cytokinesis failure (16) (Fig. S1H, Upper, and Table S2). Twenty-four hours after tetraploidization, NMuMG/vector cells displayed nuclear abnormalities, whereas NMuMG/CUX1 cells were uniformly mononucleated (Fig. S1H, Lower). NMuMG/CUX1 cells proliferated significantly faster than control cells after treatment (Fig. 2A, gray lines). This was due, at least in part, to a reduced rate of apoptosis as judged from Annexin V staining (Fig. 2B). Time-lapse microscopy revealed that most binucleated NMuMG/vector cells (63%) underwent a multipolar anaphase (Fig. 2 C and D and Movies S1 and S2). In contrast, most binucleated NMuMG/CUX1 cells (73.5%) underwent a bipolar division (P < 0.0001; Movie S3). In both cell populations, bipolar division in tetraploid cells was associated with a longer duration of mitosis (Fig. 2D; P < 0.0001), whereas mitosis was unaffected in neighboring mononucleated cells (compare Fig. 2D with Table S1). Similar experiments in U2OS cells and in the nontransformed human mammary epithelial MCF10A cells confirmed that p110 CUX1 and another isoform, p75 CUX1 (17), can promote bipolar mitoses in tetraploid cells (Fig. 2E; P < 0.0002).
Fig. 2.
p110 CUX1 expression enables bipolar mitoses in newly formed tetraploid cells. (A) Proliferation curves of NMuMG/vector (bold) and NMuMG/CUX1 (dashed) cells, with (gray) or without (black) a 12-h blebbistatin pulse treatment. Cells were counted until they ceased proliferation because of contact inhibition. (B) Percentage of apoptotic cells was measured 48 h after blebbistatin wash-off, according to Annexin V staining and FACS analysis. (C and D) Outcome of cell divisions after a blebbistatin wash-off was determined by time-lapse microscopy using frames taken every 5 min. Multipolar divisions displayed a multipolar anaphase giving rise to three or four daughter cells (Movie S1) or a tripolar anaphase followed by cytokinesis failure resulting in two cells, one of which with two nuclei (Movie S2). Neighboring mononucleated cells were used as controls. (Scale bars, 25 μm.) (E) U2OS and MCF10A cells expressing p110 CUX1 were treated with blebbistatin, and the fate of binucleated cells was determined by time-lapse microscopy. (F) NMuMG/CUX1 were pulse-treated with blebbistatin and imaged by time-lapse microscopy with increasing concentrations of MPS1-IN-1. The outcome of cell division and the duration of mitosis were scored for mono- and binucleated cells. (G) U2OS/vector cells were pulse-treated 8 h with blebbistatin. Sixteen hours later, cells were imaged by time-lapse microscopy, during which 10 μM MG132 was added for 90 min and washed away. The fate of binucleated cells entering mitosis during or just before MG132 treatment (Movie S5) was examined. Neighboring binucleated cells dividing without being exposed to MG132 were used as control (Movie S4). Alternatively, cells were fixed 1 h after MG132 treatment, and the metaphase spindle configuration was determined using centrin 3 and α-tubulin staining (Fig. S1 I–K). *P < 0.005; **P < 0.0002; ***P ≤ 1.2 × 10−5. Black bars, % bipolar division. Gray bars, % multipolar division.
Identification of CUX1 Transcriptional Targets Involved in Bipolar Mitosis.
We sought to identify transcriptional targets of p110 CUX1 that stimulate bipolar mitosis in tetraploid cells. A list of putative CUX1 transcriptional targets was first established by performing ChIP in Hs578T breast tumor cells followed by hybridization on the human HG18 promoter ChIP-on-chip oligo microarray set of NimbleGen. Independent ChIP experiments were performed in HeLa cells to validate relevant targets. We identified and validated 12 genes previously found in a genome-wide RNAi screen as being required for bipolar mitosis in cells with supernumerary centrioles [Table 1, columns 1 and 2 (from left) labeled with “*”] (18). Activation of the spindle assembly checkpoint (SAC) was proposed to favor bipolar division indirectly by extending mitosis and providing sufficient time for centrosome clustering (18). Therefore, we validated nine other SAC genes from our ChIP-on-chip dataset. The role of p110 CUX1 as a transcriptional activator of these 21 targets was demonstrated from three complementary approaches: overexpression of p110 CUX1, inducible shRNA-mediated inhibition of endogenous CUX1 expression, and reexpression of CUX1 after removal of the shRNA inducer doxycyclin (Table 1, columns 3–5). Note in the latter experiment the striking increase in the expression of all targets upon reexpression of endogenous CUX1 (Table 1, column 5). In summary, these experiments demonstrate that many genes involved in the mitotic checkpoint are directly activated by CUX1.
Table 1.
Validated transcriptional targets activated by CUX1
| mRNA expression (fold) |
||||
| Hs578t shCUX1tetON |
||||
| Gene | ChIP IgG vs. input (fold) | NmuMG (CUX1 vs. vector) | + Dox | − Dox |
| SAC | ||||
| AURKB | +3.8 | +2.6 | −1.3 | +7.0 |
| BUB1* | +3.8 | +3.6 | −2.5 | +5.6 |
| BUB3 | +3.5 | +2.4 | −2.4 | +7.5 |
| BUBR1 | +1.7 | +3.6 | −2.2 | +10.8 |
| CENPE* | +10.8 | +2.6 | −1.6 | +6.7 |
| MAD1L1 | +4.7 | +1.8 | −1.5 | +3.9 |
| MAD2L1* | +2.7 | +2.0 | −2.6 | +5.7 |
| TTK (MPS1) | +2.0 | +3.2 | −1.2 | +5.6 |
| NEK2 | +2.5 | +3.8 | −1.4 | +6.5 |
| ROD | +3.6 | +3.9 | −1.5 | +5.9 |
| ZW10 | +3.1 | +2.0 | −1.8 | +6.3 |
| ZWILCH | +2.9 | +5.0 | −2.5 | +4.3 |
| Centromeric histone | ||||
| CENPA* | +3.1 | +1.9 | −1.5 | +5.8 |
| Kinesin | ||||
| KIF2C* | +2.3 | +3.2 | −1.4 | +6.1 |
| KIFC1* | +1.9 | +2.3 | −1.8 | +9.4 |
| Other functions | ||||
| CAMK2D* | +2.9 | +1.4 | −1.9 | +2.0 |
| COG7* | +8.1 | +1.6 | −1.4 | +1.3 |
| MFAP1* | +4.3 | +1.4 | −1.7 | +3.9 |
| NUDCD1* | +3.7 | +2.6 | −1.2 | +3.8 |
| RIN2* | +4.2 | +2.3 | −1.6 | +2.4 |
| SF3B3* | +3.0 | +2.0 | −1.9 | +4.6 |
Validation of p110 CUX1 transcriptional targets relevant to the establishment of bipolar mitoses. CUX1 transcriptional targets were identified by ChIP in Hs578T breast tumor cells. From left: column 1: asterisks (*) indicate genes previously identified in an siRNA screen for bipolar mitosis (18). Column 2: validation and measurement of promoter occupancy from independent ChIP assays using HeLa cells. Columns 3, 4, and 5: fold difference in mRNA expression measured by real-time PCR between NMuMG/CUX1 and control cells (column 3), Hs578t cells bearing a doxycyclin-inducible CUX1-specific shRNA, treated or not for 5 d with doxycycline (column 4), or after endogenous CUX1 expression was permitted after removal of doxycyclin for 3 d (column 5).
p110 CUX1 Facilitates Engagement of the Spindle Assembly Checkpoint.
The role of CUX1 as a transcriptional activator of many genes involved in the control of bipolar mitosis made us consider the possibility that CUX1 overexpression might enable cells to engage more efficiently the SAC. In agreement with this notion, CUX1-expressing cells sustained an extended mitotic arrest and maintained higher cyclin B levels when the SAC was triggered by treating cells with the microtubule poison nocodazole (Fig. S2 A and B).
To verify whether efficient SAC engagement was necessary to allow bipolar division in newly formed tetraploid NMuMG/CUX1 cells, time-lapse microscopy after blebbistatin treatment was performed in the presence of MPS1-IN-1, a cell-permeable inhibitor of the SAC kinase MPS1 (19). With increasing MPS1-IN-1 concentrations the duration of mitosis was progressively shortened, and the frequency of multipolar divisions was correspondingly increased up to threefold at 5 μM MPS1-IN-1 (P < 0.0001; Fig. 2F). Note that at any drug concentration the average duration of mitosis was significantly longer for binucleated cells undergoing a bipolar division than for those undergoing a multipolar division (P < 0.0002). Importantly, these concentrations of MPS1-IN-1 did not affect the outcome nor the duration of mitosis in neighboring mononucleated cells, indicating that tetraploid cells are intrinsically more sensitive to SAC inhibition than diploid cells. These results indicate that mitotic duration and bipolar division in tetraploid NMuMG/CUX1 cells are very sensitive to SAC inhibition. Moreover, these findings suggest that CUX1 promotes bipolar divisions by allowing tetraploid cells to delay mitosis, which in turn would increase the prospect of centrosome clustering. In support of this mechanism of action, the rate of bipolar division (live cell) and bipolar spindle configuration (fixed cells) in U2OS/vector cells was increased to approximately 80% by transiently delaying anaphase onset using the proteasome inhibitor MG132 (Fig. 2G and Movies S4 and S5).
Tumorigenic Potential of p110 CUX1 Is Associated with Chromosomal Instability.
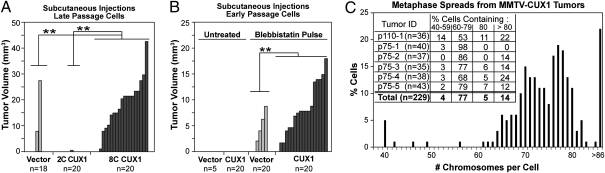
Tetraploidy and aneuploidy have previously been associated with increased tumorigenicity (2). We therefore compared the tumorigenic potential of NMuMG p110 CUX1 cells that have become aneuploid or remained diploid. We performed s.c. injections in nude mice with late-passage populations of cells carrying an empty vector, or the FACS-sorted 2C and 8C NMuMG/CUX1 cells (from Fig. 1C). Significantly more and larger outgrowths were produced by the 8C NMuMG/CUX1 cells (Fig. 3A; P < 0.0001). The fact that late-passage 2C NMuMG/CUX1 cells failed to produce outgrowths strongly suggests that the acquisition of tumorigenic potential in p110 CUX1-expressing cells is associated with chromosomal instability. We therefore directly tested whether p110 CUX1 expression enabled tumor outgrowth after cytokinesis failure. Early-passage NMuMG cells expressing p110 CUX1 or not were treated with blebbistatin before being s.c. injected into nude mice. The frequency and size of tumors were significantly higher in cells expressing p110 CUX1 than in cells carrying the empty vector (Fig. 3B; P = 0.0002). These results together with the assays performed in tissue culture indicate that p110 CUX1 promotes the survival and proliferation of tetraploid cells (Fig. 2 A and B and Fig. S2).
Fig. 3.
CUX1-induced tumorigenicity involves chromosomal instability. (A) Late-passage NMuMG/vector cells, 2C-sorted NMuMG/CUX1 cells, or aneuploid 8C-sorted NMuMG/CUX1 cells were injected s.c. in nude mice. Tumor volumes (mm3) were measured 27 d after injection. (B) Early-passage NMuMG cells expressing p110 CUX1 or not were pulse-treated with blebbistatin and injected s.c. into nude mice. (C) Chromosome number of cells from six mammary gland tumors arising in p110 or p75 CUX1 transgenic mice was determined from metaphase spreads. **P < 0.0003 for both tumor sizes and tumor numbers.
If p110 CUX1 contributes to tumorigenicity by promoting chromosomal instability, aneuploidy should be a common feature in tumors of CUX1 transgenic mouse models. We examined the ploidy on metaphase spreads of six independent mammary tumors that arose in MMTV-CUX1 transgenic mice (9). Strikingly, each tumor showed a high level of aneuploidy, with the majority of cells containing a subtetraploid chromosome content: 82% of the 229 cells scored contained between 60 and 80 chromosomes (Fig. 3C). We next verified whether CUX1 transcriptional targets identified in Table 1 were associated with tumor development in MMTV-CUX1 transgenic mice. We performed expression profiling on microdissected epithelial cells and observed a gradual increase in the expression of the 21 genes: a 1.75-fold increase between normal mammary epithelial cells from nontransgenic and transgenic mice (Fig. S3A; P = 0.014) and in transgenic mice, a 2.31-fold increase between epithelial cells from normal adjacent mammary glands and mammary tumors (P = 0.0016). The significance of these differences was confirmed independently using two other statistical approaches: the hypergeometric test and gene set enrichment analysis (GSEA) software (Fig. S3 B and C). Together, these results show that transcriptional targets of p110 CUX1 identified in cell lines are also up-regulated in the mammary glands and mammary tumors of CUX1 transgenic mice, and most cells of the mammary tumors carry a subtetraploid chromosome content.
Targets of CUX1 Predict Clinical Outcome in Human Breast Cancers.
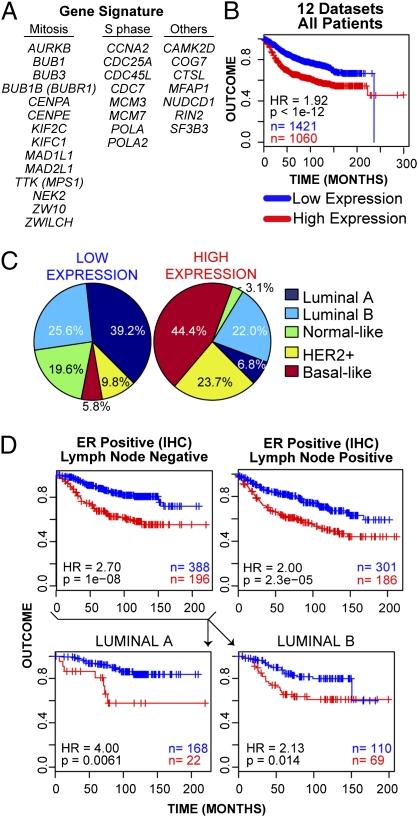
We performed a meta-analysis of 12 published gene expression datasets of breast cancer patients to analyze the expression of CUX1 transcriptional targets (SI Experimental Procedures). Most (11 of 12) datasets did not contain CUX1-specific probes. Moreover, CUX1 transcriptional activity cannot be predicted from mRNA expression because it depends on dephosphorylation and proteolytic processing (reviewed in ref. 7). Therefore, as a surrogate for CUX1 activity we used a set of eight well-defined targets of CUX1 at the G1/S transition [CCNA2, CDC7, MCM3, MCM7, CDC45L, CDC25A, POLA, and POLA2 (12)] and CTSL1. For each dataset, patients were hierarchically clustered using this gene list with the Euclidean distance metric via Ward's algorithm. The gene signature showed significant covariation across the datasets, with the majority of the 29 genes being coexpressed. This enabled us to use low and high expression of the signature to stratify patients and perform survival analysis (Fig. S4). Interestingly, high expression was associated with significantly poorer outcome when patients from all 12 datasets were combined [Fig. 4B; Cox regression P value < 10−12, Cox regression hazard ratio (HR) high vs. low = 1.92] and in 8 of 12 individual datasets (Fig. S5). The gene signature was also a strong predictor of outcome in univariate analysis (Table 2; P = 2.2 × 10−16, HR = 2.27).
Fig. 4.
CUX1 transcriptional targets are highly expressed in breast tumor subtypes with poor prognosis and identify a subset of luminal tumors with poor outcome. (A) List of 29 CUX1 transcriptional targets used in expression analysis. Targets include 20 genes validated in Table 1 (but excludes ROD, which is absent from published microarray datasets), plus eight previously characterized DNA replication genes whose expression is strongly stimulated by p110 CUX1 and CTSL1 (12). (B) Kaplan-Meier curve displaying significant segregation of good outcome vs. bad outcome patients classified into “low” and “high” groups on the basis of a 29-gene signature. (C) Breakdown and percentage of patients classified into each molecular subtype for “low” and “high” expression groups. (D) Kaplan-Meier curves comparing the outcome of low- and high-expressing groups for patients reported to be ER+/LN− and ER+/LN+. Lower: ER+/LN− patients were further classified into luminal A and luminal B subtypes.
Table 2.
Univariate and multivariate analyses
| Univariate |
Multivariate |
|||
| Variable | HR (95% CI) | P | HR (95% CI) | P |
| Grade 2 (vs. 1) | 2.26 (1.63–3.15) | 1.1 × 10−6 | 1.87 (1.34–2.61) | 2.6 × 10−4 |
| Grade 3 (vs. 1) | 2.92 (2.10–4.05) | 1.6 × 10−10 | 1.72 (1.19–2.49) | 4.0 × 10−3 |
| Lymph node | 1.63 (1.34–1.98) | 6.3 × 10−7 | 1.57 (1.29–1.90) | 5.7 × 10−6 |
| HER2 | 1.60 (1.25–2.05) | 1.6 × 10−4 | 1.20 (0.93–1.54) | 1.6 × 10−1 |
| ER | 1.57 (1.28–1.94) | 2.1 × 10−5 | 1.18 (0.93–1.49) | 1.6 × 10−1 |
| CUX1 signature | 2.27 (1.87–2.76) | 2.2 × 10−16 | 1.85 (1.48–2.32) | 7.1 × 10−8 |
Univariate and multivariate analyses were performed on patients for whom all clinical variables were reported (n = 1,474, eight datasets; Fig. S5). CI, confidence interval.
We then determined the molecular subtype distribution within the “low” and “high” expression groups as classified by the gene signature, on the basis of the correlation to centroids created from the prediction analysis of microarray method using a 50-gene signature previously described (20). The breast cancer molecular subtypes were not distributed equally between the two groups. Those subtypes with a poorer outcome, namely HER2+ and basal-like, were overrepresented in the high expression group, (7.6-fold and 2.4-fold, respectively), whereas the proportion of the good outcome-enriched luminal A subtype was reduced 5.7-fold (Fig. 4C). Furthermore, the signature identified subgroups of luminal A (72 of 557 = 11.4%, P = 0.013, HR = 1.96) and luminal B (233 of 384 = 37.8%, P = 0.0028, HR = 1.56) patients with poorer clinical outcome (Fig. S6A). When we restricted our analysis to patients with similar clinico-pathological features, high expression of the signature was associated with poor outcome regardless of grade (grade 1: P = 0.0034, HR = 2.56; grade 2: P < 10−5, HR = 1.89; grade 3: P = 0.0014, HR = 1.64), axillary lymph node (LN) status (LN+: P < 10−5, HR = 1.92; LN−: P < 10−8, HR 2.33) or estrogen receptor (ER) status (ER+: P < 10−12, HR = 2.38; ER−: P = 0.047, HR = 1.51) (Fig. S6 B–D). Importantly, multivariate Cox regression analysis confirmed that the gene signature was a significant predictor of outcome that is independent of grade, LN, HER2, and ER status (Table 2; P = 7.1 × 10−8, HR = 1.85). The signature was predictive of outcome both among patients considered at risk, like the LN+/HER2+ (Fig. S6C; 49 of 120, P = 0.0014, HR = 2.63), and among patients at a lower risk, like the ER+/LN− patients (Fig. 4D; 196 of 584 = 34%, P = 10−8, HR = 2.70). This seems to be not solely due to the enrichment for poor-outcome subtypes within this cluster, because high expression was associated with poor outcome within luminal A and luminal B patients (Fig. 4D, Lower). Thus, this expression signature is able to identify patients at risk for relapse among ER+/LN− patients.
Discussion
In the present study, we showed that p110 CUX1 contributes to the establishment of a transcriptional program that enables cells to efficiently engage SAC signaling, thus allowing the survival and proliferation of polyploid cells that evolve to become aneuploid and tumorigenic. To our knowledge, such a mechanism of oncogenic transformation by a transcription factor has not yet been described. Aneuploidy from a tetraploid precursor was thought to arise from multipolar mitoses, but recently such events were found to generate nonviable cells and to occur much less frequently than originally suspected (3). However, although centrosome clustering may succeed in producing bipolar mitosis, transient multipolar spindles were shown to increase the occurrence of merotely which, if not corrected, cause chromosome missegregation (3, 5, 6, 21). We made similar observations in cells expressing p110 CUX1. Although indirect immunofluorescence on fixed cells showed a high proportion of cells with a multipolar spindle configuration (Fig. 1E), live-cell imaging revealed that centrosome clustering to two poles was eventually achieved in virtually all of these cells (Table S1) but was accompanied by frequent chromosome segregation defects (Fig. 1 F and G).
Two lines of observations suggest that p110 CUX1 does not itself induce tetraploidization but affects its outcome once it has occurred. First, constitutive expression of p110 CUX1 did not cause a defect in mitosis or cytokinesis and was associated with tetraploidization in only two of seven cell lines expressing p110 CUX1 (Fig. 1 and Fig. S1). Second, after induction of tetraploidy with blebbistatin, p110 CUX1 enabled a greater proportion of cells to undergo a normal, bipolar, cell division (Fig. 2D and Movies S1– S3), but the protective effect of p110 CUX1 was lost in the presence of a SAC kinase inhibitor (Fig. 2F). The extended mitotic arrest in the presence of nocodazole (Fig. S2A), the delay in mitosis after blebbistatin treatment (Fig. 2 D and F), and the loss of protective effect in the presence of an inhibitor of the SAC kinase MPS1 (Fig. 2F) all concur to suggest that p110 CUX1 mediates its effects by establishing a transcriptional program that enables efficient SAC engagement, thereby allowing more time for multiple centrosomes to cluster. In addition, we must consider the possibility that p110 CUX1 directly facilitates bipolar mitosis by stimulating the expression of genes that are involved in centrosome clustering. In support of this notion, we reported that KIF2C and KIFC1 are direct targets that are activated by p110 CUX1 (Table 1). Unfortunately, it is not easy to verify a direct effect on centrosome clustering because of the close link that exists between the time spent in mitosis and successful clustering. To eliminate the effect of time, we transiently inhibited anaphase onset using MG132 and analyzed cell division in U2OS cells made tetraploid with a blebbistatin treatment (Fig. 2G). In this context, expression of p110 CUX1 did not significantly improve centrosome clustering over the already high rate observed in control cells carrying the empty vector (Fig. 2G; 80% bipolar for vector vs. 83% for CUX1). More experiments will be needed to ascertain the effect of p110 CUX1 on centrosome clustering.
Because of the unusual structure of the CUTL1 gene, currently most expression microarrays contain oligonucleotides for the Cut-alternatively spliced product, CASP, but not for CUX1. Therefore, we were forced to use a surrogate for CUX1 expression, and we chose a set of well-characterized targets that play a role in S-phase entry. Across all large-scale breast cancer gene expression datasets, CUX1 transcriptional targets that play a role in mitosis were found to cluster with established targets of CUX1 at the G1/S transition. Linking these two classes of genes produced a gene expression signature that is strongly associated with poor clinical outcome. We have thus identified a group of CUX1 targets that has predictive potential in cancer and that might be important to mediate its oncogenic activity. Not only was high expression of these genes found more frequently in breast tumor subtypes that exhibit a poor prognosis, like the basal-like and HER2+, but it also identified patients with poor outcome within the luminal A and luminal B subtypes (Fig. 4). The finding that these two sets of genes together have prognostic value could have important practical application in the clinic because the luminal subtypes represent nearly 50% of breast tumors, and there is an urgent need to identify which node-negative patients in this subtype would benefit from adjuvant therapy and which ones should be spared from the toxicities associated with these treatments. Moreover, because our gene signature includes many genes involved in mitotic processes and microtubule-based activity, it has potential to be a useful predictor of treatment response for chemotherapeutic regimens that include antimicrotubule agents. Indeed, our results using the MPS1 inhibitor clearly show that cells with centrosomal aberrations are intrinsically more sensitive than normal cells to spindle checkpoint inhibition (Fig. 2F), thus suggesting a therapeutic window for drug efficacy.
Experimental Procedures
Cell Culture.
Cell lines were generated by retroviral infection as described in SI Experimental Procedures. After blebbistatin treatment (100 μM for 12 h; Sigma) cells were allowed to recover for 4 h, during which the media was changed four times before live-cell imaging or s.c. injections. Where indicated, MPS1-IN-1 was added to the media during cell imaging, whereas 10 μM MG132 was added for 90 min and washed off during imaging. Population doubling was calculated using the formula N(t) = N(0) × ert. The proliferation rate (r) was calculated between days 0 and 4 for untreated cells, and between days 2 and 6 for blebbistatin-treated cells. Apoptosis assays were done using the Annexin V-EGFP Apoptosis Detection Kit (Genscript). Cell sorting was performed on a MoFlo cytometer (Dako), after staining nuclei with Hoechst 33342 (2 μg/mL, 1 h).
Live-Cell Imaging.
Time-lapse microscopy was performed on a Zeiss inverted microscope enclosed in a humidified chamber at 37 °C, in Leibovitz's media plus 10% FBS using 20× and 32× objectives. Frames were taken every 5 min. The duration of mitosis was measured from nuclear envelope breakdown until nuclei were visible in daughter cells.
Chromosome Spreads.
Cells were treated with 100 ng/mL colcemid for 2 h, trypsinized, washed with PBS, and swollen in 0.56% KCl for 8–12 min. Cells were centrifuged at 800 × g for 5 min, followed by two rounds of fixation in ice-cold Carnoy's fixative for 10 min at room temperature. Cells were dropped on glass slides, dried, and mounted with DAPI.
Subcutaneous Injections.
Cells (2 × 106 in 100 μL PBS) were injected contralaterally in 5-wk-old nude mice (CD1 nu/nu; Charles River Breeding Laboratories). Tumor volumes were measured approximately 30 d after injection.
Promoter Occupancy and Gene Expression Analysis.
Protocols for ChIP, ChIP-on-chip, cDNA preparation, and real-time PCR were described previously (12). Validation was done with primers encompassing a <300-bp region identified by ChIP-on-chip, with the QuantiTect SYBR Green PCR Kit (Qiagen), on a Rotor Gene 3000 real-time PCR machine (Corbett). For gene expression analysis, RNA was isolated using the RNeasy Mini kit (Qiagen), and cDNA was prepared using SuperScript II RNase H-reverse transcriptase kit (Invitrogen). Primer sequences are provided in Table S3. GSEA was performed as described elsewhere (22). The significance of enrichment was determined by performing 3,500 random gene set membership permutations.
Human Data Analysis.
Relative expression of 29 CUX1 targets (Fig. 4A) was examined in 12 publicly available gene expression datasets comprising a total of 2,481 patients with breast cancer (see SI Experimental Procedures for references). In each dataset, hierarchical clustering was performed using Euclidean distance and Ward's algorithm (Fig. S5). Kaplan-Meier survival analysis and the log–rank test were used to compare the patients in the “low” vs. the “high” classes in all five molecular subtypes and in all patients combined. The analysis was completed using R/Bioconductor (23). Because of the differences in survival characteristics of the datasets (overall survival time vs. time to relapse), the associated times were classified as time to “outcome.” The analysis of each individual dataset is shown in Fig. S6.
Supplementary Material
Acknowledgments
We thank Dr. M. Petronczki for sharing reagents; Dr. J. Gannon for anti-p21 and p53 antibodies; and Ginette Bérubé and Maria Drossos for expert technical assistance. L.S., C.V., and C.C. were supported by studentships from, respectively, the Canadian Institutes of Heath Research, the Fonds de la Recherché en Santé du Québec, and the Department of Defense Breast Cancer Research Program. This research was supported by Grant 019389 from the Canadian Cancer Society (to A.N.).
Footnotes
Conflict of interest statement: L.S., J.L., M.T.H., and A.N. are filing a patent regarding the gene expression signature.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1008403108/-/DCSupplemental.
References
- 1.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 2.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 3.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 5.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sansregret L, Nepveu A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Cadieux C, et al. Transgenic mice expressing the p75 CCAAT-displacement protein/Cut homeobox isoform develop a myeloproliferative disease-like myeloid leukemia. Cancer Res. 2006;66:9492–9501. doi: 10.1158/0008-5472.CAN-05-4230. [DOI] [PubMed] [Google Scholar]
- 9.Cadieux C, et al. Mouse mammary tumor virus p75 and p110 CUX1 transgenic mice develop mammary tumors of various histologic types. Cancer Res. 2009;69:7188–7197. doi: 10.1158/0008-5472.CAN-08-4899. [DOI] [PubMed] [Google Scholar]
- 10.Michl P, et al. CUTL1 is a target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 2005;7:521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 11.Sansregret L, et al. The p110 isoform of the CDP/Cux transcription factor accelerates entry into S phase. Mol Cell Biol. 2006;26:2441–2455. doi: 10.1128/MCB.26.6.2441-2455.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harada R, et al. Genome-wide location analysis and expression studies reveal a role for p110 CUX1 in the activation of DNA replication genes. Nucleic Acids Res. 2008;36:189–202. doi: 10.1093/nar/gkm970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santaguida M, et al. Phosphorylation of the CCAAT displacement protein (CDP)/Cux transcription factor by cyclin A-Cdk1 modulates its DNA binding activity in G(2) J Biol Chem. 2001;276:45780–45790. doi: 10.1074/jbc.M107978200. [DOI] [PubMed] [Google Scholar]
- 14.Owens RB, Smith HS, Hackett AJ. Epithelial cell cultures from normal glandular tissue of mice. J Natl Cancer Inst. 1974;53:261–269. doi: 10.1093/jnci/53.1.261. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 17.Goulet B, et al. Characterization of a tissue-specific CDP/Cux isoform, p75, activated in breast tumor cells. Cancer Res. 2002;62:6625–6633. [PubMed] [Google Scholar]
- 18.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.