Abstract
Structural chromosome aberrations and associated segmental or chromosomal aneusomies are major causes of reproductive failure in humans. Despite the fact that carriers of reciprocal balanced translocation often have no other clinical symptoms or disease, impaired chromosome homologue pairing in meiosis and karyokinesis errors lead to over-representation of translocations carriers in the infertile population and in recurrent pregnancy loss patients. At present, clinicians have no means to select healthy germ cells or balanced zygotes in vivo, but in vitro fertilization (IVF) followed by preim-plantation genetic diagnosis (PGD) offers translocation carriers a chance to select balanced or normal embryos for transfer. Although a combination of telomeric and centromeric probes can differentiate embryos that are unbalanced from normal or unbalanced ones, a seemingly random position of breakpoints in these IVF-patients poses a serious obstacle to differentiating between normal and balanced embryos, which for most translocation couples, is desirable. Using a carrier with reciprocal translocation t(4;13) as an example, we describe our state-of-the-art approach to the preparation of patient-specific DNA probes that span or ‘extent’ the breakpoints. With the techniques and resources described here, most breakpoints can be accurately mapped in a matter of days using carrier lymphocytes, and a few extra days are allowed for PGD-probe optimization. The optimized probes will then be suitable for interphase cell analysis, a prerequisite for PGD since blastomeres are biopsied from normally growing day 3 - embryos regardless of their position in the mitotic cell cycle. Furthermore, routine application of these rapid methods should make PGD even more affordable for translocation carriers enrolled in IVF programs.
Keywords: cytogenetics, chromosome aberration, translocation, IVF, PGD, fluorescence in situ hybridization (FISH), bacterial artificial chromosome (BAC), DNA probes
Introduction
The human genome is not cast in stone; it is believed to undergo changes with every generation. Besides paternal and maternal contributions to the genome of an individual human being, de novo changes may occur during the earliest steps of human reproduction, i.e., during generation of germ cells in early human development, or a later time point as we witness genetic changes in somatic cells that spur off cancer development. Looking at chromosomal changes from the standpoint of reproductive genetics, one notes that congenital anomalies which include balanced and Robertsonian translocations as well as chromosomal inversions occur in as much as 1.4% of the general population [1]. Among infertile couples and IVF patients with recurrent abortions, such structural chromosome abnormalities have been observed at even higher rates [1–2]. Stern and colleagues, for example, reported balanced translocations in 0.6% of all infertile couples, but in 3.2% and 9.2% of couples who failed more than 10 IVF cycles or experienced three or more consecutive first-trimester abortions, respectively [3].
A common consequence of balanced reciprocal translocations in carriers without clinical disease symptoms is an increased fraction of germ cells with numerical chromosome aberrations. This has been attributed to disturbed homologue pairing during meiosis or precocious chromatid separation [4–5]. As a clinical manifestation of this problem, patients suffer from reduced fertility, infertility or a history of repeated miscarriages [6]. During the course of IVF, PGD can now be offered to affected couples as an alternative to prenatal diagnosis and medically-indicated termination of pregnancies with chromosomally-unbalanced fetuses [7–9]. If a sufficient number of fertilized normal embryos is available for transfer, PGD also provides an efficient option to put an end to a familial disease [6]. However, the greatest benefit of PGD is the reduction of spontaneous abortions [10]. On the other hand, the pregnancy rates after PGD among couples carrying non-Robertsonian translocations may not improve much due to high prevalence of abnormal embryos [6,11].
The conventional cytogenetic methods, i.e., chromosome banding procedures, are challenged when dealing with very subtle chromosome rearrangements, particularly de novo abnormalities in newborns. Even more limiting, banding analysis requires cells in metaphase, but the blastomeres biopsied from day 3 embryos can be in any stage of the mitotic cell cycle.
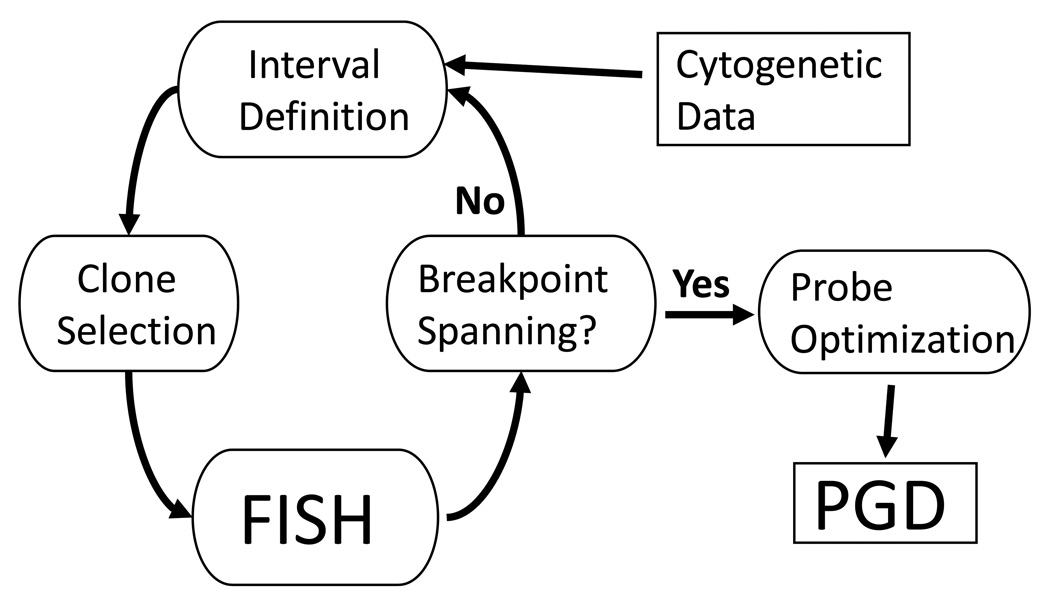
Fortunately, fluorescence in situ hybridization (FISH), a technique to mark specific DNA sequences in interphase or metaphase cells, is sensitive and specific enough to elaborate these objectives. A few years ago, we proposed to map breakpoints with yeast artificial chromosome (YAC) probes spaced in intervals of roughly 8–15 megabasepair (Mbp) along the target chromosomes [12–14]. The target interval was narrowed through repeated cycles of clone selection and hybridization until a clone had been found that spanned the breakpoint [13](Fig. 1). Breakpoint-spanning YACs and adjacent, non-chimeric clones were then assembled into larger contigs to increase FISH efficiency. Although this proved to be a straightforward approach for breakpoint mapping in some patients [12,15–16], the precise determination of breakpoint locations often became a rather time-consuming process plagued by errors in the published physical maps and YAC clone chimerisms [13,17].
Fig. 1.
The procedure to map breakpoint locations in translocation carriers begins with a rough definition of the breakpoint interval followed by cycles of probe selection, mapping on to patient metaphase chromosomes and interval refinement. The second phase, probe optimization, begins when a breakpoint-spanning probe has been identified.
The bacterial artificial chromosome (BAC) clone libraries, able to maintain DNA fragments of several hundred kb, show a very small fraction of chimeric clones, if any [18–19]. As probes in cytogenetic analyses, BACs owe their popularity to several features: relatively high stability, large DNA insert-to-vector size ratio, ease of handling and rapid growth [19–22]. Compared to the use of YACs, the latter is expected to reduce the length of each mapping cycle (Fig. 1), thus accelerating the in situ delineation of chromosomal translocation breakpoints and preparation of breakpoint-specific DNA probes [13]. Furthermore, we decided to use sets of overlapping BAC clones forming ‘contigs’ or ‘pools’ instead of single clones, since this minimizes the rates of so-called ‘FISH failures’ or uninformative results [8–9,23–25]. The present article describes the strengths of BAC clone pooling strategies expediting probe preparation for PGD.
Materials and methods
Tissue samples
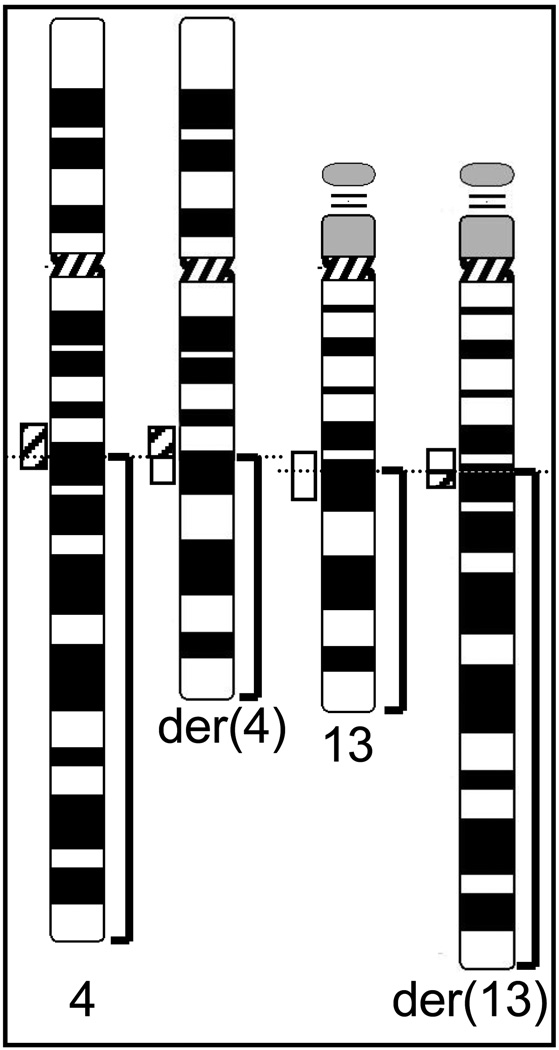
Prior to our study, lymphocytes from the 31-year old female IVF patient T-0512 were analyzed by G-banding. The karyotype 46,XX, t(4;13)(q21.3;q21.2) suggested a balanced, reciprocal translocation as shown in Figure 2.
Fig. 2.
A) Schematic diagram of the karyotypic abnormalities in case T-0512 initially reported as t(4;13)(q21.3;q21.2). Dotted horizontal lines indicate the approximate breakpoint locations at 4q22.1 and 13q21.3 as determined by FISH, and translocated parts are bracketed. The hatched and open boxes represent the breakpoint-spanning probe contigs for chromosome 4 and 13, respectively. Please note that the contigs are split unevenly by the translocation.
Metaphase spreads were made from short-term cultures of peripheral blood following published procedures [26–27]. Briefly, lymphocytes from an anonymous normal male donor or patient T-0512 were grown for 72 h in RPMI 1640 (Invitrogen, Carlsbad, CA) 10% fetal bovine serum, 1% penicillin/streptomycin and 2% phytohemagglutinin (PHA, HA-15; Abbott Molecular, Inc, Des Plaines, IL). Prior to harvest, cells were blocked in mitosis in a 30 min treatment with colcemid (0.12 µg/ml, Invitrogen, San Diego, CA) and incubated in 75 mM KCl for 15 min at 37°C. The cells were then spun down, and approximately 107 cells were incubated in 5 ml of freshly prepared fixative (acetic acid:methanol, 1:3 (vol.:vol.)). The fixation step was repeated twice, before the cells were dropped on ethanol-cleaned glass slides. Slides were aged for a minimum of 1 week in air at 20°C, then sealed in plastic bags and stored at −20 °C until used.
Fluorescence in situ hybridization
Using information in publicly available databases (http://genome.ucsc.edu/ and http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi), we selected BAC clones from the Roswell Park Cancer Institute (RPCI) library RP11 that map to the estimated breakpoint interval or to adjacent chromosome bands. For initial mapping of individual clones, BAC DNA was isolated from 10 ml bacterial overnight cultures containing 12.5 µg/ml chloramphenicol (Sigma, St. Louis, MO) using an alkaline lysis and isopropanol DNA precipitation [28]. Briefly, cell pellets resuspended in 10 ml of phosphate-buffered saline (PBS) containing 50µg/ml lysozyme (Sigma) and lyzed in sodium hydroxide (0.2 N NaOH, 1 % SDS). After neutralization by addition of 3 M NaOAc and pelleting of bacterial DNA, BAC DNA was precipitated in 2-propanol, washed in cold 70% ethanol, and resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Next, the DNA was extracted once with phenol:chloroform, precipitated with isopropanol, and resuspended in 20–40 µl sterile water. The DNA concentrations were determined by Hoechst 33342 fluorometry using a TKO100 instrument (Hoefer, San Francisco, CA) [28]. The BAC-derived probe DNA (typically 1–2 µl of DNA in a 10 µl reaction) was labeled via random priming following the instruction of the kit manufacturer (BioPrime Kit, Invitrogen) [29]. For non-isotopic, indirect labeling biotin-14-dCTP (Invitrogen), digoxigenin(dig.)-11-dUTP (Roche Molecular Systems, Indianapolis, IN), Spectrum Green or Spectrum Orange (Abbott) was incorporated into the DNA [30].
The preparation of DNA probes representing BAC pools was performed in essentially the same way with the following modification: individual BACs were grown overnight in 10 ml of broth containing 20 µg/ml chloramphenicol. Then, 5 ml from each culture was combined in the desired pool, the cells were spun down, resuspended in 10 ml PBS containing 50 µg/ml lysozyme, and DNA was isolated and labeled as described above. For an initial assessment of the breakpoint location using individual BACs, we selected 60 clones from the Roswell Park Cancer Institute (RPCI) ‘RP11’ library (Oseogawa et al. 2001) spread out over the following intervals: for chromosome 4 from 79.7 Mbp to 91.3 Mbp, i.e., from band 4q21.2 to band 4q23. Probe clones for chromosome 13 were selected to map between 57.4 Mbp and 66.8 Mbp, which corresponds to bands 13q21 and 13q23, respectively. Prior to hybridization of DNA probes to patient samples, all probes were tested on normal male metaphase spreads to ensure sufficient signal strength, correct cytogenetic map positions and absence of chimerism [17].
For delineation of the T-0512 breakpoint on the long arm of chromosome 4, we chose 13 BACs from the RP11 library that cover the region between 89.4 Mbp and 91.2 Mbp.
The BAC-derived DNA probes were combined in pools as shown in Table 1: Pool 4-1 is a five BAC contig centered on clone RP11-2I7 at 89.7 – 89.9 Mbp. Pool 4-2 binds distal of Pool 4-1 and covers the interval from 90.2 Mbp to 90.6 Mbp on chromosome 4q. Pool 4-3 is comprised of 4 clones, which map between 90.8 Mbp and 91.2 Mbp, i.e., distal of Pools 4-1 and 4-2. Pools 4-1 and 4-2 cover unique, non-overlapping regions of about 795 kb and 400 kb, respectively. The chromosome 4-specific probes or probe pools were labeled via random priming in separate reactions, then combined as needed for in situ hybridization experiments.
Table 1.
Location of BAC pools on chromosome 4q22.1.
| Region | Clone | Start point (bp)a | End point (bp) a | BAC insert size (bp)b |
|---|---|---|---|---|
| 4-1 | RP11-10L7 | 89403287 | 89513577 | 112291 |
| 4-1 | RP11-466G12 | 89513577 | 89700979 | 189402 |
| 4-1 | RP11-2I7 | 89700979 | 89802530 | 103551 |
| 4-1 | RP11-496N17 | 89802530 | 89912438 | 111908 |
| 4-1 | RP11-502A23 | 90029275 | 90198548 | 171273 |
| 4-2 | RP11-84C13 | 90198548 | 90308551 | 112003 |
| 4-2 | RP11-173C9 | 90308551 | 90427213 | 120662 |
| 4-2 | RP11-549C16 | 90427213 | 90599408 | 172195 |
| 4-2 | RP11-79M20 | 90433497 | 90599388 | 165894C |
| 4-3 | RP11-115D19 | 90755172 | 90922656 | 169484 |
| 4-3 | RP11-395B6 | 90922656 | 90939578 | 18922 |
| 4-3 | RP11-67M1 | 90939578 | 91115385 | 177807 |
| 4-3 | RP11-350B19 | 91115385 | 91242293 | 128907 |
Unique position information is estimated from the Human Genome Reference DNA Sequence, Mapviewer build 36.3 at http://www.ncbi.nlm.nih.gov/projects/mapview/.
The insert sizes were taken from information available at the NCBI Clone Registry at http://www.ncbi.nlm.nih.gov/genome/clone/clname.cgi?stype=Id&list=209311&TransHist=0
BAC size was determined via BLAST search at http://blast.ncbi.nlm.nih.gov/.
We also prepared eight chromosome 13-specific BAC pools: Pools 13-1 to 13-6 cover part of the long arm of chromosome 13 from band q21.2 to band q21.33, while Pool 13-7 comprised of 2 BACs which map in band 13q22.3 between 77.3 Mbp and 77.5 Mbp serves as a distal reference probe (Table 2).
Table 2.
Location of BAC pools on chromosome 13q.
| Pool | Region | Clone | Start point (bp)* | End point (bp)* | BAC insert size (bp) |
|---|---|---|---|---|---|
| 13-1 | 13q21.2 | RP11-524F1 | 58618242 | 58784577 | 166436 |
| 13-1 | 13q21.2 | RP11-26P21 | 59005905 | 59210355 | 206407 |
| 13-1 | 13q21.2 | RP11-218B22 | 59243134 | 59395345 | 154212 |
| 13-1 | 13q21.2 | RP11-442F12 | 59395345 | 59598822 | 209794 |
| 13-1 | 13q21.2 | RP11-430I3 | 59598822 | 59662163 | 63441 |
| 13-2 | 13q21.31 | RP11-350G11 | 60620618 | 60721182 | 102564 |
| 13-2 | 13q21.31 | RP11-310K10 | 60721182 | 60882412 | 163231 |
| 13-2 | 13q21.31 | RP11-432J3 | 60882412 | 60939684 | 59272 |
| 13-2 | 13q21.31 | RP11-210L5 | 60939684 | 61113770 | 176085 |
| 13-2 | 13q21.31 | RP11-543A19 | 61113770 | 61178155 | 66386 |
| 13-2 | 13q21.31 | RP11-179D6 | 61180150 | 61263711 | 85563 |
| 13-2 | 13q21.31 | RP11-429G17 | 61263711 | 61426542 | 164831 |
| 13-2 | 13q21.31 | RP11-71L7 | 61428536 | 61530264 | 103728 |
| 13-3 | 13q21.31 | RP11-527N12 | 62520980 | 62699203 | 178323 |
| 13-3 | 13q21.31 | RP11-282D7 | 62699203 | 62805636 | 106534 |
| 13-3 | 13q21.31 | RP11-320N6 | 62805636 | 62944551 | 139649 |
| 13-3 | 13q21.31 | RP11-67L17 | 62944551 | 63070144 | 125693 |
| 13-3 | 13q21.31 | RP11-473M10 | 63070144 | 63233263 | 163218 |
| 13-3 | 13q21.31 | RP11-394A14 | 63236873 | 63409675 | 174801 |
| 13-3 | 13q21.31 | RP11-520F9 | 63409675 | 63481486 | 73811 |
| 13-3 | 13q21.31 | RP11-205B18 | 63481486 | 63637946 | 158460 |
| 13-4 | 13q21.31 | RP11-261A1 | 64364070 | 64528551 | 166490 |
| 13-4 | 13q21.31-q21.32 | RP11-211D10 | 64561613 | 64741933 | 182321 |
| 13-4 | 13q21.32 | RP11-379K8 | 64788660 | 64966449 | 179790 |
| 13-4 | 13q21.32 | RP11-229I7 | 64966450 | 65062394 | 96045 |
| 13-4 | 13q21.32 | RP11-326D19 | 65062394 | 65222214 | 159919 |
| 13-4 | 13q21.32 | RP11-223F20 | 65233930 | 65400826 | 168897 |
| 13-4 | 13q21.32 | RP11-298H15 | 65415271 | 65575185 | 160015 |
| 13-5 | 13q21.32 | RP11-10M21 | 66264338 | 66378528 | 114290 |
| 13-5 | 13q21.32 | RP11-138D23 | 66378528 | 66543031 | 164604 |
| 13-5 | 13q21.32 | RP11-576O3 | 66543031 | 66682813 | 139881 |
| 13-5 | 13q21.32 | RP11-346A3 | 66682813 | 66750587 | 71774 |
| 13-5 | 13q21.32 | RP11-531B22 | 66750587 | 66883530 | 82944 |
| 13-5.5 | 13q21.32 | RP11-164E20 | 66882862 | 67067236 | 184374 |
| 13-5.5 | 13q21.32 | RP11-520F22 | 67006314 | 67148127 | 141913 |
| 13-5.5 | 13q21.32 | RP11-51P14 | 67148127 | 67174888 | 28761 |
| 13-5.5 | 13q21.32 | RP11-562L19 | 67174888 | 67254473 | 81586 |
| 13-5.5 | 13q21.32 | RP11-248N6 | 67254473 | 67417068 | 162695 |
| 13-6 | 13q21.33 | RP11-338L17 | 67493555 | 67560112 | 68556 |
| 13-6 | 13q21.33 | RP11-157F14 | 67560112 | 67681998 | 123887 |
| 13-6 | 13q21.33 | RP11-520F24 | 67681998 | 67841134 | 159235 |
| 13-7 | 13q22.3 | RP11-318G21 | 77297743 | 77480980 | 183337 |
| 13-7 | 13q22.3 | RP11-122N18 | 77314551 | 77489832 | 175282 |
Unique position information is estimated from the Human Genome Reference DNA Sequence, Mapviewer build 36.3 at http://www.ncbi.nlm.nih.gov/projects/mapview/.
For FISH, we combined 1 µl of each probe, 1 µl of human COT-1™ DNA (1 mg/ml, Invitrogen), 1 µl of salmon sperm DNA (10 mg/ml, Invitrogen) and 7 µl of a hybridization master mix (78.6% formamide (Invitrogen), 14.3% dextran sulfate in 1.43× SSC, pH 7.0 (20× SSC is 3 M sodium chloride, 300 mM tri-sodium citrate) [30] and denatured the mixture in a waterbath at 76 °C for 10 min. Then, the hybridization mixture was incubated for 30 min at 37 °C to pre-anneal blocking DNA with the probes, while the slides were denatured for 4 min at 76 °C in 70% formamide/2× SSC, pH 7.0, dehydrated in a 70%, 85%, and 100% ethanol series for 2 min each step, and allowed to air dry. The hybridization mixture was then pipetted on to the slides and sealed with rubber cement under a 22×22 mm2 coverslip. Following overnight hybridization at 37 °C and coverslip removal, the slides were washed twice in 50% formamide/2× SSC at 45°C for 10 min each followed by two washes in 2× SSC at 21°C. Cells were then incubated briefly in PNM (5% nonfat dry milk, 1% sodium azide in PN buffer (0.1 M sodium phosphate buffer, pH 8.0, 1% Nonidet-P40 (Sigma))) for 10 min at 21°C, before bound probes were detected with fluorescein-conjugated avidin DCS (Vector, Burlingame, CA) or anti-digoxigenin-rhodamine (Roche) [28]. Finally, the slides were mounted in 4′,6-diamidino-2-phenylindole (DAPI) (0.5 µg/ml; Calbiochem, La Jolla, CA) in antifade solution [28,30].
Image acquisition and analysis
Fluorescence microscopy was performed on a Zeiss Axioskop microscope equipped with a filter sets for observation of Texas Red/rhodamine. FITC or DAPI detection (ChromaTechnology, Brattleboro, VT). Images were collected using a CCD camera (VHS Vosskuehler, Osnabrueck, FRG) and processed using Adobe Photoshop® software (Adobe Inc., Mountain View, CA).
Results
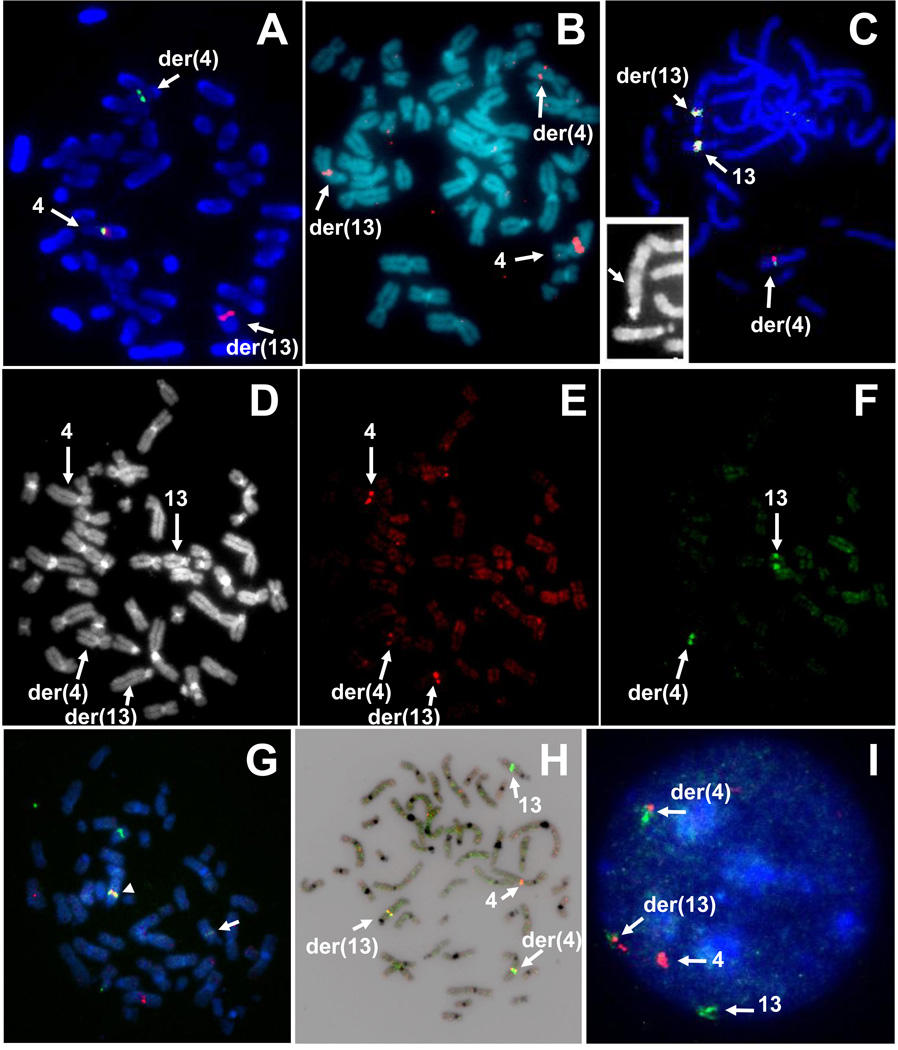
Using probes prepared from 60 individual BAC clones, hybridizations with normal or patient metaphase spreads showed disappointing results: 20 of 60 clones failed to produce informative hybridization signals (data not shown). However, clones that gave analyzable signals on t(4;13) cells suggested a breakpoint on the long arm of chromosome 4 distal of clone RP11-2I7 at 89.7–89.8 Mbp and proximal of BAC clone RP11-115D19 at 90.7–90.9 Mbp, i.e., roughly within a 1 Mbp interval in band 4q22.1 (Table 1). Based on the FISH results with individual clones, we changed our mapping strategy and replaced individual clones with selected contiguous sets of BACs. To increase hybridization efficiencies and obtain a first assessment of the location of the chromosome 4-specific breakpoint, we pooled three Spectrum Green–labeled probes (RP11-2I7, RP11-496N17 and RP11-502A23) with four Spectrum Orange-labeled probes prepared from clones RP11-115D19, RP11-395B6, RP11-67M1 and RP11-350B19 (Pool 4-3, Table 1). The FISH results demonstrated that all green fluorescent probes bound proximal of the breakpoint and gave signals on the normal homologue of chromosome 4 and the der(4) (Fig. 3A). The hybridization of our pool of Spectrum Orange-labeled probes resulted in red signals exclusively on the normal chromosome 4 and the der(13) indicating that Pool 4-3 (Table 1) mapped entirely distal of the breakpoint (Fig. 3A). Due to close proximity of green and red signals probes, the superimposed FISH images appear partially yellow in the pseudo-RGB pictures in Fig. 3. With the breakpoint on chromosome 4 in T-0512 located between clones RP11-502A23 and RP11-115D19, we prepared one large pool of dig.-labeled DNA probes comprised of all five probes that map to Region 4-1 and four clones from Region 4-2 (Table 1). The FISH results showed that this contig bound to the normal chromosome 4, as well as both derivative chromosomes, thus resulting in a probe that not only covered, but extended the chromosome 4 breakpoint (Fig. 3B).
Fig. 3.
Hybridization of BAC pools for rapid delineation of chromosome breakpoints. A) Hybridization of chromosome 4-specific BAC pools to patient metaphase cells demonstrates specific binding to the target region on the long arm of chromosome 4. The pseudo-RGB image shows the localization of proximal (green) and distal (red) BACs on DAPI counterstained metaphase chromosomes. B) Hybridization of the optimized BAC contig for chromosome 4 marks the normal homologue and both derivative chromosomes in this metaphase spread from patient T-0512. C) Hybridization of seven probe pools for chromosome 13 generates signals on the normal homologue and the der(13) as well as on the der(4) indicating that probes bind proximal and distal of the breakpoint on chromosome 13. The insert shows an enlarged, partial picture of DAPI channel with the arrow pointing at the der(13). D–F) Combined hybridization of the chromosome 4-specific BAC pools (red) and Pools 13-4 to 13-6 (green) indicated the distal position of the chromosome 13-specific probes. The DAPI, red, and green fluorescence images are shown in D, E, and F, respectively. G) Combined hybridization of Pool 13-3 (green) and the chromosome 4 contig (red) shows a strong green signal on the der(13)(arrowhead) and a weak green signal on the der(4)(arrow). H–I) Hybridization of the optimized probe sets for chromosomes 13 (green) and 4 (red) to metaphase and interphase cells. Image H) shows the superposition of probe signals with the inverted DAPI image in a T-0512 metaphase spread. In interphase nuclei (I), der(4) and der(13) can be differentiated by the strength of the red-green fluorescence signals.
The FISH mapping experiments of individual BAC clones for chromosome 13 were plagued by hybridization failures, too, but allowed to place the T-0512 breakpoint between the proximal clone RP11-16M6 at ~57.4 Mbp and the three BAC clones RP11-10M21, RP11-138D23 and RP11-346A3, which map to 66.165 Mbp – 66.753 Mbp (Table 2). This chromosome 13-specific interval measures about 9 Mbp.
A first FISH experiment to determine the chromosome 13-specific breakpoint location used BAC pools comprised of biotin-labeled DNA from Pools 13-1, 13-3, 13-5 and 13-7 and dig.-labeled probes made from Pools 13-2, 13-4 and 13-6 (Table 2). Hybridization of these ‘superpool’ DNA probes to normal male metaphase spreads showed strong, specific signals on both homologues of chromosome 13 without noticeable cross-hybridization to other chromosomes (not shown). Hybridization of the same combination of chromosome 13-specific probe pools to metaphase cell from T-0512 showed strong hybridization signals on the normal chromosome 13 and the der(13) as well as on the der(4) (Fig. 3C). Since all three hybridization domains showed green and red fluorescent signals, this first BAC pool hybridization confirmed our hypothesis, i.e., the interval covered by Pools 13-1 to 13-6 extents onto both sides of the breakpoint. It also suggested a breakpoint between Pools 13-2 and 13-6, i.e., around 60.6 – 67.8 Mbp. A second hybridization of BAC pools to patient metaphase spreads combined the two dig.-labeled pools for chromosome 4 (4-1, 4-2; Table 1) with a combination of biotinylated DNA probes prepared from Pools 13-4, 13-5, 13-6, and the newly added Pool 13-5.5 (Table 2). The chromosome 4- and 13–specific probes were detected in red and green, respectively, and all four chromosomes of interest (4, 13, der(4), der(13)) could be identified by their DAPI banding pattern and FISH signals (Fig. 3D–F). For example, the image in Fig. 3E shows three red signals: one on the normal chromosome 4 and two on derivative chromosomes der (4) and der(13) as expected for a probe pool that spans the breakpoint on chromosome 4. We also noted a strong signal on the normal copy of chromosome 4, while signals on the derivative chromosomes were of lesser strength (Fig.3E). The green fluorescent signals were found exclusively on the normal copy of chromosome 13 and the der(4)(Fig.3F), indicating that Pool 13-4 to Pool 13-6 bound distal of the breakpoint on chromosome 13.
We then decided to map Pool 13-3 knowing that the breakpoint on chromosome 13 lies proximal of Pool 13-4, but within or distal of Pool 13-2. Dual color FISH using a combination of biotinylated Pool 13-3 DNA and dig.-labeled Pools 4-1 plus 4-2 showed the expected signals on the normal non-rearranged copies of chromosomes 4 and 13 (Fig. 3G). Red and green signals were found on both derivative chromosomes indicated that Pool 13-3 spans the chromosome 13-specific breakpoint in T-0512. We also noted that the green signal on the der(4) chromosome was faint (arrow in Fig. 3G), while the green signal on the der(13) was strong (arrowhead, Fig. 3G). Thus, only a small fraction of probe from Pool 13-3 bound distal of the breakpoint, and most of this probe pool bound proximal.
In conclusion, only three overnight FISH experiments with BAC pools and patient metaphase spreads allowed us to narrow the breakpoint position on chromosome 13 to a 1.1 Mbp interval between 62.5 Mbp and 63.6 Mbp. The next step in the PGD probe preparation process was probe optimization: since the chromosome 4-specific DNA probe contigs was split about 3:2 to 2:1 (Fig. 3B), we decided to design a chromosome 13-specific BAC pool probe that will be split asymmetrically by the translocation, thus allowing unambiguous identification of derivative chromosomes in interphase cell nuclei. This was achieved easily by combining the previously prepared biotinylated probe from Pool 13-3 with DNA probes prepared from Pools 13-4, 13-5, 13-5.5, and 13-6 (Table 2) covering an interval from 62.5 Mbp to 67.8 Mbp. The FISH result showed that signals from biotinylated chromosome 13 probes were split into two differently sized parts: the signals derived from pool 13-3 BAC’s binding to the proximal long arm of chromosome 13, i.e., der(13) signals were weaker than those of probes that covered the distal part leading to green signals on the der(4) chromosome (Fig. 3H). This set of hybridization probes which extents differently on proximal and distal sites of the chromosome-specific breakpoints and a simple dual color probe detection scheme allows classification of all normal homologues and derivative chromosomes involved in this translocation in interphase cell nuclei (Fig. 3I).
Discussion
PGD is a well established procedure to identify aneuploid oocytes or chromosomally (ab-)normal embryos with the purpose to increase the chances of nidation, pregnancy and birth of a healthy baby [6–8,10]. Unlike other laboratory tests, PGD is a single cell analysis: typically, only 1–2 blastomeres are biopsied from day 3 embryos [6,9]. Interphase cell analysis is possibly the most important component of PGD, since blastomeres can be in any stage of the cell cycle. For over a decade, we and others have been using chromosome-specific DNA repeat probes, many of which are commercially available, to enumerate a limited number of chromosomes in individual interphase nuclei [8,23,31–32]. Many of these chromosome enumerator probes (CEPs) target alpha satellite DNA repeats at or near the chromosome centromeres [33,34]. However, in cells from carriers of a reciprocal translocation, in which the prevalence of unbalanced gametes carrying a partial aneusomy ranges from 50% to 70% [7,9,35], the centromeric probes miss most abnormalities.
To address this issue, we initially prepared DNA probe contigs comprised of YACs that bilaterally extended individual translocation breakpoints [12–16]. With the time constraints in IVF programs, often little time was left for probe optimization [14]. Thus, our present work focuses on choosing BAC clones rather than YACs, because the former ones have a number of significant advantages such as less chimerism or faster growth. Another aim of our study was to expedite the process of mapping translocation breakpoints by eliminating so-called hybridization failures through the pooling BAC clones.
In our FISH mapping scheme of translocation breakpoints, the normal homologues show hybridization domains in a single color. For example, red fluorescent signals delineate the hybridization target on normal homologues of chromosome 4 or green signals specifically mark chromosome 13 (Fig.3). If probe binding extents significantly on both sides of the breakpoint (i.e., it spans or extends the breakpoint region), probe signals will be found on one or both derivative chromosomes and signals are comprised of mixed colors.
Historically, breakpoint mapping is an iterative process based on the definition of the smallest interval between proximal and distal probes. Thus, many of the DNA probes prepared in breakpoint mapping experiments did not generate additional information [12–16]. Importantly, our pooling protocol for PGD probe preparation accelerates the delineation and fine mapping of translocation breakpoints without sacrificing resolution. The turnaround time for each cycle comprised of clone selection, FISH and image analysis using patient samples can now be as short as 3–4 days. Thus, with translocation breakpoints roughly determined by G-banding, large numbers of BACs can be pulled from in-house libraries and assembled in probe pools before initiation of the IVF cycle. As the example in this paper shows, only a few overnight hybridizations will be required to localize the breakpoint to a single pool and optimize the probes for single cell interphase analysis. Thus, the proposed BAC pooling strategy seems capable to provide breakpoint information as well as DNA probes suitable for interphase cell analysis in only 2–3 weeks, a significant improvement over previous methods [13]. In many instances, the costs of IVF cycles and PGD, often tens of thousands of US dollars per cycle, are borne by the patients. We believe, our approach will not only lead to reduced costs making interphase PGD more affordable to infertile couples, but also result in more reliable PDG procedures, reduce the number of failed embryo transfers and the suffering associated with failed transfers.
Acknowledgments
This work was supported in parts by NIH grants CA80792, CA88258, CA123370, and HD44313, and a grant from the Director, Office of Energy Research, Office of Health and Environmental Research, U.S. Department of Energy, under contract DE-AC02-05CH11231. JFW was supported in part by NIH grant HD45736 and a grant from the UC Discovery Program, which also supported AB. Ideograms were kindly provided by D. Adler, Ph.D., Dept. of Pathology, Univ. Washington. We acknowledge the support from staff at Reprogenetics providing metaphase spreads and mapping data. We like to express our thanks to the scientists at the Human Genome Center, California Institute of Technology, Pasadena, whose generosity made these studies possible.
Footnotes
Parts of this work have been presented at the 13th Congress of the International Federation of Societies for Histochemistry and Cytochemistry (ICHC2008), Medical University of Gdansk, Poland, August 27–30, 2008.
Disclaimer: This document was prepared as an account of work sponsored by the United States Government. While this document is believed to contain correct information, neither the United States Government nor any agency thereof, nor The Regents of the University of California, nor any of their employees, makes any warranty, express or implied, or assumes any legal responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights. Reference herein to any specific commercial product, process, or service by its trade name, trademark, manufacturer, or otherwise, does not necessarily constitute or imply its endorsement, recommendation, or favoring by the United States Government or any agency thereof, or The Regents of the University of California. The views and opinions of authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof, or The Regents of the University of California.
References
- 1.Subrt I. Reciprocal translocation with special reference to reproductive failure. Hum Genet. 1980;55:303–307. doi: 10.1007/BF00290208. [DOI] [PubMed] [Google Scholar]
- 2.Peng HH, Chao AS, Wang TH, Chang YL, Chang SD. Prenatally diagnosed balanced chromosome rearrangements: eight years' experience. J Reprod Med. 2006;51:699–703. [PubMed] [Google Scholar]
- 3.Stern C, Pertile M, Norris H. Chromosome translocations in couples with in vitro fertilisation implantation failure. Hum Reprod. 1999;14:2097–2101. doi: 10.1093/humrep/14.8.2097. [DOI] [PubMed] [Google Scholar]
- 4.Srb AM, Owen RD, Edgar RS. General Genetics. 2nd ed. San Francisco, CA: W.H. Freeman and Co; 1965. [Google Scholar]
- 5.Kalousek DK. Pathogenesis of chromosomal mosaicism and its effect on early human development. Am J Med Genet. 2000;91:39–45. doi: 10.1002/(sici)1096-8628(20000306)91:1<39::aid-ajmg7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 6.Munné S. Preimplantation genetic diagnosis of numerical and structural chromosome abnormalities. Reprod Biomed Online. 2002;4:183–196. doi: 10.1016/s1472-6483(10)61938-4. [DOI] [PubMed] [Google Scholar]
- 7.Braude P, Pickering S, Flinter F, Ogilvie CM. Preimplantation genetic diagnosis. Nat Rev Genet. 2002;3:941–953. doi: 10.1038/nrg953. [DOI] [PubMed] [Google Scholar]
- 8.Munné S, Grifo J, Cohen J, Weier H-UG. Mosaicism and aneuploidy in arrested human preimplantation embryos: a multiple probe fluorescence in situ hybridization (FISH) study. Am J Hum Genet. 1994;55:150–159. [PMC free article] [PubMed] [Google Scholar]
- 9.Sampson JE, Ouhibi N, Lawce H, Patton PE, Battaglia DE, Burry KA, Olsen SB. The role of preimplantation genetic diagnosis in balanced translocation carriers. Am J Obstet Gynecol. 2004;190:1707–1713. doi: 10.1016/j.ajog.2004.02.063. [DOI] [PubMed] [Google Scholar]
- 10.Verlinsky Y, Cohen J, Munné S, Gianaroli L, Simpson JL, Ferraretti AP, Kuliev A. Over a decade of experience with preimplantation genetic diagnosis: a multicenter report. Fertil Steril. 2004;82:292–294. doi: 10.1016/j.fertnstert.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 11.Munné S, Sandalinas M, Escudero T, Fung J, Gianaroli L, Cohen J. Outcome of preimplantation genetic diagnosis of translocations. Fertil Steril. 2000;73:1209–1218. doi: 10.1016/s0015-0282(00)00495-7. [DOI] [PubMed] [Google Scholar]
- 12.Cassel MJ, Munné S, Fung J, Weier H-UG. Carrier-specific breakpoint-spanning DNA probes for pre-implantation genetic diagnosis [PGD] in interphase cells. Hum Reprod. 1997;12:101–109. doi: 10.1093/humrep/12.9.2019. [DOI] [PubMed] [Google Scholar]
- 13.Fung J, Munné S, Duell T, Weier H-UG. Rapid cloning of translocation breakpoints: from blood to YAC in 50 days. J Biochem Mol Biol Biophys. 1998;1:181–192. [Google Scholar]
- 14.Fung J, Munné S, Garcia J, Kim U-J, Weier H-UG. Reciprocal translocations and infertility: molecular cloning of breakpoints in a case of constitutional translocation t(11;22)(q23;q11) and preparation of probes for preimplantation genetic diagnosis (PGD) Reprod Fert Dev. 1999;11:17–23. doi: 10.1071/rd98110. [DOI] [PubMed] [Google Scholar]
- 15.Munné S, Fung J, Cassel MJ, Márquez C, Weier H-UG. Preimplantation genetic analysis of translocations: case-specific probes for interphase cell analysis. Human Genetics. 1998;102:663–674. doi: 10.1007/s004390050759. [DOI] [PubMed] [Google Scholar]
- 16.Weier HUG, Munné S, Fung J. Patient-specific Probes for Preimplantation Genetic Diagnosis (PGD) of Structural and Numerical Aberrations in Interphase Cells. J Assist Reprod Genet. 1999;16:182–189. doi: 10.1023/A:1020360706317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selleri L, Eubanks JH, Giovannini M, Hermanson GG, Romo A, Djabali M, Maurer S, McElligott DL, Smith MW, Evans GA. Detection and characterization of "chimeric" yeast artificial chromosome clones by fluorescent in situ suppression hybridization. Genomics. 1992;14:536–541. doi: 10.1016/s0888-7543(05)80263-0. [DOI] [PubMed] [Google Scholar]
- 18.Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim UJ, Shizuya H, Kang HL, Choi SS, Garrett CL, Smink LJ, Birren BW, Korenberg JR, Dunham I, Simon MI. A bacterial artificial chromosome-based framework contig map of human chromosome 22q. Proc Natl Acad Sci USA. 1996;93:6297–6301. doi: 10.1073/pnas.93.13.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osoegawa K, Mammoser AG, Wu C, Frengen E, Zeng C, Catanese JJ, de Jong PJ. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liehr T, Weise A, Heller A, Starke H, Mrasek K, Kuechler A, Weier H-UG, Claussen U. Multicolor chromosome banding (MCB) with YAC/BAC-based probes and regionspecific microdissection DNA libraries. Cytogenet Genome Res. 2002;97:43–50. doi: 10.1159/000064043. [DOI] [PubMed] [Google Scholar]
- 22.Carreira IM, Mascarenhas A, Matoso E, Couceiro AB, Ramos L, Dufke A, Mazauric M, Stressig R, Kosyakova N, Melo JB, Liehr T. Three unusual but cytogenetically similar cases with up to five different cell lines involving structural and numerical abnormalities of chromosome 18. J Histochem Cytochem. 2007;55:1123–1128. doi: 10.1369/jhc.7A7244.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Alton ME, Malone FD, Chelmow D, Ward BE, Bianchi DW. Defining the role of fluorescence in situ hybridization on uncultured amniocytes for prenatal diagnosis of aneuploidies. Am J Obstet Gynecol. 1997;176:769–776. doi: 10.1016/s0002-9378(97)70600-8. [DOI] [PubMed] [Google Scholar]
- 24.Pehlivan T, Rubio C, Rodrigo L, Remohi J, Pellicer A, Simon C. Preimplantation genetic diagnosis by fluorescence in situ hybridization: clinical possibilities and pitfalls. J Soc Gynecol Investig. 2003;10:315–322. doi: 10.1016/s1071-5576(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 25.Plastira K, Maher E, Fantes J, Ramsay J, Angelopoulou R. Using BAC clones to characterize unbalanced chromosome abnormalities in interphase cells. Eur J Med Genet. 2006;49:235–246. doi: 10.1016/j.ejmg.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Fung J, Weier H-UG, Pedersen RA, Zitzelsberger HF. Spectral Imaging Analysis of Metaphase and Interphase Cells. In: Rautenstrauss B, Liehr T, editors. FISH Technology. Heidelberg, Germany: Springer Verlag; 2002. pp. 363–387. [Google Scholar]
- 27.Bayani J, Squire JA. Preparation of cytogenetic specimens from tissue samples. In: Bonifacino JS, editor. Current Protocols in Cell Biology. New York: John Wiley & Sons, Inc.; 2004. pp. 22.2.1–22.2.15. [DOI] [PubMed] [Google Scholar]
- 28.Weier HU, Rhein AP, Shadravan F, Collins C, Polikoff D. Rapid physical mapping of the human trk protooncogene (NTRK1) to human chromosome 1q21-q22 by P1 clone selection, fluorescence in situ hybridization (FISH), and computer-assisted microscopy. Genomics. 1995;26:390–393. doi: 10.1016/0888-7543(95)80226-c. [DOI] [PubMed] [Google Scholar]
- 29.Fung J, Weier H-UG, Goldberg JD, Pedersen RA. Multilocus genetic analysis of single interphase cells by spectral imaging. Hum Genet. 2000;107:615–622. doi: 10.1007/s004390000416. [DOI] [PubMed] [Google Scholar]
- 30.Lu CM, Wang M, Greulich-Bode K, Weier JF, Weier HUG. Quantitative DNA fiber mapping. In: Liehr T, editor. Springer FISH Lab Manual. Heidelberg, FRG: Springer Verlag; 2008. pp. 269–291. [Google Scholar]
- 31.Weier JF, Weier H-UG, Jung CJ, Gormley M, Yan Zhou Y, Chu LW, Genbacev O, Wright AA, Fisher SJ. Human cytotrophoblasts acquire aneuploidies as they differentiate to an invasive phenotype. Dev Biology. 2005;279:420–432. doi: 10.1016/j.ydbio.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Stumm M, Wegner R-D, Bloechle M, Eckel H. Interphase M-FISH applications using commercial probes in prenatal and PGD diagnostics. Cytogenet Genome Res. 2006;114:296–301. doi: 10.1159/000094217. [DOI] [PubMed] [Google Scholar]
- 33.Waye JS, Durfy SJ, Pinkel D, Kenwrick S, Patterson M, Davies KE, Willard HF. Chromosome-specific alpha satellite DNA from human chromosome 1: hierarchical structure and genomic organization of a polymorphic domain spanning several hundred kilobase pairs of centromeric DNA. Genomics. 1987;1:43–51. doi: 10.1016/0888-7543(87)90103-0. [DOI] [PubMed] [Google Scholar]
- 34.Baumgartner A, Weier JF, Weier H-UG. Chromosome-specific DNA repeat probes. J Histochem Cytochem. 2006;54:1363–1370. doi: 10.1369/jhc.6A6974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scriven PN, Handyside AH, Ogilvie CM. Chromosome translocation: segregation modes and strategies for preimplantation genetic diagnosis. Prenat Diagn. 1998;18:1437–1449. [PubMed] [Google Scholar]
- 36.Fung J, Munné S, Weier H-UG. Detection of chromosome translocation products in single interphase cell nuclei. In: Darzynkiewicz Z, Chrissman HA, Robinson JP, editors. Methods in Cell Biology. 3rd ed. Part B. Vol. 64. San Diego: Academic Press; 2001. pp. 98–117. Cytometry. [DOI] [PubMed] [Google Scholar]