Abstract
Bacillus subtilis two-component system DegS/U is well known for the complexity of its regulation. The cytosolic sensory kinase DegS does not receive a single predominant input signal like most two-component kinases, instead it integrates a wide array of metabolic inputs that modulate its activity. The phosphorylation state of the response regulator DegU also does not confer a straightforward “on/off” response; it is fine-tuned and at different levels triggers different sub-regulons. Here we describe serine phosphorylation of the DegS sensing domain, which stimulates its kinase activity. We demonstrate that DegS phosphorylation can be carried out by at least two B. subtilis Hanks-type kinases in vitro, and this stimulates the phosphate transfer towards DegU. The consequences of this process were studied in vivo, using phosphomimetic (Ser76Asp) and non-phosphorylatable (Ser76Ala) mutants of DegS. In a number of physiological assays focused on different processes regulated by DegU, DegS S76D phosphomimetic mutant behaved like a strain with intermediate levels of DegU phosphorylation, whereas DegS S76A behaved like a strain with lower levels of DegU phophorylation. These findings suggest a link between DegS phosphorylation at serine 76 and the level of DegU phosphorylation, establishing this post-translational modification as an additional trigger for this two-component system.
Introduction
Two-component systems are a ubiquitous means of signal transduction in bacteria [1]. The first, signal-receiving component is a sensory histidine kinase that is triggered by a stimulus binding or otherwise affecting its sensing domain. Upon activation, the histidine kinase autophosphorylates on a histidine residue, and thereafter transfers the phosphate to a specific aspartate residue of its cognate response regulator. Phosphorylation of the response regulator, in turn, triggers its regulatory role, which is in most cases transcriptional regulation via binding of a specific DNA sequence. The histidine kinases of the two-component systems are known to be highly specific, i.e. exhibiting low level of cross-talk with non-cognate response regulators [2]. Another major group of bacterial kinases involved in signal transduction is the Hanks type serine/threonine kinases [3], [4]. Hanks type kinases and two-component histidine kinases are sometimes found fusioned in a single polypeptide in Cyanobacteria [5], however, very few cases of crosstalk between these two protein families have been reported so far. Two recent studies pointed out that serine/threonine kinases can phosphorylate two-component response regulators: StkP from Streptococcus pneumoniae phosphorylates the orphan response regulator RitR [6] and serine-threonine kinase Stk1 phosphorylates and thereby abolishes the activity of the response regulator CovR in Group B Streptococcus [7]. Interestingly, a recent phosphoproteomic study in Bacillus subtilis, identified the two-component system histidine kinase DegS as being phosphorylated on the residue serine76, located in the signal sensing domain [8]. This implied the existence of a new type of crosstalk between two phosphorylation systems, namely one in which a presently unknown serine kinase would phosphorylate the two-component sensory histidine kinase.
Early mutational studies of the DegS/U two component system established that the response regulator binds DNA sequences and regulates expression of specific genes both in its phosphorylated and unphosphorylated state. This was exemplified by reciprocal effects of the two forms of DegU on exoprotease production and competence [9]. The importance of DegS/U was further underscored in two microarray experiments that independently demonstrated a total of 135 transcriptional units regulated either directly or indirectly by this two-component system [10], [11]. Only 22 transcriptional units were identified in the overlap between the two studies, which could indicate an even larger regulon. Whereas the initial studies of DegS/U in the laboratory strain 168 mainly focused on competence and exoprotease production, more recent studies using an undomesticated B. subtilis strain demonstrated that DegS/U also affects motility, complex colony and biofilm formation. The regulation was shown to depend on the discrete levels of DegU phosphorylation, in a manner far more subtle than a simple on/off switch [12], [13]. It is now well established that DegS/U system plays an important role in the transition growth phase where it receives many inputs which regulate degSU transcription or modulate the activity of synthesized DegS/U proteins. The degSU genes are transcribed as an operon and degU is itself transcribed from two additional promoters: one activated by DegU∼P and the other by nitrogen starvation [9], [13]–[16]. The signal-sensing domain of DegS interacts with the SMC-ScpA-ScpB protein complex, involved in chromosome segregation, which inhibits the kinase activity of DegS [17]. Similarly, the DNA-binding activity of DegU is inhibited by RapG and this inhibition is counteracted by PhrG in response to increased cell density [18]. DegS/U activity is further modulated by two small regulatory peptides, DegQ and DegR. DegQ enhances the phosphotransfer from DegS∼P to DegU but does not protect DegU∼P from dephosphorylation [13]. The latter is accomplished by DegR that stabilises DegU∼P [19]. DegQ is synthesized in response to quorum sensing via ComPA and has been hypothesised to be a determinant for the transition from motile to sessile-growth state [13]. degR expression is SigD dependent and peaks in late exponential phase, but the physiological role remains elusive [20].
Despite the fact that the DegS/U two-component system is submitted to an elaborate control at both transcriptional and protein level, no specific DegS-activating signal has so far been proposed. Most two-component system histidine kinases are transmembrane proteins, presumably activated by extracellular signals via their N-terminal signal-sensing domains protruding from the cell surface [21]. By contrast, DegS is a cytosolic protein and hence responds to, and integrates several cytosolic signals, some of which have been listed above. In this study, we examined the possibility that phosphorylation of DegS on serine 76 residue could represent a novel input for this regulatory system. We demonstrated that the specific phosphorylation of this residue by the Hanks kinase YbdM stimulates phosphotransfer to DegU in vitro, and a phosphomimetic mutant of DegS leads to an increased DegU∼P pool, and influences the transcription of key DegU-dependent promoters in vivo.
Results and Discussion
DegS is phosphorylated in vitro by B. subtilis Hanks-type serine/threonine kinases
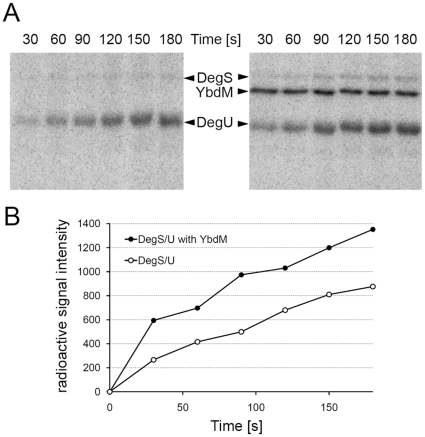
In order to characterize the serine 76 phosphorylation of DegS, we first asked the question whether this phosphorylation is auto-catalyzed or it requires another kinase. To answer this question we carried out in vitro phosphorylation experiments with purified DegS and 32P-γ-ATP. Phospho-histidine and phospho-serine can be easily distinguished, the latter being stable in acidic conditions and resistant to heat. Since the entire radioactive label present on autophosphorylated DegS was removed by acid and heat treatment (figure 1), we concluded that DegS was incapable of autophosphorylating on serine. There are a number of poorly characterized serine/threonine kinases in B. subtilis, mainly belonging to the family of Hanks-type kinases [22]. The most extensively characterized of those is the kinase PrkC that has recently been shown to participate in signalling underlying spore germination [23]. We purified the three Hanks-type kinases PrkC, YabT and YbdM, and a putative kinase PrkA to test their ability to phosphorylate DegS in vitro. PrkA (data not shown) and PrkC were unable to phosphorylate DegS, whereas both YbdM and YabT tested positive for DegS phosphorylation (figure 1). In order to verify whether serine 76 is indeed the residue phosphorylated by these kinases, we constructed a mutant protein with a non-phosphorylatable replacement DegS S76A. Phosphorylation of DegS S76A by YbdM was completely abolished, suggesting that it is the major phosphorylation site, and that the kinase YbdM is specific for this site (figure 1A). In the same assay, phosphorylation of DegS S76A by YabT was as efficient as that of the wild type (figure 1B). This situation is not unprecedented [24], and residual phosphorylation in such case could be due to a presently unknown secondary site, or the lack of specificity exhibited by the kinase under in vitro conditions. Our conclusion was that in vitro at least two different Hanks-type kinases can phosphorylate DegS, of which YbdM is specific for the residue serine 76. DegS is also known to exhibit phosphatase activity, so we tested whether serine phosphorylation of DegS could be removed by the protein itself. For this, we removed the ATP from the phosphorylation reactions catalyzed by YbdM and YabT, and allowed the dephosphorylation reaction to occur for 2 h. The radiolabel on DegS remained stable, indicating the absence of phospho-serine phosphatase activity (figure 1A and 1B, lanes 9). The B. subtilis serine/threonine kinase-encoding genes yabT, ybdM and prkA have been reported to belong to the sigma F, G and E regulon respectively [25], [26] hence linking them to sporulation specific processes. However, a recent transcriptomics study [27] identified yabT, ybdM and prkC as transcribed in the exponential growth phase, which makes it highly probable that these kinases are present in the transition phase when DegS activity is triggered. Since YbdM appeared to be the the most specific kinase for DegS serine 76, we tested the effect of YbdM-dependent DegS phosphorylation on the efficiency of phosphotransfer from DegS to DegU. To this end, we first incubated DegS with non-labelled ATP, MgCl2 and YbdM for 3 hrs, and we also prepared a control sample of DegS incubated for exactly the same time with ATP and MgCl2, but without YbdM. The DegS sample pre-incubated with YbdM was more efficient in phosphorylating DegU, especially in the first 60 s period which corresponds to the theoretical reaction time of the two-component system (figure 2).
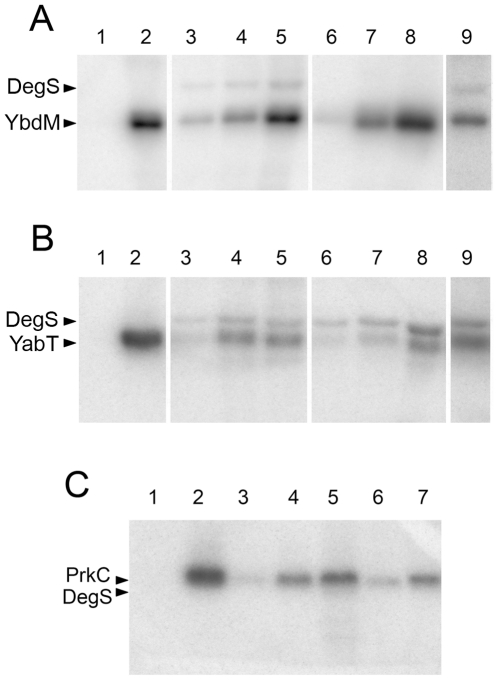
Figure 1. Phosphorylation of DegS by Hanks type kinases in vitro.
Autoradiography of SDS-Polyacrylamide gels with proteins that had been incubated with 32P-γ-ATP. Gels were treated by boiling in acid to remove phospho-histidine signals (A) Phosphorylation of DegS by the kinase YbdM. Lanes 1 and 2 are controls, with DegS alone and YbdM alone, respectively. Lanes 3–5 show phosphorylation of DegS by YbdM for 15, 30 and 60 min, respectively. Lanes 6–8 show phosphorylation of DegS S76A by YbdM for 15, 30 and 60 min, respectively. Lane 9 shows the equivalent of the reaction from lane 5, after desalting to remove the ATP and a 2 h incubation to test for phosphatase activity. (B) Phosphorylation of DegS by the kinase YabT. Lanes 1 and 2 are controls, with DegS alone and YabT alone, respectively. Lanes 3–5 show phosphorylation of DegS by YabT for 15, 30 and 60 min, respectively. Lanes 6–8 show phosphorylation of DegS S76A by YabT for 15, 30 and 60 min, respectively. Lane 9 shows the equivalent of the reaction from lane 5, after desalting to remove the ATP and a 2 h incubation to test for phosphatase activity. (C) Phosphorylation of DegS by the kinase PrkC. Lanes 1 and 2 are controls, with DegS alone and PrkC alone, respectively. Lanes 3–5 contain the reactions where PrkC concentration has been varied (2, 4 and 10 nM, respectively), and in lanes 6–7 the pH has been varied (pH 7 and pH 8, respectively).
Figure 2. Phosphorylation of DegS by YbdM stimulates phosphotransfer to DegU in vitro.
(A) Autoradiography of SDS-Polyacrylamide gels with proteins that had been incubated with 32P-γ-ATP. Gels were not treated by boiling in acid, so the phospho-histidine and phospho-aspartate signals are preserved. Efficiency of phosphotransfer of wild-type DegS to DegU (gel on the left) is compared to that of DegS that had been preincubated with YbdM, 50 µM non-labelled ATP and 5 mM MgCl2 for 3 h (gel on the right). (B) Quantification of DegU phosphorylation signals on both gels.
Phosphomimetic mutant DegS S76D exhibits increased autophosphorylation and phosphotransfer to DegU in vitro
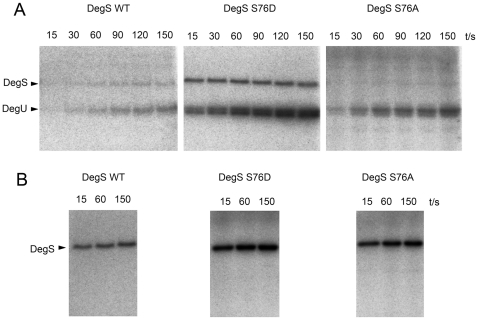
Rarely more than several percent of the target protein is phosphorylated during in vitro kinase assays, unless a specific kinase-activating signal is present [28]. In order to study the regulatory effects of phosphorylation in vivo, phosphomimetic mutants, with the phosphorylatable residue replaced by a larger, negatively charged amino acid are often employed [29]. Since our data indicated that DegS can be phosphorylated by two Hanks-type kinases, for which the specific effectors (or conditions) that trigger their activity towards DegS are unknown, it seemed particularly promising to study the effects of DegS phosphorylation in vivo using a phosphomimetic mutant DegS S76D. Before using the mutant protein in vivo, we checked whether its behaviour during an in vitro phosphorylation assay would corresponded to that of wild type DegS which had been phosphorylated by YbdM (as shown in figure 2). Purified DegS S76D showed an increase in DegU phosphorylation on aspartate (figure 3A) as well as an increase in autophosphorylation on histidine (figure 3B) compared to the wild type. The respective activites of DegS S76A were also above the wild type level, but below that of DegS S76D (figure 3). Interestingly, DegS S76D maintained a considerable level of incorporated phosphate in the presence of DegU, whereas DegS wild type and DegS S76A retained much less phosphate under these conditions. DegS/U exerts a very complex regulation of several physiological processes, some of which are affected by high and others by low levels of DegU phosphorylation. We thus hypothesized that strains where wild type DegS would be replaced by DegS S76D should be unable to activate processes that are normally stimulated by non-phosphorylated DegU, and probably overly stimulate processes that require phosphorylated DegU. We therefore decided to focus on several functions that exemplify these contrasting situations to examine the regulatory role of DegS phosphorylation on serine 76.
Figure 3. DegS mutations affect autophosphorylation and DegU phosphorylation in vitro.
Autoradiography of SDS-Polyacrylamide gels with proteins that had been incubated with 32P-γ-ATP. Gels were not treated by boiling in acid, so the phospho-histidine and phospho-aspartate signals are preserved. Time periods are indicated for each lane, for DegU phosphorylation (A) and DegS autophosphorylation (B).
DegS S76D negatively affects competence development
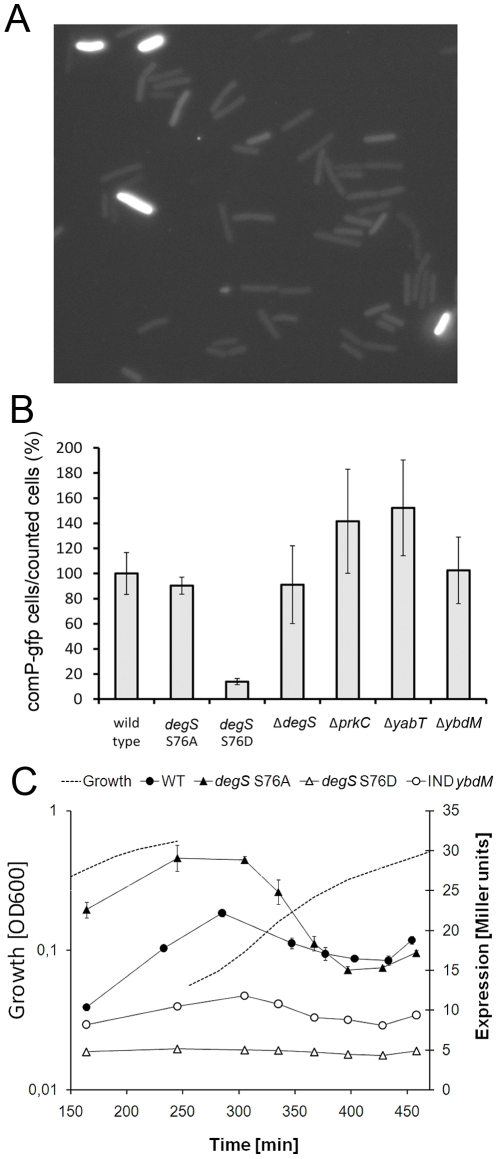
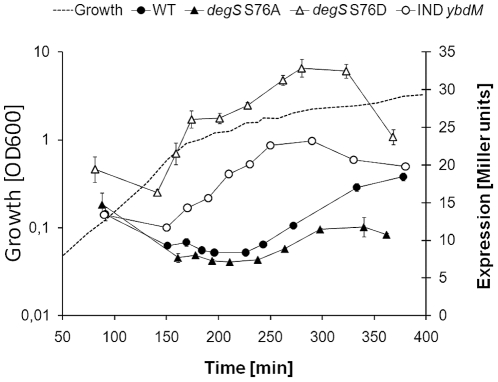
Competence development is a complex process regulated partly by DegS/U via the transcriptional activation of comK exerted by unphosphorylated DegU [30]. To test the effect of phosphorylation of DegS serine 76 on this process, strains were constructed in which the chromosomal version of degS was replaced by copies encoding either DegS S76D or DegS S76A. Competence of the resulting strains was initially compared to the wild type by using a two-step transformation protocol [31]. The competence was quantified as the number of colonies per µg of plasmid used for transformation, and normalized for the wild type (100+/−12%). The strain degS S76A (155+/−7%) was about 50% more competent than the wild type, while the degS S76D mutant exhibited an approximately 5-fold reduction in competence (18+/−0%), concurring with our working hypothesis. Next, competence development on single cell level was assayed by introducing GFP under control of the comK promoter. In this system we further assayed the effects of inactivating individual serine/threonine kinase-encoding genes prkC, ybdM and yabT (figure 4). Our hypothesis was confirmed, with an approximate 5-fold competence reduction in degS S76D, while in this set up no difference between wild type and the degS S76A strain was observed. In this setup, a mutant of the kinase phosphorylating DegS would be expected to behave as the non-phosphorylatable degS S76A. All kinase mutants had competence level identical or superior to wild-type, which is in agreement with our hypothesis, but of limited evidential value, since degS S76A was itself non-distinguishable from the wild type. Competence in B. subtilis is a bistability phenomenon that occurs in a sub-population of cells which become transiently competent during a certain window of time in the transition growth phase [32]. We thus asked the question whether the effect of degS S76D on competence could be due to a temporal shift in the window of competence or an overall decrease in DegU-dependent comK expression. In order to determine this, lacZ encoding β-galactosidase was placed under the control of the comK promoter and introduced in the amyE locus of wild type and mutant strains. Activity of the comK promoter was assayed in the two-step competence media. Expression of comK was similar in the wild type and degS S76A cells, peaking out as expected in the early transition phase. In the degS S76D mutant, expression of comK was 5-fold lower, and not induced at all in the transition phase (figure 4C). When we overexpressed ybdM from an IPTG-dependent promoter in the wild type strain, comK was still induced, but less efficiently than in the wild type. Since comK expression is known to be activated by unphosphorylated DegU, the conclusion we reached based on the presented data was that the degS S76D mutation indeed leads to an increase in the DegU∼P pool in vivo, in accordance to our phosphorylation data obtained in vitro.
Figure 4. Competence development is inhibited in degS S76D strain.
(A) Single cell analysis of competence: a representative picture demonstrating the difference in fluorescence intensity observed between competent and non-competent cells. (B) Single cell analysis of competence: numerical data. Competence of wild type, degS mutants and Ser/Thr kinase mutants normalised with respect to the wild type cells. The results (with standard deviation bars) are the average of three independent experiments. (C) comK promoter activity in wild type, degS S76A, degS S76D and a strain overexpressing ybdM grown in competence media. Wild type is shown in filled circles, degS S76A in filled triangles, degS S76D in open triangles and the strain overexpressing ybdM in open circles. Culture growth is indicated with the dotted line (broken line indicates the dilution in the new medium). The results (with standard deviation bars) are the average from three technical replicas.
DegS S76D affects complex colony formation and swarming
After confirming the effect of the phosphomimetic mutation S76D on the pool of DegU∼P using the comK promoter, activated by unphosphorylated DegU, we set out to confirm this finding from the opposite angle. We next examined the yvcA promoter, recently shown to be activated by intermediate levels of DegU∼P [12]. We used the same experimental setup with promoter-lacZ fusions introduced ectopically, and the promoter activities were determined in cells grown in LB medium (figure 5). A significant increase in yvcA promoter activity was observed in the degS S76D strain and to a lesser extent in the strain overexpressing ybdM, compared to the wild type and degS S76A strain. A gradual increase in expression from the yvcA promoter in the wild type B. subtilis compared to the degS S76A strain observed in the stationary phase might indicate a gradual increase in the level of DegS serine phosphorylation, which is completely abolished in degS S76A. These results further substantiate that the level of DegU∼P is increased in the degS S76D mutant. The yvcA promoter has been reported to be inhibited at high levels of DegU∼P, indicating that this mutation leads rather to intermediate than excessive amount of DegU∼P. Further substantiating this, protease production, known to be activated by high levels of DegU∼P, was not stimulated by the degS S76D mutant (data not shown).
Figure 5. degS S76D leads to increased yvcA promoter activity.
yvcA promoter activity in wild type, degS S76A, S76D and the strain overexpressing ybdM grown in LB medium. Wild type is shown in filled circles, degS S76A strain in filled triangles, degS S76D in open triangles and the strain overexpressing ybdM in open circles. Culture growth is indicated with the dotted line. The results (with standard deviation bars) are the average from three technical replicas.
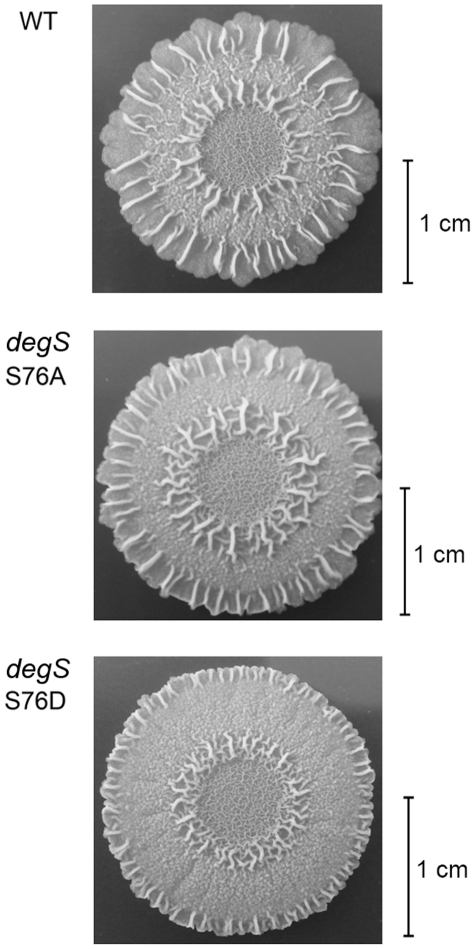
YvcA has been shown to play an important role in complex colony formation [12]. The effects on yvcA promoter activity prompted us to investigate the effects of the degS S76D mutation on this and the other social behavioural traits pellicle formation and swarming that are regulated by low levels of DegU∼P. The laboratory strain B. subtilis 168 does not readily swarm due to defects in surfactin production and a frame shift mutation in swrA [33] and we therefore tested these traits in the undomesticated strain NCIB 3610. No effect was observed on pellicle formation (data not shown). Concerning complex colony formation, all strains exhibited some variability on the MSgg medium. However, there was a subtle difference in colony morphology: the wild type and degS S76A colonies were capable of producing larger and more complex aerial structures than the strain degS S76D (figure 6).
Figure 6. Complex colony formation.
Complex colony formation of the wild type B. subtilis and mutant strains degS S76A and degS S76D grown on MSgg medium. Representative colonies are shown, together with a scale bar.
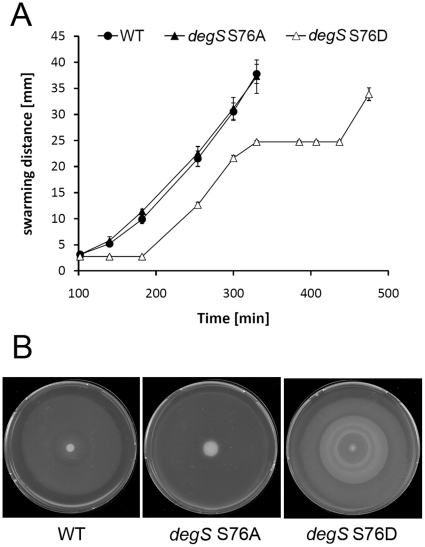
A more apparent phenotype was observed in swarming (figure 7). The wild type and degS S76A swarming pattern and speed were similar, they swarmed in a linear manner until reaching the end of the plate in just over 300 min (figure 7A). The degS S76D strain swarmed in two phases. After an initial lag (compared to the wild type) of about 60 min, it started swarming at the same speed as the wild type until reaching about one half-distance towards the end of the plate. Then it paused for about 120 min, seemingly consolidating. Finally, it continued swarming and went on to reach the end of the plate with about 180 min delay compared to the wild type. Due to this stop-and-go behaviour, the degS S76D final swarm exhibited concentric layers at various stages of consolidation (figure 7B). It is difficult to explain this altered swarming pattern in the strain degS S76D. It has previously been reported that expression of the flagella operon is inhibited at high DegU∼P levels [34], but we observed no difference in flagella amount or organisation in the three strains at the time of degS S76D consolidation (data not shown). Further, this phenotype does not seem to be related to either surfactin production or the swrA mutation since a lab strain cured for these mutations, DS155 [33], exhibited no phenotype on swarming (data not shown). Nevertheless, these data collectively support the idea that degS S76D mutation indeed leads to an increased DegU∼P level. If this mutation can mimic the phosphorylated form of DegS in vivo, it would indicate that the serine phosphorylation event could have the potential to regulate social behaviours regulated at low DegU∼P levels in B. subtilis.
Figure 7. Swarming is affected by degS S76D.
Swarming of the wild type B. subtilis and mutant strains degS S76A and degS S76D, Swarming speed (A) was followed by measuring the radii of the swarming zones on plates at designated time intervals. Wild type is shown in filled circles, degS S76A strain in filled triangles and degS S76D in open triangles. Consolidation of swarms (B) was documented one hour after the swarming reached the end of plates (367 min for wild type, 374 min for degS S76A, and 502 min for degS S76D). One of three independent experiments (all yielding similar results) is shown.
Concluding remarks
Here we describe, to the best of our knowledge, the first example of a bacterial two component sensory kinase that is regulated via serine phosphorylation of its input domain by a Hanks type Ser/Thr kinase. DegS was phosphorylated by Hanks type serine/threonine kinases YbdM and YabT in vitro. YbdM-dependent phosphorylation was specific for serine 76 and it led to increased efficiency of phosphotransfer to DegU. Moreover, ybdM overexpression led to a similar (albeit less pronounced) in vivo effect on some DegU-controlled promoters as the degS S76D phosphomimetic mutation. The degS S76D strain exhibited phenotypes corresponding to elevated levels of DegU∼P in vivo. The use of point-mutations to mimic phosphorylated and non-phosphorylatable proteins is a common tool employed in protein phosphorylation research but it has a potential pitfall. The ensuing phenotypes may be caused by conformational changes caused by the mutations, which are entirely unrelated to phosphorylation. In case of the DegS S76D mutation, the protein behaves in a similar manner as wild type DegS phosphorylated by YbdM. The fact that the mutant protein exhibits a higher DegU kinase activity is expected since it would correspond to 100% phosphorylation of DegS, which is never achieved by incubating DegS with YbdM in vitro. The non-phosphorylatable mutant DegS S76A also exhibits an increased DegU kinase activity in vitro but this does not translate to an increased pool of DegU∼P in vivo. Whether this could reflect a conformational change of the protein leading to increased kinase activity or maybe increased stability in vitro remains elusive. Knocking out the kinase YbdM did not provoke strong phenotypes, possibly due to complementation of its function by remaining Hanks kinases. The overproduction of the regulatory kinase YbdM, which possibly phosphorylates other proteins, could also lead to non-physiologically relevant phenotypes. The fact remains that there is an agreement between the observations with the phosphomimetic DegS S76D and overexpression of YbdM. If DegS S76D indeed can mimick the serine 76-phosphorylated state of DegS, it would suggest that phosphorylation of serine 76 of DegS contributes to an already very complex process of regulating the level of DegU phosphorylation in B. subtilis. The molecular mechanism by which serine phosphorylation activates DegS activity remains elusive. The fact that a phosphomimetic mutant DegS S76D behaved in a similar manner would preclude that phospho-transfer from serine to either DegS histidine or DegU aspartate is involved. It would seem plausible that phosphorylation could induce a conformational change thereby stimulating kinase activity, but due to the lack of any structural data for DegS this is merely a speculation. The residue serine 76 of DegS was found to be phosphorylated in the late exponential growth phase [8], but that study was performed on this single growth condition, and hence the temporal window of DegS serine phosphorylation is not known. Despite YbdM arguably being the most specific of the three, the other two Hanks type kinases present in B. subtilis can possibly contribute to phosphorylating DegS serine 76 in vivo to some extent. These kinases are presently almost completely uncharacterised (except PrkC) and more work will be needed to elucidate specific signals that trigger their expression, activity and substrate specificity. Our results indicate that DegS serine phosphorylation influences DegU phosphorylation in vivo, pointing towards a possible role in regulating phenomena such as motility and complex colony formation.
Materials and Methods
Bacterial strains and growth conditions
E. coli NM522 was used for plasmid propagation in cloning experiments. The chaperone overproducing strain E. coli M15 carrying pREP4-GroESL [35] was used for protein synthesis. B. subtilis strains used in this study are listed in table 1. E. coli and B. subtilis cells were grown at 37°C with shaking in LB medium. In addition, B. subtilis was grown in competence media for transformation experiments as described [31] and MSgg medium for complex colony experiments [36]. For E. coli ampicillin (100 µg/mL), kanamycin (25 µg/mL), tetracycline (8 µg/mL) and for B. subtilis erythromycin (5 µg/mL), neomycin (5 µg/mL) and tetracycline (15 µg/mL) were added as appropriate.
Table 1. List of B. subtilis strains used in this study.
| Strain | Description | Reference |
| 168 | [36] | |
| 168-degS NS | degS K9stop | This work |
| 168-PcomK | amyE::PcomK-lacZ | This work |
| 168-PcomK-gfp | amyE::PcomK-gfp | This work |
| 168-PcomK-IND ybdM | ybdM::pHT315 amyE::PcomK-lacZ | This work |
| 168-PyvcA | amyE::PyvcA-lacZ | This work |
| 168-PyvcA-IND ybdM | ybdM::pHT315 amyE::PyvcA-lacZ | This work |
| 168-degS S76A | degS S76A | This work |
| 168-S76A-PcomK | degS S76A amyE::PcomK-lacZ | This work |
| 168-S76A-PcomK-gfp | degS S76A amyE::PcomK-gfp | This work |
| 168-S76A-PyvcA | degS S76A amyE::PyvcA-lacZ | This work |
| 168-degS S76D | degS S76D | This work |
| 168-S76D-PcomK | degS S76D amyE::PcomK-lacZ | This work |
| 168-S76D-PcomK-gfp | degS S76D amyE::PcomK-gfp | This work |
| 168.S76D-PyvcA | degS S76D amyE::PyvcA-lacZ | This work |
| 168-ΔdegS PcomK-gfp | ΔdegS::pMUTIN2 amyE::PcomK-gfp | This work |
| 168-ΔprkC PcomK-gfp | ΔprkC::pMUTIN2 amyE::PcomK-gfp | This work |
| 168-ΔybdM PcomK-gfp | ΔybdM::pMUTIN2 amyE::PcomK-gfp | This work |
| 168-ΔyabT PcomK-gfp | ΔyabT::pMUTIN2 amyE::PcomK-gfp | This work |
| 3610 | sfp+ swrA+ | Bacillus Genetic Stock Center |
| 3610 degS S76A | sfp+ swrA+ degS S76A | This work |
| 3610 degS S76D | sfp+ swrA+ degS S76D | This work |
| DS155 | PY79 sfp+ swrA+ | [33] |
DNA manipulations and strain construction
B. subtilis genes degS, degU, prkA, prkC (catalytic domain), yabT (catalytic domain) and ybdM were PCR-amplified using genomic DNA from the strain 168 as template [37]. In order to improve solubility of PrkC and YabT only the cytosolic part containing the active site was used. Point mutations degS S76A and S76D were obtained using two partially overlapping mutagenic primers (table 2). The PCR products were inserted in the vector pQE30 (Qiagen) using appropriate restriction sites. For promoter-lacZ fusions the promoter regions were PCR-amplified from genomic DNA and inserted between the EcoRI and BamHI sites of pDG268-neo [38]. For comK promoter-gfp fusion, pDG268neo-PcomK was restricted with BamHI and PciI to remove lacZ, gfp was PCR-amplified using plasmid pFH2191 [39] as template, restricted and ligated with the vector. B. subtilis was transformed with the constructs yielding strains with promoter-lacZ or -gfp fusions inserted in the amyE locus using a one-step transformation method [40]. A nonsense mutation of degS (K9stop) was constructed using two partially overlapping mutagenic primers. The PCR product and pHT315 [41] were restricted with EcoRV and PvuII and the fragments ligated. The resulting vector was devoid of the Gram-positive origin of replication. Upon transformation and integration on the chromosome, the mutation was verified by sequencing. Inactivation of prkA, prkC, ybdM and yabT was done using pMUTIN2 [42]. For IPTG-inducible overexpression, ybdM was inserted in pHT315 and introduced in B. subtilis strains bearing promoter-lacZ fusions. The vector pG+host8 containing a temperature sensitive origin of replication was used to introduce degS S76A and S76D point mutations in situ, replacing the chromosomal version of degS in B. subtilis 168 [43]. BamHI-Cfr9I fragments from pQE30-degS S76A and S76D containing the mutated gene were inserted into pG+host8 between the BamHI and Cfr9I sites. B. subtilis was transformed with the constructs, plated on tetracycline-containing plates and incubated at the non-replicative temperature 37°C, which selects for integration of the vector on the chromosome by single crossing-over. The transformants were further cultured in liquid LB for loss of plasmid by the second crossing-over event and the chromosomal mutation was verified by sequencing. In our hands the second-crossing over happened with a low frequency indicating that B. subtilis, contrary to Lactococcus lactis, was not severely affected by a chromosomal copy of pG+host8 actively replicating. Point mutations in B. subtilis NCIB3610 were introduced by same method except that transformation with pG+Host8 was done by PEG treatment of protoplasts [44].
Table 2. Primers used in this study. Restriction sites are underlined and changed codons are in bold.
| Name | Sequence1 | Description |
| DegU fwd | CGCCGCGGATCCATGACTAAAGTAAACATTGTTATT | BamHI |
| DegU rev | CGCAATGGTACCTTATCTCATTTCTACCCAGCCATTTTT | KpnI |
| DegS fwd | CGCCGCGGATCCATGAATAAAACAAAGATGGATTCC | BamHI |
| DegS rev | CGCAATGGTACCTTAAAGAGATAACGGAACCTTAATCAT | KpnI |
| PrkC fwd | GAAGATCTATGCTAATCGGCAAGCGGATCAGCGGGCG | BglII |
| PrkCtrunc rev | AAAACTGCAGTTACAAAACCCACGGCCACTTTTTTCTTTTTGCCG | PstI, amplification of aa 1–333 |
| YabT fwd | GAAGATCTATGATGAACGACGCTTTGACGAGTTTGGC | BglII |
| YabTtrunc rev | AAAACTGCAGTTAGATAAGCGTTGTTTCAAATAACCCC | PstI, amplification of aa 1–321 |
| YbdM fwd | CGGGATCCATGGCATTAAAACTTCTAAAAAAACTGC | BamHI |
| YbdM rev | AAAACTGCAGTTATGTGACCGATTGAATGGCCCG | PstI |
| YbdM pHT fwd | CGCGGATCCAAAGGAGGAAAACATATGGCATTAAAACTTCTAAAAAAACTGCTATTTGACC | BamHI |
| YbdM pHT rev | AAAACTGCAGTTATGTGACCGATTGAATGGCCCGGTTTAGATCCTCG | PstI |
| PrkA fwd | CGGGATCCATGGATATATTAAAGAAAATTGAAAAGTAC | BamHI |
| PrkA rev | AAAACTGCAGTTATCGGTTCAGCAGGCTGCCG | PstI |
| RBS-gfp fwd | CGGGATCCAAAGGAGGAAAACATATGTCTAAAGGTGAAGAACTG | BamHI |
| gfp rev | CCATACATGTTTATTTATACAGCTCATGCATGC | PciI |
| degS NS1 fwd | CGGATATCATCTCGTGTTCTCCCGCTTC | EcoRV, anneals 263 bp upstream of degS |
| degS NS2 rev | GTCCAGCTGTTCATACTGCTGGCGTGACTGC | PvuII, anneals 123 bp inside degS |
| prkC_ MUT_fwd | CCCAAGCTTAAAGATCCTTTTCATCGCTACG | HindIII |
| prkC_MUT_rev | CGCCCGCGGGGTGACCGTGGCGCCTTCTTTGAC | SacII |
| yabT_MUT_fwd | CCCAAGCTTATGCAATGGAATACATAAAAGGG | HindIII |
| yabT_MUT_rev | CGCCCGCGGTTGAAGCAGCGGGTTTCCTTCG | SacII |
| ybdM_MUTfwd | CCCAAGCTTGAATTCATCATAGACGGACAGG | HindIII |
| ybdM_MUTrev | CGCCCGCGGCAGCAAGAATAACAGCGTTTCTCC | SacII |
| S76A fwd | AAACCGTTTAGCCGAGGTCAGCCGTAATTTTCA | S76A |
| S76A rev | GGCTGACCTCGGCTAAACGGTTTCTCGCATGGC | S76A |
| S76D fwd | AAACCGTTTAGACGAGGTCAGCCGTAATTTTCA | S76D |
| S76D rev | GGCTGACCTCGTCTAAACGGTTTCTCGCATGGC | S76D |
| degS NS1 rev | AATCCAGCACTTAGGAATCCATCTTTGTTTTATTC | K9Stop |
| degS NS2 fwd | GATGGATTCCTAAGTGCTGGATTCTATTTTGATG | K9Stop |
| PcomK fwd | CGGAATTCTAAAGAATCCCCCCAATGCC | EcoRI |
| PcomK rev | CGCGGATCCGTCTGTTTTCTGACTCATAT | BamHI |
| PyvcA fwd | CGGAATTCGAACGCCAAGCGGAAATGCC | EcoRI |
| PyvcA rev | CGCGGATCCCCTGTCAGGGCAAGTAATAAG | BamHI |
Protein synthesis and purification
6xHis-tagged proteins were synthesised in the chaperone-overproducing strain E. coli M15 carrying pREP4-groESL. Cultures were grown shaking at 37°C to OD600 0.5, induced with 1 mM IPTG and grown for an additional 3 hours. Cells were disrupted by sonication and 6xHis-tagged proteins were purified on Ni-NTA columns (Qiagen) according to manufacturer's instruction, desalted with PD-10 columns (GE-Healthcare) and stored in a buffer containing 50 mM Tris-Cl pH 7.5, 100 mM NaCl and 10% glycerol. Protein concentrations were estimated using the Bradford assay (Bio-Rad) with BSA as standard.
In vitro phosphorylation assay
Phosphorylation reactions were performed in a total volume of 30 µl, essentially as described [28], with 180 nM DegS, DegS S76A or DegS S76D and 15 µM DegU. For serine phosphorylation of DegS, reactions contained 10 nM of either PrkA, PrkC (catalytic domain), YabT (catalytic domain) or YbdM (unless otherwise specified in the figure legend). Besides the proteins, the reaction mixture contained 50 µM 32P-γ-ATP (20 µCi/mmol), 42.5 mM Tris-Cl (pH 7.5), 5 mM MgCl2, 85 mM NaCl and 8.5% glycerol. For PrkC, we varied the pH value of the Tris-Cl buffer to the additional pH values of 7.0, and 8.0. To measure the influence of YbdM on DegS phosphotransfer to DegU, DegS was pre-incubated with YbdM for 3 h, in exactly the same conditions as described above, only with non-labelled ATP. Reactions were started by addition of ATP, incubated at 37°C for 60 min (unless otherwise indicated in figure legends) and stopped by addition of SDS-containing loading buffer. All gels shown in the same figure have the same exposure times. For dephosphorylation assays, after the initial phosphorylation reaction described above, the DegS/YbdM and DegS/YabT reaction mixtures were desalted on a PD-10 column (to remove the ATP), lyophilized and resuspended in the identical reaction mixture as before, but without ATP, and incubated 2 hours at 37°C. The proteins were separated by SDS-PAGE (for separation of PrkC and DegS we used a Tris-tricine gel). For detection of phospho-histidine and phospho-aspartate, the gels were rinsed with water and dried directly, whereas for detection of phospho-serine they were additionally treated in boiling 0.5 M HCl for 10 min. Radioactive signals were visualised using STORM phosphoimager and quantified using the ImageQuant software (GE-healthcare).
β-galactosidase assay
150 mL LB was inoculated with overnight culture to OD600 of 0.02 and grown with shaking at 37°C. IPTG at a final concentration of 0.5 mM was added where appropriate. At time points indicated in the figures, 2 mL samples were taken, spun down (10000 g, 2 min) and cell pellets were stored at −20°C. The pellet was resuspended in 2 mL of Z-buffer (60 mM Na2HPO4·7H20, 0.04 mM NaH2PO4·H20, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol, pH 7.0) and OD600 was measured. 1 mL cell suspension was treated with 0.5 mg lysozyme for 5 min at 30°C before adding 8 µL 10% Triton X-100 and incubating for additional 5 min. Reaction was started by addition of 100 µL of 4 mg/mL ONPG and stopped by addition of 1 mL of 0.5 M Na2CO3. Miller units were calculated as described previously [45].
Competence assays
To assess the competence of B. subtilis strains cells were transformed according to the two-step protocol described by Yasbin et al. with the modification that 60 min after dilution in GM2 medium, 0.5 mL culture were transferred to three test tubes containing 1 µg DNA and incubated an additional 90 min before plating. The plasmid pDG268-neo [38] conferring neomycin resistance was used as DNA for competence experiments. Results from three biological replicates are presented as % of competence in wild type and are average and standard deviation of three transformations. For single cell analysis the cultures were incubated for 105 min after dilution in GM2 at which time cultures were concentrated 10 fold and 5 µL were deposited on a polylysine-coated glass slide (Thermo Scientific) and examined using a Xeiss Axioplan microscope equipped with a Kappa ACC 1 condenser, a Zeiss Plan Neofluor 100× objective and a Kappa DX2 HC-FW camera. Images were acquired using Kappa Imagebase Control 2.7.2 software. In the experiments 1600 to 9800 cells per strain were examined. For wild type cells about 5.4% were competent and values are given as % of wild type with standard deviation of two independent experiments.
Complex colony formation
Cells were grown in LB shaking at 37°C to OD600 0.5 at which point 5 µL culture was spotted on a dried MSgg plate (1.5% agar) and incubated for 96 hours at 28°C. Colonies were measured and photographed using a SONY Cyber-shot DSC-T20 camera with close focus enabled. For each sample, a representative image from 20 examined colonies is presented.
Swarming
Cells were grown in LB to an OD600 of 0.5 at which time LB plates (0.7% agar) dried for 20 min in a fume hood (face up) where inoculated with 5 µL culture and dried an additional 10 minutes. Petri dishes were sealed with parafilm to avoid plates drying out and incubated at 37°C. Swarm radii were measured at the times indicated in the figure. Plates were scanned about 1 hour after the swarm reached the edge of the plate using a standard HP office scanner.
Acknowledgments
We would like to thank Dan Kearns for providing the strain DS155.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Danish National Research Council (FNU) 272-07-0129, the Lundbeckfonden R17-A1535 and the Institut National de Recherche Agronomique (INRA) installation package to IM and a PhD stipend from the Technical University of Denmark (DTU) to CJ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 2.Laub MT, Goulian M. Specificity in two-component signal transduction pathways. Annu Rev Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 3.Madec E, Stensballe A, Kjellström S, Cladière L, Obuchowski M, et al. Mass spectrometry and site-directed mutagenesis identify several autophosphorylated residues required for the activity of PrkC, a Ser/Thr kinase from Bacillus subtilis. J Mol Biol. 2003;330:459–472. doi: 10.1016/s0022-2836(03)00579-5. [DOI] [PubMed] [Google Scholar]
- 4.O'Hare HM, Durán R, Cerveñansky C, Bellinzoni M, Wehenkel, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70:1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 5.Phalip V, Li JH, Zhang CC. HstK, a cyanobacterial protein with both a serine/threonine kinase domain and a histidine kinase domain: implication for the mechanism of signal transduction. Biochem J. 2001;360:639–644. doi: 10.1042/0264-6021:3600639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci. Mol Microbiol. 2009;71:382–90. doi: 10.1111/j.1365-2958.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- 7.Lin WJ, Walthers D, Connelly JE, Burnside K, Jewell KA, et al. Threonine phosphorylation prevents promoter DNA binding of the Group B Streptococcus response regulator CovR. Mol Microbiol. 2009;71:1447–1795. doi: 10.1111/j.1365-2958.2009.06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, et al. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol Cell Proteomics. 2007;6:697–707. doi: 10.1074/mcp.M600464-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Msadek T, Kunst F, Henner D, Klier A, Rapoport G, et al. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990;172:824–834. doi: 10.1128/jb.172.2.824-834.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogura M, Yamaguchi H, Yoshida K-I, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic acids res. 2001;29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mäder U, Antelmann H, Buder T, Dahl MK, Hecker M, et al. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol Genet Genomics. 2002;268:455–467. doi: 10.1007/s00438-002-0774-2. [DOI] [PubMed] [Google Scholar]
- 12.Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007;65:554–568. doi: 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007;66:395–409. doi: 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 14.Ogura M, Tsukahara K. Autoregulation of the Bacillus subtilis response regulator gene degU is coupled with the proteolysis of DegU-P by ClpCP. Mol Microbiol. 2010;75:1244–1259. doi: 10.1111/j.1365-2958.2010.07047.x. [DOI] [PubMed] [Google Scholar]
- 15.Veening JW, Igoshin OA, Eijlander RT, Nijland R, Hamoen LW, et al. Transient heterogeneity in extracellular protease production by Bacillus subtilis. Mol Syst Biol. 2008;4:184. doi: 10.1038/msb.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasumura A, Abe S, Tanaka T. Involvement of Nitrogen Regulation in Bacillus subtilis degU Expression. J Bacteriol. 2008;190:5162–5171. doi: 10.1128/JB.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dervyn E, Noirot-Gros M-F, Mervelet P, McGovern S, Ehrlich SD, et al. The bacterial condensin/cohesin-like protein complex acts in DNA repair and regulation of gene expression. Mol Microbiol. 2004;51:1629–1640. doi: 10.1111/j.1365-2958.2003.03951.x. [DOI] [PubMed] [Google Scholar]
- 18.Ogura M, Shimane K, Asai K, Ogasawara N, Tanaka T. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol Microbiol. 2003;49:1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x. [DOI] [PubMed] [Google Scholar]
- 19.Mukai K, Kawata-Mukai M, Tanaka T. Stabilization of Phosphorylated Bacillus subtilis DegU by DegR. J Bacteriol. 1992;174:7954–7962. doi: 10.1128/jb.174.24.7954-7962.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogura M, Tanaka T. Bacillus subtilis ComK negatively regulates degR gene expression. Mol Gen Genet. 1997;254:157–165. doi: 10.1007/s004380050403. [DOI] [PubMed] [Google Scholar]
- 21.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 22.Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- 23.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petranovic D, Grangeasse C, Macek B, Abdillatef M, Gueguen-Chaignon V, et al. Activation of Bacillus subtilis Ugd by the BY-Kinase PtkA Proceeds via Phosphorylation of Its Residue Tyrosine 70. J Mol Microbiol Biotechnol. 2009;17:83–89. doi: 10.1159/000206635. [DOI] [PubMed] [Google Scholar]
- 25.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, et al. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 26.Steil L, Serrano M, Henriques AO, Völker U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology. 2005;151:399–420. doi: 10.1099/mic.0.27493-0. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mijakovic I, Poncet S, Boël G, Mazé A, Gillet S, et al. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 2003;22:4709–4718. doi: 10.1093/emboj/cdg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittekind M, Reizer J, Deutscher J, Saier MH, Klevit RE. Common structural changes accompany the functional inactivation of HPr by seryl phosphorylation or by serine to aspartate substitution. Biochemistry. 1989;28:9908–9912. doi: 10.1021/bi00452a005. [DOI] [PubMed] [Google Scholar]
- 30.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 31.Yasbin RE, Wilson GA, Young FE. Transformation and transfection in lysogenic strains of Bacillus subtilis: evidence for selective induction of prophage in competent cells. J Bacteriol. 1975;121:296–304. doi: 10.1128/jb.121.1.296-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearns DB, Chu F, Rudner R, Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol. 2004;52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 34.Amati G, Bisicchia P, Galizzi A. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J Bacteriol. 2004;186:6003–6014. doi: 10.1128/JB.186.18.6003-6014.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amrein KE, Takcs B, Stieger M, Molnos J, Flint NA, et al. Purification and characterization of recombinant human p50csk protein-tyrosine kinase from an Escherichia coli expression system overproducing the bacterial chaperones GroES and GroEL. Proc Natl Acad Sci U S A. 1995;92:1048–1052. doi: 10.1073/pnas.92.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 38.Christiansen LC, Schou S, Nygaard P, Saxild HH. Xanthine Metabolism in Bacillus subtilis: Characterization of the xpt-pbuX Operon and Evidence for Purine- and Nitrogen-Controlled Expression of Genes Involved in Xanthine Salvage and Catabolism. J Bacteriol. 1997;179:2540–2550. doi: 10.1128/jb.179.8.2540-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Østergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jarmer H, Berka R, Knudsen S, Saxild HH. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol Lett. 2002;206:197–200. doi: 10.1111/j.1574-6968.2002.tb11009.x. [DOI] [PubMed] [Google Scholar]
- 41.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 42.Vagner V, Dervyn E, Ehrlich SD. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 43.Maguin E, Prévost H, Ehrlich SD, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang S, Cohen SN. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979;168:111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- 45.Miller J. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]