Abstract
Nitric oxide (NO) is an important signaling molecule that interacts with different targets depending on its redox state. NO can interact with thiol groups resulting in S-nitrosylation of proteins, but the functional implications of this modification are not yet fully understood. We have reported that treatment of RAW 264.7 cells with NO caused a decrease in levels of iron regulatory protein 2 (IRP2), which binds to iron-responsive elements present in untranslated regions of mRNAs for several proteins involved in iron metabolism. In this study, we show that NO causes S-nitrosylation of IRP2, both in vitro and in vivo, and this modification leads to IRP2 ubiquitination followed by its degradation in the proteasome. Moreover, mutation of one cysteine (C178S) prevents NO-mediated degradation of IRP2. Hence, S-nitrosylation is a novel signal for IRP2 degradation via the ubiquitin-proteasome pathway.
Physiologically, the majority of cells in vertebrates acquire iron from a well-characterized plasma glycoprotein, transferrin (Tf). Iron uptake from Tf involves the binding of Tf to the Tf receptors (TfR), internalization of Tf within an endocytic vesicle by receptor-mediated endocytosis, and the release of iron from Tf by a decrease in endosomal pH (15, 39, 41). Following iron release from Tf within endosomes, Fe2+ passes through the endosomal membrane by divalent metal transporter 1 (2, 8) and then enters the poorly characterized intracellular labile pool (LIP). Intracellular iron that exceeds the requirement for the synthesis of functional heme and nonheme iron-containing proteins is stored within ferritin (39, 41).
In general, cellular iron homeostasis is regulated posttranscriptionally by the cytoplasmic factors iron regulatory proteins 1 and 2 (IRP1 and IRP2), which “sense” iron levels in the LIP (7, 14, 35, 41). In the absence of iron in the LIP, IRPs bind to specific nucleotide sequences called iron-responsive elements (IREs), which are located in the 3′ untranslated region of TfR mRNA (36, 41) and the 5′ untranslated region of ferritin mRNA (14, 30, 41). The binding of IRPs to IREs stabilizes TfR mRNA and blocks ferritin mRNA translation. In iron-replete cells, IRP1 contains a [4Fe-4S] cluster and binds RNA with low affinity (13, 41). IRP2, however, does not have the iron-sulfur cluster and is degraded under iron-replete conditions. This degradation is dependent on a 73-amino-acid insertion, rich in cysteine, which is absent in IRP1 (11, 20). Hence, the expansion of the LIP inactivates IRP1 binding to IREs and leads to a degradation of IRP2, resulting in a rapid degradation of TfR mRNA and an efficient translation of ferritin mRNA (7, 35, 41). Importantly, IRPs can also be affected by various forms of oxidative stress and nitric oxide (NO) (6, 12, 15, 38, 40, 46).
NO is an important signaling molecule (17, 18) that interacts with different targets depending on its redox state. The reduced form of NO (the notation NO is used here as a generic expression encompassing all nitrogen monoxide species), NO·, interacts mainly with iron (39, 40) and is known to disrupt the iron-sulfur cluster in IRP1 (25), leading to an increase in IRP1 binding to IREs (6, 40, 46). On the other hand, the oxidized form of NO, NO+ (nitrosonium ion), reacts with thiol groups, resulting in S-nitrosylation (42, 43). Numerous proteins have been identified as targets for S-nitrosylation (4, 9, 23, 32, 37, 44), but the functional implications of this modification are not yet fully understood. Previously, we observed that sodium nitroprusside (SNP), a compound with NO+ character that readily promotes S-nitrosylation, decreased IRP2 protein levels (26) in RAW 264.7 cells (a macrophage cell line); the SNP-mediated degradation of IRP2 could be prevented by inhibitors of proteasome-dependent protein degradation (26). Importantly, the response to SNP was remarkably similar to the IRP2 decrease seen in lipopolysaccharide-gamma interferon-treated cells (27); these agents are known to induce NO production in macrophages (31). Specific inhibitors of inducible NO synthase prevented lipopolysaccharide-gamma interferon-mediated degradation of IRP2 (27), indicating that their effect is also caused by NO. Importantly, treatment of RAW 264.7 cells with either SNP or lipopolysaccharide-gamma interferon led to a dramatic increase in ferritin synthesis that was associated with a significant enhancement of iron incorporation into ferritin (28). The physiological relevance of NO regulation of IRP2 in macrophages is further supported by studies showing that macrophages from IRP2-deficient mice have elevated ferritin levels (E. Meyron-Holtz, A. M. Martinez, and T. A. Rouault, Proc. World Congr. Iron Metab., BioIron 2001, abstract O5, 2001). We deemed it important to define the mechanism of IRP2 degradation by NO. In this study we demonstrated that IRP2 is S-nitrosylated by SNP and that this nitrosylation leads to its degradation via the ubiquitin-proteasome pathway.
MATERIALS AND METHODS
Cells.
RAW 264.7 murine macrophages and COS1 cells were obtained from the American Type Culture Collection and grown in 100-cm2 plastic culture dishes (Life Technologies) in a humidified atmosphere of 95% air and 5% CO2 at 37°C in Dulbecco's modified Essential medium containing 10% fetal calf serum, extra l-glutamine (300 μg/ml), sodium pyruvate (110 μg/ml), penicillin (100 U/ml), and streptomycin (100 μg/ml).
Plasmid construction and transfection.
IRP2 plasmid was obtained from Elizabeth Leibold and amplified with primers containing the restriction sites of BamH1 and Xhol. This fragment was cloned into mammalian expression vector pcDNA3.1/His (Invitrogen). Mutations of IRP2 cysteines were generated with the Quick-Change site-directed mutagenesis kit (Stratagene) as described in the manufacturer's manual, followed by sequencing analysis to confirm the accuracy of mutations. Plasmids were transfected by Lipofectamine (Life Technologies) according to the manufacturer's instructions.
Preparation of 35S-labeled IRP2.
The COS1 cells overexpressing IRP2 were incubated with 50 μCi of [35S]methionine for 2 h in medium which lacked methionine, following which cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris Cl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]), and [35S]IRP2 was immunoprecipitated with anti-Xpress antibody.
In vitro ubiquitin conjugation and degradation assay.
Plasmid pcDNA3.1/His-IRP2 was linearized by XbaI and in vitro translated with a TNT-coupled wheat germ kit (Promega) in the presence of [35S]methionine. Assays of ubiquitin conjugation and degradation were performed as previously described (34).
Western blot analysis.
About 5 × 107 cells were lysed with an extraction buffer (10 mM HEPES [pH 7.5], 3 mM MgCl2, 40 mM NaCl, 5% glycerol, 1 mM dithiothreitol [DTT], 0.2% Nonidet P-40), and 60 μg of protein was resolved using SDS-6% polyacrylamide gel electrophoresis (PAGE). Protein was transferred to a nitrocellulose membrane, which was subsequently incubated with rabbit anti-IRP2 antibodies (a generous gift from E. Leibold) or antinitrosothiol (anti-SNO) antibody (Alexis Biochemicals Inc.). After 1 h of incubation, the membranes were washed and incubated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Sigma) for 1 h. The protein was then visualized with an enhanced chemiluminescence Western blotting detection system (Bio-Rad) according to the manufacturer's manual.
Isoelectric focusing.
Recombinant IRP2 was purified from yeast (generously provided by E. Leibold) and pretreated with 10 mM DTT for 10 min. After removing DTT by using Microcon (Millipore), IRP2 was treated with either 500 μM nitrosoglutathione (GSNO) or SNP for 30 min. IRP2 was treated with 35 mM iodoacetamide and subjected to isoelectric focusing using a thin gel containing pH 3.5 to 10.0 ampholyte (24). Protein was transferred to polyvinylidene difluoride membrane and detected by anti-IRP2 antibody.
Immunoprecipitation analysis.
Approximately 5 × 107 cells were incubated with [35S]methionine in methionine-deficient minimum essential medium for 1 h. The cells were then washed (three times) with cold phosphate-buffered saline (PBS). The cells were lysed with RIPA buffer, and the equal amount of protein was cleared with protein A-Sepharose (Amersham-Pharmacia). Three micrograms of antiferritin antibody was added to the lysate and incubated for 2 h at 4°C, following which 60 μl of protein A-Sepharose was added, and the beads were washed with cold RIPA (three times) and boiled with SDS loading dye. Immunoprecipitated protein was resolved using SDS-15% PAGE. The gels were dried, and the band intensity was detected by fluorography.
RESULTS AND DISCUSSION
SNP decreases the stability of IRP2.
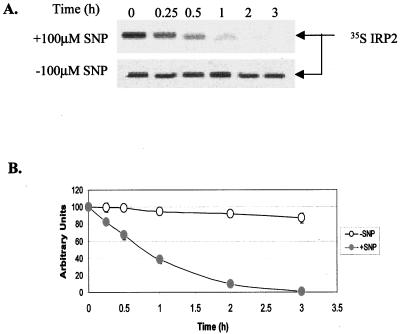
We first investigated whether the decrease in IRP2 protein levels after SNP treatment of RAW 264.7 cells is caused by the degradation of this protein. Following labeling of RAW 264.7 cells with [35S]methionine for 1 h, cells were treated with 100 μM SNP for various time intervals and the amount of IRP2 was measured by autoradiography following immunoprecipitation of IRP2 and SDS-PAGE (Fig. 1A). The levels of IRP2 started to decrease as early as 15 min of exposure of RAW 264.7 cells to SNP, and this treatment caused a rapid degradation of [35S]methionine-labeled IRP2 ([35S]IRP2) with an estimated half-life of about 45 min, whereas in control cells levels of [35S]IRP2 remained virtually unchanged during 3 h of incubation (Fig. 1).
FIG. 1.
The effect of SNP on IRP2 stability. (A) [35S]IRP2 stability in RAW264.7 cells. Following labeling of RAW264.7 cells with [35S]methionine for 1 h, cells were washed with PBS (three times) and incubated in growth medium with or without 100 μM SNP. [35S]IRP2 was immunoprecipitated using anti-IRP2 antibodies and analyzed by SDS-6.5% PAGE, followed by autoradiography. (B) Densitometric analysis of [35S]IRP2.
Because SNP is an iron complex, it may be argued that SNP could donate iron to cells, resulting in IRP2 degradation. However, it is unlikely that SNP causes IRP2 degradation by increasing iron levels in the LIP. First, the exposure of RAW 264.7 cells to SNP for 3 h does not decrease IRP1 activity (26), which would occur if intracellular iron levels increased. Second, iron-mediated deactivation of IRP1 and IRP2 occurs with similar kinetics (12, 27). Third, the treatment of 59Fe-labeled RAW 264.7 cells with SNP did not mobilize 59Fe from ferritin and other 59Fe-containing proteins (data not shown). Fourth, ferricyanide, which is structurally very similar to SNP, did not decrease IRP2 protein levels under conditions when SNP did (26).
Since SNP may under certain conditions lead to the production of peroxynitrite (ONOO−, which forms by a reaction of 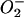 with NO·) (1, 3), we examined the effects of SIN-1 (ONOO− donor) or diethyleneaminetriamine NONO-ate (NO· donor) plus xanthine-xanthine oxidase (O2−-generating system) on the binding of IRP2 to IRE. However, peroxynitrite generated by these agents did not decrease IRP2 binding activity in RAW 264.7 cells nor its protein levels (3 h treatment; data not shown).
with NO·) (1, 3), we examined the effects of SIN-1 (ONOO− donor) or diethyleneaminetriamine NONO-ate (NO· donor) plus xanthine-xanthine oxidase (O2−-generating system) on the binding of IRP2 to IRE. However, peroxynitrite generated by these agents did not decrease IRP2 binding activity in RAW 264.7 cells nor its protein levels (3 h treatment; data not shown).
NO+ donors induce S-nitrosylation of IRP2 in vitro.
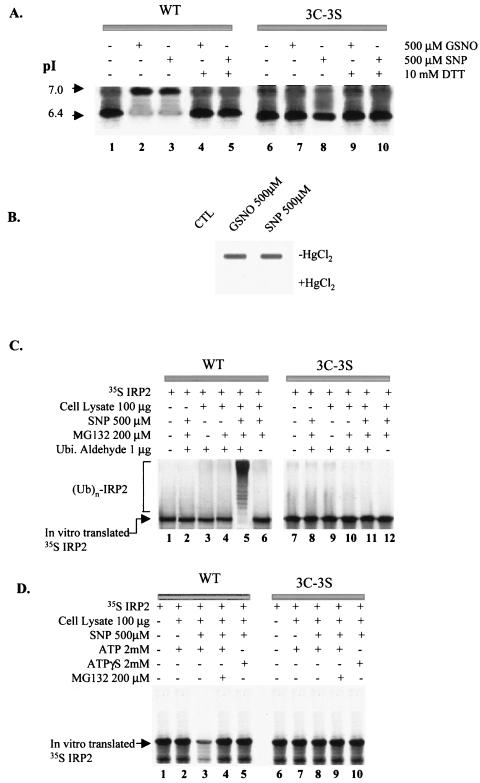
Our previous research suggested that an NO+-induced modification of IRP2 leads to the degradation of IRP2 by the proteasome (26). Despite the fact that the sequences of biochemical reactions of the ubiquitin-proteasome pathway are well understood (16), it is unclear how proteins are targeted for their ubiquitination and consequent degradation. It is reasonable to assume that this process is very specific, because numerous short-lived proteins have to be identified among all the other proteins. Degradation of a target protein seems to be mediated by specific signals, which may be found in certain domains or sequences that are required for rapid protein turnover (16). Moreover, a substrate can be modified posttranslationally after external stimuli. Examples of such modifications are phosphorylation of IκBα (5) and hydroxylation of hypoxia-inducible factor 1 (19, 22). NO+ is well known to interact with thiol groups (e.g., -SH of cysteines in protein) (23, 42, 43), causing S-nitrosylation of proteins, and this modification has been shown to regulate the activity of these proteins (9, 23, 32, 37, 44). Importantly, three cysteines in IRP2 have been reported to play an essential role in the stability of this protein (20, 21). Therefore, we examined whether IRP2 can be a target for S-nitrosylation by NO by comparing pI values of wild-type (WT) IRP2 and its triple mutant (C168, C174, and C178 to serine [3C-3S]). These cysteines reside in the unique 73-amino-acid (AA) domain of IRP2, and this domain is essential for IRP2 degradation in iron-replete cells. It has been reported that these mutations prevent iron-mediated degradation of IRP2 (20, 21), which cannot be S-nitrosylated at mutated sites. [35S]IRP2, prepared by in vitro translation in the wheat germ system, was incubated with 500 μM SNP for 30 min. Both WT (Fig. 2A, lane1) and mutant IRP2 (Fig. 2A, lane 6) showed a pI of 6.4, which is equivalent to the calculated pI value based on the amino acid composition of the protein. Importantly, neither ferricyanide nor cyanide, which are the decomposition products of SNP, caused any change in the pI of IRP2 (data not shown). Treatment with GSNO or SNP raised the pI of IRP2 from 6.4 to 7.0. When either GSNO- or SNP-treated WT IRP2 was exposed to DTT, which removes NO from S-nitrosylated proteins (33), the pI value decreased from 7.0 to 6.4 (Fig. 2A, lanes 4 and 5). In contrast, the pI value of [3C-3S] mutant IRP2 did not change following either GSNO or SNP treatment (Fig. 2A, lanes 7 and 8). In another experimental strategy, the recombinant IRP2 protein which was purified from yeast was treated without or with either GSNO or SNP. S-nitrosylation, detected by the anti-SNO antibody, occurred only in GSNO- or SNP-treated samples (Fig. 2B). Treatment of samples with HgCl2, which is known to cleave NO from SNO (32), led to a disappearance of the signal detectable by the anti-SNO antibody (Fig. 2B). Collectively, these data indicate that IRP2 is a target for S-nitrosylation after exposure to either GSNO or SNP.
FIG. 2.
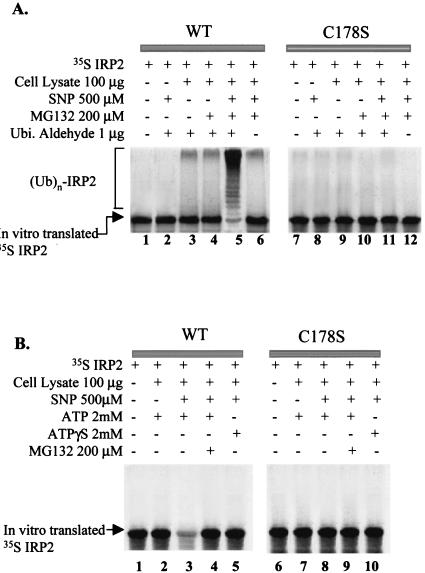
In vitro modification of IRP2 by SNP. (A) Isoelectric focusing analysis for S-nitrosylation of IRP2 and identification of target cysteines. Following treatment of in vitro-translated [35S]IRP2 with either 500 μM GSNO or SNP for 30 min, some samples (lanes 4, 5, 9, and 10) were treated with 10 mM DTT for an additional 5 min. Iodoacetate (35 mM) was added to protect sulfhydryls and to increase the negative charge on the unmodified thiol groups by its alkylation. The protein was subjected to isoelectric focusing and detected by autofluorography. (B) In vitro S-nitrosylation of recombinant WT IRP2. Recombinant IRP2 was treated without (control [CTL]) or with 500 μM concentration of either GSNO or SNP for 30 min. Following this treatment, the samples were treated without or with HgCl2 for 5 min and analyzed by slot blotting with anti-SNO antibody (Alexis Biochemicals Inc.). (C) In vitro ubiquitin conjugation assay of IRP2. In vitro-translated [35S]IRP2 was treated with or without 500 μM SNP for 30 min and then incubated (for 1.5 h) with cell lysate prepared from RAW 264.7 cells and various reagents as indicated. (D) In vitro degradation assay. In vitro-translated IRP2 was pretreated with 500 μM SNP for 30 min and incubated without or with cell lysate from RAW264.7 cells and other reagents as indicated.
S-nitrosylation of IRP2 leads to its degradation via the ubiquitin-proteasome pathway in vitro.
To examine whether S-nitrosylation is a signal for degradation by the 26S proteasome that in most cases requires ubiquitination of proteins destined for proteolysis (16), we first performed an in vitro ubiquitin conjugation assay. We compared WT IRP2 and its [3C-3S] mutant. Following the incubation of SNP-treated [35S]IRP2 in the lysate of RAW 264.7 cells in the presence of various reagents (Fig. 2C), [35S]IRP2 was analyzed by SDS-PAGE. As shown in Fig. 2C, SNP treatment substantially increased the appearance of higher-molecular-weight bands in the presence of both MG132 and ubiquitin aldehyde, which block the degradation and deubiquitination, respectively, of ubiquitinated proteins (32) (Fig. 2C, lane 5). The high-molecular-weight bands provided evidence for the accumulation of multiubiquitinated species of IRP2 whose formation was induced by SNP. However, mutation of cysteines completely prevented ubiquitination of IRP2 (Fig. 2C, lane 11).
A prominent feature of ubiquitin-dependent proteolysis is its dependence on ATP, which is required for conjugation of ubiquitin to the substrate and for degradation of the ubiquitinated protein by the proteasome (16). To further characterize the mechanism of degradation of IRP2 triggered by S-nitrosylation, we exploited ATPγS, which is a nonhydrolysable ATP analog. Pretreatment of [35S]IRP2 with 500 μM SNP for 30 min caused its degradation in the presence of ATP and cell lysate (Fig. 2D, lane 3), while in the absence of SNP, levels of [35S]IRP2 remained unchanged (Fig. 2D, lane 2). Moreover, either addition of MG132 or replacement of ATP with ATPγS blocked the SNP-induced degradation of WT IRP2 (Fig. 2D, lanes 4 and 5), indicating that IRP2 degradation induced by SNP is ATP dependent. Importantly, SNP was unable to induce the degradation of mutated IRP2 (Fig. 2D, lane 8). Taken together, these data indicate that IRP2 is a target for S-nitrosylation by SNP and that this modification leads to IRP2 degradation via a ubiquitin-proteasome pathway in vitro.
SNP causes S-nitrosylation of IRP2 and its consequent ubiquitination in vivo.
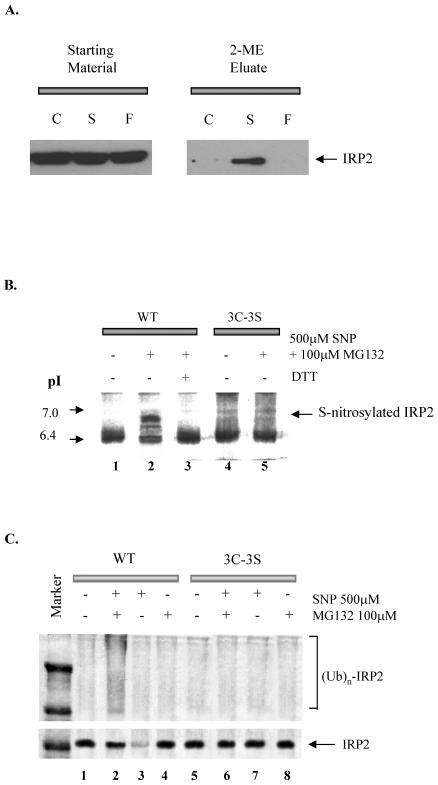
To investigate whether SNP can cause S-nitrosylation of IRP2 in vivo, RAW 264.7 cells were incubated with 100 μM SNP for 1 h and examined for S-nitrosylation of IRP2 using a novel method developed by Jaffrey et al. (23). Figure 3A shows that IPR2 is S-nitrosylated only in SNP-treated cells. Moreover, COS1 cells were transfected with an expression plasmid containing either WT or [3C-3S]IRP2, each tagged with Xpress. Following labeling of IRP2-transfected COS1 cells with [35S]methionine for 1 h, cells were treated with 500 μM SNP for 3 h, and IRP2 was analyzed by autoradiography after its immunoprecipitation and resolution by isoelectric focusing. SNP treatment of COS1 cells expressing WT IRP2 changed its pI from 6.4 to 7 (Fig. 3B, lanes 1 and 2); addition of 10 mM DTT to the lysate prepared from SNP-treated cells changed the pI to the value seen in untreated cells (Fig. 3B, lane 3). In contrast, no change in the pI of IRP2 was observed when COS1 cells transfected with [3C-3S]IRP2 were treated with SNP. These results indicate that IRP2 can be S-nitrosylated in vivo and its cysteine(s) are targets for S-nitrosylation. To examine whether SNP-mediated S-nitrosylation of IRP2 causes ubiquitination of IRPs in intact cells, COS1 cells were cotransfected with a vector expressing hemagglutinin (HA)-tagged human ubiquitin (45) and an expression plasmid harboring either WT IRP2 or the [3C-3S] IRP2 mutant, each containing the Xpress tag. Cotransfected COS1 cells were then treated with 500 μM SNP and 100 μM MG132 for 6 h, following which IRP2 was immunoprecipitated by anti-Xpress antibodies and immunoblotted using anti-HA tag antibodies. Treatment of cotransfected cells with these agents enhanced the appearance of multiubiquitinated species only in the transfectant expressing the WT IRP2 (Fig. 3C, lane 2 versus 6). These results, together with those shown in Fig. 2, indicate that IRP2 is S-nitrosylated by SNP and that this modification leads to its degradation via the ubiquitin-proteasome pathway.
FIG. 3.
In vivo modification of IRP2 by SNP. (A) In vivo S-nitrosylation of endogenous IRP2. RAW 264.7 cells were incubated without (C) or with 100 μM SNP (S) or 100 μM ferricyanide (F) for 1 h, washed, and lysed. SNO IRP2 was detected as described by Jaffrey et al. (23). Briefly, free thiols in the lysate proteins were blocked by incubation with the thio-specific methylthiolating agent methyl methanethiosulfonate. After the blocking of free thiols, nitrosothiols were selectively decomposed to thiols and subsequently reacted with a sulfhydryl-specific biotinylating reagent. All the biotinylated proteins (which were previously S-nitrosylated) were precipitated by using streptavidin beads, following which they were eluted from the beads by 2-mercaptoethanol and subjected to SDS-PAGE and Western blot analysis using anti-IRP2 antibody (2-ME Eluates). The “Starting Material” represents a Western blot analysis of IRP2 in untreated lysates. (B) In vivo S-nitrosylation of IRP2. Either WT or triple mutant [3C-3S] His-tagged IRP2 was transfected into COS1 cells. Following labeling of RAW264.7 cells with [35S]methionine for 1 h, cells were washed with PBS (three times) and incubated in growth medium with or without 500 μM SNP and 100 μM MG132. [35S]IRP2 was immunoprecipitated using anti-His antibodies. The protein was subjected to isoelectric focusing and visualized by autofluorography. (C) In vivo ubiquitination assay of [35S]IRP2. Either WT or triple mutant [3C-3S] IRP2 was transfected together with HA-tagged ubiquitin plasmid into COS1 cells. IRP2 was immunoprecipitated by using anti-IRP2 antibody and immunoblotted using HA tag antibody.
S-nitrosylation of cysteine 178 is required for IRP2 degradation.
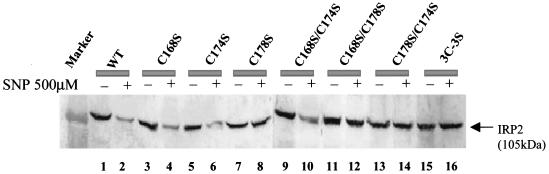
Next, we investigated the target cysteine(s) for S-nitrosylation of IRP2 by constructing a series of plasmids containing single or double substitutions of cysteines with serines in IRP2. Cells were transfected with a series of mutants and treated with 500 μM SNP for 6 h. As expected, treatment of transfected cells with SNP caused a decrease in WT IRP2 (Fig. 4, lanes 1 and 2) but did not result in the degradation of the [3C-3S] IRP2 mutant (Fig. 4, lanes 15 and 16). Moreover, SNP treatment decreased levels of IRP2 which had single cysteine mutations at positions 168 or 174 (Fig. 4, lanes 3 versus 4 and 5 versus 6), whereas the C178S IRP2 mutant did not exhibit any sign of degradation (Fig. 4, lanes 7 and 8). Importantly, SNP-induced degradation of IRP2 did not occur when the double mutants contained the C178S mutation, such as C168/C178S and C174/C178S (Fig. 4, lanes 11 to 14), while C168S/C174S IRP2 was still degraded (Fig. 4, lanes 9 and 10) in cells exposed to SNP. These data indicate that C178 is essential for IRP2 degradation following SNP treatment.
FIG. 4.
Identification of a target cysteine involved in IRP2 degradation. Western blot analysis of IRP2 after transfection of WT or mutated IRP2. Plasmids containing either WT or mutated IRP2 were transfected into COS1 cells, and 48 h after the transfection cells were treated with 500 μM SNP for 6 h. The protein was detected by anti-Xpress tag antibody.
Additional experiments (Fig. 5) further stressed the importance of cysteine 178 in IRP2 degradation via the ubiquitin-proteasome system. Using the same strategy as described for Fig. 2C, we demonstrated that the C178S mutant of IRP2 cannot be ubiquitinated (Fig. 5A, lane 11 versus 5). It is pertinent to mention that a low degree of ubiquitination of WT IRP2 occurred even in the absence of SNP, but only in the presence of the lysate prepared from RAW 264.7 cells (Fig. 5A, lanes 3 and 4). It is likely that some lysate preparations, but not all (Fig. 2C), contain molecules (e.g., GSNO) that lead to S-nitrosylation. This is not totally unexpected, since S-nitrosylating species of NO are ubiquitously present in cells (10). Moreover, in contrast to WT IRP2, the C178S mutant was not degraded in the proteasome (Fig. 5B, lane 8 versus 3).
FIG. 5.
In vitro modification of IRP2 by SNP. (A) In vitro ubiquitin conjugation assay of IRP2. In vitro-translated [35S]IRP2 (either WT or the C178S mutant) was treated with or without 500 μM SNP for 30 min and then incubated (for 3 h) with lysate prepared from RAW 264.7 cells and various reagents as indicated. (B) In vitro degradation assay. In vitro-translated [35S]IRP2 was pretreated with 500 μM SNP (lanes 3 to 5 and 8 to 10) for 30 min and incubated with cell lysate from RAW264.7 cells and other reagents as indicated.
It needs to be pointed out that NO-mediated degradation of IRP2 is associated with increased ferritin synthesis in macrophage cell lines and that IRP2-deficient mice exhibit a dramatic increase in ferritin levels in various tissues (29), including macrophages (Meyron-Holtz et al., Proc. World Congr. Iron Metab., 2001). These observations are consistent with the idea that NO regulation of IRP2 in macrophages is of physiological and pathophysiological relevance. It has been proposed that an NO-mediated increase of ferritin synthesis via IRP2 downregulation in macrophages plays a role in anemia of chronic disease (28) and that overexpression of ferritin in macrophages of IRP2-deficient mice may lead to a misperception of iron deficiency and consequently misregulation of iron export from macrophages (Meyron-Holtz et al., Proc. World Congr. Iron Metab., 2001).
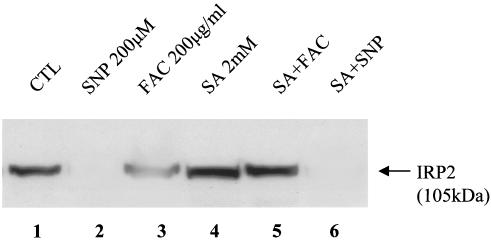
Recently, Yamanaka et al. (47) identified the ubiquitin ligase that recognizes oxidized IRP2 and also demonstrated that degradation of IRP2 by iron requires the synthesis and availability of heme. These investigators showed that succinylacetone (SA, a specific inhibitor of heme synthesis at the level of 5-aminolevulinic acid dehydratase) prevented iron-mediated degradation of IRP2. Hence, we investigated whether NO-mediated degradation of IRP2 is also a part of this pathway. COS1 cells were transfected with WT IRP2, following which the cells were pretreated with 2 mM SA for ∼12 h to ensure the depletion of heme; the cells were then treated with either 200 μM SNP or 200 μg of ferric ammonium citrate (FAC)/ml for 6 h. As expected, the treatment of cells with either FAC or SNP caused the degradation of WT IRP2 (Fig. 6, lanes 2 and 3); the treatment of cells with 2 mM SA did not significantly change the basal level of IRP2 expression. We confirmed (Fig. 6, lane 5) that the treatment of cells with SA prevented FAC-mediated degradation of IRP2 (47); however, the depletion of cellular heme did not prevent the SNP-mediated degradation of IRP2 (Fig. 6, lane 6). Moreover, we examined the effect of SA on the SNP-mediated degradation of endogenous IRP2 in RAW264.7 cells and obtained the same results as those described for IRP2-transfected COS1 cells (data not shown). These data suggest that NO-induced degradation of IRP2 does not follow the same pathway used in iron-mediated degradation of IRP2.
FIG. 6.
A plasmid containing WT IRP2 was transfected into COS1 cells, and 48 h after the transfection cells were pretreated (overnight) with 2 mM SA, following which the cells were treated with either 200 μM SNP or 200 μg of FAC/ml for 6 h. The protein was detected by anti-Xpress tag antibody.
In summary, we have demonstrated that SNP induces S-nitrosylation of IRP2, both in vitro and in vivo, and that the specific modification of C178 targets IRP2 for degradation via the ubiquitin-proteasome pathway. Hence, we have identified S-nitrosylation as a novel posttranslational modification that targets a protein for recognition by the ubiquitin-proteasome system. This modification now joins phosphorylation and hydroxylation as a family of molecular switches that can precisely activate the degradation of a protein. Moreover, our findings provide a new perspective on the molecular mechanism by which NO exerts its signaling function.
Acknowledgments
We are grateful to Elizabeth Leibold for antibodies and plasmids for IRP2. We are grateful to Sandy Fraiberg for excellent editorial assistance.
This work was supported by grants (to P.P.) and a scholarship (to S.K.) from the Canadian Institutes of Health Research.
REFERENCES
- 1.Aleryani, S., E. Milo, and P. Kostka. 1999. Formation of peroxynitrite during thiol-mediated reduction of sodium nitroprusside. Biochim. Biophys. Acta 1472:181-190. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, N. C. 1999. The iron transporter DMT1. Int. J. Biochem. Cell. Biol. 31:991-994. [DOI] [PubMed] [Google Scholar]
- 3.Arnelle, D. R., and J. S. Stamler. 1995. NO+, NO, and NO− donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 318:279-285. [DOI] [PubMed] [Google Scholar]
- 4.Beltran, B., A. Orsi, E. Clementi, and S. Moncada. 2000. Oxidative stress and S-nitrosylation of proteins in cells. Br. J. Pharmacol. 129:953-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Z., J. Hagler, V. J. Palombella, F. Melandri, D. Scherer, D. Ballard, and T. Maniatis. 1995. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9:1586-1597. [DOI] [PubMed] [Google Scholar]
- 6.Drapier, J. C., H. Hirling, J. Wietzerbin, P. Kaldy, and L. C. Kühn. 1993. Biosynthesis of nitric oxide activates iron regulatory factor in macrophages. EMBO J. 12:3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisenstein, R. S. 2000. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu. Rev. Nutr. 20:627-662. [DOI] [PubMed] [Google Scholar]
- 8.Flemming, M. D., C. C. Trenor III, M. A. Su, D. Forenzler, D. R. Beier, W. F. Dietrich, and N. C. Andrews. 1997. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat. Genet. 16:383-386. [DOI] [PubMed] [Google Scholar]
- 9.Foster, M. W., T. J. McMahon, and J. S. Stamler. 2003. S-nitrosylation in health and disease. Trends Mol. Med. 9:160-168. [DOI] [PubMed] [Google Scholar]
- 10.Gow, A. J., Q. Chen, D. T. Hess, B. J. Day, H. Ischiropoulos, and J. S. Stamler. 2002. Basal and stimulated proteins S-nitrosylation in multiple cell types and tissues. J. Biol. Chem. 277:9637-9640. [DOI] [PubMed] [Google Scholar]
- 11.Guo, B., J. D. Phillips, Y. Yu, and E. A. Leibold. 1995. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270:21645-21651. [DOI] [PubMed] [Google Scholar]
- 12.Hanson, E. S., and E. A. Leibold. 1998. Regulation of iron regulatory protein 1 during hypoxia and hypoxia/reoxygenation. J. Biol. Chem. 273:7588-7593. [DOI] [PubMed] [Google Scholar]
- 13.Hentze, M. W. 1996. Iron-sulfur clusters and oxidant stress responses. Trends Biochem. Sci. 21:282-283. [PubMed] [Google Scholar]
- 14.Hentze, M. W., S. W. Caughman, T. A. Rouault, J. G. Barriocanal, A. Dancis, J. B. Harford, and R. D. Klausner. 1987. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science 238:1570-1573. [DOI] [PubMed] [Google Scholar]
- 15.Hentze, M. W., and L. C. Kühn. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. USA 93:8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Ignarro, L. J. 1994. Regulation of cytosolic guanylyl cyclase by porphyrins and metalloporphyrins. Adv. Pharmacol. 26:35-65. [DOI] [PubMed] [Google Scholar]
- 18.Ignarro, L. J. 1996. Physiology and pathophysiology of nitric oxide. Kidney Int. Suppl. 55:S2-S5. [PubMed] [Google Scholar]
- 19.Ivan, M., K. Kondo, H. Yang, W. Ki, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 20.Iwai, K., R. D. Klausner, and T. A. Rouault. 1995. Requirements for iron regulated degradation of the RNA binding protein, iron regulatory protein 1. EMBO J. 14:5350-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwai, K., S. K. Drake, N. B. Wehr, A. M. Weissman, T. LaVaute, N. Minato, R. D. Klausner, R. L. Levine, and T. A. Rouault. 1998. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA 95:4924-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukkerji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and R. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 23.Jaffrey, S. R., H. Erdjument-Bromage, C. D. Ferris, P. Tempst, and S. H. Snyder. 2001. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3:193-197. [DOI] [PubMed] [Google Scholar]
- 24.Ji, Y., T. P. Akerboom, H. Sies, and J. A. Thomas. 1999. Gel electrofocusing method for studying protein S-nitrosylation. Methods Enzymol. 301:145-151. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, M. C., W. E. Antholine, and H. Beinert. 1997. An EPR investigation of the products of the reaction of cytosolic and mitochondrial aconitases with nitric oxide. J. Biol. Chem. 272:20340-20347. [DOI] [PubMed] [Google Scholar]
- 26.Kim, S., and P. Ponka. 1999. Control of transferrin receptor expression via nitric oxide-mediated modulation of iron-regulatory protein 2. J. Biol. Chem. 274:33035-33042. [DOI] [PubMed] [Google Scholar]
- 27.Kim, S., and P. Ponka. 2000. Effects of interferon-γ and lipopolysaccharide on macrophage iron metabolism are mediated by nitric oxide-induced degradation of iron regulatory protein 2. J. Biol. Chem. 275:6220-6226. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S., and P. Ponka. 2002. Nitrogen monoxide-mediated control of ferritin synthesis: implications for macrophage iron homeostasis. Proc. Natl. Acad. Sci. USA 99:12214-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaVaute, T., S. Smith, S. Cooperman, K. Iwai, W. Land, E. Meyron-Holtz, S. K. Drake, G. Miller, M. Abu-Asab, M. Tsokos, R. Switzer III, A. Grinberg, P. Love, N. Tresser, and T. Rouault. 2001. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 27:209-214. [DOI] [PubMed] [Google Scholar]
- 30.Leibold, E. A., and H. N. Munro. 1988. Cytoplasmic protein binds in vitro a highly conserved sequence in the 5′ untranslated region of ferritin H. Proc. Natl. Acad. Sci. USA 85:2171-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacMicking, J., Q. W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323-350. [DOI] [PubMed] [Google Scholar]
- 32.Mannick, J. B., A. Hausladen, L. Liu, D. T. Hess, M. Zeng, M., Q. X. Miao, L. S. Kane, A. J. Gow, and J. S. Stamler. 1999. Fas-induced caspase denitrosylation. Science 284:651-654. [DOI] [PubMed] [Google Scholar]
- 33.Marshall, H. E., and J. S. Stamler. 2001. Inhibition of NF-κB by S-nitrosylation. Biochemistry 40:1688-1693. [DOI] [PubMed] [Google Scholar]
- 34.Meerovitch, K., S. Wing, and D. Goltzman. 1997. Preproparathyroid hormone-related protein, a secreted peptide, as a substrate for the ubiquitin proteolytic system. J. Biol. Chem. 22:6706-6713. [DOI] [PubMed] [Google Scholar]
- 35.Mikulits, W., M. Schranzhofer, H. Beug, and E. W. Müllner. 1999. Post-transcriptional control via iron-responsive elements: the impact of aberrations in hereditary disease. Mutat. Res. 437:219-230. [DOI] [PubMed] [Google Scholar]
- 36.Müllner, E. W., B. Neupert, and L. C. Kühn. 1989. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell 58:373-382. [DOI] [PubMed] [Google Scholar]
- 37.Padgett, C. M., and A. R. Whorton. 1995. S-Nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am. J. Physiol. 269:C739-C749. [DOI] [PubMed] [Google Scholar]
- 38.Pantopoulos, K., and M. W. Hentze. 1995. Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J. 14:2917-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponka, P., C. Beaumont, and D. R. Richardson. 1998. Function and regulation of transferrin and ferritin. Semin. Hematol. 35:35-54. [PubMed] [Google Scholar]
- 40.Richardson, D. R., V. Neumannova, E. Nagy, and P. Ponka. 1995. The effect of redox-related species of nitrogen monoxide on transferring and iron uptake and cellular proliferation of erythroleukemia (K562) cells. Blood 86:3211-3219. [PubMed] [Google Scholar]
- 41.Rouault, T. A., and R. Klausner. 1997. Regulation of iron metabolism in eukaryotes. Curr. Top. Cell Regul. 35:1-19. [DOI] [PubMed] [Google Scholar]
- 42.Stamler, J. S. 1994. Redox signaling nitrosylation and related target interactions of nitric oxide. Cell 78:1-19. [DOI] [PubMed] [Google Scholar]
- 43.Stamler, J. S., D. J. Singel, and J. Loscalzo. 1992. Biochemistry of nitric oxide and its redox-activated forms. Science 258:1898-1902. [DOI] [PubMed] [Google Scholar]
- 44.Tabuchi, A., K. Sano, E. Oh, T. Tsuchiya, and M. Tsuda. 1994. Modulation of AP-1 activity by nitric oxide (NO) in vitro: NO-mediated modulation of AP-1. FEBS Lett. 351:123-127. [DOI] [PubMed] [Google Scholar]
- 45.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 46.Weiss, G., B. Goossen, W. Doppler, D. Fuchs, K. Pantopoulos, G. Werner-Felmayer, H. Wachter, and M. W. Hentze. Translational regulation via iron-responsive elements by the nitric oxide/NO-synthase pathway. EMBO J. 12:3651-3657. [DOI] [PMC free article] [PubMed]
- 47.Yamanaka, K., H. Ishikawa, V. Megumi, F. Tokunaga, M. Kanie, T. A. Rouault, I. Morishima, N. Minato, K. Ishimori, and K. Iwai. 2003. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat. Cell Biol. 5:336-340. [DOI] [PubMed] [Google Scholar]