Abstract
Proteasome-mediated protein degradation has been implicated in playing a role in nuclear receptor-mediated gene expression; inhibition of the proteasome impairs the transcriptional activity of estrogen receptor α (ERα) and most other nuclear receptors. This coincides with blockage of agonist-dependent degradation of the receptor and elevation of the steady-state levels of SRC family coactivators and CBP. Here, we examined the effects that different ERα ligands have on coactivator protein steady-state levels and demonstrate that the selective ER modulators (SERMs) 4-hydroxytamoxifen (4HT) and raloxifene are able to elevate SRC-1 and SRC-3 protein levels. Using the HeLa cell line, we show that this effect is ERα dependent. Consistent with the observed increase in coactivator protein levels, we were also able to observe an increase in the transcriptional activity of other nuclear receptors in SERM-treated cells. Information presented here demonstrates an unexpected consequence of SERM treatment, which could help further define the complex tissue responses to 4HT and raloxifene, and suggests that these ligands can have a broad biological action, stimulating the transcriptional activity of other nuclear receptors.
The estrogen receptor α (ERα), like other members of the nuclear receptor superfamily, is a ligand-activated transcription factor (24). Upon binding of the agonist ligand, 17β-estradiol (E2), ERα undergoes a conformational change that initiates a series of events that leads to the transcription of E2-regulated target genes. Transcriptional activation depends upon the recruitment of coactivators to the agonist-bound receptor's ligand-binding domain, where they serve to enhance ERα-mediated transcription through a number of mechanisms. For instance, many coactivators possess intrinsic histone acetyltransferase activity that alters the adjacent chromatin surrounding ERα-responsive genes, allowing for increased transcription. Another class of coactivators, which includes TRIP1/Sug1 (19, 35), E6-AP (28), RPF-1 (11), UBC9 (8), and Tat-binding protein (12), comprises proteins which have been identified as components of the ubiquitin-proteasome protein degradation pathway, suggesting that ubiquitin-proteasome-mediated protein degradation also plays an important role in nuclear receptor-driven gene transcription. Ubiquitin-proteasome-mediated protein degradation is a bipartate process: the first part involves the covalent attachment of ubiquitin to the protein being targeted for degradation by ubiquitin conjugases and ligases (6); in the second part the 26S proteasome, a large multisubunit protease, recognizes and degrades these ubiquitinated proteins.
Proteasome-mediated protein degradation could serve as a mechanism to allow for the temporally dynamic exchange of cofactors that would be required for efficient ERα-mediated transcription to ensue (31, 33). Consistent with this, it has been observed that inhibition of the proteasome is known to reduce the mobility of ERα, SRC-1, and glucocorticoid receptor (GR)-green fluorescent protein fusion receptors within the nucleus (5, 34). Importantly, inhibition of the proteasome also impairs ERα transcriptional activity (21), despite the fact that receptor and coactivator levels are elevated, indicating that proteasome-mediated degradation plays an obligatory role in efficient ERα-mediated gene transcription.
A number of nuclear receptors, including ERα, retinoic acid receptor-α, progesterone receptor (PR), thyroid hormone receptor, GR, and retinoic acid receptor, are degraded in a proteasome-dependent manner upon addition of their cognate ligand, concomitant with transcriptional activation (4, 17, 29, 36, 44). Our laboratory has previously shown that ligand-mediated degradation of ERα is dependent upon coactivator-binding residues located in the activation function-2 (AF-2) of the receptor, indicating that ERα degradation is integrally connected to the receptor activation process (22). Selective estrogen receptor modulators (SERMs), such as tamoxifen and raloxifene, which fail to activate ERα in most cell contexts, on the other hand are known to stabilize ERα (14, 38), consistent with their ability to block coactivator interaction with the AF-2 of the receptor.
The three members of the SRC coactivator family, SRC-1, SRC-2 (TIF-2), and SRC-3 (AIB1/RAC3/ACTR/pCIP), and CBP (24) are themselves targets of ubiquitin-proteasome-mediated protein degradation (22), but the impact that ligands have on coactivator stability has not been examined.
The spatial complexity of coactivator expression contributes to distinct biological responses in different tissues. For instance, elevated expression of SRC-1 in a uterine-derived cell line (Ishikawa) and lower expression in a breast-derived cell line (MCF-7) contribute to the SERM 4-hydroxytamoxifen (4HT) agonist and antagonist behavior, respectively, in each cell line (32). Coactivator overexpression is also likely to contribute to carcinogenesis. SRC-3/AIB1 has been identified as a protein that is overexpressed in a significant number of breast cancers (1). Other nuclear receptor coactivators such as AIB3/ASC-2 (9, 20) and TRAP220/PBP (42, 45) have also been identified which are amplified in breast malignancies (46), pointing to a role for coactivator expression in defining the qualitative and quantitative responses to steroid, thyroid, and retinoid hormones in different tissues and a role for their overexpression in malignancy. Factors which influence the protein levels of any of these coactivators could influence the tissue's potential to become carcinogenic.
Here we have examined the factors which affect the steady-state levels of SRC-1 and SRC-3 in more detail at the protein and mRNA levels. Despite their relatively constant expression level in tissues, the turnover rate of SRC-1A and SRC-3 indicate that both coactivators are unstable proteins that are continuously synthesized and degraded. Treatment with 4HT or raloxifene led to an increase in the steady-state levels of SRC-1A and SRC-3 coactivator fusion proteins and endogenously expressed coactivators in HeLa cells and an MCF-7 breast cancer-derived cell line, revealing that SERM biological action can be impacted through its influence on coactivator steady-state levels.
MATERIALS AND METHODS
Plasmids.
The expression vectors for human ERα (pCR3.1 hERα), PR (pCR3.1 hPR-B) and pGRE-E1b-LUC, have already been described (29). FLAG hERα was constructed by replacing the NheI-XmaI fragment of pCR3.1 hERα with a PCR-generated fragment containing sequence for the FLAG epitope fused in frame and upstream of the ERα cDNA. The SRC-1A luciferase fusion protein pCR3.1 SRC-1A-LUC was constructed by replacing the BsmI-XbaI fragment of pCR3.1 SRC-1A (21) with a PCR-generated fragment which removes the stop codon and adds a SalI restriction site. The luciferase cDNA from pGL3-Basic (Promega) was amplified by PCR using primers which contain SalI and XbaI restriction sites and cloned in frame into the respective sites in the modified SRC-1A expression vector. The pCR3.1 SRC-3-LUC luciferase fusion proteins were constructed by replacing the SanDI-XbaI fragment of pCR3.1 RAC3 (21) with a PCR-generated fragment which removes the stop codon and adds a SalI restriction site. The luciferase cDNA fragment described above was then inserted in frame into the modified SRC-3 vector. pSG5-KM3F2-hSRC-1 was constructed by inserting a PCR-generated full-length hSRC-1A fragment into the pSG5 vector with a modified polylinker via its NotI/XhoI restriction sites. Its 2× FLAG tag was incorporated into the carboxyl terminal end. The integrity of all vectors was confirmed by sequencing.
Cell lines and transfections.
HeLa, ts85, ZR-75-1, T47-D, and MCF-7 cells (from Richard Santen, University of Virginia, or American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum. The ubiquitin-activating enzyme temperature-sensitive cell lines were maintained at a permissive (30°C) temperature. Twenty-four hours before transfection, HeLa, ts85, and MCF-7 cells were plated at a density of 2 × 105, 9 × 105, and 6 × 105 cells per well, respectively, in six-well dishes in phenol red-free DMEM containing 5% dextran-coated charcoal-stripped serum. T47-D cells were maintained similarly, except that RPMI medium was used instead, and plated at a density of 6 × 105 cells per well. Cells were transfected with the indicated expression vector plasmids using Lipofectamine (Invitrogen) according to the manufacturer's protocol. Where indicated, cells were treated with 10−9 M E2 (Sigma), 10−7 M 4HT (Sigma), or a 10−7 M concentration of ICI 182,780 or raloxifene (Sigma) dissolved in ethanol; the ethanol vehicle alone served as a control. MG132 (Sigma) was administered to cells at a concentration of either 1 or 10 μM as indicated, dissolved in dimethyl sulfoxide (DMSO). Control cells were treated with DMSO vehicle alone. For protein turnover determinations, cells were treated with 200 μg of cycloheximide (Sigma)/ml dissolved in ethanol. For coactivator mRNA stability experiments, cells were treated with 1 μg of actinomycin D (Sigma)/ml dissolved in DMSO.
Cell extraction and assays.
At the indicated times after hormone and drug treatment, cells were harvested in TEN buffer (10 mM Tris [pH 8.0], 150 mM NaCl, 10 mM EDTA). Cell pellets were lysed in luciferase assay buffer (25 mM Tris [pH 8.0], 150 mM NaCl, 10 mM EDTA, 1 mM dithiothreitol, 5% glycerol, 0.5% Triton X-100) and passed through a 26-gauge needle 20 times. The lysate was then spun for 20 s at 21,000 × g, and the supernatant was assayed for luciferase activity. For chloramphenicol acetyltransferase (CAT) assays, cell extracts were assayed for CAT protein levels using a CAT enzyme-linked immunosorbent assay kit (Boehringer Mannheim). Luciferase and CAT activities were normalized against total cellular protein by Bradford analysis (Bio-Rad). For Western analysis, cells were extracted with luciferase assay buffer and sonicated at 4°C and then centrifuged for 5 min at 21,000 × g. Forty micrograms of total protein was resolved by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Nitrocellulose membranes were incubated in a blocking buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.5% Tween 20, 1% dried nonfat milk). The membrane was then incubated with an anti-luciferase antibody (Promega), anti-FLAG M2 antibody (Sigma), or a rabbit polyclonal anti-FLAG (Affinity Bioreagents), anti-SRC-1A (Upstate Biotechnologies), or anti-SRC-3 (BD Pharmingen) antibody, followed by the appropriate anti-goat, anti-mouse, or anti-rabbit secondary horseradish peroxidase-conjugated antibody and visualized by chemiluminescence (ECL Plus; Amersham). All experiments were repeated at least two times, and error bars represent the standard errors of the means of triplicate data points (except for the Western analysis densitometric quantitation, which is described below in the legend for Fig. 1).
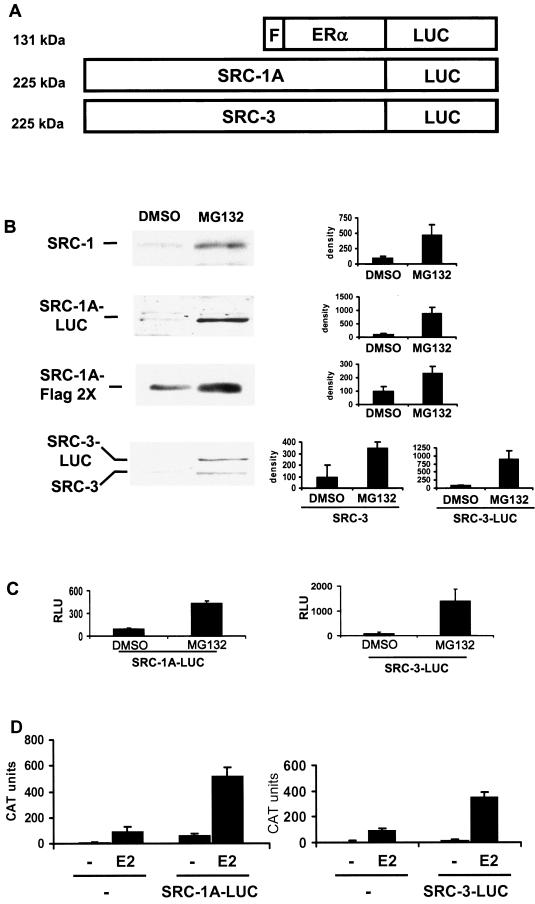
FIG.1.
SRC-1A-LUC and SRC-3-LUC fusion proteins behave as their unmodified counterparts do. (A) Schematic of luciferase fusion constructs. The luciferase (LUC) cDNA was fused to the C terminus of FLAG (F)-ERα, SRC-1A, or SRC-3. The predicted molecular masses are listed on the left. (B) Western analysis of endogenous (SRC-1) and transfected (1,000 ng of SRC-1A FLAG 2× or 1,000 ng of SRC-1A-LUC) SRC-1 in HeLa cells in the absence (DMSO) or presence of 10 μM MG132 for 24 h was performed using appropriate antibodies (see Materials and Methods). The histograms to the right represent densitometric quantitation of Western blots from two or three separate experiments. (C) The luciferase activity of 100 ng of SRC-1A-LUC or 250 ng of SRC-3-LUC transfected into HeLa cells and treated with MG132 or DMSO vehicle for 24 h. (D) SRC-1A-LUC and SRC-3-LUC can coactivate ERα. HeLa cells were transfected with 500 ng of pERE-E1b-CAT, an expression vector for wild-type ERα (10 ng of pCR3.1 hERα) along with 250 ng of the pCR3.1 empty vector (−), pCR3.1 SRC-1A-LUC, or pCR3.1 SRC-3-LUC and treated with either ethanol vehicle (−) or E2 for 24 h and assayed for CAT protein levels.
Quantitative PCR analysis.
HeLa cell total RNA was isolated from six-well culture dishes using TRIzol reagent (Invitrogen). The mRNA for SRC-1 and SRC-3 in HeLa cells was quantitated by Taqman-based reverse transcriptase PCR (RT-PCR) using the ABI Prism 7700 sequence detection system (Applied Biosystems). For SRC-1 the primer pair 5′-TGAAAGTGGAAAAGAAAGAACAGATG-3′ and 5′-GTCAAGTCAGCTGTAAACTGGC-3′ was used with a 5′-6FAM-CAAACCCACTCCTGAGGAAATAAAACTGGAGG-TAMRA-3′ probe (6FAM is 6-carboxyfluorescein, and TAMRA is 6-carboxytetramethylrhodamine). For SRC-3, the primer pair 5′-CAGCCCCAGCAGGGTTT-3′ and 5′-ATAGCCACCCTCTGTTGTCGG-3′ was used along with a 5′-6FAM-CAAAATGGTCGCCCAACGCAGC-TAMRA-3′ probe. For SGK1, the primer pair 5′-AAGCTGCCGAGGCTTTCC-3′ and 5′-GCCCTAACAGGGTTCAGAGGA-3′ and a 5′-6FAM-TTTCCTATGCGCCTCCCACGGA-TAMRA-3′ probe were used. RT-PCRs were performed using One-step RT-PCR Universal Master Mix reagents according to the manufacturer's recommendations. All mRNA quantities were normalized against 18S RNA using Taqman rRNA control reagents.
SRC-3-LUC coimmunoprecipitation.
To examine the interaction of SRC-3-LUC with PR, HeLa cells were transfected with 100 ng of pGRE-E1b-CAT, 100 ng of pCR3.1 hPR-B, 100 ng of pCR3.1hERα, or 1,000 ng of pCR3.1-SRC-3-LUC. Twenty-four hours after transfection, the cells were treated with E2 or 4HT. Twenty-four hours thereafter, the cells were treated with 10−7 M progesterone for 1 h and then harvested in the luciferase assay buffer described above and maintained at 4°C. The lysates were centrifuged 5 min, and the supernatant was transferred to a new tube and incubated with an anti-PR antibody (Santa Cruz) for 2 h. A 20-μl aliquot of 50% (wt/vol) protein A+G-Sepharose beads was then added, incubated an additional hour, and then washed three times with ice-cold luciferase assay buffer. After washing, the beads were resuspended in 100 μl of luciferase assay buffer, and a 20-μl aliquot was assayed for luciferase activity.
RESULTS
Coactivator and receptor luciferase fusion proteins behave like their wild-type equivalents. In Arabidopsis thaliana, it has been demonstrated that a fusion of the firefly luciferase protein to the AuxAII transcription factor allows for the highly sensitive and quantitative evaluation of the transcription factor's steady-state levels and stability (30). We generated similar chimeric proteins by fusing luciferase (lacking the C-terminal peroxisomal targeting sequence) to the C terminus of SRC-1A or SRC-3 (Fig. 1A). Our laboratory previously showed that luciferase is not a target of the proteasome (21) and therefore should not interfere with proteasome-mediated turnover of our coactivator fusion proteins. We also generated an ERα-luciferase fusion protein vector as an additional control, which was preferentially degraded in the presence of E2 or ICI 182,780, like the wild-type ERα (data not shown). To test these coactivator fusion proteins, their expression vectors were transfected into HeLa cells, treated with either MG132 or ligands 24 h later, and then harvested 24 h thereafter. Western analysis of the two fusion proteins indicated that both are expressed at their appropriate molecular weight of 225 (Fig. 1B) without the detection of any lower-molecular-weight fragmented proteins (data not shown). Treatment with the proteasome inhibitor MG132 resulted in an increase in the steady-state level of SRC-1A-LUC and SRC-3-LUC, indicating that these fusion proteins are targets of the proteasome like their wild-type counterparts. Luciferase assays for SRC-1A-LUC and SRC-3-LUC also indicated that they behave like their wild-type counterparts and that their observed levels by Western analysis are consistent with those seen in luciferase assays. Consequently, this method allowed accurate quantitation of cellular proteins present in low concentrations. MG132 treatment resulted in an increase in luciferase activity in cells transfected with SRC-1A-LUC or SRC-3-LUC proteins, indicating an increase in the steady-state levels of these two coactivators (Fig. 1B and C). Also, the endogenous mRNAs for SRC-1 and SRC-3 were unaffected by treatment with MG132, indicating that MG132 is not leading to an increase in coactivator levels due to alterations of the levels of their transcripts (data not shown).
Additionally, SRC-1A-LUC and SRC-3-LUC are able to perform their roles as coactivators. When transiently transfected along with a CAT-based estrogen-responsive reporter (pERE-E1b-CAT) and an expression vector for the wild-type ERα, expression of SRC-1A-LUC or SRC-3-LUC was able to coactivate ERα-mediated transcription (Fig. 1D).
Turnover rate of SRC-1A and SRC-3 coactivator proteins.
The three SRC family coactivators are targets of the proteasome, indicated by the fact that in transiently transfected HeLa cells the steady-state levels of all three coactivators increase in the presence of MG132 (21). Inhibition of the proteasome also impairs the transcriptional activity of most nuclear receptors, suggesting that the turnover of either the receptor and/or coactivators is necessary for efficient receptor-mediated transcription to ensue.
To gain further insight into the possible connection between coactivator turnover and transcription, we measured the rate of decay of SRC-1A-LUC and SRC-3-LUC in the presence of the protein synthesis inhibitor cycloheximide. Expression vectors for either coactivator fusion protein or the luciferase protein as a control were transiently transfected into HeLa cells along with an expression vector for the wild-type ERα and an estrogen-responsive CAT reporter (Fig. 2A). Twenty-four hours after transfection, cells were treated with cycloheximide (200 μg/ml) and harvested at the indicated time points following cycloheximide treatment. The decay of the coactivator fusion proteins followed apparent first-order kinetics for the first 4 h of cycloheximide treatment but not beyond this time (data not shown). MG132 was able to essentially block the decay of SRC-1A-LUC and reduced the decay rate for SRC-3-LUC, indicating that the observed decay was proteasome dependent. In contrast to that seen for the coactivator fusion proteins, luciferase degraded only minimally throughout the 4-h time course. The decay of the endogenous SRC-1 and SRC-3 proteins was also examined in the HeLa cell line (Fig. 2B). It can be seen in the figure that both coactivators displayed similar kinetics as the transfected coactivator-luciferase fusion proteins.
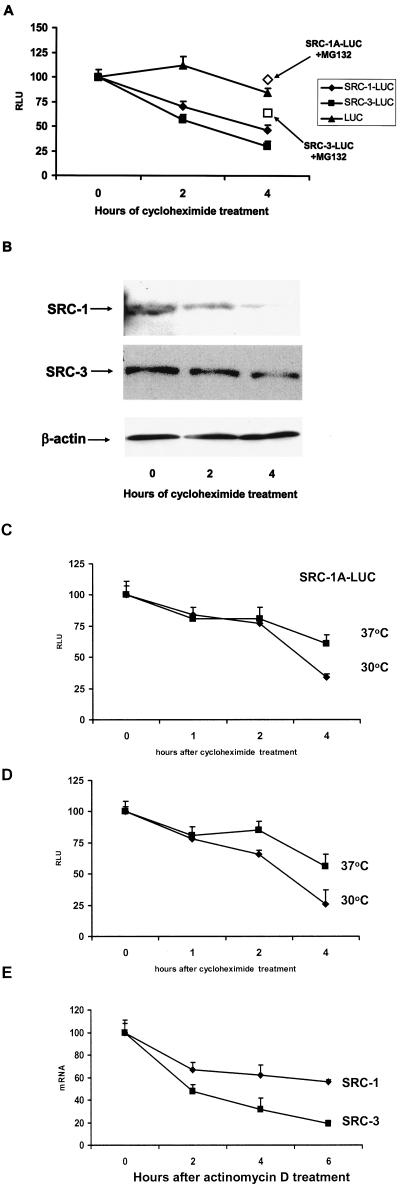
FIG.2.
Decay rate of SRC-1 and SRC-3 proteins. (A) HeLa cells were transiently transfected with expression vectors for SRC-1A-LUC, SRC-3-LUC, or luciferase along with pERE-E1b-CAT and pCR3.1 hERα. Twenty-four hours thereafter, cells were harvested just prior to treatment (0) or treated with cycloheximide (and MG132 [open boxes]; 4-h treatment time) at the zero hour time point and harvested 2 or 4 h thereafter for luciferase assays. (B) Decay of endogenous SRC-1A and SRC-3 in HeLa cells. HeLa cells were treated with cycloheximide, harvested at 0, 2, or 4 h thereafter, and visualized by Western analysis. As a loading control for the SRC-3 Western analysis, the blot was reprobed with β-actin (equal loading of SRC-1 was confirmed by the observation of nonspecific bands detected by the SRC-1 antibody [data not shown]). (C and D) SRC-1A-LUC and SRC-3-LUC are degraded in a proteasome-dependent manner in the temperature-sensitive UBA-defective ts85 cell line. Expression vectors for SRC-1A-LUC (C) and SRC-3-LUC (D) were transfected into ts85 cells. After 24 h of incubation at either the permissive (30°C) or restrictive (37°C) temperature, cells were then harvested at the time of cycloheximide treatment 1, 2, and 4 h thereafter and assayed for luciferase activity. (E) Decay rates for SRC-1 and SRC-3 in HeLa cells. HeLa cells were treated with the transcription inhibitor actinomycin D and harvested 0, 2, 4, and 6 h thereafter. SRC-1 and SRC-3 mRNA was quantitated by real-time PCR and normalized against 18S RNA.
To further explore the dependence of coactivator turnover on the ubiquitin-proteasome protein degradation pathway through another approach which does not involve proteasome inhibitor compounds, we examined the rate of decay of SRC-1A-LUC and SRC-3-LUC in the ts85 cell line, which harbors a thermo-labile ubiquitin-activating enzyme that abolishes the transfer of ubiquitin to target proteins at nonpermissive temperatures, disabling the ubiquitin-proteasome protein degradation pathway. A variety of coactivators, including SRC-1 and SRC-3, have been shown to be targets of the proteasome in this cell line (41). ts85 cells maintained at 30°C were transfected with expression vectors for SRC-1A-LUC or SRC-3-LUC. Twenty-four hours thereafter, cells were either transferred to 37°C (nonpermissive temperature) or maintained at 30°C. Twenty-four hours later, cells in either temperature regimen were treated with cycloheximide and harvested at the indicated times thereafter (Fig. 2C and D). It can be seen in the figure that the steady-state levels for both SRC-1A-LUC and SRC-3-LUC decay at a slower rate at the restrictive temperature. Both coactivators also showed appreciable decay at the nonpermissive temperature as well, suggesting that a nonproteasomal mechanism also exists to promote coactivator turnover in this cell line. These results demonstrate that the decay of SRC-1A-LUC and SRC-3-LUC is mediated primarily through the ubiquitin-proteasome protein degradation system.
SRC-1 and SRC-3 mRNAs are unstable.
Regulation of SRC-1 and SRC-3 steady-state levels could also be accomplished through regulation of their mRNA transcripts. To examine this, we assessed the stability of the mRNA for SRC-1 and SRC-3 by quantitative PCR. HeLa cells were transiently transfected with an expression vector for ERα and an ER-responsive CAT reporter and treated with the inhibitor of transcription actinomycin D (1 μg/ml). The SRC-3 transcript, like the SRC-3 protein, was less stable than that for SRC-1, having a half-life of approximately 3 h compared to approximately 5 h for SRC-1 (Fig. 2E). Ligand treatment had no effect on the decay rate for the mRNA for either coactivator (data not shown). Taken together, these results indicate that production of the message and the protein for SRC-1 and SRC-3 are being continuously synthesized and turned over.
4HT enhances protein levels of SRC-1A-LUC and SRC-3-LUC coactivator fusion proteins.
Differences in the biological activities of partial agonist-antagonists such as 4HT have been attributed to variations in the expression of coactivators, such as SRC-1. E2, thyroid hormone, and dexamethasone, which have been reported to alter coactivator mRNA expression in different cell types (10, 15, 25). Here, we have examined the SRC-1 and SRC-3 proteins in addition to their transcripts to determine if alterations in protein stability can be seen in response to different ERα ligands. HeLa cells were transfected with an ER-responsive CAT reporter and expression vectors for ERα and either SRC-1A-LUC or SRC-3-LUC, and then the cells were treated with E2, 4HT, or raloxifene as indicated. Twenty-four hours after hormone treatment, cells were harvested and assayed for luciferase activity corresponding to either coactivator (Fig. 3A). E2 treatment resulted in a moderate (∼1-fold) increase in SRC-1A-LUC or SRC-3-LUC protein, while ICI 182,780 had no significant effect (data not shown). 4HT treatment led to a much more dramatic increase in the level of SRC-1A-LUC or SRC-3-LUC protein levels. Interestingly, 4HT and raloxifene have been reported to stabilize ERα itself (14, 38), although the connection between the observed increase in SRC-1A-LUC or SRC-3-LUC and ERα is unclear. The increase in coactivator was receptor dependent and was also specific for SRC-1A-LUC and SRC-3-LUC, as the luciferase protein itself was not affected by different hormone treatments (Fig. 3A).
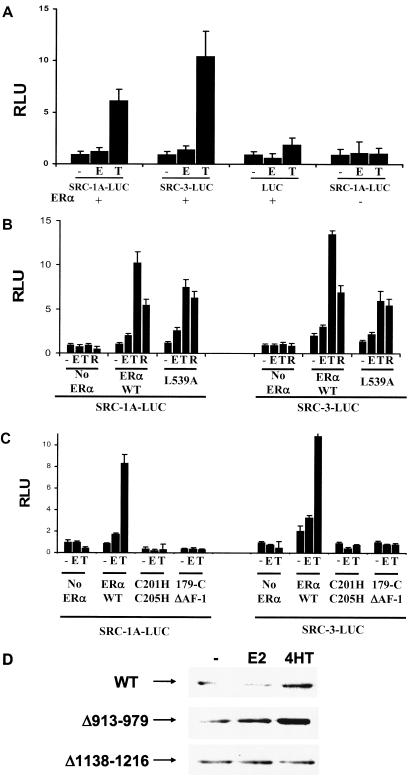
FIG. 3.
(A) 4HT can elevate the steady-state level of SRC-1A-LUC and SRC-3-LUC in HeLa cells. HeLa cells were transfected with pERE-E1b-CAT and expression vectors for the indicated coactivators along with either pCR3.1 hERα (+) or its empty vector, pCR3.1 (−). Twenty-four hours thereafter, cells were treated with ethanol vehicle (−), E2 (E), or 4HT (T) for 24 h and harvested for luciferase activity. (B) The AF-2 of ERα is dispensable for 4HT-induced elevation of SRC-1A-LUC and SRC-3-LUC protein levels. HeLa cells were transfected in a similar manner as described above, with the addition of an AF-2-defective mutant for ERα (L539A). Cells were treated with hormone as described above with the addition of raloxifene (R). (C) The AF-1 and DBD of ERα are required for 4HT-induced elevation in coactivator expression. HeLa cells were transfected and treated as described above with the cotransfection of expression vectors for the DBD mutant (C201H/C205H) or ERα with AF-1 deleted (179C ΔAF-1) instead of the wild-type receptor. (D) Deletion of amino acid residues 1138 to 1216 of SRC-1 blocks 4HT-induced elevation of the coactivator. Expression vectors for ERα and the wild-type FLAG-tagged SRC-1 or SRC-1 deletion mutants Δ913-979 and Δ1138-1216 were transfected into HeLa cells. Twenty-four hours later, cells were treated with E2, 4HT, or their ethanol vehicle for an additional 24 h and harvested for Western analysis.
Because this effect on coactivator steady-state levels was ERα dependent, we tested a number of ERα mutants to explore which part of the receptor was important for this effect. Mutation of the mouse ERα residue corresponding to the human leucine 539 disrupts coactivator binding to the ligand-binding domain of the receptor (23), leading us to test a human leucine 539-to-alanine (L539A) mutant. However, coactivator steady-state levels were still elevated in the presence of the L539A human ERα AF-2 mutant, suggesting that other parts of the receptor play the most significant role in this phenomenon (Fig. 3B). Interestingly, another partial agonist-antagonist, raloxifene, was also able to enhance the steady-state levels of SRC-1A-LUC and SRC-3-LUC. Mutation of the DNA-binding domain (DBD) (C201H/C205H) or deletion of the N-terminal AF-1 abolished the ability of 4HT to elevate the steady-state level of either coactivator-luciferase fusion protein (Fig. 3C).
We examined which portion of SRC-1 is required for the effect that 4HT has on coactivator protein levels in HeLa cells. Expression vectors for two deletions of SRC-1 were transfected along with ERα and analyzed by Western blotting to determine if deletion of the p300 interaction region (amino acids 913 to 979) of SRC-1 or another region (1138 to 1216) which encompasses two phosphorylation sites in SRC-1 and is part of the coactivator's P/CAF interaction surface would abolish 4HT-mediated elevation of the SRC-1 protein (Fig. 3D). It can be seen that the Δ913-979 SRC-1 is still affected by SRC-1, while Δ1138-1216 is now unaffected by 4HT treatment. These results point to the possibility that SRC-1 phosphorylation status plays a role in the effect that 4HT has on its stability.
To independently confirm that coactivator protein levels were elevated in 4HT-treated HeLa cells transiently transfected with ERα, the endogenous SRC-1 and SRC-3 protein levels were examined by Western analysis (Fig. 4A). These Western blots also reveal that 4HT is able to increase the steady-state levels of either coactivator, consistent with that seen for the SRC-1 and SRC-3 luciferase fusion proteins.
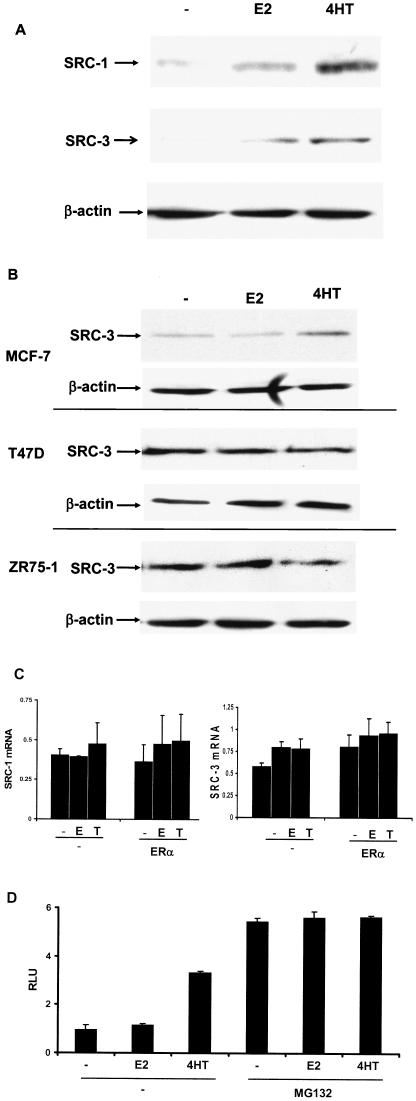
FIG.4.
4HT can elevate endogenous SRC-1 and SRC-3 in HeLa cells transiently transfected with ERα and in MCF-7 cells. (A) HeLa cells were transfected with pERE-E1b-CAT and pCR3.1 hERα, incubated 24 h, and then treated with ethanol vehicle (−), E2, or 4HT for 24 h and harvested for Western analysis. (B) 4HT is able to promote elevation of endogenous SRC-3 in MCF-7 but not T-47D or ZR-75-1 ERα-positive breast cancer cell lines. Untransfected MCF-7, T-47D, and ZR-75-1 cell lines were treated with ligands and harvested for Western analysis as described for panel A. (C) 4HT treatment has no effect on SRC-1 or SRC-3 mRNA levels in HeLa cells transfected with ERα. HeLa cells were transfected with pERE-E1b-CAT and pCR3.1 hERα and then treated for 24 h with ligands as described above. Cells were harvested for total RNA, and mRNA for SRC-1 and SRC-3 was quantitated by real-time PCR. (D) In MG132-treated HeLa cells, SRC-1A-LUC protein is not further elevated when cotreated with 4HT. HeLa cells were transfected with pERE-E1b-CAT, pCR3.1hERα, or pCR3.1 SRC-1A-LUC and treated with ethanol vehicle (−), E2, or 4HT with or without MG132 for 8 h and harvested for luciferase activity.
Next, we wanted to determine if 4HT could affect the steady-state levels of SRC-3 in cell lines where ERα and SRC-3 are endogenously expressed (Fig. 4B). In two breast cancer-derived cell lines, T47-D and ZR-75-1, 4HT was unable to elevate the steady-state level of SRC-3. However, in the MCF-7 cell line, we were able to observe an increase in SRC-3 expression in the presence of E2 or 4HT. These results indicate that the ability of 4HT to influence the steady-state level of SRC-3 is cell type specific. We also examined a variety of other ERα-positive cell lines for the ability of 4HT to influence SRC-3 protein levels. Twenty-four hours of 4HT treatment was unable to elevate SRC-3 protein in PC-3, Du145, MC3T3, or Ishikawa cells (data not shown). Further examination of SRC-3 protein levels in MCF-7 cells revealed that the greatest increase in SRC-3 protein levels was evident after 2 h of 4HT treatment. Because of this, it is still possible that a more detailed examination of other ERα-positive cell lines may reveal a modulatory effect of 4HT on coactivator protein levels.
To assess whether the elevation of SRC-1 or SRC-3 protein levels by 4HT in HeLa cells was related to up-regulation of SRC-1 or SRC-3 mRNA, either transcript was assessed by quantitative real-time PCR (Fig. 4C). HeLa cells were transfected with an estrogen-responsive reporter in the absence or presence of an expression vector for ERα and treated with ethanol vehicle, E2, or 4HT for 24 h. Cells were then harvested, and the mRNA for SRC-1 and SRC-3 was assessed by quantitative PCR. No increase in either SRC-1 or SRC-3 mRNA was observed in the presence of 4HT, indicating that the increases in SRC-1 and SRC-3 proteins were effected at the protein level.
To directly examine if the SRC-1A protein becomes more stable in the presence of 4HT, we transiently transfected HeLa cells with an estrogen-responsive reporter vector, ERα, and SRC-1A-LUC. We then treated the cells with either ethanol vehicle, E2, or 4HT, with or without MG132, and assessed whether 4HT could elevate SRC-1A-LUC levels in the presence of MG132 (Fig. 4D). Consistent with the possibility that 4HT is able to protect SRC-1A-LUC from proteasome-mediated degradation, 4HT treatment did not elevate SRC-1A-LUC levels over those for ethanol- or E2-treated cells in the presence of MG132.
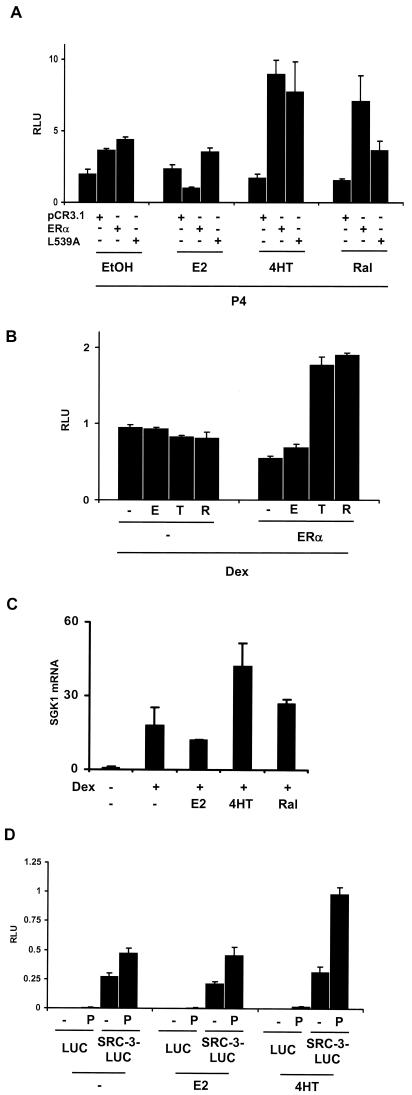
Because of the observed increase in SRC-1A-LUC and SRC-3-LUC in the presence of 4HT or raloxifene, we predicted that treatment with either SERM would also be able to enhance the transcriptional activity of other nuclear receptors in HeLa cells which have been transfected with ERα and treated with 4HT or raloxifene. This could account in part for the prior observation that tamoxifen and raloxifene are able to enhance PR-mediated transcription in the endometrial Ishikawa cell line (2). To test this, HeLa cells were transfected with a progesterone-responsive luciferase reporter (pGRE-E1b-LUC) and expression vectors for ERα or an ERα L539A mutant which is transcriptionally inactive on an ERE-containing reporter that is still sufficient to allow for SERM-mediated coactivator accumulation (Fig. 3B), along with PR-B. SERM treatment in the presence of either the wild-type ERα or ERα L539A mutant enhanced PR-B-mediated transcription by approximately fourfold compared to that seen in the absence of SERMs and ERα or L539A (Fig. 5A), suggesting that ERα- and SERM-mediated elevation of coactivator expression can likely contribute to the transcriptional activity of another nuclear receptor. Similarly, we were able to enhance transcription mediated through the endogenously expressed GR. HeLa cells were transfected with ERα and a glucocorticoid-responsive reporter. In the presence of dexamethasone, 4HT and raloxifene were able to increase GR-mediated transcription as well (Fig. 4B). These results support the idea that an increase in coactivator protein concentration in the presence of SERMs is able to affect the transcriptional potential of other nuclear receptors. To test whether SERMs could enhance nuclear hormone receptor-mediated transcription in a setting where all the relevant proteins are endogenously expressed, we examined the ability of 4HT and raloxifene to enhance GR-mediated transcription in the MCF-7 cell line. The GR target gene SGK1 (37) was quantitated by real-time quantitative PCR after 12 h of treatment with SERMs and dexamethasone (Fig. 5C). 4HT, and to a lesser extent raloxifene, were able to elevate SGK1 transcript levels when cotreated with dexamethasone.
FIG.5.
SERMs can potentiate the transcriptional activity of other nuclear hormone receptors. (A) HeLa cells were transfected with a progesterone-responsive reporter (pGRE-E1b-LUC), pCR3.1 hERα L539A, or its vector backbone, pCR3.1 (−), and a PR expression vector (pCR3.1 hPR-B). Twenty-four hours thereafter, the cells were treated with ethanol vehicle (−), E2 (E), 4HT (T), or P4 (P). (B) Cells were transfected with a GR-responsive reporter (pGRE-E1b-LUC) and ERα and then treated with 10−7 M dexamethasone (Dex) and with the ERα ligands listed above. (C) SERM-mediated potentiation of GR-mediated transcription in MCF-7 cells. Untransfected MCF-7 cells were treated with ligands as described above and harvested for total RNA 12 h thereafter. The mRNA for the GR-inducible SGK1 gene was quantitated by real-time quantitative PCR normalized against 18S RNA. (D) Treatment with 4HT promotes increased interaction between PR and SRC-3-LUC in the HeLa cell line. HeLa cells were transfected with pERE-E1b-CAT and expression vectors for ERα, PR-B and SRC-3-LUC. Twenty-four hours thereafter, the cells were treated with E2 or 4HT as indicated. After an additional 24 h, the cells were treated with progesterone (P) for 1 h and then cells were harvested and subjected to immunoprecipitation with an anti-PR antibody. The antibody-associated luciferase (LUC) or SRC-3-LUC protein was detected with a standard luciferase assay.
To further examine the possibility that an increase in coactivator levels in 4HT-treated cells leads to an increase in nuclear receptor-mediated transcription, we asked whether there would be an increased association of the luciferase fusion protein SRC-3-LUC with PR-B in HeLa cells. To test this possibility we transfected HeLa cells with the progesterone-responsive reporter, pGRE-E1b-LUC, and expression vectors for ERα, PR-B, and SRC-3-LUC or the wild-type luciferase protein as a control. Twenty-four hours after transfection, cells were treated with either E2 or 4HT and incubated an additional 24 h to allow for 4HT to stimulate an increase in SRC-3-LUC protein levels. Then, cells were treated with progesterone for an additional hour to promote the ligand-dependent interaction of PR with SRC-3-LUC and harvested for analysis in a coimmunoprecipitation assay using an anti-PR antibody (see Materials and Methods). Figure 5D demonstrates that an increased amount of SRC-3-LUC was coimmunoprecipitated with PR in 4HT-treated cells compared to cells treated with either E2 or vehicle. This result supports the possibility that the ability of 4HT to stimulate PR-mediated transcription is due to increased coactivator association with PR-B.
DISCUSSION
Coactivators have emerged as critically important proteins in potentiating nuclear receptor-mediated gene transcription. For instance, targeted disruption of the SRC-1 (39) or SRC-3 (40) genes produces a hypomorphic reproductive phenotype attributable to a reduced transcriptional output from steroid receptors. One hypothesis which is used to explain either the agonist or antagonist biological action of SERMs such as 4HT and raloxifene is that differential coactivator expression in estrogen target tissues such as breast, uterus, and bone allows for their distinct tissue-specific biological activities. Analysis of the spatial patterns of coactivator expression in different tissues has revealed a complex spatial pattern of coactivator expression which is likely to lead to a diversity of biological actions imparted through the same nuclear receptor. For instance, it has been shown that elevated expression of SRC-1 in the uterine-derived Ishikawa cell line contributes to the increased agonist action of 4HT in the uterus, while lower levels of SRC-1 in the breast-derived MCF-7 cell line allow for 4HT to function as an antagonist (32).
The difference in coactivator or corepressor expression in different tissues that has been described is based largely upon the abundance of their transcripts, which as demonstrated here may not correspond to their actual expression at the protein level in the presence of SERMs. As an additional example, protein for the corepressor NCoR was shown to be present at higher levels in a nonneuronal cell line than in a neuronal cell line, although its transcript was equally abundant (43). It was shown that the difference in protein expression levels was due to NCoR protein being targeted by the proteasome in the neuronal cell line and not in the nonneuronal cell line.
Here, we provide evidence that the SERMs 4HT and raloxifene can elevate the steady-state level of SRC-1A and SRC-3 in certain cell lines. In the cervical carcinoma-derived HeLa cell line, 4HT and raloxifene were both able to stimulate an increase in the amount of both SRC-1 and SRC-3 proteins. This effect was ERα dependent and was dependent upon the AF-1 and DBD of ERα. We were also able to see an increase in the steady-state level of the SRC-3 protein in breast-derived MCF-7 cells treated with 4HT but not in T-47D or ZR-75 cells, which are also breast derived (Fig. 3E). These data indicate that the protein expression pattern for SRC-1 or SRC-3 could be different in certain basal and 4HT-treated tissues, suggesting that a prediction of whether 4HT or raloxifene will possess agonist activity based upon the expression of SRC-1 or SRC-3 in untreated tissues may be formed on the erroneous assumption that coactivator levels remain constant upon treatment with SERM ligands. Furthermore, the influence that 4HT and raloxifene have on coactivator stability could lead to a broadened impact for these SERMs. Their ability to enhance coactivator steady-state levels could potentiate the biological actions imparted by other nuclear receptors through their respective ligands or influence transcription through other transcription factors which interact with SRC-1 or SRC-3.
We have also presented evidence here which characterizes the stability of the protein and mRNA for SRC-1 and SRC-3. Our data demonstrate that the protein and transcripts for each coactivator are turned over with half-lives of approximately 5 or 3 h for SRC-1A-LUC or SRC-3-LUC proteins, respectively (Fig. 2A), and similar decay rates were seen for endogenous SRC-1 and SRC-3 as well (Fig. 2B). Similar protein half-lives were observed for SRC-1A-LUC and SRC-3-LUC in HepG2 and MCF7 cell lines (data not shown), indicating that coactivator turnover rates are similar in cell lines derived from other (liver or breast) tissues. Cotreatment of HeLa cells used for the SRC-1A-LUC and SRC-3-LUC protein decay experiments with MG132 indicated that the decay of either coactivator was due to proteasome-mediated degradation. The transcripts for both coactivators were also unstable, decaying at similar rates as the proteins (Fig. 2E). The decay rates for both the SRC-1 and SRC-3 transcripts fell into a range that is commonly seen for other transcripts. Genome-scale analysis of the stability of transcripts in the presence inhibitors of transcription has revealed a range of transcript stabilities ranging from about 0.5 h to more than 8 h, and SRC-1 and SRC-3 mRNAs are in the middle of this range (16). The lack of effect of ERα ligands on the decay of the SRC-3 mRNA that we saw in HeLa cells agrees with that observed in MCF-7 cells (18), although the observed mRNA half-life in MCF-7 cells was reported to be 4 h. We saw a shorter SRC-3 mRNA half-life of approximately 2 h in HeLa cells, which may explain the higher steady-state SRC-3 mRNA levels that are present in the MCF-7 cell line.
The requirement for proteasome function for most nuclear receptors to be transcriptionally competent (22), coupled with the observation that coactivators are targets of the proteasome, suggests that coactivator degradation plays a role in positively influencing gene transcription. Direct evidence which demonstrates that either SRC-1, SRC-3, or another coactivator must be specifically degraded to promote nuclear receptor-mediated transcription has not yet been demonstrated, but data presented here provide kinetic information on the rate of coactivator protein, and mRNAs should be of future use in characterizing the role that their degradation plays in transcription and how regulation of coactivator protein stability leads to the observed expression levels of coactivators in individual tissues.
Other evidence exists which suggests that proteasome-mediated degradation of coactivators and nuclear receptors represents a necessary step in allowing for efficient receptor-mediated transcription. For instance, degradation of the VP16 transcription factor has been shown to depend upon its activation domain, and increases in its transcriptional potency correlate with increased transcription factor instability (26). Also, the transcriptional activation domains of a large number of transcription factors have also been shown to be the regions of those proteins responsible for directing their proteasome-mediated degradation (27). Recently, it was shown that inhibition of proteasome activity abolished androgen-dependent induction of prostate-specific antigen (PSA) transcription, concomitant with the inability of the androgen receptor to dissociate from the PSA promoter, suggesting that dynamic remodeling of the PSA promoter is necessary for transcription to ensue (13). The 19S regulatory particle of the 26S proteasome has also been reported to play a role in transcriptional elongation (7), and components of the proteasome have also been shown to interact with ligand-activated nuclear receptors (cited above). It is possible that degradation of factors which form the preinitiation complex (nuclear receptors and coactivators) allows for the release of RNA polymerase from the promoter, facilitating transcriptional elongation. However, a more precise role for the proteasome in nuclear receptor-mediated transcription has not yet been established. It has been shown that the interaction of SRC-2 with the proteasome is mediated through its activation functions (3), implying again that proteasome activity is linked to the coactivator's ability to activate transcription. However, while SERMs stabilize ERα and coactivators concomitant with transcriptional inactivity, we are still able to observe an increase in PR- and GR-mediated transcription in SERM-treated cells, suggesting that degradation of either SRC-1 or SRC-3 is not strictly necessary for transcription to ensue. It is likely that other components of the coactivator-transcriptional apparatus must be degraded for transcription to proceed. It is possible that differences in coactivator stability seen between E2- and SERM-treated cells could be due to an alternate interaction between the coactivator and receptor in the presence of 4HT. Possibly, exclusion of components of the coactivator complex, such as the ubiquitin ligases E6-AP or p300 in SERM-treated cells, may result in altered coactivator stability.
In summary, we have shown that 4HT and raloxifene can affect the steady-state level of SRC-1 and SRC-3 at the protein level. Our results shed light on a novel mechanism which is likely to play a role in the tissue-selective agonist activity of 4HT in bone, uterine, and other tissues. The biological action of SERMs will need to be reevaluated, in part, to account for their ability to stimulate transcription mediated by other nuclear receptors. Finally, our results may have some relevance to the chronic induction of tamoxifen resistance in breast tumors. If, in the presence of tamoxifen, the receptor is stabilized and coactivator levels are increased, the setting is ripe for an adaptive mutation in the tumor to cause an up-regulation of a membrane-signaling pathway that in turn would activate the receptor, take advantage of the higher concentration of coactivators, and lead to ligand-independent ERα activities.
Acknowledgments
We thank Richard Santen for providing us with MCF-7 cells. The pSG5-KM3F2-hSRC-1 expression vector was kindly provided by Dennis Dowhan, and the ts85 cell line was graciously provided by Alexander Varshavsky. We also thank Xiao Tao Li, Ming Jer-Tsai, and Andrew Dennis for proofreading and critique of the manuscript.
This work was performed with funding from the National Institutes of Health to B.W.O.
REFERENCES
- 1.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965-968. [DOI] [PubMed] [Google Scholar]
- 2.Barsalou, A., G. Dayan, S. I. Anghel, M. Alaoui-Jamali, P. Van de Velde, and S. Mader. 2002. Growth-stimulatory and transcriptional activation properties of raloxifene in human endometrial Ishikawa cells. Mol. Cell. Endocrinol. 190:65-73. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, C. T., H. Ma, R. Wolford, J. C. Reyes, P. Maruvada, C. Lim, P. M. Yen, M. R. Stallcup, and G. L. Hager. 2001. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol. Endocrinol. 15:485-500. [DOI] [PubMed] [Google Scholar]
- 4.Dace, A., L. Zhao, K. S. Park, T. Furuno, N. Takamura, M. Nakanishi, B. L. West, J. A. Hanover, and S. Cheng. 2000. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc. Natl. Acad. Sci. USA 97:8985-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deroo, B. J., C. Rentsch, S. Sampath, J. Young, D. B. DeFranco, and T. K. Archer. 2002. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol. Cell. Biol. 22:4113-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez, F., A. Delahodde, T. Kodadek, and S. A. Johnston. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548-550. [DOI] [PubMed] [Google Scholar]
- 8.Gottlicher, M., S. Heck, V. Doucas, E. Wade, M. Kullmann, A. C. Cato, R. M. Evans, and P. Herrlich. 1996. Interaction of the Ubc9 human homologue with c-Jun and with the glucocorticoid receptor. Steroids 61:257-262. [DOI] [PubMed] [Google Scholar]
- 9.Guan, X. Y., J. Xu, S. L. Anzick, H. Zhang, J. M. Trent, and P. S. Meltzer. 1996. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res. 56:3446-3450. [PubMed] [Google Scholar]
- 10.Iannacone, E. A., A. W. Yan, K. J. Gauger, A. L. Dowling, and R. T. Zoeller. 2002. Thyroid hormone exerts site-specific effects on SRC-1 and NCoR expression selectively in the neonatal rat brain. Mol. Cell. Endocrinol. 186:49-59. [DOI] [PubMed] [Google Scholar]
- 11.Imhof, M. O., and D. P. McDonnell. 1996. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol. Cell. Biol. 16:2594-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishizuka, T., T. Satoh, T. Monden, N. Shibusawa, T. Hashida, M. Yamada, and M. Mori. 2001. Human immunodeficiency virus type 1 Tat binding protein-1 is a transcriptional coactivator specific for TR. Mol. Endocrinol. 15:1329-1343. [DOI] [PubMed] [Google Scholar]
- 13.Kang, Z., A. Pirskanen, O. A. Janne, and J. J. Palvimo. 2002. Involvement of proteasome in the dynamic assembly of the androgen receptor transcription complex. J. Biol. Chem. 277:48366-48371. [DOI] [PubMed] [Google Scholar]
- 14.Kiang, D. T., R. E. Kollander, T. Thomas, and B. J. Kennedy. 1989. Up-regulation of estrogen receptors by nonsteroidal antiestrogens in human breast cancer. Cancer Res. 49:5312-5316. [PubMed] [Google Scholar]
- 15.Kurihara, I., H. Shibata, T. Suzuki, T. Ando, S. Kobayashi, M. Hayashi, I. Saito, and T. Saruta. 2002. Expression and regulation of nuclear receptor coactivators in glucocorticoid action. Mol. Cell. Endocrinol. 189:181-189. [DOI] [PubMed] [Google Scholar]
- 16.Lam, L. T., O. K. Pickeral, A. C. Peng, A. Rosenwald, E. M. Hurt, J. M. Giltnane, L. M. Averett, H. Zhao, R. E. Davis, M. Sathyamoorthy, L. M. Wahl, E. D. Harris, J. A. Mikovits, A. P. Monks, M. G. Hollingshead, E. A. Sausville, and L. M. Staudt. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol. 2:research0041.1-11. [DOI] [PMC free article] [PubMed]
- 17.Lange, C. A., T. Shen, and K. B. Horwitz. 2000. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc. Natl. Acad. Sci. USA 97:1032-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauritsen, K. J., H. J. List, R. Reiter, A. Wellstein, and A. T. Riegel. 2002. A role for TGF-beta in estrogen and retinoid mediated regulation of the nuclear receptor coactivator AIB1 in MCF-7 breast cancer cells. Oncogene 21:7147-7155. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. W., F. Ryan, J. C. Swaffield, S. A. Johnston, and D. D. Moore. 1995. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature 374:91-94. [DOI] [PubMed] [Google Scholar]
- 20.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 21.Lonard, D. M., Z. Nawaz, C. L. Smith, and B. W. O'Malley. 2000. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol. Cell 5:939-948. [DOI] [PubMed] [Google Scholar]
- 22.Lonard, D. M., and C. L. Smith. 2002. Molecular perspectives on selective estrogen receptor modulators (SERMs): progress in understanding their tissue-specific agonist and antagonist actions. Steroids 67:15-24. [DOI] [PubMed] [Google Scholar]
- 23.Mak, H. Y., S. Hoare, P. M. Henttu, and M. G. Parker. 1999. Molecular determinants of the estrogen receptor-coactivator interface. Mol. Cell. Biol. 19:3895-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna, N. J., and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 25.Misiti, S., L. Schomburg, P. M. Yen, and W. W. Chin. 1998. Expression and hormonal regulation of coactivator and corepressor genes. Endocrinology 139:2493-2500. [DOI] [PubMed] [Google Scholar]
- 26.Molinari, E., M. Gilman, and S. Natesan. 1999. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18:6439-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell. Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 28.Nawaz, Z., D. M. Lonard, C. L. Smith, E. Lev-Lehman, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1999. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 19:1182-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nawaz, Z., D. M. Lonard, A. P. Dennis, C. L. Smith, and B. W. O'Malley. 1999. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. USA 96:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos, J. A., N. Zenser, O. Leyser, and J. Callis. 2001. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, G., M. R. Hubner, R. Metivier, H. Brand, S. Denger, D. Manu, J. Beaudouin, J. Ellenberg, and F. Gannon. 2003. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11:695-707. [DOI] [PubMed] [Google Scholar]
- 32.Shang, Y., and M. Brown. 2002. Molecular determinants for the tissue specificity of SERMs. Science 295:2465-2468. [DOI] [PubMed] [Google Scholar]
- 33.Stenoien, D. L., M. G. Mancini, K. Patel, E. A. Allegretto, C. L. Smith, and M. A. Mancini. 2000. Subnuclear trafficking of estrogen receptor-α and steroid receptor coactivator-1. Mol. Endocrinol. 14:518-534. [DOI] [PubMed] [Google Scholar]
- 34.Stenoien, D. L., K. Patel, M. G. Mancini, M. Dutertre, C. L. Smith, B. W. O'Malley, and M. A. Mancini. 2001. FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nat. Cell Biol. 3:15-23. [DOI] [PubMed] [Google Scholar]
- 35.vom Baur, E., C. Zechel, D. Heery, M. J. Heine, J. M. Garnier, V. Vivat, B. Le Douarin, H. Gronemeyer, P. Chambon, and R. Losson. 1996. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 15:110-124. [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace, A. D., and J. A. Cidlowski. 2001. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J. Biol. Chem. 276:42714-42721. [DOI] [PubMed] [Google Scholar]
- 37.Webster, M. K., L. Goya, and G. L. Firestone. 1993. Immediate-early transcriptional regulation and rapid mRNA turnover of a putative serine/threonine protein kinase. J. Biol. Chem. 268:11482-11485. [PubMed] [Google Scholar]
- 38.Wijayaratne, A. L., S. C. Nagel, L. A. Paige, D. J. Christensen, J. D. Norris, D. M. Fowlkes, and D. P. McDonnell. 1999. Comparative analyses of mechanistic differences among antiestrogens. Endocrinology 140:5828-5840. [DOI] [PubMed] [Google Scholar]
- 39.Xu, J., Y. Qiu, F. J. DeMayo, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1998. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 279:1922-1925. [DOI] [PubMed] [Google Scholar]
- 40.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O'Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan, F., X. Gao, D. M. Lonard, and Z. Nawaz. 2003. Specific ubiquitin-conjugating enzymes promote degradation of specific nuclear receptor coactivators. Mol. Endocrinol. 17:1315-1331. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, J., M. G. Guenther, R. W. Carthew, and M. A. Lazar. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, J., M. Gianni, E. Kopf, N. Honore, M. Chelbi-Alix, M. Koken, F. Quignon, C. Rochette-Egly, and H. de The. 1999. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARα) and oncogenic RARα fusion proteins. Proc. Natl. Acad. Sci. USA 96:14807-14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Y., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500-25506. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, Y., C. Qi, S. Jain, M. M. Le Beau, R. Espinosa III, G. B. Atkins, M. A. Lazar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 1999. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc. Natl. Acad. Sci. USA 96:10848-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]