Abstract
Desmocollin 1 (Dsc1) is part of a desmosomal cell adhesion receptor formed in terminally differentiating keratinocytes of stratified epithelia. The dsc1 gene encodes two proteins (Dsc1a and Dsc1b) that differ only with respect to their COOH-terminal cytoplasmic amino acid sequences. On the basis of in vitro experiments, it is thought that the Dsc1a variant is essential for assembly of the desmosomal plaque, a structure that connects desmosomes to the intermediate filament cytoskeleton of epithelial cells. We have generated mice that synthesize a truncated Dsc1 receptor that lacks both the Dsc1a- and Dsc1b-specific COOH-terminal domains. This mutant transmembrane receptor, which does not bind the common desmosomal plaque proteins plakoglobin and plakophilin 1, is integrated into functional desmosomes. Interestingly, our mutant mice did not show the epidermal fragility previously observed in dsc1-null mice. This suggests that neither the Dsc1a- nor the Dsc1b-specific COOH-terminal cytoplasmic domain is required for establishing and maintaining desmosomal adhesion. However, a comparison of our mutants with dsc1-null mice suggests that the Dsc1 extracellular domain is necessary to maintain structural integrity of the skin.
Desmosomes are adhesive cell-cell junctions that are formed in epithelia and certain other tissue types (reviewed in references 22, 25, 27, 30, 40, and 60). They are particularly abundant in stratified epithelia, such as the epidermis. The cytoplasmic surface of the desmosome is connected to the intermediate filament cytoskeleton, thereby establishing a three-dimensional transcellular scaffold of filament proteins that enables tissues to withstand mechanical stress.
Impaired desmosome function has been found in patients with mutations in desmosomal genes (reviewed in references 13 and 25; see also reference 33) and patients who develop autoantibodies against desmosomal proteins (e.g., references in reference 62). Furthermore, the histopathology of the bacterium-induced skin disorders bullous impetigo and staphylococcal scaled-skin syndrome was attributed to enzymatic cleavage of the desmosomal transmembrane protein desmoglein 1 (2). In all of the examples listed above, severe skin disorders were observed, with clinical phenotypes ranging from palmoplantar keratoderma to skin blistering. Furthermore, mutations in the genes of the desmosomal plaque proteins plakoglobin and desmoplakin have been linked to cardiomyopathy (reviewed in reference 13).
Genetically modified animals with mutations in desmosomal proteins were shown to develop phenotypes consistent with tissue fragility (1, 5, 6, 14, 28, 33, 37, 38, 67; see also references 21, 23, 24, and 58), demonstrating that desmosomes are crucial for the mechanical integrity and stability of certain tissues, in particular stratified epithelia. Furthermore, at least in some tissues, the correct temporal-spatial expression pattern of desmosomal proteins seems to be a prerequisite for the development of normal tissue and organ architecture (reviewed in reference 25).
The molecular composition of desmosomes has been extensively analyzed in the last few years. At least five components appear to be necessary to form a simple desmosome: desmoglein(s) (Dsg), desmocollin(s) (Dsc), and the plaque proteins plakoglobin, desmoplakin, and plakophilin(s) (PKP) (25-27, 35).
The transmembrane core of desmosomes is formed by both Dsg and Dsc. These type I transmembrane glycoproteins belong to the cadherin superfamily of calcium-dependent cell adhesion receptors. It is thought that heterophilic interactions between Dsc and Dsg establish the intercellular connection (e.g., see references 17, 26, 43, 63, and 64), although homophilic interactions between Dsc2 molecules in vitro have been reported (63). Plakoglobin, PKP(s), and desmoplakin bind to the cytoplasmic domains of desmosomal transmembrane receptors (directly or indirectly), thereby establishing the electron-dense plaque and the connection to the intermediate filament cytoskeleton (reviewed in references 9, 18, 25, 27, and 48).
Three Dsc isoforms (Dsc1 to Dsc3) have been identified in mammals (reviewed in references 26, 27, and 35) that show tissue- and cell type-specific expression patterns 3, 15, 32, 49-51). All Dsc are synthesized in the epidermis, where they show very complex and overlapping expression patterns. Dsc1 is the predominant Dsc isoform synthesized in terminally differentiating keratinocytes of stratified epithelia (3, 32, 51). This protein is also present in the hair follicle, in particular the companion cell layer of the outer root sheath and the inner root sheath (e.g., see reference 42).
All Dsc proteins are synthesized in two forms (a and b), the corresponding mRNAs being generated by differential splicing (reviewed in reference 26). The a and b variants differ only with respect to the COOH-terminal cytoplasmic amino acid sequences. The smaller b variants end with short amino acid sequences that are unique to these variants (11 amino acids in the case of Dsc1b [32]). The a variants have longer COOH termini (65 amino acids for Dsc1a) that show sequence homologies to classical cadherins, such as E-cadherin. These sequence elements at the COOH termini of the a variants, termed cadherin-type sequences (35), allow binding of plaque proteins and are therefore thought to be crucial for desmosome assembly (reviewed in references 25 to 27).
Transfection experiments done by Troyanovsky and colleagues demonstrated that the COOH-terminal domain of Dsc1a, but not Dsc1b, can recruit plakoglobin and desmoplakin to the plasma membrane of epithelial cells and serve as a nucleation site for the assembly of an electron-dense plaque with attached intermediate filaments (65, 66). Given these results, it appears that the a variants play a central role in the assembly of desmosomes. It is not known in which way the b variants contribute to desmosome assembly and/or function.
The initial goal of the present study was to analyze the role of the Dsc1a variant in vivo. Given the results of in vitro studies summarized above, one would expect that absence of Dsc1a would affect desmosome assembly, cell-cell adhesion, and/or proper tissue differentiation. We decided to investigate this hypothesis by generating mice that are deficient in this protein. Unexpectedly, however, we generated mice that synthesize a truncated Dsc1 protein that lacks both the Dsc1a- and Dsc1b-specific COOH-terminal cytoplasmic domains. Mutant mice showed a subclinical phenotype, suggesting that the splice variant-specific cytoplasmic domains are essential for neither proper epidermal development nor maintenance of tissue integrity.
MATERIALS AND METHODS
Animal protocols.
Mice were kept under standard housing conditions at Baylor College of Medicine. The experiments described in this report were approved by the animal review committee of Baylor College of Medicine.
Vector construction, ES cell targeting, and generation of mutant mice.
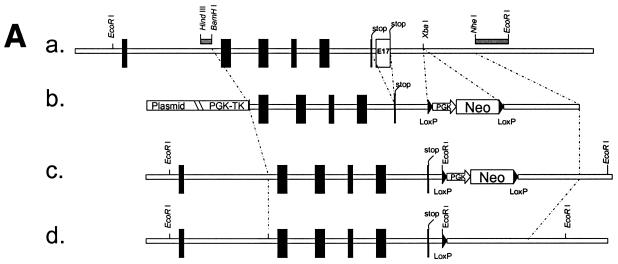
We have isolated a BAC clone from an Sv/129 BAC library (Genome Systems) that contained the entire dsc1 gene (data not shown). Exon 17 and flanking DNA sequences were isolated from the recombinant BAC and cloned into conventional cloning vectors. In the final targeting construct (TDSC1aΔE17; Fig. 1A), intron 16 and exon 17 were deleted, and the 3′ end of exon 16 was fused to the 3′ UTR (untranslated region) of the gene. In TDSC1aΔE17, exon 16 is flanked by 5.4-kb 5′ sequences and 4.2-kb 3′ sequences. A neomycin resistance minigene (PGKneoBpA; provided by Allan Bradley, Baylor College of Medicine), flanked by loxP sites, was inserted into an XbaI site 0.23 kb downstream of exon 16. A herpes simplex virus thymidine kinase minigene (provided by John Lydon, Baylor College of Medicine) was inserted into the targeting construct as well. Both antibiotic resistance cassettes contained a phosphoglycerate kinase I promoter. The targeting vector was linearized and electroporated into AB2.2 ES cells (provided by Allan Bradley). ES cells were selected with G418/FIAU, and drug-resistant colonies were tested for successful recombination by using a mini Southern procedure (56, 57). Three probes were used: a 5′ probe (HindIII-BamHI fragment, Fig. 1A) and a 3′ probe (NheI-EcoRI fragment, Fig. 1A) to confirm targeting of the dsc1 locus and a neomycin probe to ensure that no additional copy of the targeting vector had integrated into the ES cell genome (data not shown). Recombinant ES cell clones were transiently transfected with a Cre expression plasmid (CMV-Cre; provided by Allan Bradley) to remove the neomycin resistance cassette from the ES cell genome (data not shown). Randomly selected clones from these electroporations were tested by Southern blot analysis to confirm excision of the neo cassette. ES cell clones that were heterozygous for the ΔE17LoxP allele were injected into C57BL/6 blastocysts, a service provided by the Transgenic and Mutant Mouse Core at the Baylor College of Medicine. Chimeric mice were obtained from three ES cell clones. One of these lines showed germ line transmission of the mutation. Heterozygous mice (dsc1a+/−ΔE17LoxP) were intercrossed to obtain homozygous mutant mice. Mice were genotyped by PCR with tail DNA. The primers used were DSC1-m3 (exon 16 specific; GAATCCATTAGAGGACAC) and dsc1-m23 (3′ UTR specific; GGAGCTATGATTGGTAA). In wild-type mice, these primers amplify a genomic DNA fragment that contains exon 16, intron 16, exon 17, and the 3′ UTR. The fragment sizes are 0.5 kb (wild-type locus) and 0.23 kb (dsc1aΔE17LoxP locus), respectively.
FIG. 1.
Strategy used to generate dsc1−/−ΔΕ17LoxP mutant mice. (A) Schematic representation of the targeting strategy used in this study. (a) The 3′ end of the mouse dsc1 gene locus is shown. Exons are represented by vertical bars. Exons 16 and 17 contain stop codons. Probes used to identify recombinant ES cell clones are shown as horizontal bars (HindIII/BamHI and NheI/EcoRI fragments). (b) Targeting construct TDSC1aΔE17. Intron 16 and exon 17 were deleted in this construct. A neomycin resistance minigene with flanking loxP sequences was inserted downstream of exon 17. The targeting vector also contained a thymidine kinase (TK) cassette. (c) Homologous recombination in ES cells generated the dsc1ΔE17Neo gene locus, in which intron 16 and exon 17 were deleted. (d) The neomycin resistance minigene was deleted through transient expression of CRE recombinase, generating the dsc1ΔE17LoxP allele. (B) RNase protection assays to detect Dsc1 RNA. Desmoglein 3 (left panel) and β-actin (right panel) were used as internal controls. Probes derived from dsc1 exon 17 (E17) and the 5′ end of the Dsc1 mRNA (5′), respectively, were used. Homozygous dsc1−/−ΔE17LoxP mutants express a Dsc1 RNA that does not contain exon 17 sequences. The genotype of the samples is indicated above the lanes (MT, dsc1−/−ΔE17LoxP mutant; +/+, wild type). Note that the expression levels in homozygous mutant mice are slightly lower than those in wild-type mice. (C) Western blot analysis using whole-skin extracts from wild-type (+/+) and homozygous mutant (MT/MT) mice with antibody gp899 (Dsc1). The positions of Dsc1a and Dsc1b in the wild-type sample are indicated. Note the absence of the Dsc1a band in the mutant sample.
RNase protection assays (RPAs).
Newborn mouse epidermis was isolated as previously described (31). Tissue lysates were prepared with the reagents from the Direct Protect Lysate RPA kit (Ambion). Biotinylated cRNA probes were synthesized with the MAXIscript II kit (Ambion) by using biotin-16-UTP (Roche). The following DNA templates were used to synthesize cRNA: dsc1 exon 1 (nucleotide positions 125 to 345; accession number X97986), dsc1 exon 17, dsc2 (nucleotide positions 79 to 385; accession number L33779), dsc3 (nucleotide positions 4 to 360; accession number Y11169), dsg1 (exon 15; generously provided by Mŷ Mahoney, Thomas Jefferson University, Philadelphia, Pa.), Dsg2, Dsg3 (clone B9) (38), β-actin (pTRI-Actin M; Ambion), and cyclophilin (pTRI-Cyclophilin M; Ambion).
RPA products were analyzed as previously described (38). To determine the expression levels of the various desmosomal genes, RPA signals on Kodak films were scanned with a flat-bed scanner and analyzed with the QuantiScan software package (Biosoft). β-Actin and cyclophilin were used as internal standards in the RPA reactions.
RT-PCR amplification of Dsc1 mRNA, cloning, and sequence analysis.
Total RNA was isolated from newborn epidermis with the RNeasy Mini kit (Qiagen). Reverse transcription (RT)-PCR was performed with the One Step RT-PCR kit (Qiagen) by using primers DSC1-m41 (GAAGAAGTGACGGAAGCCAAT; derived from the coding sequences of exons 14 and 15) and DSC1-m19 (TTAATTTTTAATCAGACTGTGTCCTC; derived from exon 16). PCR products were gel purified with the Zymoclean Gel DNA Recovery kit (Zymo Research) and then cloned into the pCR2.1TOPO vector (Invitrogen). The cDNA insertions were sequenced with M13 reverse and T7 primers following standard procedures.
Antibodies.
cDNA sequences encoding portions of the cytoplasmic domains of mouse Dsc1, Dsc2, and Dsc3, respectively, were cloned into bacterial expression vector pMAL-c2X (pMAL Protein Fusion and Purification System; BioLabs). The following polypeptide sequences were used (amino acid positions are given with respect to the mature protein): Dsc1, C579 to P683; Dsc2, C581 to L715; Dsc3, C578 to E704. The fusion proteins were purified with amylose resin (BioLabs). Guinea pigs were immunized with these proteins (a service provided by Rockland Immunochemicals, Gilbertsville, Pa.), and the resulting antibodies were affinity purified with Affi-Gel 10 and Affi-Gel 15 columns (Bio-Rad), to which the Dsc fusion proteins had been coupled. The antibodies generated were gp899 (Dsc1), gp2295 (Dsc2), and gp2280 (Dsc3).
The additional antibodies used were DG3.10 (Dsg1 and Dsg2; Research Diagnostics), plakoglobin (Santa Cruz Biotechnology), plakophilin 1 (PKP1), PKP3 (generously provided by Werner W. Franke and Lutz Langbein, German Cancer Research Center, Heidelberg, Germany), repetin (generously provided by Daniel Hohl, CHUV/DHURDV, Lausanne, Switzerland), and β-catenin (Santa Cruz Biotechnology). Antibodies against filaggrin, loricrin, K14, K10, and K6 were generously provided by Dennis Roop (Baylor College of Medicine).
Western blotting.
Newborn skin samples were pulverized in liquid nitrogen and then incubated for 10 min in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer (0.0625 M Tris-HCl, 5% SDS, 10% glycerol, 20% β-mercaptoethanol, pH 6.8) at 95°C (whole-tissue lysate). In certain experiments, newborn skin was extracted with ice-cold TX buffer (1% Triton X-100 [TX], 50 mM Tris-HCl, 100 mM NaCl, pH 7.4) in the presence of protease inhibitors (COMPLETE protease inhibitor cocktail; Roche). Tissue lysates were centrifuged twice for 15 min each time at 4°C and 10,000 × g. TX-insoluble pellets were washed twice with phosphate-buffered saline (supplemented with protease inhibitors [see above]) and then denatured in SDS-PAGE loading buffer (cytoskeletal fraction). The TX-soluble proteins were precipitated with methanol-chloroform and then denatured in SDS-PAGE loading buffer (TX-soluble fraction). SDS-PAGE, blotting, and antibody detections were done essentially as previously described (38).
Histology, immunofluorescence microscopy, and low-temperature immunoelectron microscopy.
For conventional histological analysis, tissue sections were fixed overnight at 4°C in phosphate-buffered saline-buffered formalin, transferred to 70% ethanol, and then embedded in paraffin. Sections were stained with hematoxylin and eosin.
Tissues for deconvolution microscopy were embedded in O.C.T. compound (Tissue-TekII; Lab-Tek Products). Cryosections (6 μm) were stained as previously described (34). The sections were photographed with a Zeiss AxioVert S100 TV microscope and a DeltaVision Restoration Microscopy System (deconvolution microscopy; Applied Precision, Inc.).
Samples for low-temperature immunoelectron microscopy were isolated from the oral mucosa of mutant and wild-type control mice, and postembedding immunogold electron microscopy was performed as described previously (29). Ultrathin sections were incubated with gp899 (diluted 1:100) and then with 5-nm colloidal gold-conjugated goat anti-guinea pig immunoglobulin G (heavy and light chains; diluted 1:40; BBInternational). For low-power electron microscopy, the colloidal gold probes were amplified with the IntenSE silver enhancement kit (Amersham International) as described previously (61). Double immunolabeling was performed with a primary antibody mixture containing gp899 (1:100) and rabbit anti-desmoplakin (1:40; Research Diagnostics) or gp899 (1:100) and rabbit anti-plakoglobin (1:40; Santa Cruz Biotechnology) as previously described (44).
In situ hybridization.
Nonradioactive Dsc2 sense and antisense probes (see above) were synthesized with a digoxigenin labeling kit from Roche and hybridized to formalin-fixed tissue sections in hybridization buffer (Ambion) by following standard protocols. Probes were detected with anti-digoxigenin-alkaline phosphatase Fab fragments (Roche) and stained with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Roche).
Yeast two-hybrid assays.
Yeast two-hybrid vectors encoding the Gal4 DNA binding (pLP-GBK) or transcription activation (pLP-GAD) domain were purchased from BD Biosciences Clontech. Plakoglobin and PKP1 head (amino acids 1 to 286) constructs were generated as previously described (10, 41). The Dsc1a cytoplasmic domain and the COOH-terminally truncated Dsc1 cytoplasmic domain constructs were subcloned into the Creator system donor vector (BD Biosciences Clontech), followed by recombination subcloning into acceptor vector pLP-GAD. All constructs were then verified by sequencing. To assay for interactions between proteins, plasmid DNA was transformed into yeast strain AH109 (BD Biosciences Clontech) and double transformants were selected by using growth in the absence of leucine and tryptophan. Expression of the histidine- and adenine-encoding reporter genes was analyzed by monitoring colony growth on plates lacking histidine, adenine, leucine, and tryptophan.
RESULTS
Generation of mice with a deletion of exon 17.
The dsc1 gene consists of 17 exons and encodes two gene products (Dsc1a and Dsc1b) that differ only in their COOH-terminal amino acid sequences. The Dsc1a and Dsc1b mRNAs are generated by differential splicing of a single precursor RNA. The COOH terminus of Dsc1b is encoded by exon 16, while that of Dsc1a is encoded by exon 17 (see references in reference 26). The strategy for generating mice that do not synthesize Dsc1a was to delete exon 17. By following the steps outlined in Fig. 1A, heterozygous mutant mice (dsc1+/−ΔE17LoxP) were obtained and intercrossed to generate homozygous mutants (dsc1−/−ΔE17LoxP). All three genotypes (wild type and heterozygous and homozygous mutants) were obtained in the expected Mendelian ratio. Newborn and adult mutant mice were morphologically indistinguishable from wild-type littermates. In particular, we did not observe any abnormalities with respect to birth weight, growth rate, or appearance of the coat (data not shown). A histological analysis indicated that Dsc1-expressing tissues, including skin, tongue, and forestomach, developed normally in mutant mice (data not shown).
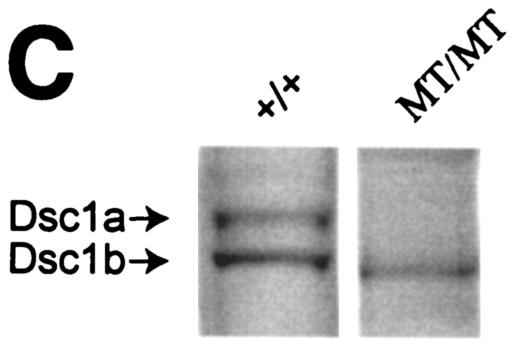
Homozygous mutant mice did not contain exon 17, as demonstrated by PCR with genomic DNA as a template (see Materials and Methods). Furthermore, RPAs with an exon 17 probe confirmed that homozygous mutant mice did not express Dsc1a mRNA (Fig. 1B). As expected, a 5′ probe detected a Dsc1 transcript, demonstrating that the mutant dsc1 gene locus is transcriptionally active (Fig. 1B). Finally, Western blot analysis with polyclonal Dsc1 antibodies confirmed that Dsc1a was not synthesized in homozygous mutant mice (Fig. 1C).
Mutant mice express a truncated Dsc1 receptor that lacks the Dsc1a- and Dsc1b-specific COOH-terminal domains.
The dsc1ΔE17LoxP allele was designed to encode a mutant transcript in which exon 16 is the last coding exon. To verify that exon 16 was correctly spliced to exon 15, thereby generating the Dsc1b mRNA, we performed RT-PCR analysis (Fig. 2). RT-PCR products from three mutant and three wild-type samples were cloned and sequenced. As expected, we identified the Dsc1b mRNA in wild-type samples (Fig. 2A). Surprisingly, the mutant samples yielded an RT-PCR product that was much larger than predicted (Fig. 2A). cDNA sequencing of the cloned RT-PCR products revealed that the mutant Dsc1 mRNA retained intron 15. Our sequence analysis predicts that the mutant protein lacks the 11 amino acids encoded by exon 16. Instead, the COOH terminus of the mutant protein contains an unrelated sequence of 13 amino acids encoded by intron 15 (Fig. 2B). We did not find mutations in exon 15, intron 15, or exon 16 of the dsc1ΔE17LoxP allele. This indicates that the abnormal splicing is not due to a mutation in splice sites located in intron 15.
FIG. 2.
Characterization of dsc1 transcripts in newborn epidermis of wild-type (WT) and homozygous mutant (MT; dsc1−/−ΔE17LoxP) mice by RT-PCR. The 5′ primer was derived from the junction between exons 14 and 15, i.e., designed to suppress amplification of genomic DNA. The 3′ primer was derived from exon 16. (A) Agarose gel with RT-PCR products derived from three wild-type and three mutant samples. The PCR products were sequenced. The exon-intron organization of the product is schematically shown. (B) dsc1 gene structure and mRNA in wild-type and mutant mice. Note that exons 16 and 17 both contain stop codons. In mutant mice, we detected a single transcript that was aberrantly spliced, i.e., retained intron 15. DNA sequence analysis predicted that the mutant (MT) transcript encodes a protein in which the Dsc1b-specific sequence ESIRGHTLIKN is replaced by the sequence VSAQSSSAHSVQC. The sequence RLGE is encoded by the 3′ end of exon 15. Note that we have not confirmed the presence of the aberrant amino acid sequence at the COOH terminus of mutant Dsc1 by protein sequencing.
We have repeated the RT-PCR analysis with various primer pairs, all of which confirmed that intron 15 is present in the mutant dsc1 transcript (data not shown). Furthermore, RPAs with a probe consisting of exon 15 and exon 16 sequences failed to detect correctly spliced transcripts in mutant mice (data not shown).
The COOH-truncated Dsc1 receptor does not bind desmosomal plaque proteins plakoglobin and PKP1.
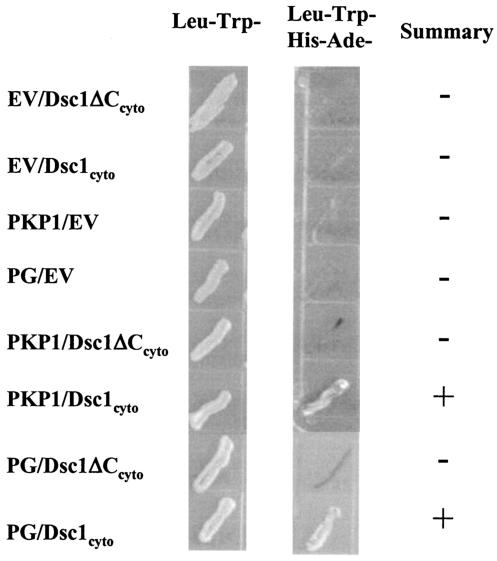
The Dsc1 receptor synthesized by dsc1−/−ΔE17LoxP mutant mice lacks COOH-terminal cytoplasmic amino acid sequences that have previously been shown to bind plakoglobin and PKP (16, 65, 66). To confirm that the mutant Dsc1 receptor does not bind these proteins, we carried out a yeast-two-hybrid analysis. As shown in Fig. 3, the cytoplasmic domain of wild-type Dsc1a binds plakoglobin and PKP, whereas the mutant receptor does not.
FIG. 3.
Molecular interactions of the cytoplasmic domains of Dsc1a (Dsc1cyto) and the truncated Dsc1 receptor (Dsc1ΔCcyto) synthesized in dsc1−/−ΔE17LoxP mutant mice. Yeast two-hybrid experiments were carried out to test for direct interactions by using growth in the absence of histidine and adenine (middle column) as a reporter for interactions between the cytoplasmic domains of the two Dsc1 molecules and several components of the desmosomal plaque. The cytoplasmic domain of Dsc1a, but not the COOH-terminally truncated Dsc1 mutant, interacts directly with the head domain of PKP1 and plakoglobin. Both Dsc polypeptides (Dsc1cyto and Dsc1ΔCcyto) were expressed as fusion proteins with the Gal4 activation domain and tested for interactions with either empty DNA binding domain vector (EV), the head domain of PKP1, or plakoglobin (PG) cloned into the Gal4 DNA binding domain vector.
The Dsc1 receptor synthesized in dsc1−/−ΔE17LoxP mutant mice integrates into desmosomes.
We have generated polyclonal antibodies against the cytoplasmic domain of mouse Dsc1. These antibodies recognize epitopes in a sequence of 107 amino acids that is present in Dsc1a, Dsc1b, and the predicted protein synthesized in dsc1−/−ΔE17LoxP mutant mice. In Western blot assays with wild-type samples, the Dsc1 antibodies recognized two bands with the predicted molecular weights of Dsc1a and Dsc1b (Fig. 1D). The Dsc1 band recognized in protein extracts of mutant samples represents the truncated Dsc1 receptor and not Dsc1b.
Consistent with published data regarding the distribution of Dsc1 in mouse skin (see Introduction), these antibodies stained the suprabasal layers of the epidermis and the inner root sheath of anagen hair follicles. To determine whether the mutant protein was incorporated into desmosomes of dsc1−/−ΔE17LoxP mutant mice, we costained epidermis of newborn pups with the Dsc1 antibodies and antibodies against desmoplakin. As shown in Fig. 4A, the mutant protein colocalized with desmoplakin, suggesting that neither the Dsc1a-specific nor the Dsc1b-specific COOH-terminal domain is required for assembly into the desmosomal protein complex. The desmosomal localization of the mutant receptor was confirmed by low-temperature immunoelectron microscopy (Fig. 4B). Furthermore, conventional transmission electron microscopy revealed no gross abnormalities in the structure and abundance of desmosomes in the granular layers of mutant epidermis (data not shown).
FIG. 4.
Subcellular localization of Dsc1 in newborn epidermis of wild-type (WT) and mutant (MT) mice. (A) Deconvolution microscopy. The sections were stained with antibody gp899 (Dsc1; red) and a monoclonal antibody against desmoplakin (green). Colocalization resulted in yellow fluorescence. Note that the mutant protein coassembles with desmoplakin into desmosomes. Identical results were obtained by staining with gp899 and a monoclonal antibody against Dsg1 and Dsg2 (DG3.10; data not shown). (B) Low-temperature immunoelectron microscopy. Mutant (a, c, e, g) and wild-type (b, d, f, h) samples were incubated with various antibodies. (a, b) gp899 (silver enhancement; see Materials and Methods). (c, d) Higher magnification of desmosomes stained with gp899. (e, f) Costaining with gp899 (5-nm gold particles) and desmoplakin antibodies (15-nm gold particles). (g, h) Costaining with gp899 (5-nm gold particles) and plakoglobin antibodies (15-nm gold particles). Note that the mutant Dsc1 receptor is integrated into desmosomes and that the staining patterns of desmoplakin and plakoglobin are not affected by the mutation. Bars: a and b, 1 μm; c to h, 0.1 μm.
Normal expression of differentiation markers in mutant epidermis.
In order to determine whether the dsc1 mutation affected epidermal differentiation, we tested the tissue distribution and expression levels of various differentiation markers. Immunofluorescence microscopy of newborn skin of mutants and wild-type controls showed normal epidermal staining with antibodies against desmosomal proteins (Dsc3, Dsg1, Dsg2, plakoglobin, PKP1, and PKP3), keratins (K14, K10, and K6), a keratin-associated protein (filaggrin), and cornified cell envelope components (repetin and loricrin) (data not shown).
We also determined the proliferation index in the skin of mutant and wild-type control mice that were injected with bromodeoxyuridine. We did not find a significant difference in the number of bromodeoxyuridine-positive keratinocytes in mutant and control samples (data not shown).
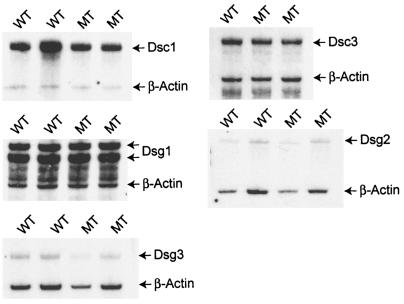
Western blot assays with antibodies against desmosomal proteins that are coexpressed with Dsc1 in the epidermis did not show significant changes in their expression levels (examples are shown in Fig. 5). Antibody DG3.10, which we used in our experiments, cross-reacts with Dsg1 and Dsg2 (36, 39). Therefore, we confirmed the protein expression data for these desmogleins by RPA (Fig. 6). No differences were found in the expression levels of Dsg1 and Dsg2 in mutant mice.
FIG. 5.
Western blot analysis of total-tissue lysates from back skin of newborn wild-type (WT) and dsc1−/−ΔE17LoxP mutant (MT) mice. Equal amounts of protein were blotted with antibodies against Dsg1 and Dsg2 (DG3.10), Dsg3, Dsc3 (gp2280; see Materials and Methods), plakoglobin (Pg), PKP1 (PP1), PKP3 (PP3), and β-catenin (β-cat). No significant differences in the expression levels of the analyzed proteins in wild-type and mutant samples were observed.
FIG. 6.
RNase protection assays to determine the expression levels of desmosomal cadherins in wild-type (WT) and dsc1−/−ΔE17LoxP mutant (MT) mice. RNA was isolated from the back epidermis of newborn mice. β-Actin served as an internal control in each experiment. Normalization of the expression data revealed that none of the markers showed a significant change (>50%) in their expression levels due to the dsc1 mutation. Note that the Dsg1 probe yielded two bands, which is probably due to a mouse strain polymorphism. The Dsg1 probe was derived from C57BL/6 genomic DNA. The mutant and wild-type samples tested were derived from mice on a segregating C57/BL6 and 129/SV background.
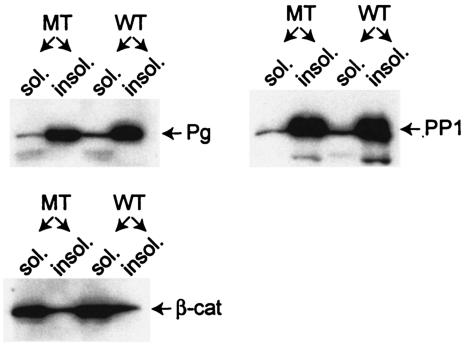
Next, we extracted mutant and wild-type skin samples with TX-containing buffers and subjected the protein fractions that were soluble and insoluble in this detergent, respectively, to Western blot analysis. Insolubility in TX is a characteristic of proteins assembled into desmosomes (52-54). We were particularly interested in possible changes in the cytoplasmic pools of PKPs and plakoglobin in mutant epidermis. Both plaque proteins bind to Dsc1a and are found in the nuclei, where they are thought to influence gene expression (4, 7, 9, 11, 12, 25, 46, 47, 59). As shown in Fig. 7, the distribution of plakoglobin and PKP1 in the TX-soluble and -insoluble pools was not affected by the mutation. In mutant and wild-type samples, these two plaque proteins were mainly found in the TX-insoluble (cytoskeletal) protein fraction.
FIG. 7.
Western blot analysis of the TX-soluble (sol.) and -insoluble (insol.) protein fractions from the back skin of newborn wild-type (WT) and dsc1−/−ΔE17LoxP mutant (MT) mice. Note that plakoglobin (Pg) and PKP1 (PP1) are mainly found in the insoluble fraction, which is characteristic for junction-associated proteins. β-Catenin, on the other hand, is present predominantly in the soluble cytoplasmic pool. Again, no significant difference was observed between wild-type and mutant samples.
Abnormal expression of Dsc2 in mutant mice.
Surprisingly, the only desmosomal gene tested that showed a significant change in its expression level in dsc1−/−ΔE17LoxP mutant mice was dsc2 (Fig. 6 contains expression data on other desmosomal cadherins). RPA analysis showed a dramatic up-regulation of this gene in mutant mice. In the examples shown in Fig. 8A, the Dsc2 mRNA levels were increased by a factor of 38. In situ hybridization experiments confirmed this finding and demonstrated that Dsc2 is expressed mainly in the suprabasal layers of the newborn epidermis (Fig. 8B). Unfortunately, we were not able to assess the tissue distribution of Dsc2 with our antibody, because it did not stain formalin-fixed (paraffin-embedded) or unfixed (frozen) skin sections.
FIG. 8.
Effects of the dsc1 mutation on the expression of Dsc2. (A) RPA analysis of epidermal RNA from newborn wild-type (WT) and dsc1−/−ΔE17LoxP mutant (MT) samples. Note the dramatic increase in Dsc2 expression (approximately 38-fold) in the two mutant samples. β-Actin was used as an internal standard to normalize the expression data. (B) In situ hybridization with Dsc2 antisense probes. Note the strong suprabasal expression of Dsc2 in the mutant sample. The signal obtained with wild-type samples was barely above the background. A sense probe did not yield a signal on mutant or wild-type samples (data not shown). The dotted line indicates the position of the basement membrane. (C) Western blot analysis using total skin extracts from newborn mice with antibody gp2295. Note that Dsc2a and Dsc2b are expressed at similar levels in wild-type and homozygous mutant samples.
Surprisingly, Western blot assays did not indicate a concomitant increase in Dsc2 protein synthesis in mutant mice (Fig. 8C).
DISCUSSION
We have generated mutants that synthesize a truncated Dsc1 protein that contains neither the a nor the b variant-specific COOH-terminal (cytoplasmic) amino acid sequence. The truncated adhesion receptor consists of the extracellular domain of Dsc1, as well as the intracellular sequence (107 amino acids) that is common to both Dsc1a and Dsc1b. The mechanism that leads to aberrant splicing of the mutant Dsc1 pre-mRNA is not clear. The deletion of intron 16 or the introduction of an artificial XhoI site at the 3′ end of exon 16, which was necessary to fuse that exon to the 3′ UTR of the gene, could have contributed to this phenomenon. On the basis of our sequence analysis of RT-PCR products, we can rule out the possibility that a mutation in intron 15 contributed to the aberrant splicing.
Our microscopy data indicate that the truncated Dsc1 protein integrates into desmosomes in the upper spinous and granular layers of the epidermis. Given the fact that this protein does not bind plakoglobin or PKP1, it is tempting to speculate that this integration is mediated through direct interactions with desmogleins, either in cis (within the plane of the plasma membrane) or in trans (with desmogleins of an adjacent cell). We have also tested the head domain of desmoplakin for interaction with our Dsc constructs in yeast two-hybrid assays. However, this construct (consisting of amino acid positions 1 to 585; DPNTP in reference 8) did not directly bind to either Dsc construct (data not shown).
It remains to be seen whether the truncated Dsc1 receptor binds other cytoplasmic proteins and whether these interactions are required for desmosomal localization.
The observation that our mutant mice show no apparent phenotype was surprising. Mouse strains with null mutations in desmosomal cadherin genes that are expressed in skin (e.g., dsc1, dsg3, and dsg4) (14, 33, 37, 38, 55) show pathological phenotypes; i.e., functional compensation by desmosomal cadherins has not been observed previously.
Furthermore, it appears that loss of cytoplasmic function (e.g., plakoglobin and PKP binding in our mutant receptor) is better tolerated than loss of extracellular functions (see below).
The most likely explanation for the occurrence of functional desmosomes in our mouse model is that Dsc2 and Dsc3, which are both expressed throughout the suprabasal layers of the epidermis (references 15, 26, and 51 and our own data), are sufficient to establish and maintain the structural integrity of desmosomes.
An analysis of a large set of epidermal differentiation markers failed to reveal abnormal expression patterns in mutant epidermis by RPAs and Western blot assays, with one exception; the expression of Dsc2 mRNA was dramatically increased in mutant skin. This phenomenon was consistently observed in mutant mice. Nevertheless, Western blot assays indicated that the amount of Dsc2 protein synthesized in mutants was normal. It is noteworthy that the mRNA expression levels of all other desmosomal cadherins were not significantly changed in mutant mice.
It is possible that the increase in Dsc2 expression constitutes a stress response, i.e., subtle defects in the mutant epidermis triggering the increase in transcription. However, the lack of K6 induction in the epidermis of mutants suggests that this is not the case. K6 is synthesized in the interfollicular epidermis in response to mechanical and biochemical insults, as well as abnormal differentiation (see references in references 19 and 68).
Fail-safe mechanisms must exist that prevent increased synthesis of Dsc2. It has been shown that ectopic overexpression of at least one desmosomal cadherin (Dsg3) in transgenic mice can cause abnormal epidermal differentiation (20, 45). It is therefore reasonable to assume that the expression levels of desmosomal cadherins are tightly controlled in vivo. Future studies will have to identify the molecular mechanisms by which Dsc2 protein levels are controlled.
Chidgey and colleagues recently generated dsc1-null mice, i.e., animals that synthesize neither Dsc1a nor Dsc1b (14). Newborn dsc1-null mice showed localized acantholysis in the granular layer of the epidermis. Furthermore, the epidermis showed hyperproliferation and interfollicular expression of stress proteins K6 and K16. Parakeratosis (retention of nuclei in the cornified cell layer of the epidermis) suggested abnormal differentiation of keratinocytes. Localized skin barrier defects were found in newborn mutant mice. It is possible that some of the abnormal features of dsc1-null epidermis are due to these barrier defects caused by localized acantholysis. Older mice developed ulcerating lesions and hair loss. Interestingly, Chidgey and colleagues did not observe a change in the synthesis of the Dsc2 and Dsc3 proteins, which is consistent with the data presented in the present report.
On the basis of the Dsc1-null phenotype, it is safe to conclude that the dsc1ΔE17LoxP allele does not represent a null mutation. Obviously, this mutation does not create a dominant-negative allele either. Nevertheless, a comparison of the two animal models suggests that the extracellular domain of Dsc1 and maybe the cytoplasmic domain that is common to Dsc1a, Dsc1b, and the truncated Dsc1 receptor are necessary and sufficient for normal epidermal development and homeostasis.
Acknowledgments
We thank the following coworkers and colleagues: Mitsi Blount, Mary Ann Longley, Donna Wang, and Stephanie Purdum for technical assistance; Susan Berget for helpful discussions about splicing of the Dsc1 RNA in our mutant mice; Werner W. Franke and Lutz Langbein (German Cancer Research Center), Daniel Hohl (CHUV/DHURDV, Lausanne, Switzerland), and Dennis Roop for the generous gifts of antibodies; and John Sundberg (The Jackson Laboratory) for help with analyzing histology slides.
This work was supported in part by a grant (AR47343) from the National Institutes of Health (to P.J.K.) and a Career Development Award from the Dermatology Foundation (to P.J.K.).
REFERENCES
- 1.Allen, E., Q. C. Yu, and E. Fuchs. 1996. Mice expressing a mutant desmosomal cadherin exhibit abnormalities in desmosomes, proliferation, and epidermal differentiation. J. Cell Biol. 133:1367-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amagai, M., N. Matsuyoshi, Z. H. Wang, C. Andl, and J. R. Stanley. 2000. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat. Med. 6:1275-1277. [DOI] [PubMed] [Google Scholar]
- 3.Arnemann, J., K. H. Sullivan, A. I. Magee, I. A. King, and R. S. Buxton. 1993. Stratification-related expression of isoforms of the desmosomal cadherins in human epidermis. J. Cell Sci. 104(Pt. 3):741-750. [DOI] [PubMed] [Google Scholar]
- 4.Barker, N., and H. Clevers. 2000. Catenins, Wnt signaling and cancer. Bioessays 22:961-965. [DOI] [PubMed] [Google Scholar]
- 5.Bierkamp, C., K. J. McLaughlin, H. Schwarz, O. Huber, and R. Kemler. 1996. Embryonic heart and skin defects in mice lacking plakoglobin. Dev. Biol. 180:780-785. [DOI] [PubMed] [Google Scholar]
- 6.Bierkamp, C., H. Schwarz, O. Huber, and R. Kemler. 1999. Desmosomal localization of beta-catenin in the skin of plakoglobin null-mutant mice. Development 126:371-381. [DOI] [PubMed] [Google Scholar]
- 7.Bonne, S., J. van Hengel, F. Nollet, P. Kools, and F. van Roy. 1999. Plakophilin-3, a novel armadillo-like protein present in nuclei and desmosomes of epithelial cells. J. Cell Sci. 112(Pt. 14):2265-2276. [DOI] [PubMed] [Google Scholar]
- 8.Bornslaeger, E. A., C. M. Corcoran, T. S. Stappenbeck, and K. J. Green. 1996. Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 134:985-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borrmann, C. M., C. Mertens, A. Schmidt, L. Langbein, C. Kuhn, and W. W. Franke. 2000. Molecular diversity of plaques of epithelial-adhering junctions. Ann. N. Y. Acad. Sci. 915:144-150. [DOI] [PubMed] [Google Scholar]
- 10.Calkins, C. C., B. L. Hoepner, C. M. Law, M. R. Novak, S. V. Setzer, M. Hatzfeld, and A. P. Kowalczyk. 2003. The armadillo family protein p0071 is a VE-cadherin- and desmoplakin-binding protein. J. Biol. Chem. 278:1774-1783. [DOI] [PubMed] [Google Scholar]
- 11.Charpentier, E., R. M. Lavker, E. Acquista, and P. Cowin. 2000. Plakoglobin suppresses epithelial proliferation and hair growth in vivo. J. Cell Biol. 149:503-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, X. Y., S. Bonne, M. Hatzfeld, F. van Roy, and K. J. Green. 2002. Protein binding and functional characterization of plakophilin 2—evidence for its diverse roles in desmosomes and beta-catenin signaling. J. Biol. Chem. 277:10512-10522. [DOI] [PubMed] [Google Scholar]
- 13.Chidgey, M. 2002. Desmosomes and disease: an update. Histol. Histopathol. 17:1179-1192. [DOI] [PubMed] [Google Scholar]
- 14.Chidgey, M., C. Brakebusch, E. Gustafsson, A. Cruchley, C. Hail, S. Kirk, A. Merritt, A. North, C. Tselepis, J. Hewitt, C. Byrne, R. Fassler, and D. Garrod. 2001. Mice lacking desmocollin 1 show epidermal fragility accompanied by barrier defects and abnormal differentiation. J. Cell Biol. 155:821-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chidgey, M. A., K. K. Yue, S. Gould, C. Byrne, and D. R. Garrod. 1997. Changing pattern of desmocollin 3 expression accompanies epidermal organisation during skin development. Dev. Dyn. 210:315-327. [DOI] [PubMed] [Google Scholar]
- 16.Chitaev, N. A., R. E. Leube, R. B. Troyanovsky, L. G. Eshkind, W. W. Franke, and S. M. Troyanovsky. 1996. The binding of plakoglobin to desmosomal cadherins: patterns of binding sites and topogenic potential. J. Cell Biol. 133:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chitaev, N. A., and S. M. Troyanovsky. 1997. Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell-cell adhesion. J. Cell Biol. 138:193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coulombe, P. A. 2002. A new fold on an old story: attachment of intermediate filaments to desmosomes. Nat. Struct. Biol. 9:560-562. [DOI] [PubMed] [Google Scholar]
- 19.Coulombe, P. A., and M. B. Omary. 2002. 'Hard' and 'soft' principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 14:110-122. [DOI] [PubMed] [Google Scholar]
- 20.Elias, P. M., N. Matsuyoshi, H. Wu, C. Lin, Z. H. Wang, B. E. Brown, and J. R. Stanley. 2001. Desmoglein isoform distribution affects stratum corneum structure and function. J. Cell Biol. 153:243-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eshkind, L., Q. Tian, A. Schmidt, W. W. Franke, R. Windoffer, and R. E. Leube. 2002. Loss of desmoglein 2 suggests essential functions for early embryonic development and proliferation of embryonal stem cells. Eur. J. Cell Biol. 81:592-598. [DOI] [PubMed] [Google Scholar]
- 22.Franke, W. W., P. J. Koch, S. Schafer, H. W. Heid, S. M. Troyanovsky, I. Moll, and R. Moll. 1994. The desmosome and the syndesmos: cell junctions in normal development and in malignancy. Princess Takamatsu Symp. 24:14-27. [PubMed] [Google Scholar]
- 23.Gallicano, G. I., C. Bauer, and E. Fuchs. 2001. Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development 128:929-941. [DOI] [PubMed] [Google Scholar]
- 24.Gallicano, G. I., P. Kouklis, C. Bauer, M. Yin, V. Vasioukhin, L. Degenstein, and E. Fuchs. 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143:2009-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrod, D. R., A. J. Merritt, and Z. Nie. 2002. Desmosomal adhesion: structural basis, molecular mechanism and regulation (review). Mol. Membr. Biol. 19:81-94. [DOI] [PubMed] [Google Scholar]
- 26.Garrod, D. R., A. J. Merritt, and Z. Nie. 2002. Desmosomal cadherins. Curr. Opin. Cell Biol. 14:537-545. [DOI] [PubMed] [Google Scholar]
- 27.Green, K. J., and C. A. Gaudry. 2000. Are desmosomes more than tethers for intermediate filaments? Nat. Rev. Mol. Cell Biol. 1:208-216. [DOI] [PubMed] [Google Scholar]
- 28.Isac, C. M., P. Ruiz, B. Pfitzmaier, H. Haase, W. Birchmeier, and I. Morano. 1999. Plakoglobin is essential for myocardial compliance but dispensable for myofibril insertion into adherens junctions. J. Cell. Biochem. 72:8-15. [PubMed] [Google Scholar]
- 29.Ishiko, A., H. Shimizu, A. Kikuchi, T. Ebihara, T. Hashimoto, and T. Nishikawa. 1993. Human autoantibodies against the 230-kD bullous pemphigoid antigen (BPAG1) bind only to the intracellular domain of the hemidesmosome, whereas those against the 180-kD bullous pemphigoid antigen (BPAG2) bind along the plasma membrane of the hemidesmosome in normal human and swine skin. J. Clin. Investig. 91:1608-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamora, C., and E. Fuchs. 2002. Intercellular adhesion, signalling and the cytoskeleton. Nat. Cell Biol. 4:E101-E108. [DOI] [PubMed] [Google Scholar]
- 31.Jarnik, M., T. Kartasova, P. M. Steinert, U. Lichti, and A. C. Steven. 1996. Differential expression and cell envelope incorporation of small proline-rich protein 1 in different cornified epithelia. J. Cell Sci. 109:1381-1391. [DOI] [PubMed] [Google Scholar]
- 32.King, I. A., T. J. O'Brien, and R. S. Buxton. 1996. Expression of the “skin-type” desmosomal cadherin DSC1 is closely linked to the keratinization of epithelial tissues during mouse development. J. Investig. Dermatol. 107:531-538. [DOI] [PubMed] [Google Scholar]
- 33.Kljuic, A., H. Bazzi, J. P. Sundberg, A. Martinez-Mir, R. O'Shaughnessy, M. G. Mahoney, M. Levy, X. Montagutelli, W. Ahmad, V. M. Aita, D. Gordon, J. Uitto, D. Whiting, J. Ott, S. Fischer, T. C. Gilliam, C. A. Jahoda, R. J. Morris, A. A. Panteleyev, V. T. Nguyen, and A. M. Christiano. 2003. Desmoglein 4 in hair follicle differentiation and epidermal adhesion: evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 113:249-260. [DOI] [PubMed] [Google Scholar]
- 34.Koch, P. J., P. A. de Viragh, E. Scharer, D. Bundman, M. A. Longley, J. Bickenbach, Y. Kawachi, Y. Suga, Z. Zhou, M. Huber, D. Hohl, T. Kartasova, M. Jarnik, A. C. Steven, and D. R. Roop. 2000. Lessons from loricrin-deficient mice. Compensatory mechanisms maintaining skin barrier function in the absence of a major cornified envelope protein. J. Cell Biol. 151:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koch, P. J., and W. W. Franke. 1994. Desmosomal cadherins: another growing multigene family of adhesion molecules. Curr. Opin. Cell Biol. 6:682-687. [DOI] [PubMed] [Google Scholar]
- 36.Koch, P. J., M. D. Goldschmidt, M. J. Walsh, R. Zimbelmann, and W. W. Franke. 1991. Complete amino acid sequence of the epidermal desmoglein precursor polypeptide and identification of a second type of desmoglein gene. Eur. J. Cell Biol. 55:200-208. [PubMed] [Google Scholar]
- 37.Koch, P. J., M. G. Mahoney, G. Cotsarelis, K. Rothenberger, R. M. Lavker, and J. R. Stanley. 1998. Desmoglein 3 anchors telogen hair in the follicle. J. Cell Sci. 111(Pt. 17):2529-2537. [DOI] [PubMed] [Google Scholar]
- 38.Koch, P. J., M. G. Mahoney, H. Ishikawa, L. Pulkkinen, J. Uitto, L. Shultz, G. F. Murphy, D. Whitaker-Menezes, and J. R. Stanley. 1997. Targeted disruption of the pemphigus vulgaris antigen (desmoglein 3) gene in mice causes loss of keratinocyte cell adhesion with a phenotype similar to pemphigus vulgaris. J. Cell Biol. 137:1091-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch, P. J., M. J. Walsh, M. Schmelz, M. D. Goldschmidt, R. Zimbelmann, and W. W. Franke. 1990. Identification of desmoglein, a constitutive desmosomal glycoprotein, as a member of the cadherin family of cell adhesion molecules. Eur. J. Cell Biol. 53:1-12. [PubMed] [Google Scholar]
- 40.Kowalczyk, A. P., E. A. Bornslaeger, S. M. Norvell, H. L. Palka, and K. J. Green. 1999. Desmosomes: intercellular adhesive junctions specialized for attachment of intermediate filaments. Int. Rev. Cytol. 185:237-302. [DOI] [PubMed] [Google Scholar]
- 41.Kowalczyk, A. P., M. Hatzfeld, E. A. Bornslaeger, D. S. Kopp, J. E. Borgwardt, C. M. Corcoran, A. Settler, and K. J. Green. 1999. The head domain of plakophilin-1 binds to desmoplakin and enhances its recruitment to desmosomes. Implications for cutaneous disease. J. Biol. Chem. 274:18145-18148. [DOI] [PubMed] [Google Scholar]
- 42.Kurzen, H., I. Moll, R. Moll, S. Schafer, E. Simics, M. Amagai, M. J. Wheelock, and W. W. Franke. 1998. Compositionally different desmosomes in the various compartments of the human hair follicle. Differentiation 63:295-304. [DOI] [PubMed] [Google Scholar]
- 43.Marcozzi, C., I. D. Burdett, R. S. Buxton, and A. I. Magee. 1998. Coexpression of both types of desmosomal cadherin and plakoglobin confers strong intercellular adhesion. J. Cell Sci. 111(Pt. 4):495-509. [DOI] [PubMed] [Google Scholar]
- 44.Masunaga, T., H. Shimizu, A. Ishiko, T. Fujiwara, T. Hashimoto, and T. Nishikawa. 1995. Desmoyokin/AHNAK protein localizes to the non-desmosomal keratinocyte cell surface of human epidermis. J. Investig. Dermatol. 104:941-945. [DOI] [PubMed] [Google Scholar]
- 45.Merritt, A. J., M. Y. Berika, W. Zhai, S. E. Kirk, B. Ji, M. J. Hardman, and D. R. Garrod. 2002. Suprabasal desmoglein 3 expression in the epidermis of transgenic mice results in hyperproliferation and abnormal differentiation. Mol. Cell. Biol. 22:5846-5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mertens, C., I. Hofmann, Z. Wang, M. Teichmann, C. S. Sepehri, M. Schnolzer, and W. W. Franke. 2001. Nuclear particles containing RNA polymerase III complexes associated with the junctional plaque protein plakophilin 2. Proc. Natl. Acad. Sci. USA 98:7795-7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mertens, C., C. Kuhn, and W. W. Franke. 1996. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J. Cell Biol. 135:1009-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.North, A. J., W. G. Bardsley, J. Hyam, E. A. Bornslaeger, H. C. Cordingley, B. Trinnaman, M. Hatzfeld, K. J. Green, A. I. Magee, and D. R. Garrod. 1999. Molecular map of the desmosomal plaque. J. Cell Sci. 112(Pt. 23):4325-4336. [DOI] [PubMed] [Google Scholar]
- 49.North, A. J., M. A. Chidgey, J. P. Clarke, W. G. Bradsley, and D. R. Garrod. 1996. Distinct desmocollin isoforms occur in the same desmosome and show reciprocally graded distribution in bovine nasal epidermis. Proc. Natl. Acad. Sci. USA 93:7701-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nuber, U. A., S. Schafer, A. Schmidt, P. J. Koch, and W. W. Franke. 1995. The widespread human desmocollin Dsc2 and tissue-specific patterns of synthesis of various desmocollin subtypes. Eur. J. Cell Biol. 66:69-74. [PubMed] [Google Scholar]
- 51.Nuber, U. A., S. Schafer, S. Stehr, H. R. Rackwitz, and W. W. Franke. 1996. Patterns of desmocollin synthesis in human epithelia: immunolocalization of desmocollins 1 and 3 in special epithelia and in cultured cells. Eur. J. Cell Biol. 71:1-13. [PubMed] [Google Scholar]
- 52.Pasdar, M., and W. J. Nelson. 1988. Kinetics of desmosome assembly in Madin-Darby canine kidney epithelial cells: temporal and spatial regulation of desmoplakin organization and stabilization upon cell-cell contact. I. Biochemical analysis. J. Cell Biol. 106:677-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasdar, M., and W. J. Nelson. 1989. Regulation of desmosome assembly in epithelial cells: kinetics of synthesis, transport, and stabilization of desmoglein I, a major protein of the membrane core domain. J. Cell Biol. 109:163-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Penn, E. J., I. D. Burdett, C. Hobson, A. I. Magee, and D. A. Rees. 1987. Structure and assembly of desmosome junctions: biosynthesis and turnover of the major desmosome components of Madin-Darby canine kidney cells in low calcium medium. J. Cell Biol. 105:2327-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pulkkinen, L., Y. W. Choi, A. Simpson, X. Montagutelli, J. Sundberg, J. Uitto, and M. G. Mahoney. 2002. Loss of cell adhesion in Dsg3bal-Pas mice with homozygous deletion mutation (2079del14) in the desmoglein 3 gene. J. Investig. Dermatol. 119:1237-1243. [DOI] [PubMed] [Google Scholar]
- 56.Ramirez-Solis, R., A. C. Davis, and A. Bradley. 1993. Gene targeting in embryonic stem cells. Methods Enzymol. 225:855-879. [DOI] [PubMed] [Google Scholar]
- 57.Ramirez-Solis, R., J. Rivera-Perez, J. D. Wallace, M. Wims, H. Zheng, and J. D. Bradley. 1992. Genomic DNA microextraction: a method to screen numerous samples. Anal. Biochem. 201:331-335. [DOI] [PubMed] [Google Scholar]
- 58.Ruiz, P., V. Brinkmann, B. Ledermann, M. Behrend, C. Grund, C. Thalhammer, F. Vogel, C. Birchmeier, U. Gunthert, W. W. Franke, and W. Birchmeier. 1996. Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J. Cell Biol. 135:215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt, A., L. Langbein, M. Rode, S. Pratzel, R. Zimbelmann, and W. W. Franke. 1997. Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res. 290:481-499. [DOI] [PubMed] [Google Scholar]
- 60.Schwarz, M. A., K. Owaribe, J. Kartenbeck, and W. W. Franke. 1990. Desmosomes and hemidesmosomes: constitutive molecular components. Annu. Rev. Cell Biol. 6:461-491. [DOI] [PubMed] [Google Scholar]
- 61.Shimizu, H., T. Masunaga, A. Ishiko, T. Hashimoto, D. R. Garrod, H. Shida, and T. Nishikawa. 1994. Demonstration of desmosomal antigens by electron microscopy using cryofixed and cryosubstituted skin with silver-enhanced gold probe. J. Histochem. Cytochem. 42:687-692. [DOI] [PubMed] [Google Scholar]
- 62.Stanley, J. R., T. Nishikawa, L. A. Diaz, and M. Amagai. 2001. Pemphigus: is there another half of the story? J. Investig. Dermatol. 116:489-490. [DOI] [PubMed] [Google Scholar]
- 63.Syed, S. E., B. Trinnaman, S. Martin, S. Major, J. Hutchinson, and A. I. Magee. 2002. Molecular interactions between desmosomal cadherins. Biochem. J. 362:317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Troyanovsky, S. M. 1999. Mechanism of cell-cell adhesion complex assembly. Curr. Opin. Cell Biol. 11:561-566. [DOI] [PubMed] [Google Scholar]
- 65.Troyanovsky, S. M., L. G. Eshkind, R. B. Troyanovsky, R. E. Leube, and W. W. Franke. 1993. Contributions of cytoplasmic domains of desmosomal cadherins to desmosome assembly and intermediate filament anchorage. Cell 72:561-574. [DOI] [PubMed] [Google Scholar]
- 66.Troyanovsky, S. M., R. B. Troyanovsky, L. G. Eshkind, R. E. Leube, and W. W. Franke. 1994. Identification of amino acid sequence motifs in desmocollin, a desmosomal glycoprotein, that are required for plakoglobin binding and plaque formation. Proc. Natl. Acad. Sci. USA 91:10790-10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vasioukhin, V., E. Bowers, C. Bauer, L. Degenstein, and E. Fuchs. 2001. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076-1085. [DOI] [PubMed] [Google Scholar]
- 68.Wojcik, S. M., D. S. Bundman, and D. R. Roop. 2000. Delayed wound healing in keratin 6a knockout mice. Mol. Cell. Biol. 20:5248-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]