Abstract
A large number of extracellular polypeptides bound to their cognate receptors activate the transcription factor STAT3 by phosphorylation of tyrosine 705. Supplemental activation occurs when serine 727 is also phosphorylated. STAT3 deletion in mice leads to embryonic lethality. We have produced mice with alanine substituted for serine 727 in STAT3 (the SA allele) to examine the function of serine 727 phosphorylation in vivo. Embryonic fibroblasts from SA/SA mice had ∼50% of the transcriptional response of wild-type cells. However, SA/SA mice were viable and grossly normal. STAT3 wild-type/null (+/−) animals were also normal and were interbred with SA/SA mice to study SA/− mice. The SA/− mice progressed through gestation, showing 10 to 15% reduced birth weight, three-fourths died soon after birth, and the SA/− survivors reached only 50 to 60% of normal size at 1 week of age. The lethality and decreased growth were accompanied by altered insulin-like growth factor 1 (IGF-1) levels in serum, establishing a role for the STAT3 serine phosphorylation acting through IGF-1 in embryonic and perinatal growth. The SA/− survivors have decreased thymocyte number associated with increased apoptosis, but unexpectedly normal STAT3-dependent liver acute phase response. These animals offer the opportunity to study defined reductions in the transcriptional capacity of a widely used signaling pathway.
The utility of hypomorphic mutations in delineating signaling pathways has been amply proven: such mutations are widely used in detecting gene interactions which intensify or ameliorate a mutant phenotype. Many, perhaps most, extracellular ligand-activated signaling pathways report to a more limited number of transcription factors (7). To learn the quantitative importance of a given transcription factor in a particular cellular or developmental event, it would be valuable to introduce mutations with known stepwise reductions in transcription factor activity. We have constructed a mouse strain with the aim of producing reduced STAT3 activity.
The STAT (signal transducers and activators of transcription) proteins are latent transcription factors activated through receptors that bind cytokines, growth factors, or peptides, triggering intracellular tyrosine phosphorylation of the STATs (23). STAT3 is ubiquitously expressed and transiently activated by a large number of different ligands, including cytokines of the interleukin-6 (IL-6) family that also includes ciliary neurotrophic factor (CNTF), oncostatin M (OSM), and leukemia inhibitory factor (LIF), as well as noncytokine ligands such as the growth factors epidermal growth factor, platelet-derived growth factor, hepatocyte growth factor, and granulocyte colony-stimulating factor (1, 24). Many genes are known to be induced in response to STAT3 activation. The STAT3 gene is the only member of this family that results in embryonic lethality when removed in mice (43). Tissue-specific removal has revealed a broad spectrum of biological functions of STAT3 in adult tissues based on disturbed cell growth, suppression or induction of apoptosis, and cell differentiation (1, 23, 24). STAT3 has also gained attention because it is persistently active in a high proportion of all human cancers and is demonstrably required for survival in a number of cultured tumor cells (8).
In addition to the obligatory requirement for tyrosine phosphorylation to activate all the STATs, a single serine phosphorylation (residue 727 in both STAT1 and STAT3) in the transcriptional activation domain is required for full transcriptional activation of STATs 1, 3, 4, 5A, and 5B (13, 23, 46). In the case of STAT3 S727A, where the serine 727 was replaced by alanine, it was estimated that the mutant was about 50% as effective as wild type in transcriptional stimulation (46). Serine phosphorylation most likely increases STAT activity through association with other proteins. For example, serine phosphorylation of STAT1 increases association with MCM5 and CaMKII (30, 48). Several different kinases have been implicated in serine phosphorylation, implying an interaction between STAT signaling and serine kinase signaling pathways. However, the precise role of STAT serine phosphorylation, especially in animals, remains unclear (13).
We have therefore replaced the wild-type STAT3 allele in mice with the STAT3 S727A mutant (hereafter referred to as the SA mutant). The effects of diminished STAT3 transcriptional activity of the SA mutant were examined in both cultured fibroblasts and the whole animal. There was a definite stepwise reduction of STAT3 transcriptional activation in fibroblasts of the genetic series +/+, SA/SA, and SA/−. While SA/SA mice appeared normal, ∼75% perinatal lethality was found in SA/− mice. Both of the two major polypeptides that stimulate growth, insulin-like growth factor 1 (IGF-1) and growth hormone (GH), can activate STAT3 (35, 49; reviewed in reference 18), and IGF-1 is most important for growth prenatally and perinatally (5, 15). We have found slow pre- and perinatal growth and altered IGF-1 levels in SA/− mice. Thus, full STAT3 activation based on serine phosphorylation acting through IGF-1 is required for normal fetal and early perinatal growth. Also, decreased thymocyte number and increased thymocyte apoptosis were found in the SA/− mice. The animals produced in this study will be valuable in dissecting between the STAT pathway and serine kinase pathways and determining the effects of predictably reduced STAT3 activity in a wide variety of specific developmental decisions.
MATERIALS AND METHODS
Construction of the targeting vector and generation of STAT3(S727A) mice.
An 18.5-kb genomic clone containing STAT3 exons 12 to 24 was isolated from a 129/Svj genomic library (Stratagene) using a STAT3 cDNA probe (36). To construct the targeting vector, the ∼10-kb ClaI-SpeI fragment (exons 16 to 24) was subcloned into pBluescript KS(+). The serine 727 code that locates in exon 23 was mutated into alanine code, and the mutation was confirmed by DNA sequencing. A PacI site was introduced by mutation into the 3′ untranslated region at ∼90 bp downstream of the poly(A) site. A loxP-flanked neo cassette was introduced into the PacI site. Embryonic stem (ES) cells were electroporated with the SalI-linearized targeting vector and selected in the presence of G418. Correctly targeted ES clones were screened by Southern blot analysis using a 240-bp probe generated by PCR with the following two primers: forward primer, 5′ CTCAGTCTATGGCAGCAGTTGGCTG, and reverse primer, 5′ ACATGCCCAGGAAAGCAGGGGTTTA. To select the targeted clones that included the serine mutation, PCR analysis (Expanded Long PCR; Roche Biochemicals) using primer a1 (corresponding to the wild-type serine allele) or a2 (corresponding to a mutated alanine allele), in pair with primer b (which locates outside the targeting vector), was performed to amplify a 4.5-kb (wild-type) or a 6.5-kb (mutant) fragment, respectively. The primers (shown below in Fig. 1) were A1 (5′ ATACCATTGACCTGCCGATGT), A2 (5′ ATACCATTGACCTGCCGATGG), and B (5′ ACATGCCCAGGAAAGCAGGGGTTTA).
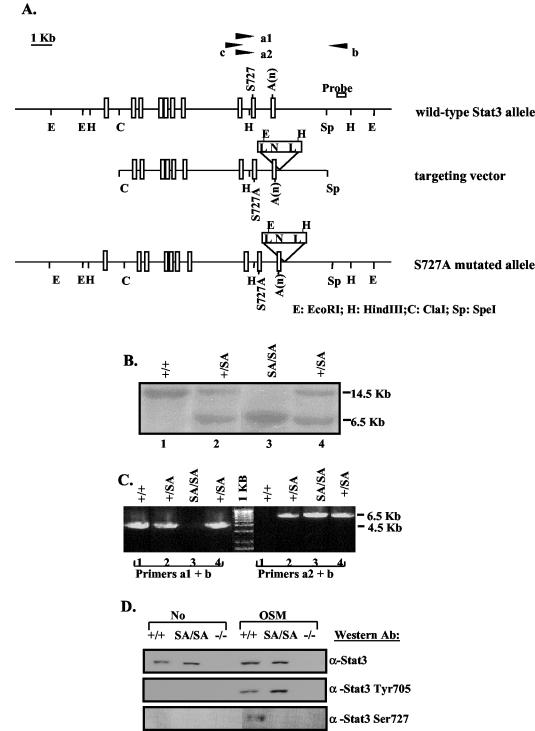
FIG. 1.
Generation of the Stat3 S727A mutant mice. (A) Schematic diagrams of the mouse endogenous wild-type Stat3 allele, S727A targeting vector, and the resulting S727A mutant allele after homologous recombination. Exons are shown as rectangles. Primers a1 and a2 were matched to wild-type serine 727 or mutated alanine sequences, respectively, and were used in pairs with primer b to perform PCR screening for mutation. Primers b and c were used to PCR amplify the fragment in between, which was sequenced to confirm the S727A mutation. N, Neo cassette; A(n), polyadenylation sequence; L, loxP sequence. (B) Southern blot analysis of EcoRI-digested genomic DNA from +/+, SA/+, and SA/SA mice using the probe shown in panel A. The probe detects a 14.5-kb band from the wild-type allele and a 6.5-kb band from the mutant S727A allele. (C) PCR analysis of the genomic DNA analyzed in panel B. Primers a1 and b detect the ∼4.5-kb band from the +/+ allele, and primers a2 and b detect the ∼6.5-kb band from the mutant S727A allele. (D) Western blot analysis to check serine phosphorylation. Whole-cell extracts prepared from +/+, SA/SA, or −/− MEFs untreated or treated with OSM for 30 min were immunoprecipitated with an anti-STAT3 antibody, followed by Western blotting using an antibody against phospho-STAT3 (Ser727). The same extracts were also straightly probed with an anti-phospho-STAT3 (Tyr705) antibody or stripped and reprobed with an anti-STAT3 antibody.
Four STAT3 clones carrying the S727A mutation were microinjected into C57BL/6 blastocytes to generate chimeric mice. Germ line transmission was obtained from three of these clones. Two distinctive clones were selected to cross back to C57BL/6 mice. After verifying they were both fertile, mice derived from one clone were chosen for further studies.
STAT3SA-neo mice generated as described above were crossed with transgenic adenovirus EIIA-Cre mice (22) to delete the neo cassette. The progeny were crossed back to C57BL/6, and the offspring that did not possess the Cre transgene were selected, backcrossed to C57BL/6, and used for subsequent studies. The primers used to detect Cre transgene were Cre1 (5′ TCCAATTTACTGACCGTACA) and Cre2 (5′ TCCTGGCAGCGATCGCTATT). STAT3+/− mice on a C57BL/6 background were described before (36).
Preparation of embryonic fibroblasts and MEF transfection.
The mouse embryonic fibroblasts (MEFs) were prepared from embryos 13.5 days postcoitum according to the standard protocol (19) and maintained in Dulbecco's modified Eagle's medium in 10% bovine calf serum. The STAT3−/− MEFs were derived from STAT3+/− MEFs by Cre-loxP-mediated recombination of the floxed STAT3 allele through retroviral expression of the Cre recombinase in STAT3+/− MEFs (36). The MEFs were transfected with an M67-luciferase reporter construct that contains four STAT binding sites, using Effectene transfection reagent (Qiagen) following the manufacturer's protocol. Typically, 400 ng of M67 and 200 ng of β-galactosidase control plasmids were transfected into cells on 24-well plates using 4.8 μl of enhancer and 3 μl of Effectene. Twenty-four hours after transfection, cells were treated with human IL-6 (12.5 ng/ml; SERVA) and recombinant human IL-6 receptor (IL-6R; 50 ng/ml; R&D Systems) or OSM (25 ng/ml; R&D Systems) for 6 h before cells were harvested for the luciferase assay. Luciferase activity was normalized to β-galactosidase activity.
Whole-cell extract preparation and Western blot analysis.
MEFs treated or untreated with OSM (25 ng/ml) were trypsinized, washed in phosphate-buffered saline (PBS), and extracted in radioimmunoprecipitation (RIPA) buffer (1% NP-40, 0.5% Na-deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM EDTA in PBS, with 100 mM Na3VO4, 5 mM NaF, 1 mM phenylmethylsulfonyl fluoride). The protein concentration was determined by Bradford assay according to the manufacturer's protocol (Bio-Rad). Ten micrograms of the proteins was separated on an SDS-7% polyacrylamide gel electrophoresis (PAGE) and blotted with phospho-STAT3 (Tyr705) antibody (Cell Signaling Technology) or stripped and reprobed with the control STAT3 antibody (Upstate Biotechnology). To detect the STAT3 serine 727 phosphorylation, 400 μg of the same extract proteins was incubated with 4 μg of anti-STAT3 antibody (Upstate Biotechnology) at 4°C overnight, bound to 40 μl of protein A-agarose beads (Upstate Biotechnology), washed in RIPA, low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 500 mM NaCl), and PBS. The bound proteins were eluted from beads by boiling in 2× SDS sample buffer and separated on an SDS-7% PAGE and blotted with phospho-STAT3 (Ser727) antibody (6E4; Cell Signaling Technology).
RT-PCR.
Total RNA was prepared from tissues snap-frozen on dry ice or cells using the TRIzol reagent (Gibco BRL). Reverse transcription (RT) reaction and subsequent PCR analysis were performed as described in reference 40. The PCR primers used were the following: SOCS3-forward, 5′ GGGGGAGGCAGGAGGTGATGGA; SOCS3-reverse, 5′ GGGCGGGCTGGAGGTGGATT; Fos-forward, 5′ CATTCCCCAGCCGACTCCTTCTCC; Fos-reverse, 5′ TTGCCCCTTCTGCCGATGCTCTG; JunB-forward, 5′ GACCTGCACAAGATGAACCACGTGACGCC; JunB-reverse, 5′ GTGCCTCCGGTACGGTCTGCGGTTCC; IRF1-forward, 5′ GCACTGTCACCGTGTGTCGTCAGCAGC; IRF1-reverse, 5′ TGTCCCCTCGAGGGCTGTCAATCTCTGG; GAPDH-forward, GGTGAAGGTCGGTGTGAACGGATTTGG; GAPDH-reverse, TGTGCCGTTGAATTTGCCGTGAGTGG; SAA-forward, 5′ GCCTGGTCTTCTGCTCCCTGCTC; SAA-reverse, 5′ TCAGGCAGTCCAGGAGGTCTGTAG; AGP-forward, 5′ TTGCAGAGACCTGATAGTCTTGAAGATGC; AGP-reverse, 5′ TGTGGCAGAGACAGAATCAAAGTGCAC; SAP-forward, 5′ CACCCTTCATTGTCATCCAAGGCACATA; SAP-reverse, 5′ GAAAGGTCACTGTAGGTTCGGAAACACAGTG; HPX-forward, 5′ GGACCAAGCCAGATTCAGATGTACCC; HPX-reverse, 5′ CACTGTCAGGACCACGGAATGCAGC; Fibrogen-α-forward, 5′ GGTCCTGCAGCAGGGCTGTAAACCGTG; Fibrogen-α-reverse, 5′ TCACTTCCAGACCCGGAGTTCCCAG; Fibrogen-γ-forward, 5′ CATCCCACAACGGCATGCACTTCAGTAC; Fibrogen-γ-reverse, 5′ GGAGAGTCTGTTGAAGGGAATTATCTTCATG; HP-forward, 5′ TCGCTGCCGACAGTTCTACAGACTAC; and HP-reverse, 5′ CTCGGTTACAAGCACCCTCTGCTTGAG.
Growth analysis.
Mice were weighed using a Scout II scale (Ohaus, N.J.) daily from postnatal day 3 (P3) until 28 days and then weekly until 8 weeks of age. The body length was measured from the tip of the nose to the anus. The tail length was measured from the beginning of the base of the tail to the tip.
IGF-1 and GH measurement.
IGF-1 levels in serum from cardiac punctures or decapitation were determined using an enzyme immunoassay kit (rat IGF DSL-10-2900; Diagnostic Systems Laboratories) according to the manufacturer's instructions. GH was measured by using an enzyme immunoassay kit (rat growth hormone Biotrak RPN2561; Amersham Biosciences).
Thymocyte preparation and apoptosis assay.
Thymi from mice at the indicated age were minced and washed gently in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Thymocytes were counted and analyzed for apoptosis using Annexin V-fluorescein isothiocyanate (BD PharMingen) and propidium iodide (Sigma) staining followed by fluorescence-activated cell sorter analysis.
Treatment of animals.
To induce acute phase response, 8- to 10-week-old STAT3SA/− mice and littermate control mice were injected subcutaneously with 100 μl of turpentine (Sunnyside Corp.) per mouse or intraperitoneally with recombinant mouse IL-6 (carrier-free 406-ML-025/CF; R&D Systems) dissolved in sterile pyrogen-free saline (Sigma) at a dose of 5 μg/mouse. As negative controls for injection, sterile pyrogen-free saline was also injected. Twelve hours after turpentine or 6 h after IL-6 injection, mice were sacrificed and livers were removed immediately and frozen on dry ice before further processing.
Statistical analysis.
Results were analyzed by a two-tailed Student's t test. A P value of less than 0.05 was considered to be statistically significant.
RESULTS
Generation of STAT3SA mutant mice.
In order to generate SA/SA mice, a targeting vector that contains 10 kb of the STAT3 genomic DNA including exons 16 to 24 was constructed as shown in Fig. 1A. In this targeting vector, the residues encoding serine 727, which is located in exon 23, were mutated to encode alanine, and a lox-P-flanked neo cassette was inserted ∼90 bp downstream of the polyadenylation site, so that the insertion of the Neo cassette would be unlikely to affect STAT3 expression. ES cells were electroporated with the linearized targeting vector, and transfected cells were selected in the presence of G418. Homologous recombination was identified by Southern blot detection of a 6.5-kb fragment using the indicated probe after EcoRI digestion of the genomic DNA (Fig. 1B). Thirty-three clones out of 400 G418-resistant clones were positive for homologous recombination. To screen for recombinations that included the S727A mutation, a long PCR approach was used to screen for the mutation, which is ∼1.5 kb upstream of the 5′ end of the Neo cassette (Fig. 1A). In this PCR analysis, the primer a1 corresponding to the wild-type serine or a2 corresponding to the mutated alanine sequence was paired with the primer b to amplify either a 4.5-kb fragment from the wild-type allele or a 6.5-kb fragment from the SA STAT3 allele (Fig. 1C). Since primer b is located outside the targeting vector, the 6.5-kb fragment could only be generated from the correct recombination of the SA allele. In addition, the serine mutation was confirmed by sequence analysis of PCR-amplified product from the genomic DNA. Five out of the 33 positive clones had the S727A mutation. These appropriately targeted ES cell clones were injected into blastocysts of C57BL/6 mice. Germ line transmission was obtained for three clones and, after confirming the viability and fertility of the strains from these clones, animals derived from one clone were used for subsequent breeding and analysis.
To further confirm the disruption of serine phosphorylation in the SA mutation, the heterozygous SA/+ mice were crossed to generate SA/SA mice. MEFs were derived from +/+ or SA/SA littermate embryos. These MEFs were treated with OSM, which activates STAT3, and analyzed for STAT3 tyrosine and serine phosphorylation (Fig. 1D). While the STAT3 tyrosine phosphorylation was intact in the SA/SA MEFs (Fig. 1D, anti-STAT3 Tyr705 blot), the serine phosphorylation observed in the +/+ extract was absent in the SA/SA extract (Fig. 1D, anti-STAT3 Ser727 blot), confirming that the S727A mutation did abolish the serine phosphorylation. There was no difference in any of the cells in DNA binding tested by electrophoretic mobility shift assay (data not shown).
Normal development and apparent normalcy of STAT3SA/SA mice.
Crosses of heterozygous SA/+ mice showed normal Mendelian segregation of all expected genotypes, including the SA/SA genotype. Both SA/+ and SA/SA mice developed, were fertile, and appeared normal compared to their +/+ littermates. Several tissues where STAT3 has an established function were then tested in SA/SA mice. The liver acute phase responses (3), liver regeneration (10, 11), endotoxin-induced septic shock (41), wound healing (38), and doxorubicin-induced cardiomyopathy (21) all occurred equally in SA/SA and wild-type mice (data not shown).
Transcriptional responses in mutant fibroblasts.
To examine whether the serine mutation does affect transcription, MEFs were derived from +/+, SA/+, and SA/SA littermate embryos. A luciferase reporter gene containing four STAT3 binding sites was transiently transfected into these MEFs (Fig. 2A). The IL-6-dependent luciferase activity was reduced approximately 25% in the SA/+ cells and further to ∼50% of wild-type level in the SA/SA cells. Thus, at least in MEFs, each SA allele subtracts about 25% of the transcriptional efficiency of activated (tyrosine-phosphorylated) STAT3.
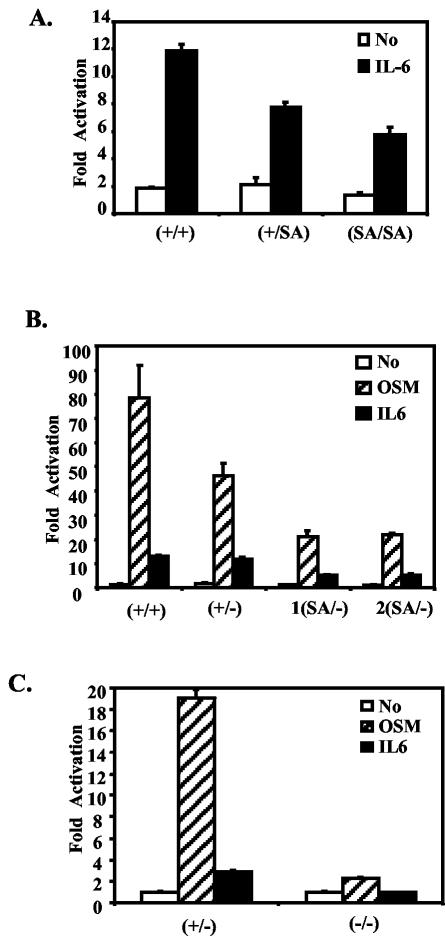
FIG. 2.
The STAT3SA mutation decreases STAT3 transactivation activity in MEFs. (A) MEFs were prepared from E13.5 STAT3+/+, STAT3+/SA, and STAT3SA/SA littermate embryos. MEFs were transiently transfected with an M67 luciferase reporter that contains four copies of the STAT binding site in its promoter, left untreated (no), or treated with IL-6 and IL-6R (IL-6) for 6 h and then harvested for a luciferase assay. Shown are average results from two, three, and three lines of MEFs derived from STAT3 +/+, +/SA, and SA/SA animals, respectively (triplet samples were tested for each cell line). (B) As for panel A, MEFs were prepared from STAT3+/+, +/−, and two SA/− littermate embryos. Transiently transfected MEFs were left untreated (no) or treated with IL-6 and IL-6R (IL-6) or OSM for 6 h. (C) As for panel B, transfections were done in STAT3+/− or −/− MEFs. (D) MEFs prepared from STAT3+/+, +/−, and two SA/− littermate embryos were treated with OSM (25 ng/ml) or IFN-γ (10 ng/ml) for the indicated times, and the indicated gene expression was analyzed by RT-PCR. For the OSM treatment, SOCS3, Fos, and JunB expression was also quantified and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by phosphorimager analysis; data are shown in the graphs underneath (SOCS3/GAPDH, Fos/GAPDH, and JunB/GAPDH).
A mouse strain that is STAT3 wild type/null (referred to as +/−) has been generated and is essentially normal (36). Since no apparent phenotype was detected in SA/SA mice, we crossed SA/+ mice with +/− mice to generate the SA/− mouse, so as to decrease the STAT3 transcription potential even farther. As discussed below, SA/− mice were able to survive through embryogenesis, and it was possible to obtain fibroblasts from fetuses of the SA/− genotype. Replacement of one SA allele by a null allele further reduced the transcriptional response compared to two SA alleles (Fig. 2B). Thus, fibroblasts from the most compromised viable mutant, SA/−, had no more than about 25% of the STAT3 transcription activation potential when cells were stimulated with IL-6 or OSM in transfection experiments. As expected, almost no response of the reporter gene to OSM remained in STAT3−/− fibroblasts (Fig. 2C).
The induction of endogenous chromosomal genes in SA/− fibroblasts by OSM, a STAT3 activator, or gamma interferon (IFN-γ), a STAT1 activator, was also tested. Both the SOCS3 and IRF-1 genes in SA/−, +/−, or +/+ cells gave similar induced responses to IFN-γ (Fig. 2E, IRF1 and SOCS3). However, for at least three potential STAT3 target genes, Fos, SOCS3, and JunB, the SA/− cells responded either less strongly or somewhat later than +/+ or +/− cells (Fig. 2D), confirming the limited transcriptional potential of SA/−.
Partial lethality of STAT3SA/− mice.
The genotypes of the offspring from the +/− by SA/+ crosses were determined at 21 days of age by PCR analysis of the tail DNA (Table 1). Mendelian segregation would yield 25% each of the +/+, SA/+, +/−, and SA/− mice. Similar numbers of +/+, SA/+, and +/− mice were found, consistent for these genotypes with a Mendelian ratio (61, 68, and 62 out of 208 offspring, respectively). In contrast, only 17 mice were of the SA/− genotype, corresponding to about 75% lethality before 21 days.
TABLE 1.
Partial perinatal lethality of STAT3SA/− mice
| Parental cross and offspring genotype | No. of mice | Ratio of total (%) |
|---|---|---|
| STAT3+/− × STAT3+/SAa | ||
| +/+ | 61 | 29 |
| +/SA | 68 | 33 |
| +/− | 62 | 30 |
| SA/− | 17 | 8 |
| Total | 208 | 100 |
| STAT3+/− × STAT3SA/SAb | ||
| +/SA | 105 | 79 |
| SA/− | 28 | 21 |
| Total | 133 | 100 |
| STAT3+/− × STAT3SA/SAc | ||
| +/SA | 11 | 46 |
| SA/− | 13 | 54 |
| Total | 24 | 100 |
Thirty-one litters were genotyped at day P21; 25% of each of the +/+, SA/+, +/−, and SA/- genotypes was expected according to the Mendelian ratio.
Twenty-five litters were genotyped at day P21; 50% of each of the +/SA and SA/− genotypes was expected according to the Mendelian ratio.
Three litters were genotyped at day E18.5; 50% SA/− mice were expected.
This experiment was repeated in offspring from the +/− by SA/SA crosses, where 50% of the offspring should be SA/−. As shown in Table 1, of the 133 offspring examined, 105 were SA/+ and only 28 were SA/− (corresponding to 21% of the total offspring). Again, about three-fourths of the SA/− pups were missing (i.e., [105 − 28]/105 × 100%).
To determine more precisely when the SA/− pups were dying, genotypes of embryonic day 18.5 (E18.5) embryos were determined. SA/− embryos were present in the expected Mendelian ratio (Table 1), indicating that SA/− mice survived to term and that one copy of a defective STAT3 gene was sufficient to overcome the early embryonic block of the STAT3−/− genotype. When the offspring of the +/− to SA/SA breeding were carefully watched and counted on a daily basis, most of the STAT3SA/− mice that died did so within 48 h after birth, usually within the first 24 h. If they survived the first 2 days they usually survived for >28 days.
Gross external examination did not show any apparent difference between SA/− newborn pups and their control littermates, except they were smaller in size (Fig. 3) (see below). Histologic evaluation (hematoxylin-eosin stain) of all major organs of surviving SA/− mice revealed no defects. Pups that died within the first postnatal day were also evaluated. Almost no milk was found in the stomachs of the dead SA/− pups. Also, in some cases the lungs were not fully inflated; however, there were patches of uninflated lung tissue in some SA/+ animals. Thus, no regular and significant abnormality was found in any major organ.
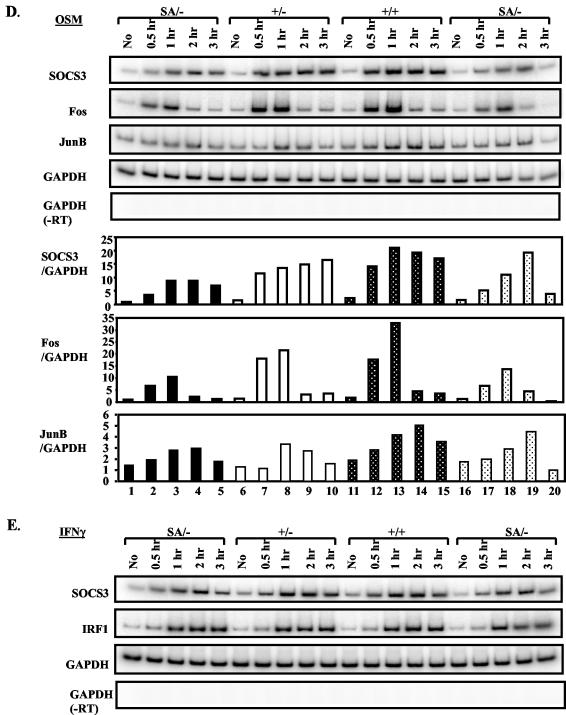
FIG.3.
Growth retardation of STAT3SA/− mice. (A) Representative male littermates at day P24. The mouse on top is SA/−, and the one on the bottom is SA/+. (B) Growth curves with standard deviations of STAT3+/SA (filled squares; total of 12 animals) and STAT3SA/− (filled circles; total of 8 animals) male mice from day P3 to 8 weeks of age. P was <0.0001 in the 2 to 6 weeks. (C) Representative female littermates at day P24. The top animal is SA/−, and the one on the bottom is SA/+. (D) Growth curves with standard deviations of STAT3+/SA (filled squares; total of 13 animals) and STAT3SA/− (filled circles; total of 6 animals) female mice from day P3 to 8 weeks of age. P was <0.0001. (E) A litter of E18.5 embryos showing reduced weight of SA/− mice during embryogenesis. The ones marked with an M are SA/−, and the rest are SA/+. (F) Weight of STAT3+/SA (11 pups) and STAT3SA/− (13 pups) embryos at E18.5. Three asterisks indicate that P was <0.001. (F) Body and tail lengths of STAT3+/SA and STAT3SA/− mice at 24 days of age. Top, males; bottom, females. Filled bar, SA/+; open bar, SA/−. Three asterisks indicate that P was <0.001. (G) Organ weights of STAT3+/SA (total of six animals) and STAT3SA/− (total of four animals) male mice at 24 days of age. The weight of each organ of the SA/+ mice was assigned as 100%. Filled bar, SA/+; open bar, SA/−. For liver and spleen, P < 0.01; for the rest, P < 0.001. (F) As for panel G but for female mice. Total of five SA/+ and six SA/− mice. Spleen, P = 0.016; for the rest, P < 0.0001.
Because STAT3 was recently reported to activate the promoter of surfactant protein B (SP-B), an essential postnatal lung protein (47), and because the SP-B null mutation causes lethal perinatal respiratory distress (9, 32), we examined the SA/− pups for the mRNA for SP-B. However, SP-B expression level was normal in SA/− mice (data not shown).
Other possible reasons for immediate mortality in 75% of the SA/− animals were also examined. No significant alteration was found in the blood glucose level. Brain and spinal cord appeared normal, and a neuronal analysis showed no apparent differences in facial and hypoglossal motor neurons of the SA/− mice (data not shown). This was in contrast to the lethal phenotypes of the JAK1−/− (37), CNTFR−/− (12), and LIFR−/− mice (25). In these three cases (where STAT3 activation would be depressed), a neuronal defect is the presumptive cause for mortality. Finally, using the technique of gene deletion in some but not all embryonic cells (mosaic deletion of STAT3 [4]), a phenotype similar to SA/− mice was observed. Particularly, these STAT3 mutant mice did not breathe or feed well. When the neuronal cells from the nodose ganglia were explanted, they underwent apoptosis at an enhanced rate. Therefore, it is possible that some lethal neuronal defect too subtle to be detected in our examinations exists in SA/− mice.
Growth retardation of STAT3SA/− mice.
We also followed the SA/− animals who survived. Compared to SA/+ animals, both male and female SA/− mice weighed only 50 to 60% of wild type through the first 28 days (Fig. 3B and D) (P < 10−5). Also, SA/− mice at day E18.5 weighed ∼13% less than their control littermates (P = 0.0004) (Fig. 3F and E). In addition, a number of fetal organs at E18.5 were also smaller. Thus, a growth defect in SA/− animals started during late fetal development. Although smaller in size, the STAT3SA/− mice appeared normal, and by 8 weeks of age they were only ∼10% smaller than their control littermates, exhibiting the catch-up growth phenomenon (Fig. 3B and D) (15). No significant difference was found between +/+, SA/+, or +/− mice (data not shown). The growth retardation of the SA/− mice was evident in a decrease of skeletal size, as reflected in the reduced body length and tail length (Fig. 3G). Lower body weight of the SA/− mice was associated with reduced weight of major visceral organs for both males and females (Fig. 3H and I). Statistically smaller size for liver, heart, lung, kidney, thymus, spleen, and testis compared to control mice was observed.
Growth defects based on altered IGF-1.
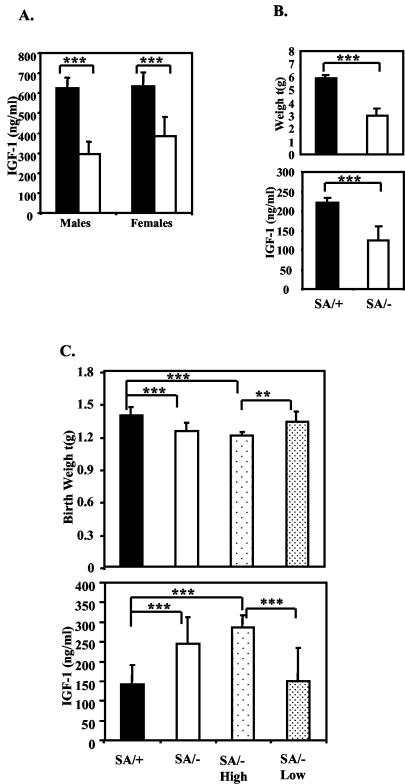
The late fetal growth and perinatal growth defect of the SA/− mice were accompanied by altered serum IGF-1 levels. Decreased IGF-1 was found in both males and females at day P24 (Fig. 4A) (male SA/+, 625 ± 52 ng/ml; SA/−, 295 ± 63 ng/ml; female SA/+, 634 ± 70 ng/ml; SA/−, 383 ± 98 ng/ml [mean ± standard deviation]), as well as at 8 days (Fig. 4B) (SA/+, 224 ± 13 ng/ml; SA/−, 141 ± 21 ng/ml). Serum IGF-1 levels were assayed also in SA/− and SA/+ newborn pups. In contrast to the P8 pups, the average newborn IGF-1 level in SA/− pups was higher than in the SA/+ pups. The SA/− pups fell into two groups based on serum IGF-1 level (Fig. 4C). Approximately 75% (16 out of 22) of the SA/− pups had ∼2-fold higher IGF-1 levels than the SA/+ pups (+/SA, 141 ± 51 ng/ml; SA/− High, 287 ± 66 ng/ml; P = 2 × 10−8). The pups with the highest IGF-1 levels, however, had ∼13% reduced weight (+/SA, 1.39 ± 0.08 g versus SA/− High, 1.21 ± 0.08 g; P = 10−7). In contrast, the remainder of the SA/− pups (6 out of 22, i.e., ∼25%) had IGF-1 levels and birth weights similar to SA/+ pups. The proportion that will survive is, therefore, the same as the proportion that has near-normal IGF-1 levels. Indeed, a few of the dying SA/− pups were associated with reduced birth weight and higher IGF-1 levels. It seems possible that the overabundance of IGF-1 may be correlated with perinatal lethality, but we have no speculation to why this should be.
FIG. 4.
Altered IGF-1 levels in STAT3SA/− mice. (A) Serum IGF-1 level of 24-day-old littermate SA/+ and SA/− males and females. Four to six mice of each genotype were analyzed. Filled bar, +/SA; open bar, SA/−. Three asterisks indicate that P was <0.001. (B) Serum IGF-1 levels and weight of 8-day-old littermate SA/+ and SA/− mice. Filled bar, +/SA; open bar, SA/−. SA/+, n = 8; SA/−, n = 4. Three asterisks indicate that P was <0.001. (C) Serum IGF-1 level and weight of SA/+ and SA/− newborns. SA/+, total of 21 pups; SA/−, total of 22 pups; SA/− High, 16 SA/− pups grouped according to higher IGF-1 levels; SA/− Low, 6 SA/− pups grouped according to lower IGF-1 levels. Two asterisks indicate that P was <0.01; three asterisks indicate that P was <0.001.
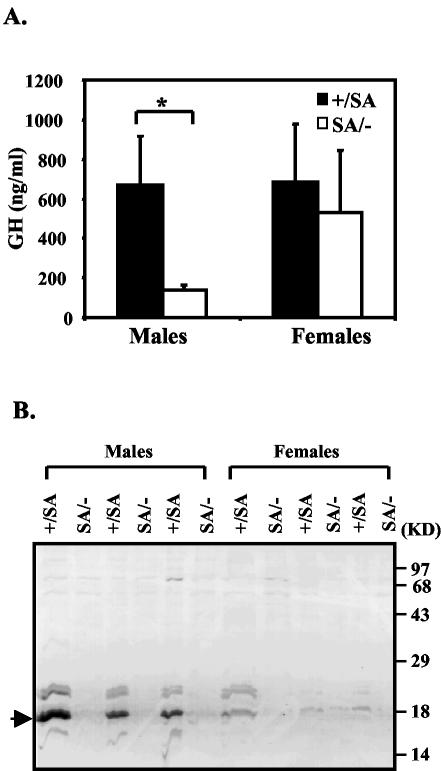
Later in perinatal life (after ∼P15), GH mediates growth through induction of IGF-1, but GH is thought to become important only around P15 (15, 27). Since in the first two postnatal weeks IGF-1 expression is independent of GH, decreased GH function should not account for the reduced IGF-1 level we found in the perinatal SA/− mice. Therefore, we measured GH at 24 days and found it was reduced in SA/− males but was approximately normal in females (Fig. 5A). Major urinary protein (MUP) is a GH-dependent liver secretory protein that comprises the major protein component of mouse urine (33). Less MUP was detected in both male and female SA/− mice, with a greater reduction in males, in accord with decreased GH function (Fig. 5B).
FIG. 5.
GH pathway in SA/− mice. (A) Serum GH levels in 24-day-old males and females at the time of sacrifice. Filled bar, SA/+; open bar, SA/−. P = 0.011 (one asterisk indicates that P was <0.05). (B) MUP levels in SA/+ and SA/− mice. Urine was collected from 24-day-old littermate animals at the time of sacrifice and checked by SDS-15% PAGE. Samples from the SA/− animals were loaded adjacent to those from the littermate SA/+ animals. The MUP is indicated by the arrow.
Defective thymocyte survival in STAT3SA/− mice.
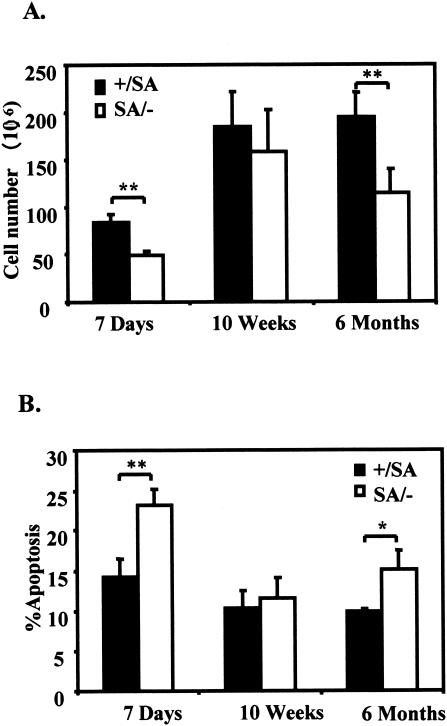
Cre-loxP deletion of STAT3 in T cells and in thymus epithelial cells in mice leads to defects in thymocytes (39, 42). Therefore, we harvested thymocytes from STAT3SA/− mice and control mice. Flow cytometric analysis revealed normal CD4+/CD8+ cell ratios, indicating normal development of the T cells present. However, the total thymocyte number was significantly reduced in SA/− mice from either preadolescent mice (7 days) or older mice (6 months) (Fig. 6A). Since more apoptotic cells were found in the SA/− thymocytes compared with the control mice (Fig. 6B), the decrease in thymocyte number can be logically attributed to defective thymocyte survival.
FIG. 6.
Decreased thymocyte survival in STAT3SA/− mice. (A) Thymocyte numbers of littermate STAT3+/SA and STAT3SA/− mice at ages of 7 days (three mice each), 10 weeks (four mice each), and 6 months (five mice each). The values shown are the mean cell number per thymus with the standard error. Two asterisks indicate that P was <0.01. (B) Thymocyte apoptosis of the littermate STAT3+/SA and STAT3SA/− mice. Thymocytes from the mice examined in panel A were stained with fluorescein isothiocyanate-Annexin V and propidium iodide; the mean percentages of the Annexin V-positive cells are shown with standard errors. One asterisk indicates that P was <0.05; two asterisks indicate that P was <0.01.
STAT3SA/− mice show normal skin and liver phenotypes.
We also tested whether there was any skin defect in the SA/− mice, as has been described for targeted STAT3 removal in epithelial cells of skin (38). The SA/− mice had a normal hair cycle (evaluated by histologic analysis). The SA/− mice also were comparable to the control mice in a wound healing test conducted after skin biopsy. Finally, later-stage phenotypes (sparse hair and spontaneous ulcers) that occurred in the epithelial STAT3-deficient mice (38) were not seen in the STAT3SA/− mice at a later age (data not shown).
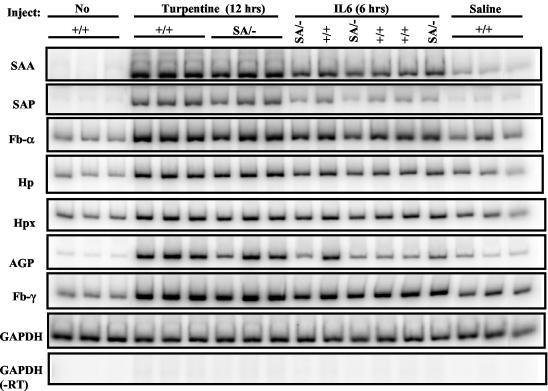
Chemical or infectious damage to the liver evokes a rise in so-called acute phase proteins in plasma (28). This response is impaired in IL-6−/− mice (2, 16, 20) or in mice with liver-specific STAT3 removal (3). We verified such increases by turpentine injection (induces local tissue damage) or intraperitoneal and intravenous injection (data not shown) of IL-6 with an array of classical mouse acute phase genes in +/+ mice, but there was unexpectedly no defect in response in SA/− mice (Fig. 7).
FIG. 7.
Induction of liver acute phase response gene expression in STAT3SA/− mice. Littermate STAT3+/+ and STAT3SA/− mice were injected with turpentine subcutaneously or recombinant IL-6 intraperitoneally. After 12 h of turpentine injection or 6 h of IL-6 injection, liver total RNA was prepared from each animal and analyzed by RT-PCR. As negative controls, mice were either left untreated or injected with sterile pyrogen-free saline. SAA, serum amyloids A; SAP, serum amyloids P; FB, fibrinogen; HP, haptoglobin; HPX, hemopexin; AGP, α1-acid glycoprotein.
DISCUSSION
The original STAT3 deletion with consequent early embryonic death in mice (43) obscured any possible subsequent necessity for the protein later in development. Recently, tissue-specific removal of STAT3 has revealed requirements for the protein in many adult tissues, in accord with the widespread distribution of the protein and its activation by a host of different ligands (reviewed in reference 23). The present study was undertaken to determine how a compromised transcriptional potential of STAT3 would affect embryogenesis and adult function. We first documented the effect of the S727A mutation in MEFs. Replacement of serine 727 with alanine (SA allele) reduced transcriptional potential in cultured MEFs by ∼50%, and pairing the SA allele with the null allele (SA/−) reduced the cytokine response for some chromosomal or transfected genes even farther (Fig. 2). Thus, at least some of the cells in the STAT3SA/− animals should have had no more than ∼25% of the STAT3-directed transcriptional potential of wild-type cells, and any genes that specifically required phosphorylated S727 would have been specifically affected. Of course, we have no way of knowing at present what the actual level of STAT3-dependent transcriptional potential is in any specific cell type, nor do we know which of the many STAT3 target genes might be most affected by this mutation. Phosphorylation of STAT3 is due to different kinases in different cell types and after different ligand stimulation. Thus, the need for serine-phosphorylated STAT3 implies a cooperation in interaction between the activating ligand for tyrosine phosphorylation and the activating serine kinase.
Compromised STAT3 activity in the SA/− mouse allows apparently near-normal embryonic development (attested to by the Mendelian ratio of 18.5-day SA/− fetuses in the SA/SA by +/− cross). However, the importance of an irreducible level of transcriptional activity by STAT3 (possibly including serine 727-dependent transcription) is clear from the perinatal death of ∼75% STAT3SA/− animals compared to the normal survival of +/− mice. Despite an intensive search, we have not uncovered a distinct cause for the perinatal lethality, although it seems likely that a neuronal defect results from the STAT3 insufficiency in SA/− mice. Targeted knockouts of JAK1 (37), CNTFR (12), and LIFR mice (25), all of which involve STAT3 activation, and neuronally deficient STAT3 mosaic mice (4) all exhibit perinatal lethality thought to be due to neuronal defects.
There is, however, a likely connection between the perinatal death of the SA/− mice and IGF-1 levels. For example, animals deficient in IGF-1 or IGF-1R exhibit perinatal lethality without a described cause (26). IGF-1 is established as the major growth factor in late embryogenesis and postnatal growth (5). The pattern of IGF-1 expression in the SA/− mice was interesting. At both P8 and P24, all the SA/− mice assayed had lower serum IGF-1 levels than the SA/+ mice. However, newborns, 75% of which were destined to die within a day or so, had higher IGF-1 levels than SA/+ newborns. It may be fortuitous, but it seems possible that the mice scheduled to die had overexpressed IGF-1. While GH is required for IGF-1 synthesis in the liver after the perinatal period, IGF-1 is also produced in local tissues that are not GH dependent (31). We do not know what triggers IGF-1 synthesis during late fetal or perinatal times, but it is presumably not GH. A second known connection between IGF-1 and STAT3 exists: STAT3 is activated by IGF-1 treatment of cultured cells or several tissues in vivo (34, 49). Thus, the STAT3 activation through IGF-1R may normally require S727 phosphorylation, since the mitogen-activated protein kinase pathway also is induced through IGF-1R (14). Thus, in the nonsurviving fraction of the newborns which may have the highest levels of IGF-1, a necessary compensation is not achieved. In the surviving fraction of SA/− animals, a deficiency of IGF-1 persists, but slow growth is allowed rather than lethality.
Although IGF-1 does not activate STAT5, GH activates STAT1, STAT3, and STAT5 (18, 35). STAT1-deficient mice do not show any growth phenotype (29). A comparison of the effects of the SA/− phenotype with STAT5A- and -5B-deficient mice shows considerable differences and emphasizes the different roles of IGF-1 and GH as well as STATs 3 and 5 in growth control. STAT5B null animals are viable but show male growth deficiency exhibited at puberty and normal female growth. STAT5A deficiency alone does not affect survival or growth. However, STAT5A/5B deficiency causes early lethality in one-third of the animals without a known cause and growth retardation in both male and female animals (44, 45). The growth defect of STAT5−/− mice is mainly imposed through lack of response to GH, while as discussed above, the STAT3 effect may be more involved in IGF-1 pathway. Nevertheless, STAT3 is activated by GH and also may be required for normal growth of both males and females.
Many adult requirements for STAT3 as detected by tissue-specific STAT3 removal were not seen in the STAT3SA/− animals. The acute phase response in the liver, wound healing, and hair growth all appeared normal, whereas targeted STAT3 removal would block these responses (23). Thus, studies of the STAT3SA/− mice would be critical to dissect the contribution of STAT3 serine phosphorylation to STAT3 activity in different cellular and signaling contexts in vivo. We did, however, detect in the SA/− animals increased thymocyte cell death leading to fewer thymocytes; however, the remaining cells differentiated normally. This contrasts with targeted removal of STAT3 in T-cell precursors, which did not decrease T-cell number (42) but rendered the T-cell precursors more susceptible to apoptosis outside the animal. In contrast, removal of STAT3 in the epithelium of the thymus did destroy thymus architecture, reduce its size, and affect differentiation of T cells (39). Thus, the thymic phenotype of the SA/− mice suggests both T-cell precursors and epithelial cell function may be faulty.
That a three- to fourfold decrease in transcriptional activity allowed near-normal development is somewhat surprising in view of the effects of manipulation of STAT92E activity by negative-acting proteins in Drosophila melanogaster. There, increased PIAS protein or an increase in a dominant-negative form of STAT92E upsets various developmental events (6, 17). Thus, we conclude that for many but not all purposes ∼25% STAT3 activity must be adequate. Finally, once a STAT3SA/− animal has survived the immediate postnatal period, viability is most likely assured.
.
Acknowledgments
We thank Chingwen Yang for assistance in gene targeting and Markus Stoffel, David Shih, Ann Kato, Patricia Morris, Paul Feinstein, James Krueger, and Michel Nussenzweig for many useful suggestions and aid in various phenotypic tests. Peter Mombaerts is acknowledged for the EIIa-Cre transgenic mice from Heiner Westphal. We thank Sharon White and Peter Shapiro for excellent technical assistance and Domenico Accili, Honghua Li, and Darnell lab members for discussions. We are grateful to Lois Cousseau for preparation of the manuscript and William Xin for photographic assistance.
This work was supported by National Institutes of Health grants AI34420 and AI32489 to J.E.D. Y.S. was partially supported by a postdoctoral fellowship from the Norman and Rosita Winston Foundation.
REFERENCES
- 1.Akira, S. 2000. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19:2607-2611. [DOI] [PubMed] [Google Scholar]
- 2.Alonzi, T., E. Fattori, M. Cappelletti, G. Ciliberto, and V. Poli. 1998. Impaired Stat3 activation following localized inflammatory stimulus in Il-6-deficient mice. Cytokine 10:13-18. [DOI] [PubMed] [Google Scholar]
- 3.Alonzi, T., D. Maritano, B. Gorgoni, G. Rizzuto, C. Libert, and V. Poli. 2001. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol. Cell. Biol. 21:1621-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonzi, T., G. Middleton, S. Wyatt, V. Buchman, U. A. Betz, W. Muller, P. Musiani, V. Poli, and A. M. Davies. 2001. Role of STAT3 and PI 3-kinase/Akt in mediating the survival actions of cytokines on sensory neurons. Mol. Cell. Neurosci. 18:270-282. [DOI] [PubMed] [Google Scholar]
- 5.Baker, J., J. P. Liu, E. J. Robertson, and A. Efstratiadis. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73-82. [PubMed] [Google Scholar]
- 6.Betz, A., N. Lampen, S. Martinek, M. W. Young, and J. E. Darnell, Jr. 2001. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. USA 98:9563-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brivanlou, A. H., and J. E. Darnell, Jr. 2002. Signal transduction and the control of gene expression. Science 295:813-818. [DOI] [PubMed] [Google Scholar]
- 8.Bromberg, J. F. 2001. Activation of STAT proteins and growth control. Bioessays 23:161-169. [DOI] [PubMed] [Google Scholar]
- 9.Clark, J. C., S. E. Wert, C. J. Bachurski, M. T. Stahlman, B. R. Stripp, T. E. Weaver, and J. A. Whitsett. 1995. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl. Acad. Sci. USA 92:7794-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cressman, D. E., R. H. Diamond, and R. Taub. 1995. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology 21:1443-1449. [PubMed] [Google Scholar]
- 11.Cressman, D. E., L. E. Greenbaum, R. A. DeAngelis, G. Ciliberto, E. E. Furth, V. Poli, and R. Taub. 1996. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 274:1379-1383. [DOI] [PubMed] [Google Scholar]
- 12.DeChiara, T. M., R. Vejsada, W. T. Poueymirou, A. Acheson, C. Suri, J. C. Conover, B. Friedman, J. McClain, L. Pan, N. Stahl, et al. 1995. Mice lacking the CNTF receptor, unlike mice lacking CNTF, exhibit profound motor neuron deficits at birth. Cell 83:313-322. [DOI] [PubMed] [Google Scholar]
- 13.Decker, T., and P. Kovarik. 2000. Serine phosphorylation of STATs. Oncogene 19:2628-2637. [DOI] [PubMed] [Google Scholar]
- 14.Dupont, J., and D. LeRoith. 2001. Insulin and insulin-like growth factor I receptors: similarities and differences in signal transduction. Horm. Res. 55(Suppl. 2):22-26. [DOI] [PubMed] [Google Scholar]
- 15.Efstratiadis, A. 1998. Genetics of mouse growth. Int. J. Dev. Biol. 42:955-976. [PubMed] [Google Scholar]
- 16.Fattori, E., M. Cappelletti, P. Costa, C. Sellitto, L. Cantoni, M. Carelli, R. Faggioni, G. Fantuzzi, P. Ghezzi, and V. Poli. 1994. Defective inflammatory response in interleukin 6-deficient mice. J. Exp. Med. 180:1243-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriksen, M. A., A. Betz, M. V. Fuccillo, and J. E. Darnell, Jr. 2002. Negative regulation of STAT92E by an N-terminally truncated STAT protein derived from an alternative promoter site. Genes Dev. 16:2379-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrington, J., L. S. Smit, J. Schwartz, and C. Carter-Su. 2000. The role of STAT proteins in growth hormone signaling. Oncogene 19:2585-2597. [DOI] [PubMed] [Google Scholar]
- 19.Hogan, B., R. Beddington, F. Costantini, and E. Lacy (ed.). 1994. Manipulating the mouse embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Kopf, M., H. Baumann, G. Freer, M. Freudenberg, M. Lamers, T. Kishimoto, R. Zinkernagel, H. Bluethmann, and G. Kohler. 1994. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature 368:339-342. [DOI] [PubMed] [Google Scholar]
- 21.Kunisada, K., S. Negoro, E. Tone, M. Funamoto, T. Osugi, S. Yamada, M. Okabe, T. Kishimoto, and K. Yamauchi-Takihara. 2000. Signal transducer and activator of transcription 3 in the heart transduces not only a hypertrophic signal but a protective signal against doxorubicin-induced cardiomyopathy. Proc. Natl. Acad. Sci. USA 97:315-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakso, M., J. G. Pichel, J. R. Gorman, B. Sauer, Y. Okamoto, E. Lee, F. W. Alt, and H. Westphal. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 93:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 24.Levy, D. E., and C. K. Lee. 2002. What does Stat3 do? J. Clin. Investig. 109:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., M. Sendtner, and A. Smith. 1995. Essential function of LIF receptor in motor neurons. Nature 378:724-727. [DOI] [PubMed] [Google Scholar]
- 26.Liu, J., J. Baker, A. S. Perkins, E. J. Robertson, and A. Efstratiadis. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75:59-72. [PubMed] [Google Scholar]
- 27.Lupu, F., J. D. Terwilliger, K. Lee, G. V. Segre, and A. Efstratiadis. 2001. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 229:141-162. [DOI] [PubMed] [Google Scholar]
- 28.Mackiewicz, A., I. Kushner, and H. Baumann (ed.). 1993. Acute phase proteins. CRC Press, Boca Raton, Fla.
- 29.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 30.Nair, J. S., C. J. DaFonseca, A. Tjernberg, W. Sun, J. E. Darnell, Jr., B. T. Chait, and J. J. Zhang. 2002. Requirement of Ca2+ and CaMKII for Stat1 Ser-727 phosphorylation in response to IFN-γ. Proc. Natl. Acad. Sci. USA 99:5971-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakae, J., Y. Kido, and D. Accili. 2001. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr. Rev. 22:818-835. [DOI] [PubMed] [Google Scholar]
- 32.Nogee, L. M., G. Garnier, H. C. Dietz, L. Singer, A. M. Murphy, D. E. deMello, and H. R. Colten. 1994. A mutation in the surfactant protein B gene responsible for fatal neonatal respiratory disease in multiple kindreds. J. Clin. Investig. 93:1860-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norstedt, G., and R. Palmiter. 1984. Secretory rhythm of growth hormone regulates sexual differentiation of mouse liver. Cell 36:805-812. [DOI] [PubMed] [Google Scholar]
- 34.Prisco, M., F. Peruzzi, B. Belletti, and R. Baserga. 2001. Regulation of Id gene expression by type I insulin-like growth factor: roles of Stat3 and the tyrosine 950 residue of the receptor. Mol. Cell. Biol. 21:5447-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ram, P. A., S. H. Park, H. K. Choi, and D. J. Waxman. 1996. Growth hormone activation of Stat 1, Stat 3, and Stat 5 in rat liver. Differential kinetics of hormone desensitization and growth hormone stimulation of both tyrosine phosphorylation and serine/threonine phosphorylation. J. Biol. Chem. 271:5929-5940. [DOI] [PubMed] [Google Scholar]
- 36.Raz, R., C. K. Lee, L. A. Cannizzaro, P. d'Eustachio, and D. E. Levy. 1999. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 96:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodig, S. J., M. A. Meraz, J. M. White, P. A. Lampe, J. K. Riley, C. D. Arthur, K. L. King, K. C. Sheehan, L. Yin, D. Pennica, E. M. Johnson, Jr., and R. D. Schreiber. 1998. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell 93:373-383. [DOI] [PubMed] [Google Scholar]
- 38.Sano, S., S. Itami, K. Takeda, M. Tarutani, Y. Yamaguchi, H. Miura, K. Yoshikawa, S. Akira, and J. Takeda. 1999. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18:4657-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sano, S., Y. Takahama, T. Sugawara, H. Kosaka, S. Itami, K. Yoshikawa, J. Miyazaki, W. van Ewijk, and J. Takeda. 2001. Stat3 in thymic epithelial cells is essential for postnatal maintenance of thymic architecture and thymocyte survival. Immunity 15:261-273. [DOI] [PubMed] [Google Scholar]
- 40.Shen, Y., and J. E. Darnell, Jr. 2001. Antiviral response in cells containing Stat1 with heterologous transactivation domains. J. Virol. 75:2627-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeda, K., B. E. Clausen, T. Kaisho, T. Tsujimura, N. Terada, I. Forster, and S. Akira. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10:39-49. [DOI] [PubMed] [Google Scholar]
- 42.Takeda, K., T. Kaisho, N. Yoshida, J. Takeda, T. Kishimoto, and S. Akira. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 161:4652-4660. [PubMed] [Google Scholar]
- 43.Takeda, K., K. Noguchi, W. Shi, T. Tanaka, M. Matsumoto, N. Yoshida, T. Kishimoto, and S. Akira. 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94:3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teglund, S., C. McKay, E. Schuetz, J. M. van Deursen, D. Stravopodis, D. Wang, M. Brown, S. Bodner, G. Grosveld, and J. N. Ihle. 1998. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell 93:841-850. [DOI] [PubMed] [Google Scholar]
- 45.Udy, G. B., R. P. Towers, R. G. Snell, R. J. Wilkins, S. H. Park, P. A. Ram, D. J. Waxman, and H. W. Davey. 1997. Requirement of Stat5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl. Acad. Sci. USA 94:7239-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen, Z., Z. Zhong, and J. E. Darnell, Jr. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82:241-250. [DOI] [PubMed] [Google Scholar]
- 47.Yan, C., A. Naltner, M. Martin, M. Naltner, J. M. Fangman, and O. Gurel. 2002. Transcriptional stimulation of the surfactant protein B gene by STAT3 in respiratory epithelial cells. J. Biol. Chem. 277:10967-10972. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, J. J., Y. Zhao, B. T. Chait, W. W. Lathem, M. Ritzi, R. Knippers, and J. E. Darnell, Jr. 1998. Ser727-dependent recruitment of MCM5 by Stat1α in IFN-γ-induced transcriptional activation. EMBO J. 17:6963-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zong, C. S., J. Chan, D. E. Levy, C. Horvath, H. B. Sadowski, and L. H. Wang. 2000. Mechanism of STAT3 activation by insulin-like growth factor I receptor. J. Biol. Chem. 275:15099-15105. [DOI] [PubMed] [Google Scholar]