Abstract
IFNs play a critical role in innate immunity against viral infections. Melanoma differentiation-associated protein 5 (MDA5), an RNA helicase, is a key component in activating the expression of type I IFNs in response to certain types of viral infection. MDA5 senses noncellular RNA and triggers the signaling cascade that leads to IFN production. Synthetic double-stranded RNAs are known activators of MDA5. Natural single-stranded RNAs have not been reported to activate MDA5, however. We have serendipitously identified a viral mRNA from parainfluenza virus 5 (PIV5) that activates IFN expression through MDA5. We provide evidence that the signaling pathway includes the antiviral enzyme RNase L. The L mRNA of PIV5 activated expression of IFN-β. We have mapped the RNA to a region of 430 nucleotides within the L mRNA of PIV5. Our results indicate that a viral mRNA, with 5′-cap and 3′-poly (A), can activate IFN expression through an RNase L-MDA5 pathway.
Keywords: RIG-I, RNA polymerase
IFNs play a critical role in innate immune responses to viral infections. Viruses trigger expression of IFN-β in infected cells, and IFN-β can lead to activation of IFN-α expression through phosphorylation of IFN regulatory factor 7 (1, 2). IFNs induce an antiviral state in cells that inhibits the spread of infection. MDA5 (melanoma differentiation-associated gene 5), an RNA helicase, plays an essential role in the activation of IFN expression (3). MDA5 is involved in the cytoplasmic sensing of infections by some RNA viruses (4). Recognition of RNA molecules generated during viral infections by MDA5 leads to activation of IFN-β promoter stimulator (IPS)-1, NF-κB, and IFN expression (5). How MDA5 differentiates between self and nonself RNA is unclear. It has been reported that stable, long, double-stranded (ds) RNA structures greater than 2 kb in size, presumably with 5′-triphosphates, generated during RNA virus infection (not typical of self RNA) may serve as a distinguishing factor for MDA5-specific recognition (6). Long, synthetic dsRNA polymers of poly(I):poly(C) are often used as a surrogate for the putative activator of MDA5 (7). A natural single-stranded (ss) RNA trigger for MDA5 has not been identified.
The role of MDA5 in regulating IFN expression was first reported in studies of parainfluenza virus 5 (PIV5), formerly known as simian virus 5 (3, 8). PIV5 is a prototypical paramyxovirus in a family of nonsegmented, negative-stranded RNA viruses that includes many important human and animal pathogens, including mumps virus, measles virus, Nipah virus, and respiratory syncytial virus (9). The viral RNA-dependent RNA polymerase, minimally consisting of the L protein and the P protein, transcribes the nucleocapsid protein (NP or N)-encapsidated viral genome RNA into 5′ capped and 3′ polyadenylated mRNAs (10). The V protein of PIV5, a component of PIV5 virions (∼350 molecules per virion) is a multifunctional protein with important roles in viral pathogenesis. The V protein C-terminal domain contains seven cysteine residues, resembling a zinc finger domain, and binds atomic zinc (11). A recombinant virus lacking the C terminus of the V protein of PIV5 (rPIV5VΔC) induces a higher level of IFN expression compared with WT virus, indicating that the V protein plays an essential role in blocking IFN production in virus-infected cells (12, 13). Andrejeva et al. (3) found that the V protein interacts with MDA5, resulting in a blockade of IFN-β expression. They also found that knocking down the expression of MDA5 reduces IFN expression induced by poly(I):poly(C), indicating that MDA5 plays an essential role in the induction of IFN expression by dsRNA. In this work, we investigated the activation of IFN by rPIV5VΔC infection and have identified a viral mRNA with 5′-cap as an activator of IFN expression through an MDA5-dependent pathway that includes RNase L.
Results
Region II of the L Gene Activates NF-κB Independent of AKT1.
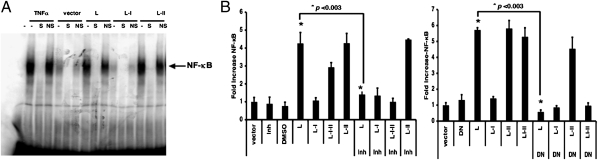
Previous work indicated that the portion of the L gene containing the conserved regions I and II (L-I-II) together is sufficient to activate NF-κB (14). Further deletion mutagenesis analysis of the L gene by EMSA showed that region II, which contains 144 amino acid residues, was sufficient for the activation of NF-κB (Fig. 1A). This finding, which was confirmed by a reporter gene assay (Fig. S1A), was somewhat surprising, given that the previous study showed that activation of NF-κB by the L gene requires AKT1 and region I (L-I), which binds to AKT1 (14). We reexamined the interaction between AKT1 and L-II and confirmed that L-II does not bind to AKT1 (Fig. S1B). Interestingly, an AKT1 inhibitor (AKTIV) and an AKT1 dominant-negative (DN) mutant had no effect on the activation of NF-κB by L-II (Fig. 1B), indicating that the L-II region activates NF-κB through an AKT1-independent mechanism.
Fig. 1.
Activation of NF-κB by region II of the L gene in an AKT-independent manner. (A) Detection of activation of NF-κB by L region-expressing plasmids using EMSA. Nuclear extracts from cells transfected with empty vector or plasmids encoding L, L-I, or L-II were prepared and incubated with 32P-labeled NF-κB probe and appropriate competitors, and then were resolved on a 6% polyacrylamide gel. Treatments were as follows: TNF-α, nuclear extracts from cells treated with 20 ng/mL of TNF-α for 3 h; NF-κB DNA primers labeled with 32P; S (specific competitor), unlabeled NF-κB probe (20-fold excess); NS (nonspecific competitor), unlabeled mutant NF-κB probe (20-fold excess). (B) Activation of NF-κB by the L-II region was independent of AKT1. (Left) A dual-luciferase assay, in which BSR-T7 cells were transfected with a plasmid encoding a firefly luciferase gene (F-Luc) under the control of NF-κB–responsive elements and a plasmid encoding PIV5 L, L-II, L-II mut, or L-I-II proteins, along with a plasmid encoding an R-Luc as an indicator of transfection efficiency, was performed in the presence of an AKTIV inhibitor (Inh) (0.5 μM; Calbiochem) or vehicle (DMSO). Ratios of F-Luc to R-Luc serve as an indicator of reporter gene activity. These ratios were normalized to the activity of the vector alone. All transfections were carried out in replicates of four. Error bars represent SD. All P values were calculated using the paired t test and are shown. (Right) Inhibition of L-activated NF-κB activity by AKT1 DN.
RNA of Region II of the L Gene Activates NF-κB.
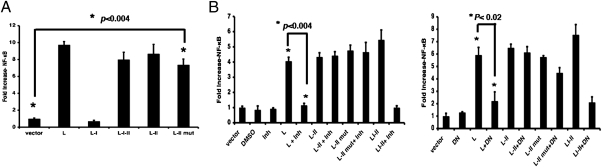
Given that RNA can activate NF-κB (15), we speculated that the RNA sequence within the L-II region, and not the amino acid residues encoded by the L-II region, might be responsible for NF-κB activation. We mutated the start codon of L-II into a stop codon (L-II mut) and found that the L-II mut did not express protein, although the expression levels of RNAs were similar in L-II and L-II mut (Fig. S2 A, B, and C). Interestingly, this mRNA generated from the plasmid pCAGGS, which is under the control of a pol-II promoter (16), activated NF-κB (Fig. 2A), as confirmed by EMSA (Fig. S2D), suggesting that an mRNA of viral origin can activate NF-κB. Consistent with previous observations (Fig. 1B), this activation was not inhibited by AKTIV or an AKT1 DN mutant (Fig. 2B).
Fig. 2.
L-II RNA activates NF-κB. (A) Activation of NF-κB by the L-II mut. The L-II mutant contains a stop codon in place of the start codon of L-II. A reporter gene assay was performed as described in Fig.1A. (B) Activation of NF-κB by L-II RNA is independent of AKT1. A dual-luciferase experiment was performed using AKT1 inhibitor (Inh) (Left) or AKT1 DN (Right), along with L, L-II, L-II mut, or L-I-II plasmids as described in Fig. 1B. All transfections were carried out in replicates of four. Error bars indicate SD.
RNA of Region II of the L Gene Activates IFN-β Expression.
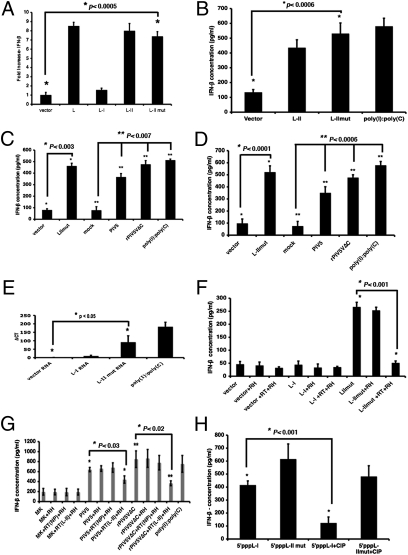
Because activation of NF-κB can lead to activation of IFN expression, we examined the ability of this RNA to activate IFN expression using a plasmid containing a reporter gene (F-Luc) under the control of an IFN-β promoter. As shown in Fig. 3A, the plasmid expressing the L-II mut RNA activated IFN-β promoter-driven reporter gene expression, suggesting that the RNA activates the IFN promoter. We evaluated the amount of IFN-β in the medium of cells transfected with plasmids encoding L-II or L-II mut mRNAs by ELISA. The plasmid encoding the L-II mut induced IFN-β production equivalent to that seen with the positive control, poly(I):poly(C) (Fig. 3B). To confirm that the RNA, not plasmid DNA, activated IFN-β expression, the RNAs from transfected cells were purified and transfected into fresh cells, and levels of IFN-β in the medium of the RNA-transfected cells were measured after 1 d. As shown in Fig. 3C, the RNAs from the L-II mut-transfected cells produced a higher level of IFN-β compared with the RNAs from vector-transfected cells. Interestingly, RNAs from both WT and rPIV5VΔC-infected cells induced IFN-β expression, indicating that RNAs capable of activating IFN-β expression exist in virus-infected cells as well (Fig. 3C).
Fig. 3.
L-II RNA activates IFN-β expression. (A) Activation of IFN-β promoter by L-II mut. A dual-luciferase assay was performed as described in Fig. 1A. A plasmid containing F-Luc under control of an IFN-β promoter was used in place of the NF-κB–containing promoter described in Fig. 1A. (B) Induction of IFN-β production by L-II RNA. Plasmids encoding L-II or L-II mut were transfected into 293T cells, and the amount of IFN-β in the media was measured by ELISA at 1 d posttransfection. Each graph showing concentrations of IFN-β using ELISA is the average of three independent experiments. Error bars represent SD. (C) IFN-β production induced by purified RNA. Vero cells were transfected with empty vector or plasmids containing L-II mut, or were infected with WT PIV5 or rPIV5VΔC, mock-infected, or transfected with poly(I):poly(C). Total RNAs were purified from transfected or infected cells. The purifed RNAs were then transfected into 293T cells, and concentrations of IFN-β were measured in the media using ELISA after 1 d. (D) Induction of IFN-β by purified mRNA. Vero cells were transfected with empty vector or plasmid containing L-II mut, infected with WT PIV5 or rPIV5VΔC, or mock-infected. mRNAs were purified and transfected into 293T cells, and IFN-β concentrations after 1 d were measured using ELISA. (E) Induction of IFN-β by L-II in the presence of CHX. The 293T cells in six-well plates were transfected with 1 μg of RNA or 250 ng of poly(I):poly(C) and then incubated with CHX (20 μg/mL) for 16 h. The total RNAs were purified and subjected to RT, followed by real-time PCR analysis. ΔCT was calculated using actin from each sample as a control. (F) Lack of production of IFN-β in the absence of L-II mRNA. The purified L-II RNA was reverse-transcribed using an L-II sequence-specific primer and RT. The product and/or purified L-II RNA were treated or untreated with RNase H (RH). The purified products were then transfected into 293T cells, and IFN-β concentrations after 1 d were determined by ELISA. (G) Lack of production of IFN-β in the absence of L mRNA. The same experiment as in Fig. 3F was performed using RNAs purified from infected cells. RT(NP), RT using NP-specific primer; RT(L-II), RT using L-specific primer. The graph shows the average of three independent experiments. Error bars represent SD. (H) Induction of IFN-β production by in vitro transcribed L-II RNA. The L-I and L-II RNA were synthesized in vitro using the Riboprobe in vitro transcription system (Promega). The RNA transcripts were treated or untreated with CIP to remove 5′-triphosphate and then transfected into 293T cells. At 1 d posttransfection, IFN-β concentrations in the media were measured by ELISA.
To further confirm that it was the mRNA that activated IFN-β expression, mRNAs from cells transfected with plasmids encoding L-II mut were purified and then transfected into fresh cells. The amounts of IFN-β in the medium of cells transfected with mRNAs from cells transfected with a plasmid expressing L-II mut mRNA were similar to that of those stimulated with poly(I):poly(C) (Fig. 3D), indicating that the mRNAs activate expression of IFN-β.
The finding that L-II RNA activated IFN-β transcription in the presence of the protein synthesis inhibitor cyclohexamide (CHX) (Fig. 3E) suggests that the activation of IFN-β does not require new protein synthesis, and that L-II RNA activates IFN-β expression at the mRNA level. To validate the role of L-II mRNA in activating IFN-β expression, the mRNA was removed from the total RNA purified from the L-II plasmid-transfected cells through a reverse-transcription (RT) reaction using a L-II–specific primer, followed by treatment of the RT products with RNase H, which digests RNA in an RNA–DNA hybrid. This L-II mRNA-depleted mRNA did not stimulate production of IFN-β, indicating that L-II mRNA is essential for the activation of IFN-β expression (Fig. 3F). A similar experiment was carried out using RNA purified from virus-infected cells (Fig. 3G). RT using a L-specific primer, but not an NP-specific primer, reduced the production of IFN-β, indicating that the L mRNA in viral infections is responsible for activating the expression of IFN-β. Furthermore, a plasmid expressing a mutant L gene, with two stop codons placed downstream in-frame from its start codon, induced activation of NF-κB and IFN-β, confirming that the L mRNA is capable of activating IFN-β expression (Fig. S3). To determine whether the L-II RNA is capable of activating IFN-β expression by itself, we generated L-II RNA by in vitro transcription using T7 RNA polymerase (Fig. S4). RNAs from both the L-I and L-II regions activated expression of IFN-β (Fig. 3H), as expected given that T7 RNA polymerase transcripts contain 5′-triphosphate, a known activator of IFN through RIG-I. Interestingly, although removing 5′-triphosphate with calf intestinal phosphatase (CIP) reduced activation of IFN-β by L-I RNA, this had a minimal impact on the effect of L-II RNA, confirming that L-II RNA can activate IFN-β expression in its own right (Fig. 3H).
RNA of Region II of the L Gene Activates IFN-β Expression Through an MDA5-Dependent Pathway.
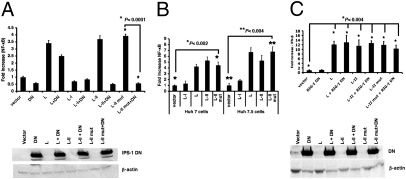
Two known cytoplasmic proteins sense noncellular RNA: RIG-I and MDA5 (17). Both of these proteins activate IFN expression through IPS-1 protein (18, 19). Expression of a DN mutant of IPS-1 was found to block the activation of NF-κB by plasmids expressing L-II RNA (Fig. 4A), implying that RIG-I and/or MDA5 may play a role in L-II–induced NF-κB activation. To examine whether these two proteins are involved in the activation of NF-κB by L-II RNA, we transfected plasmids encoding the L-II RNA into Huh7 or Huh7.5 cells, the latter of which have a defective RIG-I gene (20). We found no difference in NF-κB activation between the two cell lines, suggesting that RIG-I does not play a role in NF-κB signaling in response to L-II (Fig. 4B). Furthermore, RIG-I DN demonstrated no effect on NF-κB activation (Fig. 4C), confirming that RIG-I does not play a role in L gene-induced activation of NF-κB.
Fig. 4.
Role of RIG-I in the activation of NF-κB and IFN-β by L-II RNA. (A) Effect of IPS-1 DN on activation of NF-κB by L RNA. A dual-luciferase experiment was performed as described in Fig. 1A, using IPS-1 DN with a Flag tag (500 ng/μL). Immunoblot analysis was performed to examine the expression of IPS-1 DN using anti-Flag and anti–β-actin antibodies. All transfections were carried out in replicates of four. Error bars represent SD. (B) Activation of NF-κB by L-II RNA was independent of RIG-I. At 18–20 h after transfection, a dual-luciferase assay was performed using lysate from Huh7 or Huh7.5 cells (RIG-I defective due to a T to I mutation at amino acid residue 55) transfected with vector, L, L-I, L-II, or L-II mut. (C) Effect of RIG-I DN on activation of NF-κB by L-II RNA. A reporter gene assay was performed using a plasmid expressing RIG-I DN with a Flag tag (500 ng/μL) along with the plasmids indicated. Immunoblot analysis was performed to examine the expression of RIG-I DN using anti-Flag and anti–β-actin antibodies. All transfections were carried out in replicates of four. Error bars represent SD.
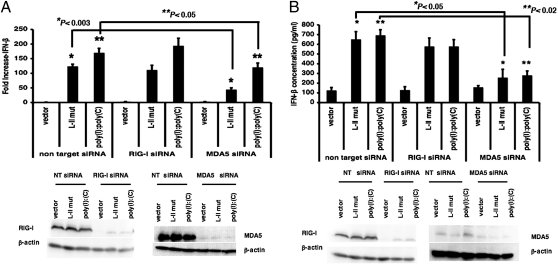
To investigate the role of MDA5, we reduced MDA5 expression using siRNA and found that this resulted in reduced NF-κB activation (Fig. S5), indicating that MDA5 plays a critical role in NF-κB activation. To further characterize the roles of RIG-I and MDA5 in activating IFN-β expression, we investigated the effects of siRNA targeting RIG-I or MDA5 on IFN-β promoter activation. We found that siRNA targeting MDA5, but not RIG-I, reduced IFN-β promoter activation by the plasmid expressing L-II mut RNA (Fig. 5A). This was further confirmed by examining IFN-β production by cells transfected with plasmids expressing L-II RNA after treatment with siRNA targeting RIG-I or MDA5. As shown in Fig. 5B, MDA5 siRNA reduced IFN-β production after plasmid transfection, whereas RIG-I siRNA had no significant effect, indicating that MDA5, but not RIG-I, plays a role in activating IFN-β expression by L-II RNA.
Fig. 5.
MDA5 plays a critical role in the activation of IFN-β by viral mRNA. (A) The roles of RIG-I and MDA5 in the activation of the IFN-β promoter by viral mRNA. The 293T cells were transfected with siRNA targeting RIG-I or MDA5 or with NT siRNA. At 48 h after transfection of siRNA, the cells were transfected with vector, L-II mut, or poly(I):poly(C), along with the luciferase reporter plasmids. Luciferase activity was measured at 18–20 h posttransfection. (B) The roles of RIG-I and MDA5 in induction of IFN-β production by viral mRNA. siRNA transfection was performed as described in Fig. 5A in 293T cells, and at 48 h after transfection with siRNA, the cells were transfected with vector, L-II mut, or poly(I):poly(C). After 18–20 h, the amount of IFN-β in the medium was measured by ELISA. Expression levels of RIG-I, MDA5, and β-actin were examined by immunoblot analysis.
RNA of Region II of the L Gene Activates IFN-β Expression Through an RNase L-Dependent Pathway.
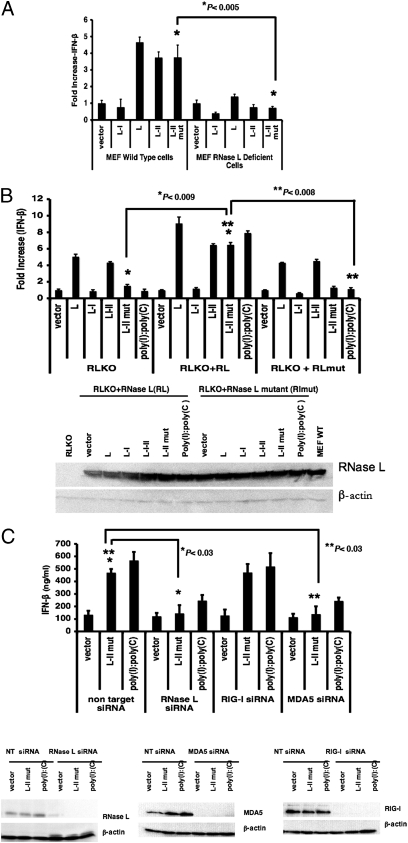
RNase L has been reported to play an important role in IFN expression (21). To examine the role of RNase L, we used mouse embryonic fibroblast (MEF) cells from mice expressing RNase L (WT) or deficient in RNase L (RLKO). We found that plasmids expressing L-II RNA activated the NF-κB and IFN-β promoters in WT MEFs, but not in RLKO MEFs (Fig. 6A and Fig. S6), suggesting that RNase L plays an important role in the activation of IFN-β by this viral mRNA. To confirm these results, we transfected WT RNase L or a defective RNase L lacking ribonuclease activity into RLKO MEFs. We found that the plasmid expressing L-II RNA activated IFN-β in MEFs in the presence of WT RNase L, but not in the presence of defective RNase L (Fig. 6B). To confirm the role of RNase L in the activation of IFN-β expression by L-II RNA, we examined the effects of siRNA targeting RNaseL. As shown in Fig. 6C, siRNA targeting RNaseL reduced the production of IFN-β.
Fig. 6.
RNase L plays a critical role in the activation of NF-κB and IFN-β by viral mRNA. (A) The role of RNase L in activating the IFN-β promoter. A dual-luciferase assay for IFN-β promoter activation was performed as described in Fig. 3A using WT or RLKO MEFs. (B) Restoration of IFN-β activation in RLKO MEFs. IFN-β activation after complementation with RNase L cDNA in RLKO MEFs was examined by a dual-luciferase experiment. RLKO MEFs were transiently transfected with RNase L cDNA or inactive RNase L mutant (R667A) cDNA. At 18 h after transfection, the cells were transfected with 1 μg/μL of vector, L, L-I, L-I-II, or L-II mut plasmids, along with reporter plasmids. At 1 d posttransfection, the luciferase assay was performed. RNase L and β-actin levels were measured by immunoblot analysis. All transfections were carried out in replicates of four. Error bars represent SD. (C) The role of RNase L in activating IFN-β expression. Then 293T cells were transfected with siRNA targeting RIG-I, MDA5, RNase L, or control siRNA. IFN-β production in response to vector, L-II mut, or poly(I):poly(C) was measured by ELISA. The expression of RIG-I, MDA5, and RNase L was examined by immunoblot analysis, with β-actin as a loading control.
Discussion
RIG-I and MDA5 are two well-known sensors of virus infection for induction of IFN expression, and 5′-triphosphate is an activator for RIG-I. Viral genomic RNA of negative-stranded viruses, such as influenza virus and Sendai virus, which have a 5′-triphosphate, can activate IFN expression via a RIG-I–dependent pathway (17, 22–24). A natural activator for MDA5 has not yet been reported, however. Because MDA5 is required for induction of IFN by the positive-stranded RNA virus encephalomyocarditis virus (EMCV), whose 5′ RNA is covalently linked to VPg, a polypeptide, thereby avoiding detection by RIG-I, MDA5 is thought to recognize a viral RNA different from the 5′-triphosphate RNA that is recognized by RIG-I (17). Because dsRNA, such as poly(I):Poly(C), can activate IFN expression through MDA5, it is thought that the activator for MDA5 is a dsRNA. Unlike the negative-stranded influenza virus and Sendai virus, PIV5, a negative-stranded RNA virus, was found to activate IFN-β expression through a viral mRNA-induced, RNase L/MDA5-dependent pathway, consistent with previous reports indicating that PIV5 and some paramyxoviruses activate IFN expression via MDA5 (7, 25). PIV5 replicates entirely in the cytoplasm (9), and viral mRNA can be readily accessed by RNase L/MDA5 proteins. Because dsRNA is a known trigger of MDA5, it is speculated that dsRNA generated during viral genome replication might activate MDA5. Viral genomic RNA of paramyxovirus is tightly encapsidated by nucleocapsid protein, resistant to RNase digestion, and inaccessible to other host proteins, such as MDA5 (9). Thus, it is not surprising that no dsRNA was detected in paramyxovirus infection using dsRNA-recognizing antibody (26). In addition, because the nascent genome RNA is encapsidated, the ds region of the viral RNA genome within the replication/transcription complex is small (>2 kb is thought to be an MDA5 trigger). Pichlmair et al. (27) reported that long dsRNA was not sufficient to activate IFN expression through a MDA5-dependent pathway, and that higher-order RNA structures containing both dsRNA and ssRNA are activators of MDA5-dependent IFN expression. The nature of this higher-order RNA structure remains unidentified. Our identification of a viral mRNA that activates IFN-β expression through MDA5 is consistent with those results.
We propose that RNase L plays a role in sensing viral RNA by MDA5, leading to activation of IFN-β. RNase L is an antiviral protein activated by 2′-5′ oligoadenylate (2-5A). This 2-5A is produced by 2′,5′ oligoadenylate synthetase, the expression of which is induced by IFN (28). Activated RNase L cleaves viral mRNA and prevents viral replication (28). RNase L has recently been reported to amplify MDA5-dependent IFN expression through the cleavage of cellular RNA (21). We speculate that RNase L recognizes the viral mRNA and processes it into an activator of MDA5, leading to expression of IFN, given that siRNAs targeting RNase L and MDA5 were seen to decrease the activation of IFN-β expression by viral mRNA. Alternatively, it is possible that viral mRNA activates MDA5-dependent IFN expression, and that RNase L plays a role in amplifying IFN production.
WT PIV5 is known to induce low levels of IFN expression, and rPIV5VΔC infection produces high levels of IFN (12, 13). Interestingly, RNAs purified from cells infected with PIV5 and rPIV5VΔC induced high expression of IFN-β. This finding is consistent with previous reports that the V protein of PIV5 can block induction of IFN induced by PIV5 infection (12, 13).
We have mapped the L RNA sequence to a 432-nt-long region. Preliminary analysis using RNA structure prediction programs indicates potential secondary structures within the sequence. Further detailed structure and function analysis of the sequence will define the sequence element and structure(s) within the viral mRNA that activate IFN-β expression through RNase L and MDA5. This sequence and structure(s) may serve as a prototype for other natural triggers of MDA5. This work has not only identified a trigger for MDA5, but also may lead to the discovery of small RNA molecules capable of activating IFN-β expression, which might be useful in antiviral therapy.
Materials and Methods
Cells and Plasmids.
BSR-T7, HeLa, Vero, 293T, Huh 7.0, Huh 7.5, and MEF cells were cultured as described previously (14, 20, 21, 29). Plasmids encoding WT RNase L, RNase L mutant (R667A), L-I, L-I-II, PIV5 L, AKT1 with a Flag tag, the DN mutant of AKT (pMT2-AH-AKT1, which contains residues 1–147 of AKT with a Myc antigen tag), the RIG-I DN (Flag-tagged RIG-I consisting of residues 218–925), the IPS-1 DN (with deletion of the CARD domain), phTK-RL, pNF-kB-TATA-F-Luc, and a plasmid containing F-Luc under control of an IFN-β promoter have been described previously (13, 14, 18, 19, 21, 30–33). Plasmids L-II (consisting of domain II) and L-II mut (consisting of a stop codon instead of a start codon in an L-II background) with an antigenic tag (HA) in expression vector pCAGGS, were generated using standard molecular cloning techniques. Plasmids were prepared using the Maxi Prep Kit (Qiagen). Endotoxin concentration was measured using the LAL Endotoxin Assay Kit (GenScript). The endotoxin concentrations of all plasmids were <0.1 EU/μg of DNA.
EMSA.
BSR-T7 cells were transfected with a vector or a plasmid encoding L, LI, or L-II, and nuclear extracts were prepared using a nuclear extraction kit (Marligen Biosciences). Nuclear extracts from TNF-α–treated BSR-T7 cells were used as a positive control. The cells were treated with 20 ng/mL of TNF-α for 2 h. EMSA was performed as described previously (14).
Dual Luciferase Assay.
Cells were transfected in 24-well tissue culture plates at 80–90% confluency. Transfection was performed using Plus and Lipofectamine (Invitrogen) for BSR-T7 cells and Lipofectamine 2000 (Invitrogen) for 293T and HeLa cells. Vector plasmid (pCAGGS) was used to maintain a constant total plasmid DNA per well. Plasmid amounts were 2.5 ng of phRL-TK, 60 ng of pNF-κB-TATA-F-Luc, and 240 ng of pIFN-Luc. A range of concentrations up to 1,500 ng of plasmids encoding L, L-I, L-II, L-I-II, and L-II mut were used. The AKT DN, pMT2-AH-AKT, was used at 800 ng, and plasmid C-RIG (RIG-I DN) and IPS-1 DN were used at 500 ng. At 18–24 h after transfection, cells were lysed in 100 μL of passive lysis buffer (Promega) for 30–45 min. Then 20 μL of lysate from each well was used for a dual-luciferase assay with a Luminometer (Promega) following the manufacturer's protocol. To examine the effect of AKT inhibitor on L-activated NF-κB, 0.5 μM of AKTIV was added to BSR-T7 cells at 4 h after transfection.
Coimmunoprecipitation.
To examine the interaction between L-II and AKT1, coimmunoprecipitation was performed as described previously (14). In brief, BSR-T7 cells transfected with a plasmid encoding AKT1 were immunoprecipitated with anti-AKT1 antibody, and then the precipitated AKT1 was used for further immunoprecipitation with L-I and L-II. L-I and L-II with an HA tag were synthesized in vitro using the TNT coupled transcription/translation system (Promega) with 35S[methionine/cysteine] labeling, as described previously (14).
Immunoprecipitation and Immunoblot Analysis.
Cells transfected with plasmids encoding L-I, LII, LI-II, LII mut with an HA tag, or vector were metabolically labeled with 35SMet and 35SCys for 3 h at 24 h posttransfection. The cell lysates were precipitated with anti-HA antibody. The precipitated proteins were resolved by 15% SDS/PAGE and visualized using a Storm PhosphorImager (Molecular Dynamics). For immunoblotting, lysates from the luciferase assays were diluted 1:1 with protein lysis buffer [2% SDS, 62.5 mM Tris-HCl (pH 6.8), 2% DTT] and sonicated. Up to 100 μL of the lysate was resolved in 10% SDS/PAGE, and immunoblot analysis was performed using respective antibodies (3).
RNA Purification and Transfection.
The 293T or Vero cells were transfected with empty vector or a plasmid encoding L, L-I, L-II, LI-II, or L-II mut. HeLa or Vero cells were infected with WT PIV5 or rPIV5VΔC or mock-infected. At 18–20 h after transfection or infection, total RNA was isolated using a Qiagen RNeasy Kit or mRNA was isolated using a Qiagen Oligotex Direct mRNA Purification Kit, following the manufacturer's instructions. The purified total RNA (1 μg/μL per well of a 24-well plate) or mRNA (200 ng/μL per well of a 24-well plate) was transfected into 293T cells using Lipofectamine 2000. Poly(I):poly(C) (500 ng/μL per well of a 24-well plate) was used as a positive control. At 1 d posttransfection, IFN-β production was determined using a human IFN-β ELISA kit (PBL InterferonSource), following the manufacturer's instructions.
Northern Blot Analysis.
The RNA samples purified from BSR-T7 cells that were transfected with empty vector (pCAGGS) or with a plasmid encoding L-I, L-II, or L-II mut for 18 h were electrophoresed on a 1.2% agarose gel in the presence of 0.44 M formaldehyde, transferred to a positively charged nylon membrane (Roche Diagnostics), fixed by UV cross-linking, and analyzed by hybridization with digoxigenin (DIG)-labeled RNA probes that were generated by in vitro transcription using the DIG Northern Starter kit (Roche Applied Sciences). The hybridized probes were detected with anti-DIG AP Fab fragments and were visualized using CDP-Star (Roche) chemiluminescence substrate on x-ray films. The DNA templates for generating DIG-RNA probes were prepared by PCR with a gene-specific sense oligomer and an antisense oligomer with a T7 RNA polymerase promoter sequence. The amplified PCR fragments were purified using a PCR purification column and gel purification kit (GenScript). A DIG-labeled RNA molecular weight marker (Roche) was used to indicate the size of RNA.
RNase H Treatment.
The purified RNA from L-II–transfected cells or infected cells was used for an RT reaction using an L-II–specific primer or an NP-specific primer. The RT products were treated with RNase H and purified using a Qiagen RNeasy column. The treated or untreated RT products or L-II RNA were transfected into 293T cells, and the concentration of IFN-β in the medium was measured by ELISA at 1 d posttransfection.
In Vitro RNA Transcription.
DNA containing region I or II of the L gene was amplified by PCR with a sequence-specific sense primer containing a T7 polymerase promoter sequence and an antisense primer, using plasmid containing L-I or L-II as a template. The L-I or L-II RNA fragments were synthesized in vitro using the Riboprobe In Vitro Transcription System (Promega). The synthesized fragments were treated with DNase I to remove the DNA template and were then purified using an RNeasy column. The in vitro synthesized RNA fragments were treated with CIP for 2 h and then purified. The purified CIP-treated in vitro RNA transcripts (200 ng) were transfected into 293T cells using Lipofectamine 2000. IFN-β production was measured with a human IFN-β ELISA kit (PBL InterferonSource) at 1 d posttransfection.
siRNA.
The siRNA experiments were performed as described previously (14, 31). Cells in 24-well plates at 30–50% confluency were transfected with 100 nM of siRNA purchased from Dharmacon [nontarget (NT) siRNA pool, MDA5 siRNA] and Santa Cruz Biotechnology (RNaseL, RIG-I siRNA) with the use of Oligofectamine (Invitrogen). At 48 h after siRNA transfection, the cells were transfected with empty vector; with plasmids expressing L, LII, or LII mut (1 μg/μL); or poly(I):poly(C) (500 ng/mL) using Lipofectamine 2000, along with phRL-TK and pNF-κB-TATA-F-Luc or pIFN-Luc, as described previously. Dual-luciferase assays and immunoblot analyses were performed at 1 d posttransfection.
ELISA for IFN-β.
Medium was collected and centrifuged to remove cell debris. Then 50 μL of the cleared medium or the IFN-β standard were used in duplicate for detection of IFN-β using a human IFN-β ELISA Kkit (PBL IFN Source), following the manufacturer's instructions.
Real-Time PCR.
The 293T cells in six-well plates were transfected with 1 μg of purified RNA (vector, L-I, or L-II mut) or poly(I):Poly(C) (250 ng) in Opti-MEM (Invitrogen) using Lipofectamine 2000 for 4 h. After transfection, the medium was changed to complete medium (10% FBS, 1% penicillin-streptomycin, and DMEM) with DMSO or CHX (20 μg/mL). After 16 h of incubation, the total RNA was isolated using the RNeasy Mini Kit. RT was performed with 11 μL of total RNA for each sample, using SuperScript III reverse transcriptase (Invitrogen) with oligo (dT)15 according to the manufacturer's protocol. The cDNA (4 μL of 1:20 diluted cDNA) from each sample was used for a real-time PCR on a Step One Plus Real-Time PCR System (Applied Biosystems) using Taqman Universal PCR Master Mix (Applied Biosystems) and Taqman Gene Expression 1 Assays (Applied Biosystems) for the IFN-β gene with FAM dye and for the β-actin gene with VIC dye. Results were analyzed to obtain Ct values. Relative levels of IFN mRNA and β-actin mRNA were determined by calculating ΔCt values. Each sample was run in three replicates.
Supplementary Material
Acknowledgments
We thank Dr. Richard Randall for providing the IFN-β Luc reporter plasmid; Dr. Craig Cameron for providing the Huh7 and Huh7.5 cells; Dr. Michael Teng and Kim Teng for helping with the Northern blot analysis and providing the IPS-1 and RIG-I plasmids; members of the B.H. laboratory for providing comments, suggestions, and technical help; and Dr. Kaori Sakamoto for carefully reading the manuscript. This work was supported by National Institutes of Health Grants K02 065795 and R56 AI081816 (to B.H.), National Institutes of Health/National Cancer Institute Grant CA044059 (to R.H.S.), and the Georgia Research Alliance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012409108/-/DCSupplemental.
References
- 1.Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato M, et al. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 6.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatziandreou N, et al. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5) J Gen Virol. 2004;85:3007–3016. doi: 10.1099/vir.0.80200-0. [DOI] [PubMed] [Google Scholar]
- 9.Lamb RA, Kolakofsky D. Paramyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 4th Ed. Philadelphia: Lippincott, Williams & Wilkins; 2001. [Google Scholar]
- 10.Emerson SU, Yu Y-H. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paterson RG, Leser GP, Shaughnessy MA, Lamb RA. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology. 1995;208:121–131. doi: 10.1006/viro.1995.1135. [DOI] [PubMed] [Google Scholar]
- 12.He B, et al. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: The multifunctional V protein blocks both interferon-β induction and interferon signaling. Virology. 2002;303:15–32. doi: 10.1006/viro.2002.1738. [DOI] [PubMed] [Google Scholar]
- 13.Poole E, He B, Lamb RA, Randall RE, Goodbourn S. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-β. Virology. 2002;303:33–46. doi: 10.1006/viro.2002.1737. [DOI] [PubMed] [Google Scholar]
- 14.Luthra P, Sun D, Wolfgang M, He B. AKT1-dependent activation of NF-κB by the L protein of parainfluenza virus 5. J Virol. 2008;82:10887–10895. doi: 10.1128/JVI.00806-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 16.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 17.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 18.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF-3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA–induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 20.Sumpter R, Jr, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichlmair A, et al. RIG-I–mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 23.Loo YM, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Yount JS, Gitlin L, Moran TM, López CB. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J Immunol. 2008;180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 26.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pichlmair A, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchholz UJ, Finke S, Conzelmann KK. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling Z, Tran KC, Teng MN. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J Virol. 2009;83:3734–3742. doi: 10.1128/JVI.02434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun M, et al. Akt plays a critical role in replication of nonsegmented negative-stranded RNA viruses. J Virol. 2008;82:105–114. doi: 10.1128/JVI.01520-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun M, et al. Conserved cysteine-rich domain of paramyxovirus simian virus 5 V protein plays an important role in blocking apoptosis. J Virol. 2004;78:5068–5078. doi: 10.1128/JVI.78.10.5068-5078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y, et al. Inhibition of interleukin-6 expression by the V protein of parainfluenza virus 5. Virology. 2007;368:262–272. doi: 10.1016/j.virol.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.