Abstract
EGFR overexpression is associated with resistance to chemotherapy and radiotherapy. EGFR modulates DNA repair after radiation-induced damage through association with the catalytic subunit of DNA protein kinase (DNA-PKcs). We investigated the role of EGFR nuclear import and its association with DNAPKcs on DNA repair after exposure to cisplatin or ionizing radiation (IR). The model system was based on EGFR-null murine NIH3T3 fibroblasts where EGFR expression was restored with isoforms that were wild-type, derived from human cancers (L858R, EGFRvIII), or mutated in the nuclear localization signal (NLS) sequence. In cells expressing wtEGFR or EGFRvIII there was complete unhooking of cisplatin-induced interstrand crosslinks and repair of IR-induced strand breaks. In contrast, cells expressing L858R or NLS mutations, showed reduced unhooking of interstrand crosslinks and repair of strand breaks. Immunoprecipitation demonstrated wtEGFR and EGFRvIII binding to DNA-PKcs, increasing 2-fold 18 hours after cisplatin. Confocal microscopy and proximity ligation assay showed this interaction in the cytoplasm and nucleus was associated with increased DNA-PK activity. Cells expressing the EGFR L858R mutation, which has constitutive kinase activity, exhibited reduced DNA repair without nuclear localization. EGFR-NLS mutants showed impaired nuclear localization and DNA-PKcs association with reduced DNA repair and DNA-PK kinase activity. In summary, EGFR nuclear localization was required for modulation of cisplatin and IR-induced repair of DNA damage, and EGFR-DNAPKcs binding was induced by cisplatin or IR, but not by EGFR nuclear translocation per se. Our findings show how EGFR subcellular distribution can modulate DNA repair kinetics, with implications for design of EGFR-targeted combinational therapies.
INTRODUCTION
The epidermal growth factor receptor (EGFR) promotes the activation of survival signalling pathways including RAS/MAPK, PI3/AKT, JAK/STAT (1, 2). Increased EGFR activation and overexpression is strongly associated with tumorigenesis and cancer progression (3). EGFR is an important target for cancer therapies including antibodies disrupting ligand/receptor interactions such as cetuximab (4, 5), and small molecules inhibiting EGFR kinase activity including gefitinib (6, 7) and erlotinib (8). There has been extensive investigation of the mechanisms by which EGFR inhibition modulates the activity of chemotherapy and radiation (3). Combinations of the monoclonal antibody cetuximab with cisplatin or radiation have been useful clinically in the treatment of head and neck and colon cancer (9, 10). In contrast, despite effects in vitro (11), only small benefits have been obtained combining small molecules inhibiting EGFR with conventional treatment. Several studies have demonstrated association of EGFR with the catalytic subunit of DNA-dependent protein kinase (DNAPKcs), a central component of the non-homologous end joining (NHEJ) pathway involved in the repair of DNA strand breaks (12, 13).
Recently, it has been reported that a fraction of intracellular EGFR is located within the nucleus where it may activate transcription of genes associated with cell proliferation and the nitric oxide pathway, including cyclin D1, iNOS, c-myb and COX-2 (14-17). Evidence for the expression and activity of nuclear EGFR has been found using a variety of techniques including fractional immunoblotting, confocal microscopy, electron microscopy, reporter assays and chromatin immunoprecipitation. Nuclear EGFR has been shown to correlate with worse prognosis in a variety of malignancies including breast, head and neck and ovarian cancer (16, 18). The intracellular localization of EGFR may therefore have profound effects on response both to chemotherapy and to novel therapies inhibiting the EGFR pathway.
EGFR cellular distribution is dictated by several regulatory motifs within the juxtamembrane domain (19). Two basolateral signals control EGFR re-sorting to the transmembrane (20), whereas the lysosomal signal (accessible following EGF receptor activation) determines EGFR degradation (21). The nuclear localization sequence (NLS), comprising 13 amino acids 645-657 (RRRHIVRKLLRR) (22), has a dual role. It allows nuclear translocation via sequence recognition and binding to Importin β (23), and mediates EGFR allosteric conformational change and dimer stabilisation, which are indispensable for the receptor activation (24, 25).
Studies on the somatically acquired, constitutively active EGFR mutant L858R, found in certain non-small cell lung cancers, have shown impaired nuclear localization and DNAPKcs binding (26). This suggests that EGFR activation and nuclear translocation are related and that nuclear localization may modulate DNA repair.
Nuclear translocation of EGFR following ionising radiation has been shown to result in increased repair of DNA stand breaks (12). The effects of nuclear translocation on repair of chemotherapy-induced DNA damage are less clear. We therefore investigated the significance of nuclear localization for the repair of cisplatin and ionizing radiation (IR)-induced DNA damage, using EGFR constructs with mutations in the NLS, as well as mutations found in human cancers (EGFRvIII, L858R). Cells expressing EGFR with mutations impairing nuclear transport demonstrated reduced repair of DNA stand breaks following IR, and reduced unhooking of interstrand crosslinks following treatment with cisplatin, as compared with cells expressing wild type EGFR. Immunoprecipitation experiments confirmed association of EGFR with DNAPKcs following treatment with cisplatin or IR. Confocal microscopy confirmed that cells with mutations in the NLS failed to translocate to the nucleus following IR and cisplatin treatment. These findings confirm the importance of nuclear translocation of EGFR in mediating effects on DNA repair and emphasise the significance of subcellular EGFR expression in determining responses to therapy.
Materials and Methods
Materials
Cisplatin (DBL 1mg/ml) was obtained from Mayne Pharma PLC. EGF was obtained from Sigma Aldrich.
Irradiation condition
Cells were plated at a concentration of 1 × 105/ml. Following 48 hours transfection, cells were serum starved for 24 hours and irradiated with 4Gy using the A.G.O. HS 321kV X-ray system.
Cell Lines and Culture Conditions
NIH3T3 mouse fibroblast cell lines (obtained from CR-UK London Research Institute) were grown in Dulbecco's Minimal Essential Medium (DMEM) (Autogen Bioclear). Transfected NIH3T3 cells were grown in the same medium containing G418-selective agent (Sigma-Aldrich) at a concentration of 1 mg/mL. All cells were supplemented with 10% FCS and 1% glutamine and incubated at 37°C in 5% CO2.
Plasmids and Site directed mutagenesis
The plasmid DNA used was the pUSEamp vector (Upstate Cell Signaling Solutions, NY, USA).
The wtEGFR and the L858R constructs were kindly provided by Dr. Daphne Bell and Matthew Meyerson from the MGH Cancer Centre, Harvard Medical School, Boston, USA.
The NLS mutant constructed used the wtEGFR and the L858R plasmid as template. The two designed and SDS purified mutagenic primers (Forward 5′cctcttcatggcagcggcccacatcgttgcgaaggccacgctggcggcgctgctgcagg3′ and Reverse 5′cctgcagcagcgccgccagcgtggccttcgcaacgatgtgggccgctgccatgaagagg3′) were utilized according to the Site directed mutagenesis XL kit protocol (Stratagene) to change the EGFR NLS sequence 645-RRRHIVRKRTLRR-657 into 645-AAAHIVAKATLAA-657.
EGFRvIII was kindly provided by Prof. William Gullick from the Department of Biosciences University of Kent, Canterbury, UK.
EGFR M1, M12, KMT, ΔNLS encoding point mutations of the EGFR NLS sequence (M1: AAAHIVRKRTLRR, M12: AAAHIVAAATLRR), the deletion of the NLS sequence (ΔNLS) and a mutation within the kinase domain (KMT: K821A) were kindly obtained from Dr. M.C. Hung (MD Anderson Cancer Center, USA).
Plasmid Transfection
Cells were plated at 1×105/ml and following 2 hours, cells were transfected according to the Genjuice transfection reagent protocol (Novagen EMD Bioscience). Cells were then treated 48 hours following transfection or serum starved for 24 hours and then treated.
Alkaline Single-Cell Gel Electrophoresis (Comet) Assay
Measurement of DNA interstrand crosslinks was performed as previously described (27).
DNA strand breaks repair in cells irradiated with 15 Gy was measured using the comet assay as previously described (28).
Data were presented as a percentage of tail moment i.e. as a percentage of the amount of strand breaks resulting immediately following IR treatment.
Statistical Analysis
The 2 way ANOVA, Bonferroni post tests and Student's t test were used for calculating the significance of the differences in repair and DNA-PK kinase activity. All the cell lines were considered individually and compared to the wtEGFR expressing cell line. Statistical values of P<0.01 were considered significant.
Immunoprecipitation
Stably expressing NIH3T3 cell lines were plated at 2 × 105/ml and left overnight before treatment. Cells were then incubated in growing medium, or treated with 50μM cisplatin for one hour in serum-free media and then left in drug-free medium for 18 hours, or serum starved for 24-36 hours or serum starved for 24-36 hours and treated with EGF 100ng/ml or treated with 4Gy IR and incubated 20 minutes in serum-free media. Approximately 5×106 cells were lysed on ice in 500μl of CelLytic™M Cell lysis reagent (Sigma) supplemented with Protease and phosphatase inhibitor (Roche) and Benzonase (Merck) according to manufacturer's protocol. 1.5 mg of protein sample was incubated with 2μg of anti-EGFR antibody (clone R19/48 Invitrogen) and left rotating at 4 °C for 2.5 hours. Immunoprecipitation was performed as previously described (29).
Western Blotting
Western blotting was performed as previously described (27). Proteins were probed using anti-EGFR (Cell Signalling 1:1000), anti-DNAPKcs (AbCam 1:400) anti-PY20 (Santa Cruz 1:1000). Finally, the primary antibody was probed with horseradish peroxidase–conjugated polyclonal antibodies for chemiluminescence detection (ECL System, Amersham Biosciences).
Immunofluorescence Staining
2 ×104 cells were plated on 13mm glass cover slips (VWR). Cells were then treated with cisplatin and IR as detailed above and stained as previously described (26). Respective primary antibodies were added as follows: anti-rabbit EGFR (1:50, clone 15F8 cell signaling) and anti-mouse DNAPKcs (1:50 AbCam). Then, secondary fluorescent conjugated Antibodies were added as follows: 1:100 Alexa Fluor 647 goat anti-rabbit and 1:100 Alexa Fluor 488 goat anti-mouse (Molecular probes and Invitrogen Life Technologies). Cells were visualized by confocal microscopy (objective × 40, Leica TCS SP2). Nuclear slice images were acquired by sequential scanning using the LAS AF Lite programme.
DNA-PK Functional Assay
DNA-PK activity was detected using the Promega (Madison, Wisconsin, USA) SignaTECT DNA-PK assay system, according to the manufacturer's protocol. The enzymatic activity of DNA-PK was analysed by scintillation counting and expressed as a percentage change of control DNA-PK activity as measured in untreated cells.
Proximity Ligation assay
Proximity ligation was performed according to the manufacturer's protocol using the Duolink Detection Kit (Cambridge BioScience Ltd, Ca,bridge UK). NIH3T3 cells were grown on 13 mm glass cover slips (VWR) and treated with cisplatin or IR as detailed above. Immunofluorescence staining protocol was carried out until the primary antibody incubation. Cy3 signal amplification was utilised for the assay. Cells were examined with a confocal microscope (objective × 40, Leica TCS SP2).
RESULTS
EGFR NUCLEAR TRANSLOCATION MODULATES DNA REPAIR
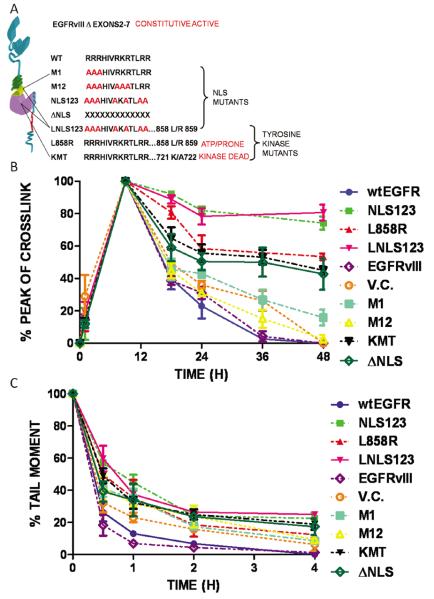
The synergistic effects of both cisplatin and IR with EGFR inhibition have been well described (12, 23). However, the role and consequences of EGFR nuclear translocation and kinase activation in the repair of drug-induced DNA interstrand crosslinks, or radiation-induced DNA strand breaks, have not been fully examined. To probe the role of EGFR in modulating DNA repair, the EGFR negative cell line, NIH3T3, was transfected with each of ten plasmids (wtEGFR, NLS123, L858R, LNLS123, EGFRvIII, M1, M12, KMT, ΔNLS, vector control) and therapy-induced DNA damage and its repair was assessed. The resulting transfectants express either wtEGFR or mutations within the NLS sequence (NLS123, LNLS123, M1, M12, ΔNLS), kinase domain (L858R, KMT) or extracellular domain (EGFRvIII) of EGFR (Figure 1A). Their expression was confirmed over a period of 72 hours following transfection (supplementary data S1). Transfected cells were incubated with 50 μM cisplatin for 1 hour, or treated with 15 Gy IR. Formation and repair of cisplatin-induced interstrand crosslinks, critical cytotoxic lesions produced following drug treatment, were measured over a 48 hour period using a modification of the comet assay as previously described (27) (Figure 1B). Repair of IR-induced strand breaks was measured over a 4 hour period (Figure 1C).
Figure 1.
Effects of EGFR modulation in repair of interstrand crosslinks and DNA strand breaks. (A) Graphic representation of the EGFR constructs employed in the study. (B-C) Measurement of drug-induced DNA interstrand crosslinks or strand breaks in NIH3T3 cells transfected with wtEGFR, NLS123, L858R, LNLS123, EGFRvIII, M1, M12, KMT, ΔNLS, vector control were treated with (B) 50μM cisplatin alone, (C) 15 Gy IR. Interstrand crosslink formation is represented as a percentage decrease of the peak of crosslink and strand breaks are showed as a percentage of tail moment.
There was no alteration in the formation of the peak of DNA interstrand crosslinks by cisplatin in cells expressing any of the constructs (supplementary figure S2, table 1). However, cells expressing mutations of the nuclear localization signal (NLS) sequence (NLS123, LNLS123) and of the kinase domain (L858R, KMT, ΔNLS) clearly showed reduction in repair (unhooking) of interstrand crosslinks (Figure 1B). Unhooking of cisplatin–induced interstrand crosslinks was >95% by 36 hours and complete by 48 hours in cells expressing wtEGFR and EGFRvIII. In contrast, cells expressing mutations NLS123 and LNLS123 showed only 26 ± 4.07 % and 19.3 ± 4.8 % unhooking at 48 hours, respectively. Intermediate levels of unhooking were observed for the L858R (46.57 ± 2.13%), KMT (54.95 ± 2,56%) and ΔNLS (57.21 ± 9.72%) mutants, whereas M1 and M12-expressing cell lines showed 84.09 ± 4.87% and 97.41 ±2.73% unhooking of interstrand crosslinks, respectively, at 48 hours following cisplatin treatment. Statistical analysis demonstrated significance (P value < 0.01) at the 48 hours time points and/or at earlier time points among the different mutants (Table 2A supplementary data).
The effect of the EGFR constructs on repair of DNA strand breaks induced by IR was also investigated. Repair of IR-induced DNA strand breaks is shown as percentage of the IR-induced tail moment calculated from the comet assay data (Figure 1C). Decrease of tail moment was 100% in both wtEGFR and EGFRvIII-expressing cell lines at 4 hours following treatment indicating complete repair of strand breaks. Significant differences (P value <0.001) in repair kinetics between wtEGFR and the mutant EGFR-expressing were found at 30 minutes following IR (Table 2B supplementary data). At 4 hours, cells expressing NLS123 and LNLS123 showed significant delay in repair of strand breaks with 22.48 ± 3.72 % and 24.94 ± 1.45 % unrepaired strand breaks. Intermediate levels of repair were observed for cells expressing L858R (12.33 ±1.00 %), KMT (18.86±3.45%), ΔNLS (17.38±5.06 %), M1 (8.51±1.12%) and M12 (9.28±2.26%) plasmids.
WILD TYPE EGFR AND EGFRVIII ASSOCIATE WITH DNAPKcs FOLLOWING TREATMENT WITH IR OR CISPLATIN
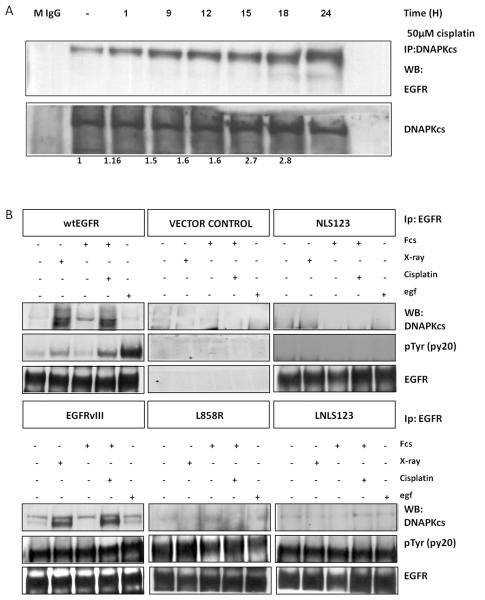
Previous studies have demonstrated association of EGFR and DNAPKcs following IR (12, 30, 31). However, this association, and its significance, have not been described following cisplatin treatment. NIH3T3 cells were transfected with wtEGFR and treated with 50 μM cisplatin for 1 hour. Cells were then collected at various time points up to 24 hours following treatment, protein extracts were prepared, immunoprecipitated using an anti-DNAPKcs monoclonal antibody and blotted with an anti-EGFR antibody (Figure 2A). There was a time-dependent association of EGFR and DNAPKcs resulting in a 2.7 fold increase at 18 hours following the cisplatin treatment.
Figure 2.
(A) wtEGFR Transfected NI3T3 cells were treated with 50μM cisplatin for 1 hour in serum free media. Cells were then lysed 1,9,12,15,18,24, hours following treatment. 750μg of protein lysate were immunoprecipitated using anti DNAPKcs and blotted with anti EGFR and anti DNAPKcs. EGFR pull-down was quantified by 2D densitometric analysis and shown as a binding fold compared to the untreated control. Mouse unrelated antibody (M IGg) was used as negative control. (B) Stable NIH3T3 cells expressing wtEGFR, NLS123, L858R, LNLS123, EGFRvIII, and Vector control were treated with 50μM cisplatin or 4 G IR, or treated with 100ng/ml EGF as described in the materials and methods. 1.5mg of protein lysate was then immunoprecipitated using anti-EGFR monoclonal antibody and blotted with anti DNAPKcs, anti-PY20 and anti-EGFR.
To determine the levels of EGFR-DNAPKcs association and whether it is induced by cisplatin or IR, we compared the levels of this association following cisplatin treatment, IR and EGF treatment. Experiments were carried out using extracts from cells expressing wtEGFR, NLS123, L858R, LNLS123 and EGFRvIII (Figure 2B) immunoprecipitated using an anti-EGFR antibody and blotted using an anti-DNAPKcs antibody. Western blot analysis showed that in cells expressing wtEGFR and EGFRvIII, there is association with DNAPKcs following treatment with IR or cisplatin. However, cells expressing NLS123, L858R and LNLS123 showed no interaction between EGFR and DNAPKcs. The immunoprecipitated samples were also blotted with a pan-phosphotyrosine antibody to determine activity of EGFR. Cells expressing wtEGFR showed maximal activation of the receptor following EGF treatment. Intermediate levels of activation were detected following IR or cisplatin. L858R, LNLS123 and EGFRvIII expressing cells showed a constitutive activation of the receptor whereas NLS123 showed no receptor activation. Therefore, the EGFR-DNAPKcs binding is triggered by cisplatin or IR and not by the EGFR nuclear translocation per se.
DNAPKcs AND EGFR LOCALIZE IN THE SAME CELLULAR COMPARTMENTS FOLLOWING IR OR CISPLATIN
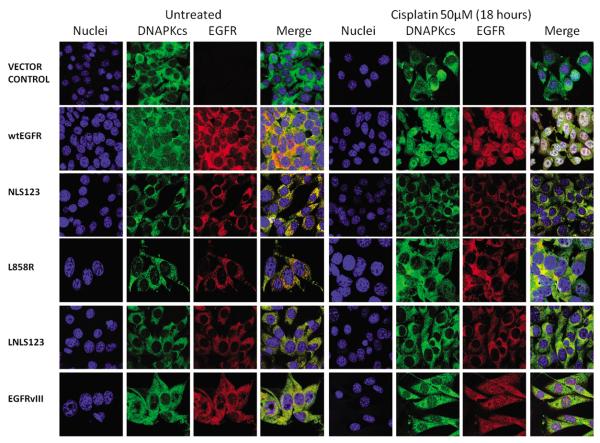
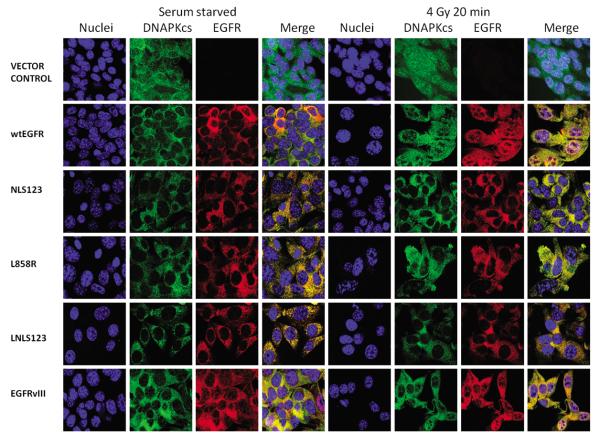
Having established an association between EGFR and DNAPKcs following either cisplatin or IR treatment, we investigated their cellular localization by confocal microscopy following IR and cisplatin treatment. Cell transiently transfected with wtEGFR and EGFRvIII showed clear EGFR nuclear expression following IR (Figure 3) or cisplatin (Figure 5) treatment. In contrast cells transiently transfected with NLS123, L858R, LNLS123, KMT and ΔNLS showed lack of EGFR nuclear accumulation. M1 and M12 transfected cells showed only reduced EGFR nuclear expression following IR or cisplatin treatment. Next, stably transfected cells were utilised to investigate this pattern. Cells expressing wtEGFR and EGFRvIII showed nuclear expression of both EGFR and DNAPKcs following IR (Figure 4) or cisplatin treatment (Figure 6). In contrast, L858R-expressing cells showed impaired EGFR nuclear localization following either treatment. Expression of EGFR and DNAPKcs was exclusively cytosolic in NLS123 and LNLS123 expressing cell lines following either IR (Figure 4) or cisplatin treatment (Figure 6). Therefore, the NLS123 mutation inhibits EGFR nuclear localization and also indirectly inhibits DNAPKcs sub-cellular localization following IR or cisplatin. Nuclear translocation of EGFR was verified via cellular fractionation following cisplatin and IR treatment (data not shown).
Figure 3.
EGFR and DNAPKcs cellular localisation following cisplatin treatment. NIH3T3 transiently transfected with wtEGFR, NLS123, L858R, LNLS123, EGFRvIII, M1, M12, KMT, ΔNLS and Vector control, were treated with 50μM cisplatin for one hour in serum free media and then fixed with 4%PFA 18 hours following treatment. Cells were stained with Goat anti-Rabbit Alexa fluor 647 (EGFR), Goat anti-Mouse Alexa fluor 488 (DNAPKcs), and Dapi (nucleus).
Figure 5.
EGFR and DNAPKcs cellular localisation following IR treatment. NIH3T3 transiently transfected with wtEGFR, NLS123, L858R, LNLS123, EGFRvIII, M1, M12, KMT, ΔNLS and Vector control were serum starved for 24 hours, treated with 4Gy Ionising radiation and then fixed with 4%PFA 20 minutes following treatment. Cells were stained with Goat anti-Rabbit Alexa fluor 647 (EGFR), Goat anti-Mouse Alexa fluor 488 (DNAPKcs), and Dapi (nucleus).
Figure 4.
EGFR and DNAPKcs cellular localisation following cisplatin treatment. Stable NIH3T3 cells expressing wtEGFR, NL123, L858R, LNLS123, EGFRvIII, Vector control were treated with 50μM cisplatin for one hour in serum free media and then fixed with 4%PFA 18 hours following treatment. Cells were stained with Goat anti-Rabbit Alexa fluor 647 (EGFR), Goat anti-Mouse Alexa fluor 488 (DNAPKcs), and Dapi (nucleus).
Figure 6.
EGFR and DNAPKcs cellular localisation following IR treatment. Stable NIH3T3 cells expressing wtEGFR, NL123, L858R, LNLS123, EGFRvIII, Vector control were serum starved for 24 hours, treated with 4gy Ionising radiation and then fixed with 4%PFA 20 minutes following treatment. Cells were stained with Goat anti-Rabbit Alexa fluor 647 (EGFR), Goat anti-Mouse Alexa fluor 488 (DNAPKcs), and Dapi (nucleus).
EGFR AND DNAPKcs ASSOCIATION FOLLOWING CISPLATIN OR IR TREATMENT
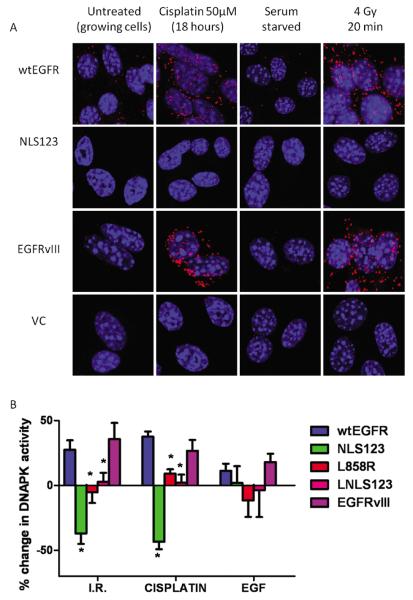
Previous studies have demonstrated binding between EGFR and DNAPKcs in the nucleus following EGFR nuclear translocation (23, 32). To investigate the co-localization of EGFR and DNAPKcs following treatment with cisplatin or IR, we performed a Duolink proximity assay. This assay allows visualisation of the interaction between two proteins in fixed cells. Each interaction is represented via a single red fluorescent dot (Figure 7A). In cells expressing vector and NLS123 constructs, no interaction was detectable. In contrast, cells expressing wtEGFR and EGFRvIII showed subcellular interaction between EGFR and DNAPKcs following either IR or cisplatin.
Figure 7.
(A) EGFR-DNAPKcs complex cellular localization. Stable NIH3T3 cells expressing wtEGFR, NLS123, EGFRvIII and Vector control were treated with 50μM cisplatin for one hour in serum free media and then fixed with 4%PFA 18 hours following treatment or 4 Gy and then fixed with 4%PFA 20 minutes follwowing radiation. Cells were then immuno blocked with anti-rabbit EGFR and anti-mouse DNAPKcs. Interacting complexes were then visulised via the duo link proximit assay. Each red spot represents a single interaction. (B) EGFR modulation of DNA-PK kinase activity. Stable NIH3T3 cells expressing wtEGFR, NLS123, L858R, LNLS123 and EGFRvIII were treated with 50μM cisplatin for one hour or 4gy or 100ng/ml EGF in serum free media. 18 hours following the treatment with cisplatin, 20 minutes following the treatment with IR and at 1 hour following EGF incubation samples prepared for the DNAPK Kinase assay. The graph shows the percentage change in DNAPK activity following each treatment compared to untreated. Stars indicate statistical significance.
EGFR MODULATION OF DNA-PK ACTIVITY
We previously demonstrated that the association of EGFR and DNAPKcs resulted in stimulation of DNAPKcs activity (31). The experiments detailed above demonstrated that nuclear translocation of EGFR is required for association with DNAPKcs. To investigate whether this association resulted in alteration of enzyme activity, we investigated the effects of expression of different EGFR constructs on DNA-PK kinase activity. As compared with individual untreated control, cells expressing wtEGFR showed a 27.5 ± 7.22% increase in DNA-PK activity following IR (4Gy) and a 37.52 ± 4.01 % increase following 50μM cisplatin treatment for 1 hour (Figure 7B). In cells expressing EGFRvIII there was a 32.42 ± 16.58 % increase of DNA-PK activity following IR and a 26.6 ± 8.49% increase following cisplatin treatment. In contrast, no significant change in DNA-PK activity compared to controls was found in L858R and LNLS123 expressing cells following IR (−5.29 ± 8.27% and 2.72 ± 7.06%) or cisplatin (9.04 ±3.46% and 2.25 ±6.07%). Results are shown in Table 2C supplementary data. Only cells expressing NLS123 showed a clear decrease in DNA-PK kinase activity compared to control following IR (−36.95± 8.08 %) or cisplatin (−43.30 ± 5.82 %). Therefore, nuclear localization is required for EGFR-induced stimulation of DNA-PK activity. EGF treatment did not induce a significant change in DNA-PK kinase activity (P value > 0.05) in NLS123, LNLS123 and EGFRvIII expressing cell lines; only L858R showed borderline significant differences (P value <0.05) compared to the wtEGFR expressing cells.
DISCUSSION
Inhibitors of EGFR play a major role in cancer therapeutics. However, the activity of these agents as monotherapies is low and it is important to investigate regimens with optimal combinations using chemotherapy and radiation (3). There is evidence of the effects of EGFR inhibition on DNA repair following irradiation. In this study, we demonstrate the importance of nuclear EGFR in modulating the repair of DNA damage following cisplatin chemotherapy or radiation.
Expression of EGFR in the nucleus is well-established but the implications on effects of therapy are not clear. According to recent reports, EGFR nuclear translocation requires receptor dimerization and activation as, following internalization, mature and active EGFR may become a poor substrate for lysosomal degradation (33, 34). This allows indirect sorting of the receptor through the Golgi, or direct sorting through the endoplasmic reticulum (ER). Subsequent association with sec61 results in retro-translocation to the cytosol where EGFR is stabilized following association with HSP70 (33). Binding of Importin β, mediated through the NLS sequence, translocates the receptor to the nucleus (22, 33). Inactive receptors (or those without an active conformation) are usually sent back to the plasma membrane via the recognition of a basolateral signal in the JX domain (19).
The interaction of EGFR with DNAPKcs has been demonstrated in several studies (12)The DNAPK complex plays a key role in non-homologous end joining, the major method of repair of DNA strand breaks following IR. Interaction of EGFR with DNAPKcs has been shown to contribute to the repair of DNA strand breaks. Inhibition of EGFR, by cetuximab or gefitinib, inhibits repair of IR-induced DNA strand breaks and impairs EGFR-DNAPKcs interaction (4, 11).
It is well known that interstrand crosslinks contribute significantly to cisplatin cytotoxicity and that unhooking of interstrand crosslinks may be used to determine clinical sensitivity (35). In previous studies we demonstrated that the unhooking of cisplatin DNA interstrand crosslinks was inhibited by gefitinib, and that this is mediated through the DNA-PK pathway (31). This pathway has been demonstrated to have relevance in the repair of cisplatin-induced DNA damage (36).
In this study, we show that EGFR nuclear expression in transfected EGFR-null cells modulates repair of DNA damage through the DNA-PK pathway. Cells expressing EGFR constructs with mutated NLS sequence are inhibited in their ability to unhook DNA interstrand crosslinks (35). Abrogation of nuclear expression of EGFR results in significant delay in repair of interstrand crosslink in these cells. This correlates with reduced association of EGFR with DNAPKcs. Cells expressing constructs which do not translocate to the nucleus showed increased sensitivity to cisplatin (supplementary figure S3). Previous studies have shown that EGFR nuclear translocation correlated with repair of IR induced strand breaks.
It has been shown previously that the association between DNAPKcs and EGFR peaks at 20 minutes following IR (37), but association following cisplatin treatment has not been described. Here we show that binding peaks at 18 hours following cisplatin treatment. The different timing following cisplatin and IR likely reflect different types of DNA lesions and repair mechanisms. There was reduced repair of DNA IR-induced strand breaks in cells expressing EGFR constructs which do not translocate to the nucleus. Repair of DNA strand breaks following IR has been shown to be modulated by EGFR (37), and the less marked effects of impaired EGFR nuclear translocation, as compared with the repair of cisplatin-induced interstrand crosslinks, may be secondary to the activation of other DNA repair pathways (12).
In this study, the nuclear translocation of EGFR was demonstrated by confocal microscopy. Interestingly, these experiments suggest that there is co-localization of EGFR and DNAPKcs both within the nucleus and the cytoplasm. Although the primary location of DNAPKcs is in the nucleus in the formation of complexes on damaged DNA, cytoplasmic expression of DNAPKcs has been demonstrated in several studies (38-40). The results of the Proximity Ligation Assay demonstrate physical proximity of DNAPKcs and EGFR following DNA damage by cisplatin or IR in cells with intact nuclear localization. This was not shown in cells expressing EGFR constructs deficient in nuclear localization.
There is contradictory evidence regarding nuclear expression of EGFRvIII. While EGFRvIII and STAT3 co-localization within the nucleus was demonstrated in some studies (17, 41), other studies have reported lack of nuclear expression in glioma models (42). Cells expressing EGFRvIII show elevated activation of DNA-PKcs and enhancement of DNA strand breaks repair. In this study we show that EGFRvIII and wtEGFR undergo nuclear translocation and binding with DNAPKcs following cisplatin or IR treatment.
Although cells expressing kinase-dead EGFR showed lack of nuclear translocation, expression of the L858R mutant also resulted in impaired nuclear expression despite constitutive kinase activity. This resulted in reduction of DNA strand breaks repair which is consistent with the observation that non-small cell lung cancer lines expressing L858R show increased sensitivity to IR (43) and reduced nuclear expression (26). Moreover, the difference in repair between cells expressing L858R (kinase active but with impaired nuclear localization) and M1or M12 (kinase active and expressed in the nucleus) suggests that it is the impaired nuclear EGFR accumulation (which is a consequence of the lack of allosteric activation) and not kinase activity per se that determines the reduced DNA repair in these models.
The impaired kinase activity shown by the NLS123 mutant supports the previously described allosteric mode of activation of EGFR and the importance played by the third cluster of arginines (646-RR-647) within the NLS sequence in adopting an α-helical conformation which is indispensable for EGFR activation (25, 34). The impairment of other EGFR functions, as a result of mutations in the NLS sequence, has not been excluded. Interestingly, the LNLS123 mutant shows kinase activation despite bearing the same NLS mutation that renders the NLS123 kinase-dead. This suggests that the L858R mutation, which has been shown to thermodynamically stabilise EGFR (44, 45), allows receptor activation that does not require the allosteric conformational change. Further work will be required to investigate whether the L858R inhibition of nuclear translocation is therefore due to a structurally hidden NLS sequence.
EGFR inhibition by gefitinib has been shown to suppress DNA repair following treatment with radiation and cisplatin (31). Similarly, in this study, the kinase dead mutant KMT (mutation K721A), shows no nuclear localization suggesting that the targeting of the EGFR kinase domain interferes with nuclear translocation and consequently with repair. Maximal effect of gefitinib on inhibition of interstrand crosslinks was observed only in EGFR constructs translocating to the nucleus (supplementary figure S4). A recent study demonstrated that cisplatin resistance and DNA repair was dependent on nuclear translocation (23). Here, we have shown that a variety of EGFR mutants deficient in nuclear expression show impaired repair and that nuclear accumulation is a major determinant of repair of cisplatin induced interstrand crosslinks. Additionally we recently demonstrated that expression of HER2 modulated repair of cisplatin-induced interstrand crosslinks and that this also requires nuclear expression (27). Other factors may contribute to the interaction between cisplatin and the EGFR pathway in therapy including ubiquitination of EGFR induced by cisplatin in head and neck cancer cells (46).
There has been extensive study on the interaction of the EGFR and DNAPK pathways. Following the initial observation that cetuximab treatment inhibits EGFR/DNAPKcs interaction (47), the role of this interaction in modulation of DNA repair has been confirmed. Cells expressing specific EGFR mutations found in human cancer such as the L858R in non-small cell lung cancer have been found to show sensitivity to IR. This includes delayed DNA repair kinetics, defective IR-induced arrest in DNA synthesis and increased apoptosis (26). Here we show that inhibition of EGFR nuclear translocation alters DNAPKcs cellular distribution.
Stimulation of DNA-PK kinase activity was associated with nuclear expression and binding. There are likely other factors apart from EGFR-DNAPKcs binding which may influence the effect of EGFR on DNA repair. EGF induces nuclear translocation of EGFR but this is not associated with EGFR-DNAPKcs interaction. Receptor kinase activation may influence, indirectly, DNA-PK possibly by activation of the AKT pathway which has been shown to be a kinase of DNAPKcs (48, 49).
These results suggest that nuclear expression of EGFR plays a significant role in response to cisplatin and radiation. The intracellular localization of EGFR may play a critical part in response to therapies combining inhibitors of the EGFR pathway with chemotherapy or radiation. Understanding of the mechanisms by which nuclear expression modulates therapeutic effects of these modalities will optimise design of clinical studies in the future.
Supplementary Material
Acknowledgements
Gianmaria Liccardi was supported by CRUK for the studentship (C2259/A7475). This work was also funded by CRUK through a programme grant to John A. Hartley and Daniel Hochhauser (C2259/A9994).
REFERENCES
- 1.Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol. 2007;19:124–34. doi: 10.1016/j.ceb.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–25. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 3.Nyati MK, Morgan MA, Feng FY, Lawrence TS. Integration of EGFR inhibitors with radiochemotherapy. Nat Rev Cancer. 2006;6:876–85. doi: 10.1038/nrc1953. [DOI] [PubMed] [Google Scholar]
- 4.Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 2005;76:157–61. doi: 10.1016/j.radonc.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–13. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–46. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 8.Ng SS, Tsao MS, Nicklee T, Hedley DW. Effects of the epidermal growth factor receptor inhibitor OSI-774, Tarceva, on downstream signaling pathways and apoptosis in human pancreatic adenocarcinoma. Mol Cancer Ther. 2002;1:777–83. [PubMed] [Google Scholar]
- 9.Bonner JA, Raisch KP, Trummell HQ, Robert F, Meredith RF, Spencer SA, et al. Enhanced apoptosis with combination C225/radiation treatment serves as the impetus for clinical investigation in head and neck cancers. J Clin Oncol. 2000;18:47S–53S. [PubMed] [Google Scholar]
- 10.Herbst RS, Kim ES, Harari PM. IMC-C225, an anti-epidermal growth factor receptor monoclonal antibody, for treatment of head and neck cancer. Expert Opin Biol Ther. 2001;1:719–32. doi: 10.1517/14712598.1.4.719. [DOI] [PubMed] [Google Scholar]
- 11.Friedmann B, Caplin M, Hartley JA, Hochhauser D. Modulation of DNA repair in vitro after treatment with chemotherapeutic agents by the epidermal growth factor receptor inhibitor gefitinib (ZD1839) Clin Cancer Res. 2004;10:6476–86. doi: 10.1158/1078-0432.CCR-04-0586. [DOI] [PubMed] [Google Scholar]
- 12.Rodemann HP, Dittmann K, Toulany M. Radiation-induced EGFR-signaling and control of DNA-damage repair. Int J Radiat Biol. 2007;83:781–91. doi: 10.1080/09553000701769970. [DOI] [PubMed] [Google Scholar]
- 13.Wang SC, Hung MC. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009;15:6484–9. doi: 10.1158/1078-0432.CCR-08-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2007;96(Suppl):R16–20. [PubMed] [Google Scholar]
- 15.Xia W, Lau YK, Zhang HZ, Xiao FY, Johnston DA, Liu AR, et al. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res. 1999;5:4164–74. [PubMed] [Google Scholar]
- 16.Hadzisejdic I, Mustac E, Jonjic N, Petkovic M, Grahovac B. Nuclear EGFR in ductal invasive breast cancer: correlation with cyclin-D1 and prognosis. Mod Pathol. 23:392–403. doi: 10.1038/modpathol.2009.166. [DOI] [PubMed] [Google Scholar]
- 17.Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 8:232–45. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–7. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choowongkomon K, Carlin CR, Sonnichsen FD. A structural model for the membrane-bound form of the juxtamembrane domain of the epidermal growth factor receptor. J Biol Chem. 2005;280:24043–52. doi: 10.1074/jbc.M502698200. [DOI] [PubMed] [Google Scholar]
- 20.He C, Hobert M, Friend L, Carlin C. The epidermal growth factor receptor juxtamembrane domain has multiple basolateral plasma membrane localization determinants, including a dominant signal with a polyproline core. J Biol Chem. 2002;277:38284–93. doi: 10.1074/jbc.M104646200. [DOI] [PubMed] [Google Scholar]
- 21.Bao J, Alroy I, Waterman H, Schejter ED, Brodie C, Gruenberg J, et al. Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J Biol Chem. 2000;275:26178–86. doi: 10.1074/jbc.M002367200. [DOI] [PubMed] [Google Scholar]
- 22.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–40. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 23.Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am J Transl Res. 2009;1:249–58. [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson JP, Berger MB, Lin CC, Schlessinger J, Lemmon MA, Ferguson KM. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25:7734–42. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hubbard SR. The juxtamembrane region of EGFR takes center stage. Cell. 2009;137:1181–3. doi: 10.1016/j.cell.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 26.Das AK, Chen BP, Story MD, Sato M, Minna JD, Chen DJ, et al. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–74. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 27.Boone JJ, Bhosle J, Tilby MJ, Hartley JA, Hochhauser D. Involvement of the HER2 pathway in repair of DNA damage produced by chemotherapeutic agents. Mol Cancer Ther. 2009;8:3015–23. doi: 10.1158/1535-7163.MCT-09-0219. [DOI] [PubMed] [Google Scholar]
- 28.Hartley JM, Spanswick VJ, Gander M, Giacomini G, Whelan J, Souhami RL, et al. Measurement of DNA cross-linking in patients on ifosfamide therapy using the single cell gel electrophoresis (comet) assay. Clin Cancer Res. 1999;5:507–12. [PubMed] [Google Scholar]
- 29.Rodriguez-Viciana P, McCormick F. Characterization of interactions between ras family GTPases and their effectors. Methods Enzymol. 2006;407:187–94. doi: 10.1016/S0076-6879(05)07016-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedmann BJ, Caplin M, Savic B, Shah T, Lord CJ, Ashworth A, et al. Interaction of the epidermal growth factor receptor and the DNA-dependent protein kinase pathway following gefitinib treatment. Mol Cancer Ther. 2006;5:209–18. doi: 10.1158/1535-7163.MCT-05-0239. [DOI] [PubMed] [Google Scholar]
- 32.Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–72. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, et al. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wynne P, Newton C, Ledermann JA, Olaitan A, Mould TA, Hartley JA. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. Br J Cancer. 2007;97:927–33. doi: 10.1038/sj.bjc.6603973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durant S, Karran P. Vanillins--a novel family of DNA-PK inhibitors. Nucleic Acids Res. 2003;31:5501–12. doi: 10.1093/nar/gkg753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, et al. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–9. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- 38.Huston E, Lynch MJ, Mohamed A, Collins DM, Hill EV, MacLeod R, et al. EPAC and PKA allow cAMP dual control over DNA-PK nuclear translocation. Proc Natl Acad Sci U S A. 2008;105:12791–6. doi: 10.1073/pnas.0805167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J, Tamaskovic R, Yang Z, Brazil DP, Merlo A, Hess D, et al. Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J Biol Chem. 2004;279:35510–7. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- 40.Toulany M, Kehlbach R, Florczak U, Sak A, Wang S, Chen J, et al. Targeting of AKT1 enhances radiation toxicity of human tumor cells by inhibiting DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Ther. 2008;7:1772–81. doi: 10.1158/1535-7163.MCT-07-2200. [DOI] [PubMed] [Google Scholar]
- 41.de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–62. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–9. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das AK, Sato M, Story MD, Peyton M, Graves R, Redpath S, et al. Non-small-cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res. 2006;66:9601–8. doi: 10.1158/0008-5472.CAN-06-2627. [DOI] [PubMed] [Google Scholar]
- 44.Yang S, Park K, Turkson J, Arteaga CL. Ligand-independent phosphorylation of Y869 (Y845) links mutant EGFR signaling to stat-mediated gene expression. Exp Cell Res. 2008;314:413–9. doi: 10.1016/j.yexcr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLoS Comput Biol. 2009;5:e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahsan A, Hiniker SM, Ramanand SG, Nyati S, Hegde A, Helman A, et al. Role of epidermal growth factor receptor degradation in cisplatin-induced cytotoxicity in head and neck cancer. Cancer Res. 70:2862–9. doi: 10.1158/0008-5472.CAN-09-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bandyopadhyay D, Mandal M, Adam L, Mendelsohn J, Kumar R. Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J Biol Chem. 1998;273:1568–73. doi: 10.1074/jbc.273.3.1568. [DOI] [PubMed] [Google Scholar]
- 48.Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–13. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 49.Lu D, Huang J, Basu A. Protein kinase Cepsilon activates protein kinase B/Akt via DNA-PK to protect against tumor necrosis factor-alpha-induced cell death. J Biol Chem. 2006;281:22799–807. doi: 10.1074/jbc.M603390200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.