Abstract
The phytoplankton community in the oligotrophic open ocean is numerically dominated by the cyanobacterium Prochlorococcus, accounting for approximately half of all photosynthesis. In the illuminated euphotic zone where Prochlorococcus grows, reactive oxygen species are continuously generated via photochemical reactions with dissolved organic matter. However, Prochlorococcus genomes lack catalase and additional protective mechanisms common in other aerobes, and this genus is highly susceptible to oxidative damage from hydrogen peroxide (HOOH). In this study we showed that the extant microbial community plays a vital, previously unrecognized role in cross-protecting Prochlorococcus from oxidative damage in the surface mixed layer of the oligotrophic ocean. Microbes are the primary HOOH sink in marine systems, and in the absence of the microbial community, surface waters in the Atlantic and Pacific Ocean accumulated HOOH to concentrations that were lethal for Prochlorococcus cultures. In laboratory experiments with the marine heterotroph Alteromonas sp., serving as a proxy for the natural community of HOOH-degrading microbes, bacterial depletion of HOOH from the extracellular milieu prevented oxidative damage to the cell envelope and photosystems of co-cultured Prochlorococcus, and facilitated the growth of Prochlorococcus at ecologically-relevant cell concentrations. Curiously, the more recently evolved lineages of Prochlorococcus that exploit the surface mixed layer niche were also the most sensitive to HOOH. The genomic streamlining of these evolved lineages during adaptation to the high-light exposed upper euphotic zone thus appears to be coincident with an acquired dependency on the extant HOOH-consuming community. These results underscore the importance of (indirect) biotic interactions in establishing niche boundaries, and highlight the impacts that community-level responses to stress may have in the ecological and evolutionary outcomes for co-existing species.
Introduction
The open ocean is the largest biome on the surface of the earth, but due to its distance from coastal and deep ocean sediments, is also one of the most oligotrophic. Nutrient scarcity is especially prevalent in the surface mixed layers of highly stratified systems, with inputs of nutrients often restricted to new production (e.g. nitrogen fixation) or atmospheric (dust) deposition [1]. Microbes dominate biomass in this “wet desert” [2] and the most abundant phytoplankter in the tropics and subtropics (∼40°N to 40°S latitude) is the unicellular cyanobacterium, Prochlorococcus [3]. This oligotrophic specialist has the smallest cell size (0.4 to 1.2 µm in diameter) and genome (1.7–2.5×106 bp) [4], [5] of any known photoautotroph. The small cell size results in a superior surface to volume ratio that is believed to provide a key advantage in nutrient scavenging versus larger competitors [6]. The small genome size, together with a reliance on sulfo- rather than phospholipids [7], greatly diminishes its cellular P quota, providing an additional advantage over larger competitors. As a result of these and other adaptations, Prochlorococcus populations span the entire euphotic zone, often exceeding 105 cells mL−1, and due to this numerical dominance have been credited for roughly half of all photosynthesis in the oceans [8]–[12].
Genetically distinct ecotypes of Prochlorococcus partition the oligotrophic euphotic zone niche with respect to depth and latitude, in response to gradients of light and temperature, respectively. The upper euphotic zone is dominated by two closely-related ecotypes, eMED4 (“e” for ecotype, “MED4” for the type strain of the lineage) and eMIT9312, which are high-light adapted (HL), while the lower euphotic zone is dominated by low-light adapted (LL) ecotypes that include eNATL2A, eMIT9313, and eSS120 [13]–[17]. In seasonally-stratified regions of the subtropics, deep mixing events facilitate an invasion of the surface mixed layer by the LL ecotype eNATL2A [17], [18], which in certain instances may lead to numerical dominance of the LL ecotypes in the mixed layer [19]. The high irradiances found near the ocean surface restrict the growth of eNATL2A [17] as well as the other LL ecotypes [16] but eNATL2A appears unique amongst the LL ecotypes in its ability to survive temporary exposures to high light, such as would be experienced during deep vertical mixing events [18]. The two HL ecotypes further partition the upper euphotic zone by latitude: the eMIT9312 ecotype dominates the middle band from 30° N to 30° S latitude, while eMED4 dominates the higher latitudes at the extremes of Prochlorococcus' distribution [20]. This latitudinal niche partitioning is driven primarily by ocean temperature [17]–[21], and is consistent with the growth properties of cultured ecotype representatives [20].
The pattern of diversification within the Prochlorococcus lineage is consistent with an evolutionary progression from LL ancestors restricted to the deep euphotic zone, towards HL strains able to exploit the high light niche in the surface mixed layer. The eMIT9313 ecotype is the earliest branching lineage from the last common ancestor of Prochlorococcus and Synechococcus, and like all Prochlorococcus contains pigments that optimize the cells for the utilization of the blue light wavelengths that penetrate deepest into the euphotic zone [22]. The LL lineages are polyphyletic, with the eNATL2A ecotype the most recently derived. Based on reconstructions of Prochlorococcus evolution from molecular phylogenies along with ecological observations, the emergence of the eNATL2A lineage coincided with the ability of Prochlorococcus to invade the surface mixed layer, albeit only in instances of deep vertical mixing that minimize the lengths of exposure to high light [17]. Acquisition of a DNA photolyase and an elevated number of high light inducible proteins may have been responsible for this adaptation for high light tolerance [23]. With the emergence of the eMED4 and eMIT9312 “true” high light ecotypes – the most derived lineages – Prochlorococcus gained a sustained presence in the mixed layer, irrespective of mixed layer depth. The HL ecotypes have a common “core” of ∼100 genes not found in the LL ecotypes, and many of these may be responsible for the exploitation of high light near the surface [23].
In addition to the high light and low nutrient stresses associated with the oligotrophic surface mixed layer, the invasion of the mixed layer by Prochlorococcus may have also involved elevated oxidative stress. While hydrogen peroxide (HOOH) is ubiquitous in the ocean, the photooxidation of dissolved organic carbon (DOC) by sunlight [24], particularly by light in the UV range [25], results in near-surface HOOH maxima. Along with iron, HOOH is enriched in rainwater [26]–[28], and the combination of the two may cause periodic oxidative stress in marine organisms via the generation of highly reactive hydroxyl radicals (OH•) by the Fenton reaction [29], [30]. HOOH has been shown to inhibit the growth of diverse marine algae, including cyanobacteria, macro- and microscopic chlorophytes, diatoms, and coral-associated zooxanthellae [31]–[36], albeit at levels higher than those typically measured in pelagic waters.
In prior work, we demonstrated that Prochlorococcus was dependent on “helper” heterotrophic bacteria to thrive in dilute laboratory cultures [37]. Many heterotroph strains were capable of helping Prochlorococcus, including members of the α- and γ-Proteobacteria and the Bacteriodetes cluster, suggesting the mechanism(s) are common bacterial activities. In these experiments, the helping phenomenon occurred when Prochlorococcus and heterotrophic bacteria were inoculated at ecologically relevant concentrations, suggesting that the helping mechanism might also play an important role in natural communities. Preliminary evidence suggested the helper phenotype was associated with protection from HOOH, compensating for Prochlorococcus' conspicuously diminished suite of antioxidant genes (e.g. catalase) compared to other aerobes [4], [38]. In this study, we were driven by the following questions regarding the heterotroph/Prochlorococcus interactions: is the removal of HOOH from the medium necessary and sufficient for the helping phenotype? Does this mechanism of protection have relevance to natural communities in the open ocean? How universal is the dependency on helpers for the growth of the Prochlorococcus lineages? Finally, what does the apparent loss of endogenous HOOH-protection mechanisms imply regarding the genomic streamlining of the Prochlorococcus genus?
Results
HOOH removal is necessary and sufficient for the helping phenotype
From our prior work, we hypothesized that heterotrophs help Prochlorococcus by the removal of HOOH from the medium [37]. To test this hypothesis we began by analyzing the impacts of Prochlorococcus and heterotrophic growth on the medium chemistry. Standard Pro99 medium prepared with filtered, autoclaved seawater was found to contain approximately 0.2±0.02 µM HOOH. Axenic cultures of Prochlorococcus UH18301 (a novel isolate of the eMIT9312 ecotype, Table S1) depleted medium HOOH either very slowly or not at all, depending on the initial inoculum (Figure S1). Compared to these axenic controls, however, co-cultures with the “helper” strain Alteromonas sp. EZ55 rapidly depleted this HOOH to below detection (<0.01 µM) (Figure S1). In contrast, pH, dissolved oxygen, bicarbonate, and dissolved organic carbon were not significantly different in axenic versus mixed cultures of Prochlorococcus during the first week of culture (t-tests for individual data points, df = 4, p>0.05, Figure S2). While CO2 concentration was different in mixed versus axenic Prochlorococcus cultures, this potential carbon source for Prochlorococcus was less abundant in co-cultures as a consequence of slightly elevated pH (Figure S2, C and E). While EZ55 on its own had the predicted effect of lowering O2 concentrations, and raising inorganic carbon concentrations, this change was relatively minor, short-lived, and not reflected in a difference between axenic UH18301 and co-cultures with EZ55 (Figure S2). Thus, of all the potential means by which helpers may benefit Prochlorococcus through their modification of the bulk medium, whether by adding a nutrient or removing a growth inhibitor, the removal of HOOH was the only one consistent with a beneficial effect for Prochlorococcus.
All confirmed helpers are catalase-positive [37], but the helping phenotype could not be unambiguously assigned to the catalase activity of these bacteria. While insertional inactivation of catalase in Alteromonas sp. EZ55, Vibrio fischeri ES114, and Silicibacter lacuscaerulensis ITI-1157 resulted in the loss of ability to help Prochlorococcus growth on agar media ([37], and data not shown), the results in liquid media were less clear. Catalase mutants of EZ55 and ES114 were still capable of helping dilute liquid cultures of Prochlorococcus, but they also showed negligible loss of HOOH scavenging ability in liquid (Table S2). Redundant HOOH scavenging pathways are common in aerobic heterotrophs [39], [40], and these are likely to maintain the heterotrophs in a help-competent state, at least for experiments in liquid (semisolid agar media may have higher HOOH levels that do require active catalase). In contrast, the S. lacuscaerulensis catalase mutant was unable to help Prochlorococcus in liquid, but by an unknown mechanism actually raised HOOH levels in culture media relative to sterile controls (Table S2). Studies with purified bovine liver catalase likewise provided ambiguous results. While catalase provided some benefit to UH18301 in comparison to untreated, axenic controls, it was clearly inferior in comparison to co-culture with bacterial helpers (Table S2). Catalase is photo-inactivated ([41] and Figure S3), and heat-killed catalase resulted in higher Prochlorococcus mortality than unamended controls (Table S2). Thus, light-driven degradation of catalase to toxic end products may account for these ambiguous results.
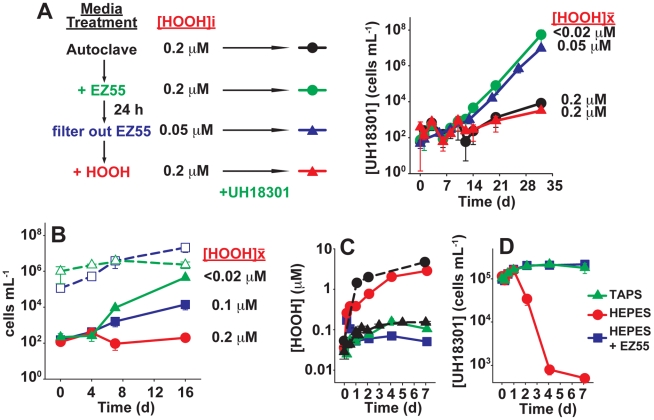
Because complications obscured the results of the catalase studies, we confirmed the role of HOOH scavenging in the helping phenotype by another line of experimentation, which was to manipulate the HOOH and heterotroph activities in media prior to inoculation with Prochlorococcus. Dilute (∼100 cells mL−1) axenic Prochlorococcus UH18301 was unable to lower the initial HOOH of the autoclaved medium, resulting in a mean daily exposure (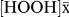 ) of 0.2 µM, and consequently exhibited very poor growth over this period (Figure 1A). In contrast, pre-inoculation of the medium with the EZ55 helper strain resulted in robust growth of UH18301. This significant growth improvement was associated with a much lower
) of 0.2 µM, and consequently exhibited very poor growth over this period (Figure 1A). In contrast, pre-inoculation of the medium with the EZ55 helper strain resulted in robust growth of UH18301. This significant growth improvement was associated with a much lower 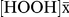 (<0.02 µM) as a result of the activity of EZ55. The activity of EZ55 was sufficient to pre-condition the medium for Prochlorococcus growth, as evidenced by the robust UH18301 growth in media pre-incubated with EZ55 for 24 hours, followed by removal of the EZ55 cells via filtration prior to inoculation of the Prochlorococcus cells. As for the co-culture treatment, this pre-conditioning treatment also resulted in a significantly lower [HOOH]
(<0.02 µM) as a result of the activity of EZ55. The activity of EZ55 was sufficient to pre-condition the medium for Prochlorococcus growth, as evidenced by the robust UH18301 growth in media pre-incubated with EZ55 for 24 hours, followed by removal of the EZ55 cells via filtration prior to inoculation of the Prochlorococcus cells. As for the co-culture treatment, this pre-conditioning treatment also resulted in a significantly lower [HOOH]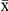 for Prochlorococcus. Filtration of the EZ55 did not appear to release intracellular components into the medium, as no detectable hydroperoxidase or alkaline phosphatase activity remained in the filtrates, in contrast to the unfiltered cultures themselves (Figure S4). Finally, if HOOH was added back to the EZ55-conditioned medium, restoring the HOOH concentration to 0.2±0.02 µM, the enhanced growth effect of the EZ55 preconditioning step on Prochlorococcus growth was lost (Figure 1A). This indicates that any activity of EZ55 other than the removal of HOOH is not sufficient to explain the helping phenotype of these cells.
for Prochlorococcus. Filtration of the EZ55 did not appear to release intracellular components into the medium, as no detectable hydroperoxidase or alkaline phosphatase activity remained in the filtrates, in contrast to the unfiltered cultures themselves (Figure S4). Finally, if HOOH was added back to the EZ55-conditioned medium, restoring the HOOH concentration to 0.2±0.02 µM, the enhanced growth effect of the EZ55 preconditioning step on Prochlorococcus growth was lost (Figure 1A). This indicates that any activity of EZ55 other than the removal of HOOH is not sufficient to explain the helping phenotype of these cells.
Figure 1. Removal of HOOH from media facilitates Prochlorococcus growth.
A) Effects of pre-treatment of Pro99 medium on growth of dilute (initially ≈100 cells mL−1) axenic Prochlorococcus UH18301 cultures grown under low light. Prior to inoculation of UH18301, medium was autoclaved (black circles), pre-inoculated with 106 cells ml−1 EZ55 (green circles), pre-incubated for 24 hours followed by removal of EZ55 via filtration (blue triangles), and then supplemented with HOOH to restore its initial concentration (red triangles). [HOOH]i, HOOH concentration at the time of inoculation; [HOOH]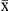 , mean daily HOOH exposure, obtained by integrating [HOOH] over the first 7 d and dividing by the elapsed time. B) Growth kinetics of dilute Prochlorococcus VOL1 under high light in axenic cultures (red circles) and in co-culture with more concentrated Prochlorococcus VOL7 (solid blue squares) or EZ55 (solid green triangles). Also shown are the kinetics of the co-cultured VOL7 (open blue squares, dashed line) and EZ55 (open green triangles, dashed line). Error bars are the standard errors of 3 biological replicates. C) HOOH accumulation in high light-exposed UH18301-inoculated seawater containing 3.75 mM HEPES either with (blue squares) or without (red circles) EZ55, compared to an axenic control grown in media with 3.75 mM of the non-HOOH producing buffer TAPS (green triangles). Dashed lines, sterile media containing HEPES (black circles) or TAPS (black triangles). D) Cell concentrations over time in buffer-treated cultures. Symbols represent the same cultures as in 1C.
, mean daily HOOH exposure, obtained by integrating [HOOH] over the first 7 d and dividing by the elapsed time. B) Growth kinetics of dilute Prochlorococcus VOL1 under high light in axenic cultures (red circles) and in co-culture with more concentrated Prochlorococcus VOL7 (solid blue squares) or EZ55 (solid green triangles). Also shown are the kinetics of the co-cultured VOL7 (open blue squares, dashed line) and EZ55 (open green triangles, dashed line). Error bars are the standard errors of 3 biological replicates. C) HOOH accumulation in high light-exposed UH18301-inoculated seawater containing 3.75 mM HEPES either with (blue squares) or without (red circles) EZ55, compared to an axenic control grown in media with 3.75 mM of the non-HOOH producing buffer TAPS (green triangles). Dashed lines, sterile media containing HEPES (black circles) or TAPS (black triangles). D) Cell concentrations over time in buffer-treated cultures. Symbols represent the same cultures as in 1C.
The ability of axenic Prochlorococcus to grow in concentrated but not dilute cultures [37] suggests that Prochlorococcus can help itself in a density-dependent manner. To confirm this, we conducted co-culture experiments between two strains of Prochlorococcus distinguishable by quantitative PCR. Axenic VOL1 (a streptomycin-resistant [SmR] mutant of MIT9215, Table S1) had a minimal influence on the medium [HOOH] and was incapable of growth from 100 cells mL−1 (Figure 1B and [37]). However, when co-cultured with 105 cells mL−1 of VOL7 (a SmR mutant of MED4, Table S1), VOL1 survived and grew at approximately the same rate as VOL7 (Figure 1B). VOL1 grew slower in co-culture with VOL7 compared to the heterotroph, EZ55, and consistently, the ability of VOL7 to remove HOOH from the medium was also substantially lower than EZ55 (Figure 1B). These observations support the hypothesis that Prochlorococcus is a poor scavenger of exogenous HOOH, but by mass-action at high densities can draw down HOOH to sub-lethal levels.
The experiments described above involved exposing dilute Prochlorococcus cultures to the elevated HOOH of autoclaved media. However, the poor HOOH scavenging capacity of Prochlorococcus and its dependency on help is also apparent when ecologically-relevant concentrations of cells (∼105 cells mL−1) are challenged with HOOH that accumulates gradually in the medium. To avoid potential influences of the concentrated Fe, N and P in growth media, we performed these experiments in sterile unamended seawater. In the presence of light, HEPES buffer generates HOOH [42], [43], and under high light (∼250 µmol quanta m−2 s−1), the rate of HOOH generation in sterile seawater containing 3.75 mM HEPES was approximately 0.4 µM d−1 (Figure 1C). Axenic Prochlorococcus UH18301 was unable to prevent HOOH accumulation to lethal levels (Figure 1C), resulting in a rapid decline in cell counts (Figure 1D). The same outcome was observed even when the HEPES concentration was lowered to reduce the rate of HOOH production by 75% (Figure S5). However, if the Prochlorococcus cultures also contained EZ55, or if the HEPES buffer was substituted with the buffer TAPS, which generates much less HOOH (Figure 1C), HOOH concentrations were maintained at or below 0.1 µM, and subsequently Prochlorococcus cells survived, doubling about once before running out of nutrients in this unamended seawater medium (Figure 1D).
Ecologically relevant levels of HOOH inhibit axenic Prochlorococcus
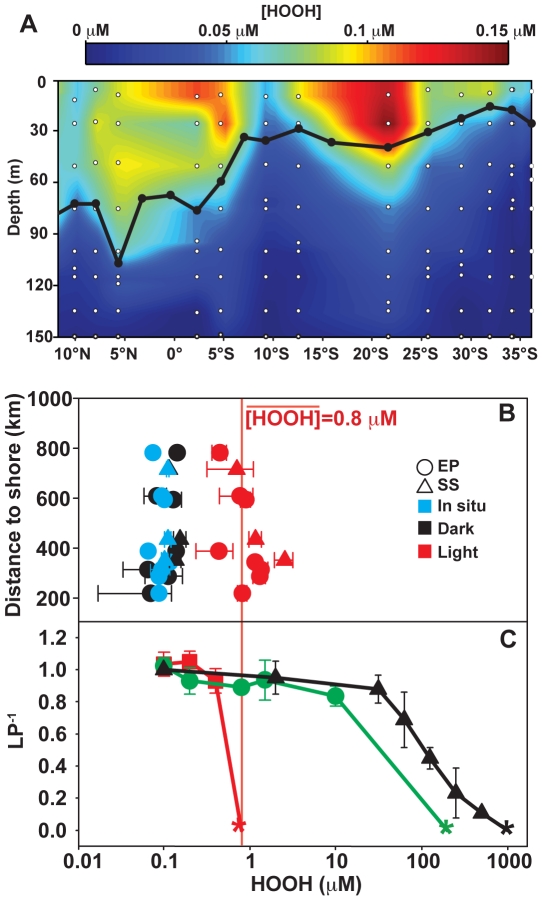
For the experiments described above, initial HOOH concentrations were between ∼0–0.2 µM. Importantly, this is well within the range observed in situ in the ocean euphotic zone [26], [44]–[49], and as these experiments also involved ecologically-relevant concentrations of Prochlorococcus and helper, our results may reflect important interactions that occur in natural communities. The primary sources of HOOH in the ocean are rainfall events [26]–[28], [32] and photochemical (especially UV) degradation of dissolved organic carbon [25], [50], [51]. Accordingly, the highest concentrations of HOOH in the oceans are in the upper euphotic zone, which we confirmed in a 2007 meridional transect in the Pacific Ocean (Figure 2A and Table S3). With a few exceptions, the concentration of HOOH in the surface mixed layer ranged from 0.075–0.15 µM, whereas the concentration rapidly dropped below the mixed layer and fell below the limit of detection (<0.01 µM). From these measurements we conclude that the greatest threat of HOOH-mediated oxidative damage for Prochlorococcus is within the mixed layer.
Figure 2. Removal of HOOH is necessary for Prochlorococcus survival at the ocean's surface.
A) [HOOH] in the euphotic zone along a transect from Hawai'i to Australia, Jan-Feb 2007. Sampled depths (open circles) used in the linear interpolation of HOOH, and surface mixed layer depth (black line) are reported. B) Effect of solar irradiation on [HOOH] of seawater. “Light” and “Dark” indicate HOOH concentrations after 24 h incubations as described in Methods. Vertical line represents the mean [HOOH] in light incubations, represented by the SMC value as described in the text. EP, Eastern Pacific stations; SS, Sargasso Sea stations. C) Effect of HOOH exposure on the long-term growth of Prochlorococcus UH18301 cultures. Lag phase duration, expressed as the inverse ratio of each sample to a no added HOOH control (LP−1) as a function of HOOH dosage with (green circles) or without (red squares) EZ55. Black triangles, LP−1 of EZ55 in YT3 medium; *, all replicate cultures failed to show detectable growth after 60 d. Error bars are the standard error of three biological replicates. Cultures were grown under low light with an initial inoculum of ∼105 cells mL−1.
HOOH concentrations in the ocean are instantaneous measurements that reflect competing rates of production and removal. While sunlight is the primary source of HOOH, the primary sink is the microflora [52], [53]. To investigate the role that the extant microbial community plays in maintaining HOOH concentrations at the surface within the tolerable range for Prochlorococcus, we examined the effect that removal of this primary sink had on the local concentrations of HOOH. In total, 11 Pacific and Atlantic open ocean stations were chosen for analysis (Table S3), each with mixed layer HOOH concentrations of ∼0.1 µM (Figure 2B) and Prochlorococcus present in typical abundance (104–105 cells mL−1, data not shown). Extant microbiota were removed from surface seawater via 0.2 µM filtration, and samples were assessed for gross HOOH abiotic production in the absence of the sink. After 24 hours incubation, dark controls were not significantly different than the in situ values (paired t-test, p>0.05), whereas the experimental samples incubated at approximately 80% of surface intensity sunlight (visible + UV), simulating exposure at 5 m depth, showed dramatic increases in HOOH (Figure 2B). This maximal concentration varied from station to station but had a mean of 0.8 µM±0.6 µM. In the experiments that follow we refer to this HOOH concentration as the surface monoculture (SMC) challenge, as it represents the hypothetical situation where Prochlorococcus is exposed to naturally generated HOOH in the surface mixed layer in the absence of HOOH-removing helpers.
In the absence of helpers, the SMC concentration of HOOH was lethal to all three of the mixed layer ecotypes. While concentrations of HOOH within the range of observed field values (<0.2 µM) had minimal impact on axenic cultures at ecologically relevant cell concentrations of 105 cells mL−1, exposure to the SMC concentration of 0.8 µM HOOH was lethal to representative strains of the eMIT9312 (UH18301 and VOL1), eMED4 (VOL7) and eNATL2A (VOL3) ecotypes (Figure 2C and Figure S6). This lethality occurred for all these strains under both high (250 µmol quanta m−2 s−1, data not shown) and low (40 µmol quanta m−2 s−1, Figure 2C and Figure S6) light conditions. In contrast, when co-cultured with ecologically relevant concentrations of EZ55 (106 cells mL−1) these strains were able to tolerate HOOH exposures at least an order of magnitude higher than the SMC value (Figure 2C and Figure S6). Indeed, the resistance of these strains in co-cultures approached that of the EZ55 helpers themselves (Figure 2C and Figure S6), well beyond any naturally occurring HOOH concentration observed in the open ocean.
HOOH stress results in catastrophic loss of cell integrity and photosynthetic capacity
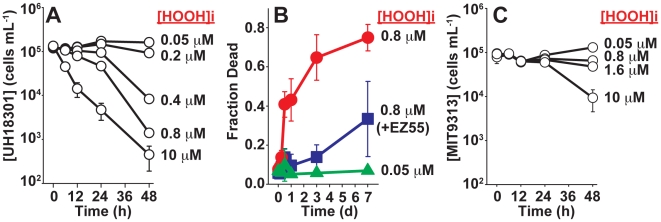
The lethal effects of the SMC challenge were further characterized for axenic UH18301 in unamended seawater. Cell concentrations (detected by flow cytometry) remained constant or increased slightly during 48 h exposure to HOOH dosages up to 0.2 µM, but exposures of 0.4 µM and greater resulted in significant loss of cells (Figure 3A). At the SMC concentration of 0.8 µM, >99% of the initial UH18301 cell population was undetectable after 48 h. Moreover, as time progressed, an increasing proportion of the remaining cells stained positive for the membrane-impermeable vital stain Sytox Green (Figure 3B), strongly suggesting that cell loss is due to lysis. Sytox Green uptake was greatly attenuated in co-cultures with EZ55 (Figure 3B), suggesting that helpers protect Prochlorococcus against HOOH-mediated membrane permeabilization.
Figure 3. Prochlorococcus cell viability during HOOH exposure in unamended sterile seawater under low light.
A) Change in cell concentration following exposure to the indicated [HOOH]i for axenic UH18301 in low light. B) Proportion of Sytox Green-positive (i.e. dead) UH18301 cells following exposure to the indicated [HOOH]i. Green triangles, axenic UH18301 with no added HOOH; red circles, axenic UH18301 with added HOOH; blue squares, co-culture of UH18301 with EZ55 and added HOOH. C) As in A, but for MIT9313.
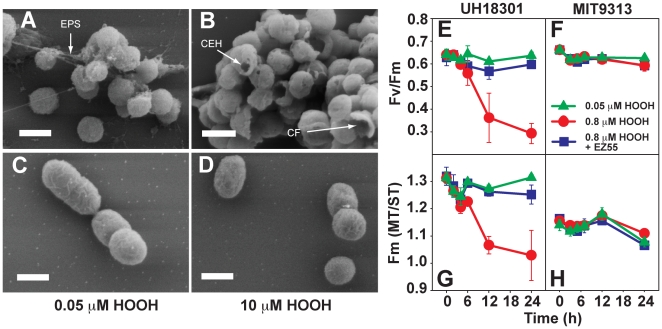
Consistent with the Sytox Green results, scanning electron microscopy revealed pronounced cell envelope damage in HOOH-stressed Prochlorococcus cells. Untreated (≤0.05 µM HOOH) axenic UH18301 cells had the typical spherical morphology of HL Prochlorococcus strains, and this strain appeared to produce an extracellular matrix (Figure 4A). Damaged cells were rare, and showed signs of lysis due to osmotic stress during the fixation process. In contrast, after 24 h of exposure to HOOH, UH18301 exhibited profound envelope damage (Figure 4B). While cell size and basic shape were unchanged, cell population density was greatly diminished in these samples (e.g. Figure 3A), and no matrix of any kind was visible. Many of the intact cells had holes in their membranes, and partial fragments of cell envelopes were also observed (Figure 4B). These images strongly suggest that the decline in cell concentration upon HOOH exposure (Figure 3A) is due to a catastrophic loss of cell integrity, such that the cells lose the light-scattering and chlorophyll-based fluorescence properties used in their detection.
Figure 4. Cell envelope and photophysiological effects of HOOH on Prochlorococcus grown under low light.
SEM of UH18301 (A and B) and MIT9313 (C and D) after 24 h exposure to 0.05 µM (A and C) or 10 µM (B and D) HOOH. All scale bars = 500 nm. EPS, extracellular polymeric substances; CEH, cell envelope hole; CF, cell fragment. Cells were exposed to ∼10 fold higher HOOH to compensate for the ∼100 fold higher cell concentrations necessary to recover sufficient biomass for imaging. Photophysiological parameters Fv/Fm and Fm(MT/ST) (see text) of Prochlorococcus UH18301 (E and G) and MIT9313 (F and H) were also measured during 24 h exposure to the indicated [HOOH] in sterile unamended seawater.
Concurrent with the decline in cell concentration, Prochlorococcus cells exposed to the SMC concentration of HOOH also suffered in their capacity for photosynthesis. Fv/Fm, a measure of photosynthetic efficiency and health, declined steadily in UH18301 cultures during 24 h of HOOH exposure, but was rescued by the presence of EZ55 helpers (Figure 4E and Figure S7). Similar results were observed for another eMIT9312 strain (VOL1), as well as representative eMED4 (VOL7) and eNATL2A (VOL3) strains, under high or low light conditions (Figure S7). Fv/Fm of control cultures (no added HOOH) was also significantly lower in cells grown under high light than under low (Figure S7). While the decrease in Fv/Fm resulting from elevated HOOH was largely eliminated by the presence of helpers, the decrease resulting from elevated light levels was not (Figure S7), which may indicate that distinct mechanisms of inhibition are at play, with the high-light associated mechanism not involving a diffusible reactive oxygen intermediary that can be removed by helpers.
The FIRe protocol used in these studies measures two distinct types of variable fluorescence. Fm(ST) is obtained using a rapid series of short flashlets to induce a single turnover of the PSII pool, whereas Fm(MT) is measured during a much longer series of flashlets, during which multiple turnovers of the system completely reduce the plastoquinone (PQ) pool. The ratio of these two values, Fm(MT/ST), thus allows an estimate of the ratio of PQ and/or other downstream components of the photosynthetic electron transport chain to photosystem II reaction centers (RCIIs) [54]. Fm(MT/ST) was significantly greater in UH18301 cultures grown in low light than in high light (Figure 4G), suggesting that under low light conditions, PQ availability increases, perhaps to maximize the efficiency of harnessing of scarce excitation energy. Following HOOH exposure, however, Fm(MT/ST) of low light acclimated cultures dropped to the level of high light acclimated cultures (Figure 4G and Figure S7). The target of this HOOH-mediated oxidative damage is not known, but whether it is the RCII itself and/or some downstream element (PQ, cytochrome b6f, etc.), this damage appears to result in diminished PQ re-oxidation rates and depression of Fm(MT) in relation to Fm(ST). Like Fv/Fm, however, the HOOH-mediated decline in Fm(MT/ST) is suppressed in co-cultures with EZ55 (Figure 4A). Like UH18301, all other strains assayed that are able to grow at high light (i.e., all except MIT9313) had higher Fm(MT/ST) in low light conditions. Interestingly, HOOH exposure eliminated this difference in all strains except VOL7 (Figure S7), suggesting that this strain may have unique, currently unknown, protective mechanisms.
Phylogenetic distribution of helper dependency
Interestingly, while the eMIT9313 ecotype has never been observed at high abundance in the surface mixed layer [17] - most likely due to its sensitivity to high light [16] - strain MIT9313 was significantly more resistant to HOOH than strains of the three ecotypes (eMIT9312, eMED4, and eNATL2A) that are found in the HOOH-enriched surface mixed layer. MIT9313 was uniquely capable of growth at the SMC HOOH concentration (Figure S6), and this concentration had minimal impact on photosynthetic efficiency (Figure 4F, 4H, and Figure S3) or cell integrity (Figure 3C and Figure 4C–D). Importantly, none of the strains tested were able to significantly lower the HOOH concentration of the medium within the time frame of the photophysiology experiments (24 h, Figure S1), suggesting that the greater resistance of MIT9313 relative to the other strains is not achieved via higher rates of HOOH scavenging.
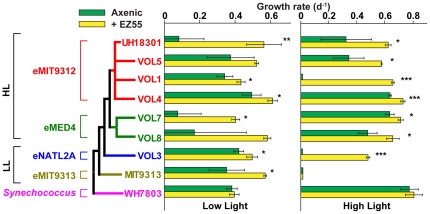
The results with the SMC HOOH concentrations suggested that the genetically distinct ecotypes may have unique responses to oxidative stress. To explore this possibility at a finer scale, we compared their growth properties at a HOOH concentration permissive to all strains, and under a high and low light intensity that may be experienced within the surface mixed layer. Altogether, eight axenic strains of Prochlorococcus representing two HL and two LL ecotypes, as well as one strain of marine Synechococcus were tested. Invariably, axenic Prochlorococcus cultures inoculated at 100 cells mL−1 into autoclaved Pro99 (containing ∼0.2 µM HOOH) grew poorer relative to co-cultures with EZ55, with higher variability in growth rate (Figure 5) and lag time (data not shown) between replicates. In contrast, there was no difference in the growth rate or lag of Synechococcus WH7803 in the presence or absence of EZ55. Light intensity had variable effects on the EZ55 helping phenotype for the Prochlorococcus strains (Figure 5). Amongst the HL ecotypes, eMED4 strains VOL7 and VOL8 were more reliant on help in low light, whereas the members of the eMIT9312 ecotype did not share a single light preference: UH18301 was less dependent on help in high light, while VOL1 was incapable of axenic growth at this light intensity. The LL eNATL2A strain VOL3 was incapable of axenic growth under high light, whereas growth under high light was permissible in the presence of EZ55. While it exhibited relatively high HOOH resistance in low light (Figure 3C), the LL strain MIT9313 exhibited no growth after 30 days in high light from an inoculum of 100 cells mL−1, with or without EZ55, supporting our assertion in the prior section that photoinhibition of Prochlorococcus [16] includes a mechanism that is independent of diffusible HOOH.
Figure 5. Growth at 0.2 µM HOOH as a function of helpers and light intensity for cultures with very dilute (∼100 cells mL−1) initial cell concentrations.
Growth rates for strains representing 4 Prochlorococcus ecotypes and a marine Synechococcus ecotype with (yellow bars) or without (green bars) EZ55 helpers in low or high light (40 or 250 µmol quanta m−2 s−1, respectively). Consensus (100% support in 1000 bootstrap replicates) neighbor-joining tree was prepared using the entire rDNA operon (≈5.5 kb) and rooted on the Synechococcus branch. Only topology is shown; branch lengths do not represent genetic distance. Significance levels of t-tests (df = 4): *, P<0.05; **, P<0.01; ***, P<0.001. HL, high light adapted ecotypes; LL, low light adapted ecotypes.
Discussion
HOOH removal is the primary mechanism responsible for the helper phenotype
In this work we have confirmed our initial report [37] that heterotrophic bacteria help dilute Prochlorococcus cultures grow via removal of HOOH from the environment. HOOH diffuses freely across cellular membranes [55], and hence the heterotrophic HOOH degradation could occur in the cytoplasm, periplasm, or extracellular milieu, perhaps with the same overall effect. Density-dependence of HOOH resistance has also been observed in Escherichia coli, and catalase-positive E. coli has been shown to cross-protect more vulnerable catalase-negative mutants in co-culture [56]. Inter-species cross-protection from oxidative stress has been noted for heterotrophic bacterial communities involved in aromatic degradation [57] and in oral biofilms [58], as well as in a benthic mat-forming cyanobacterium [59], but to our knowledge this is the first demonstration that such helping can occur between planktonic microbes of different trophic levels.
While HOOH removal is sufficient to explain the helping phenotype of heterotrophic bacteria (at least for the EZ55 strain), other types of interactions may also contribute. For instance, heterotrophs can help eukaryotic algae by improving carbon fixation, either via increasing DIC concentrations [60] or lowering oxygen concentrations (thus diminishing the competing oxygenase reaction of Rubisco [61]). However, the addition of heterotrophic helpers to the Prochlorococcus cultures either led to no observable difference in bulk bicarbonate, CO2, or oxygen, or a change in the opposite direction to that expected for a beneficial role (Figure S2). In other cases heterotrophs help by supplying an essential metabolite (e.g. vitamin B12 [62] or indole acetic acid [63]) or trace nutrient (e.g., by producing iron-binding siderophores [64]). While we cannot rule out involvement of such cross-feeding interactions, our results suggest that any nutritional mechanism must play a secondary role relative to oxidative stress reduction, as adding HOOH back to helper treated media was alone sufficient to restore the growth limitation of dilute Prochlorococcus cultures (Figure 1A). Other than inorganic C, N, P, and metals, Prochlorococcus has no known nutritional requirements for growth [65], [66], and in fact grows better when dense versus dilute, contrary to expectations for an organism with a nutritional deficiency. Additionally, that a dense strain of Prochlorococcus can help a dilute strain (Figure 1B) argues against a requirement for a nutrient that Prochlorococcus itself cannot produce from the medium.
With the exception of MIT9313, exposure of all axenic cultures of Prochlorococcus to SMC levels of HOOH results in catastrophic loss of cell envelope integrity (Figure 3B and Figure 4B). Concomitant with this envelope damage is the loss of photosynthetic efficiency (Figure 4E,G), and both of these effects may be consequences of lipid peroxidation. The slow decline of HOOH in sterile, light exposed media (Figure S1) may be the result of the Fenton reaction, where photochemically produced Fe(II) reacts with HOOH to produce highly reactive OH• [67]. In turn, OH• can attack polyunsaturated fatty acids such as those found in most biological membranes, producing relatively stable lipid peroxide radicals that can spread via radical propagation if not countered by antioxidant defenses [55]. Both the cytoplasmic and the photosynthetic membranes may be susceptible to lipid peroxidation, and this may account for the coincident loss of cell envelope integrity and photosynthetic capacity we observed. Of note, dependence of HOOH-induced lethality on free Fe(II) could explain the greater resistance of MIT9313 to SMC challenge, as this is the only strain of Prochlorococcus to express the Fe-binding dpsA gene [4] which is linked to oxidative stress tolerance in other organisms [68]. Interestingly, the Synechococcus PCC7942 DpsA has a weak catalase activity [69] and if similar, the DpsA homolog of MIT9313 may be responsible for the cell's ability to eventually deplete the HOOH after 2 weeks under the SMC treatment (data not shown).
HOOH may also affect photosynthesis by impinging on the turnover of RCII protein D1. D1 proteins are continuously degraded in illuminated photosynthetic membranes, and cells must repair or replace these proteins to maintain photosynthetic activity [70]. In the freshwater cyanobacteria Synechococcus PCC7942 and Synechocystis PCC6803, exogenous HOOH inhibits protein synthesis by specific inactivation of elongation factor G [71], leading to net loss of photosystems containing functional D1. Hence, two non-mutually-exclusive mechanisms – lipid peroxidation and interruption of D1 turnover – may be responsible for the HOOH-dependent loss of photosynthetic efficiency in Prochlorococcus, and future studies should be aimed at testing these hypotheses.
Impact of helpers on Prochlorococcus ecology
Our results suggest that Prochlorococcus depends on the HOOH-degrading members of the microbial community to grow in the surface mixed layer of the open ocean. Opposing photochemical production reactions [24], [25] and microbial degradation reactions [52], [53] maintain the HOOH concentration within the permissive range (<0.2 µM) for the three ecotypes of Prochlorococcus that exploit the mixed layer niche. Absent the mixed layer microbial community, photochemical production of HOOH yields concentrations that are lethal to all three ecotypes (Figure 2B–C). The surface monoculture (SMC) challenge (Figure 3 and Figure S6) demonstrated that, absent a counteracting microbial community, HOOH in surface seawater from the open ocean climbs to concentrations that ecologically relevant cell concentrations of these ecotypes are unable to survive. Curiously, the one strain that can tolerate the SMC challenge, MIT9313, belongs to an ecotype that is restricted from the mixed layer [17], probably due to its sensitivity to high light [16]. While the SMC challenge reflects a worst case scenario for un-helped Prochlorococcus (e.g. sustained presence within 5 m of the surface), we note that even modest increases of HOOH above the steady state mixed layer concentrations (e.g. 0.2 µM, Figure 2A) are enough to significantly impact growth (Figure 1, Figure 3A, and Figure 5), suggesting that the entire Prochlorococcus mixed layer population, not just cells at the very surface, benefit from the activity of helpers. Hence, an important but previously unrecognized role of the microbial community is to make the surface mixed layer permissive for the growth of Prochlorococcus, and by doing so, facilitate the expansion of its habitat range. Whether this relationship between Prochlorococcus and its HOOH-consuming neighbors is commensal or mutualistic remains to be determined, but as the numerically-dominant primary producer in the surface mixed layer, it is certainly conceivable that the HOOH consumers may benefit indirectly through the release of organic carbon during the growth [72] or lysis of Prochlorococcus.
Our study contributes to a growing body of evidence that interspecies interactions can contribute significantly to niche expansion via stress reduction. For instance, land plants in high altitude alpine sites subjected to high abiotic stress (e.g., low moisture, low temperature, strong winds) experience pronounced losses in productivity as well as increased mortality in the absence of other members of the plant community, whereas this helping effect does not occur in otherwise similar communities in less stressful, lower altitude habitats [73]. Likewise, lichenized algae and fungi gain vastly improved resistance to desiccation and oxidative stress in symbiosis, expanding the range of each to include subaerial habitats [74]. Similarly, cnidarians (e.g., corals) and zooxanthellae tolerate temperature and hypoxia extremes together that neither can tolerate separately [75]. Collectively, these studies support the broader “stress-gradient hypothesis” that cooperation should be stronger in more stressful environments [76], and emphasize the need to assess stress responses at the community level in order to understand their impacts on biological distributions in nature.
The microbial community in the surface mixed layer is genetically diverse [2], and identifying the microbes contributing most significantly to HOOH degradation is a major challenge for future studies. We know that Prochlorococcus degrades HOOH poorly (Figure S1) and thus is not likely to contribute significantly to HOOH decomposition in the ocean. Intriguingly, the same may be true for the most abundant heterotroph in the oligotrophic ocean as well, as the genome of Pelagibacter ubique, the first cultured representative of the SAR11 cluster, also lacks homologs of catalase and other antioxidant defenses and [77]. The lack of robust HOOH scavenging pathways in both the numerically dominant autotroph and heterotroph implies that this critical function may be performed by less abundant “keystone” species in the mixed layer. Given the high catalytic rate of catalase [78] and the efficiency of HOOH removal observed in culture (e.g. Figure S1), it is conceivable that a relatively small number of catalase-expressing organisms could protect the entire surface mixed layer community from solar-generated HOOH. We note that many of the confirmed helpers of Prochlorococcus [37] are catalase-positive members of the alpha and gamma proteobacteria whose genetic signatures appear at reasonable frequencies in the open ocean mixed layer [2], [79], and are thus potential candidates for these keystone microbes. However, there are clearly other biological mechanisms for removing HOOH (e.g., the residual HOOH scavenging abilities of catalase mutants, Table S2), any one of which may contribute to the helper phenotype of surface mixed layer communities.
Evolutionary implications of the helper-Prochlorococcus interaction
Our results present an apparent paradox regarding the evolutionary history of the Prochlorococcus lineage: the eMIT9313 ecotype, which is restricted from the HOOH-enriched mixed layer by its sensitivity to high light, is more resistant to HOOH than the ecotypes found in high abundance in the mixed layer. eMIT9313 is the earliest branching lineage from the last common ancestor of Prochlorococcus and Synechococcus [23], [80], and although it lacks catalase, may share other ROS defense mechanisms with Synechococcus (e.g. dpsA). Indirect defense mechanisms may also be involved in eMIT9313. For example the peptidoglycan synthesis genes of MIT9313 are more similar to those of Synechococcus than those of the HL strain MED4, and the MIT9313 cell wall is intermediate in thickness between those of Synechococcus and MED4 [81]. A thicker cell wall may confer enhanced resistance by limiting HOOH diffusion into cells and/or preventing cell lysis (e.g., Figure 4B versus Figure 4D). It is not clear why eMIT9313 has retained the relatively high resistance to HOOH; perhaps the genes conferring resistance are also involved in other cellular functions that remain under selection. However, what is clear is that for the more recently derived lineages, including the LL ecotype eNATL2A, the net genomic reduction that has occurred relative to eMIT9313 [23] has coincided with a loss of HOOH resistance. Thus, the evolution of the genus leading to tolerance of temporary exposures to high light (eNATL2A) and true high light adaptation (eMED4 and eMIT9312) that allowed this lineage to exploit the surface mixed layer habitat did not coincide with a greater resistance to HOOH; in fact, the opposite occurred.
We believe that this paradox is resolved because the HOOH-consuming microbes present at the ocean's surface made the HOOH resistance genes in Prochlorococcus dispensable. Prochlorococcus evolved in the context of an extant HOOH-scavenging community, and thus as it developed tolerance to high light and possibly other environmental stresses in the surface mixed layer, it had no selection pressure to maintain the high level of resistance to HOOH. As the oligotrophic environment imposed pressure to reduce genome size [5], [22], [23], [82], [83], genes encoding these resistance mechanisms were amongst the pool of expendable genes, and were eventually lost. Similar mechanisms may have been at play during the genome streamlining of the ocean's most abundant heterotroph, P. ubique [84], although it is unknown as yet whether or not this organism benefits from co-culture with helpers. It is intriguing to note that both Prochlorococcus and P. ubique are conspicuously difficult to cultivate, leading to speculation that other, as-yet-uncultured organisms may be similarly dependant on helpers in nature (e.g., [64]). Hence, this study emphasizes the importance of community-level stress responses not only for the ecology, but also the evolutionary history of free-living microbes.
Finally, we note that while the catalase gene could have been lost from the Prochlorococcus lineage (either prior to or after the divergence from Synechococcus) as a consequence of neutral genetic drift, it may also have been lost as a selectively favorable event. One reason may be economic: as hypothesized for the reduction of peptidoglycan in the HL strain MED4 [81] and the reduction of phospholipids in favor of sulfolipids throughout the Prochlorococcus lineage [7], the loss of catalase may have been selected to lower the cell quota for scarce nutrients in the oligotrophic ocean. The catalase-peroxidase found in most cyanobacteria is a large dimeric enzyme (160 kilodaltons) that has 4 iron-containing heme co-factors [78]. If we make certain assumptions about expression levels based on published data (see Methods), we may estimate that loss of catalase-peroxidase would result in a reduction of cell quotas for Fe, P, and N by 0.2%, 0.14%, and 0.05%, respectively, for Prochlorococcus MED4 (Table S4). As the primary selective pressure implicated in the genomic reductions for this oligotrophic lineage is to lower the cost of such scarce nutrients for cell production [5], [22], [23], [82], [83], it is reasonable that such a gene would be lost in the absence of selective pressure for its retention. Additionally, catalases in keratinocytes have been shown to generate an unidentified form of reactive oxygen species when exposed to UV-B [85]. If similar side reactions occur for bacterial catalases, cells in the UV-exposed surface mixed layer may experience a tradeoff, degrading HOOH while also producing another form of activated oxygen; under these conditions the negative consequences may outweigh the positive ones for Prochlorococcus. In the future, development of robust genetic tools for Prochlorococcus should allow us to ectopically express catalase and determine if it indeed has a positive or negative impact under mixed layer conditions.
Methods
Field sites and methods
HOOH levels in the euphotic zone were measured on three cruises (Table S3): Jan-Feb 2007 in the Western Pacific (WP2), Jun-Jul 2008 in the Eastern Pacific (DCM08), and May-Jun 2009 in the Sargasso Sea (BC09). Seawater was collected in 20 L Niskin bottles and dispensed into lightproof polypropylene bottles rinsed three times with sample water. Care was taken to shield these samples from light prior to laboratory analysis. In situ HOOH levels were measured within 30 minutes of sample retrieval. HOOH measurements for WP2 were conducted using an acridinium ester chemiluminescence flow-injection method [86] implemented with a FeLume (Waterville Analytical, Waterville, ME). HOOH measurements for DCM08 and BC09 were performed using a similar protocol modified for use in an Orion-L microplate luminometer (Berthold Detection Systems, Pforzheim, Germany) (see Methods S1), and restricted to surface mixed layer (≤10 m) depths. Flow cytometry or quantitative PCR [20] confirmed that Prochlorococcus was abundant (≥104 cells ml−1) in all mixed layer samples analyzed (data not shown).
To measure rates of HOOH production in sterile seawater, approximately 15 ml of the above samples were passed through 0.22 µm Millex GV syringe filters (Millipore, Billerica, MA) into custom-built UVT acrylic cuvettes, and sealed with threaded nylon caps. These cuvettes permit about 90% transmission of photosynthetically active radiation, UV-A, and UV-B, as assessed using PAR (QSL2100, Biospherical Instruments, San Diego, CA) and UV sensors (Mannix UV340, General Tools, New York, NY). Both the cuvettes and the caps were sterilized prior to use using a UV germicidal lamp. Samples in cuvettes were exposed to ambient light in a UVT acrylic deck incubator, and HOOH levels were measured at 0 and 24 h. Samples received between 60–80% of surface irradiance (i.e., ∼1200–1600 µmol quanta m−2 s−1 on clear days), and temperature was stabilized by continuous flow of surface water through the incubator. Control cuvettes were treated identically, only shielded from light.
Cultures and culture conditions
Strains used in this study are listed in Table S1. Prochlorococcus cultures were grown in Pro99 natural seawater medium [66] prepared using water collected from 15 m at station R2 in the Georgia Bight (31°22.35′ N, 80°33.68′ W). Strain UH18301 was isolated from Station ALOHA (22°45′N, 158°W) in the North Pacific Subtropical Gyre, at 125 m on July 14, 2006. Seawater was diluted onto 75% Pro99 plates pre-seeded with ∼106 cells mL−1 of helper UH06001 (Alteromonas sp. and a close relative of EZ55) and incubated at 22°C in a natural diel light cycle incubator with a 12∶12 light:dark cycle with a maximum irradiance of 50 µmol quanta m−2 s−1. A green Prochlorococcus colony that formed after ∼2 months of incubation was picked and grown in Pro99 liquid medium and this culture was later rendered clonal and axenic through the antibiotic selection method described below.
Axenic MIT9313 [87] was a generous gift of L. R. Moore, whereas the other strains (Table S1) were rendered axenic by a modified version of our previously described method [37]. Spontaneously SmR mutants of the Prochlorococcus strains were diluted to extinction in Falcon Microtest U-bottom 96-well microtiter plates (#35-3077, BD, Franklin Lakes, NJ) containing 200 µL Pro99 and approximately 2×105 cells of the streptomycin sensitive helper Alteromonas sp. EZ55. These plates were incubated at 24°C in a “Sunbox” growth chamber (Percival, Perry, IA) designed to simulate a natural light regime by gradually raising light level from darkness to a “noon” maximum of 40 µmol quanta m−2 s−1 and back to darkness again over a 12/12 light/dark photoperiod [88]. Microtiter cultures exhibiting growth of Prochlorococcus were tested for purity (apart from the streptomycin-sensitive helper) by placing aliquots into 1/10 ProAC broth [37] containing streptomycin, followed by incubation in the dark. If this purity broth did not become turbid within a week, the Pro99 culture was diluted into fresh Pro99, grown until visibly green, and treated with 100 µg mL−1 streptomycin to remove the helpers. Purity of cultures was routinely confirmed by diluting aliquots of the culture into 1/10 ProAC or PLAG media [37]. Cultures were considered axenic if no turbidity formed after 60 d in purity broth, and no DAPI-stained cells other than Prochlorococcus were observed via fluorescence microscopy and flow cytometry.
Heterotrophic bacteria were grown in YT3 medium (per L, 10 g tryptone, 5 g yeast extract, 1 mM TAPS buffer pH 8.0, dissolved in Turk's Island Sea Salts [66] and filter sterilized with a 0.22 µm Supor filter [Pall]). Helpers were harvested by centrifugation from 2 d cultures and washed twice with sterile Pro99 prior to addition to Prochlorococcus cultures.
All experiments were performed at 24°C in a Sunbox (see above). “Low light” experiments were conducted in a Sunbox fitted with blue theater gels (Roscolux #69, Rosco, Stamford, CT) and set to produce a noon maximum of about 40 µmol quanta m−2 s−1; “high light” experiments used unfiltered white light with a noon maximum of about 250 µmol quanta m−2 s−1. Prochlorococcus strains were acclimated to growth conditions over at least two passages into fresh media prior to the start of experiments. All growth experiments used inocula taken from mid-exponential phase cultures (between 107 and 108 cells mL−1). Unless otherwise specified, all chemicals were Sigma Ultra grade (Sigma-Aldrich, St. Louis, MO).
Quantification of Prochlorococcus and helpers
Prochlorococcus concentrations were determined by flow cytometry, either on a Cytopeia Influx instrument (BD, Franklin Lakes, NJ) or a Guava EasyCyte 8HT (Millipore), using established protocols [20], [89]. For some experiments, growth was tracked by bulk chlorophyll fluorescence using either a TD700 fluorometer (Turner Designs, Sunnyvale, CA, USA) equipped with an in vivo chl a filter set (excitation 340–500 nm; emission ≥665 nm) or a Synergy HT-1 96-well plate fluorometer (BioTek, Winooski, VT). In experiments using co-cultures of different Prochlorococcus strains, concentrations of the different populations were distinguished using quantitative PCR (qPCR) as previously described [15]. Helper cell viable counts were determined on YTSS plates containing 50 U ml−1 bovine liver catalase.
Exponential growth rates (µ) were calculated as the slope of the natural logarithms of at least three points during exponential phase; the highest of these measurements is Cend, taken at time tend. Lag phase duration (LPD) was calculated using the equation
| (1) |
where C0 is the initial measurement at time 0. LPD data was then expressed as an inverse lag proportion (LP−1), given by the ratio of LPD for a control culture (LPD[HOOH]0) with <0.05 µM HOOH and a culture with an elevated HOOH concentration (LPD[HOOH]x):
| (2) |
such that cultures that did not grow (LPD[HOOH]x = infinity) could be assigned a plottable value of ∼0.
Prochlorococcus viability was determined by staining with 5 µM Sytox Green (Invitrogen, Carlsbad, CA) for at least 10 minutes prior to flow cytometry, following manufacturer's instructions. Dead cells were defined as events in the Prochlorococcus gate (forward angle light scatter and red fluorescence) that exhibited green fluorescence values similar to a glutaraldehyde-killed control.
HOOH exposure experiments
The effects of elevated HOOH (i.e., different from the concentration obtained by simply autoclaving seawater) on Prochlorococcus were tested using cultures initially containing approximately 105 cells mL−1 of Prochlorococcus, either with or without 106 cells mL−1 of the helper heterotroph EZ55. These concentrations represent typical levels of Prochlorococcus and total bacteria, respectively, observed in the field [2], [3]. Seawater for the Pro99 media was prepared by ultrafiltration with a Labscale tangential flow filtration device fitted with a Pellicon XL 30 kilodalton cutoff regenerated cellulose cartridge (Millipore), followed by passage through a washed 0.22 µm Supor filter. This was necessary to produce a medium with a sufficiently low initial HOOH concentration to serve as a low HOOH control. Both autoclave and microwave sterilization led to similar HOOH levels of ∼0.2 µM (e.g., Figure S1). While filter-sterilized seawater had much lower HOOH levels (∼40±1 nM), the possible presence of Prochlorococcus phage in natural seawater necessitated either some form of heat sterilization prior to preparation of culture media, or else the ultrafiltration to remove virus size-class particles. Prochlorococcus was inoculated into Pro99 media made from this ultrafiltered seawater either unamended (<0.05 µM HOOH) or amended with HOOH (0.1 to 200 µM), and if no growth was detectable after 60 d, a culture was determined to be dead.
Experiments involving exposure to accumulating HOOH were designed using the buffer HEPES that facilitates steady, light-driven production of HOOH [42], [43]. All cultures were set to pH 8.0 and contained 3.75 mM buffer, using a combination of HEPES and TAPS [(a non-HOOH generating buffer, see Figure 1C)]) to produce a range of HOOH fluxes in a constant buffering capacity background. Cultures were acclimated to the presence of 3.75 mM buffer in TAPS-containing media before being transferred into media containing HEPES.
Chemical and enzymatic assays
All HOOH measurements in the laboratory were performed using the Orion-L luminometer and an acridinium ester chemiluminescence protocol (Methods S1). Bulk water chemistry measurements followed standard protocols [90] using samples filtered through washed 0.22 µm Millex GV filters. Dissolved inorganic carbon (DIC) concentrations were determined by calculation based on measurements of alkalinity (by the bromcresol green endpoint method) and pH (using an UltraBasic probe, Denver Instrument, Bohemia, NY). Dissolved O2 (DO) was measured using an Orion 4star probe (Fisher, Waltham, MA) calibrated in water-saturated air. Dissolved organic carbon (DOC) levels were measured using a TOC-VCP analyzer (Shimadzu, Kyoto, Japan). Alkaline phosphatase activity was measured colorimetrically, following the increase in absorbance at 420 nm after addition of p-nitrophenyl phosphate (final concentration 0.04% w/v) using a BioTek Synergy HT-1 plate reader [91]. Total hydroperoxidase activity was measured by adding 0.8 µM HOOH to a sterile sample and measuring HOOH concentrations every 2 h for 6 h.
Effects of HOOH on Prochlorococcus photophysiology and morphology
Photophysiological parameters were assessed using a Fast Induction and Relaxation (FIRe) fluorometer (Satlantic, Halifax, Nova Scotia) using the methodology described by Johnson [92]. Variable photosystem II fluorescence (Fv/Fm) was determined by curve fitting in MATLAB using both a rapid single turnover (ST) flash series and a longer multiple turnover (MT) series [93]. Photophysiological diel measurements were all performed beginning at the same point in the diel cycle (∼3 h after subjective “sunrise”). Initial decreases in photophysiological parameters (Figure 4E–H) were observed in all cultures, including controls, and likely represent normal diel cycling of these characteristics, as observed in Prochlorococcus PCC 9511 [94].
Cell morphology was assessed by scanning electron microscopy (SEM). 20 mL cultures at approximately 108 cells ml−1 were concentrated by centrifugation at 6500 g for 15 min. Pellets were resuspended in 0.1 M cacodylate-buffered 3% glutaraldehyde for 60 min, and then washed three times with buffer water for 10 min. Cells were then post-fixed in 0.1 M cacodylate-buffered 2% osmium tetroxide for 60 min. After fixation cells were washed three times with water; during the final wash cells were allowed to settle onto a clean silicon chip. Samples were dehydrated in a graded ethanol series at 15 min intervals. Following dehydration samples were critical point dried with liquid CO2 in a critical point dryer (Ladd, Williston, VT). Dried samples were sputtered with a thin layer of gold before examination with a LEO 1525 scanning electron microscope (Carl Zeiss SMT Inc, Peabody, MA). As higher concentrations of cells (108 cells ml−1) were required for the preparation of the SEM samples, we challenged the cells with 10 µM HOOH rather than the usual 0.8 µM SMC value. That an effect of this HOOH treatment was observed for the UH18301 cells but not for the more HOOH-resistant MIT9313 cells (Figure 4) indicates that the concentrations used was appropriate for these assays.
Calculation of cell quota effects for catalase loss
Cell quotas for C, N, and P for Prochlorococcus MED4 were obtained from [95]. Fe quotas were obtained from [96]. By calculation the Synechococcus WH 7803 KatG polypeptide is 80 kD in size and contains two heme cofactors. This enzyme's active configuration is dimeric [78], so the holoenzyme is 160 kD and contains 2054 N and 4 Fe atoms. In E. coli there is an average of 2 mRNA copies/OTU and about 1270 copies of each expressed protein [97]. The volume of an average E. coli cell is 1.1 µm3 [98]; assuming that MED4 is spherical and 0.5 µm in diameter, its volume is 0.065 µm3, or about 1/17 that of E. coli. If we assume that the abundance ratio of an average Prochlorococcus protein to an average E. coli protein is proportional to the volume ratio of these organisms, then the average protein would be present in about 75 copies in MED4. If we further assume that catalase is present at average abundance, and that a MED4 cell contains a single chromosomal katG and 2 mRNA's, then loss of the katG gene from a putative catalase-positive MED4 ancestor would give the savings indicated in Table S4.
Supporting Information
Measurement of HOOH using a microplate luminometer.
(DOC)
Strains used in this study.
(DOC)
Results of helper assays in liquid Pro99. Prochlorococcus was inoculated at 100 cells mL−1 in media with the indicated treatment.
(DOC)
Field sampling stations.
(DOC)
Estimated effects of katG loss from a hypothetical catalase positive ancestor of Prochlorococcus MED4 on cell quotas for selected nutrients.
(DOC)
HOOH removal by Prochlorococcus spp. and Alteromonas sp. EZ55. Prochlorococcus was added to autoclaved Pro99 media at the indicated initial inoculum (cells mL−1) either with or without 106 cells mL−1 EZ55. Changes in [HOOH] were followed using acridinium ester chemiluminescence (Methods S1).
(TIF)
Influence of Prochlorococcus UH18301 and Alteromonas EZ55 on Pro99 medium chemistry. A) EZ55 cell concentration; B) Prochlorococcus UH18301 cell concentration; C) pH; D) HCO3 − concentration; E) CO2 concentration, representing the sum of dissolved CO2 and H2CO3; F) dissolved O2 concentration; G) Total organic carbon in 0.22 µm-filtered media. Error bars are the standard error of three biological replicates. Black circles, sterile media; green triangles, axenic UH18301; red circles, axenic EZ55; blue squares, co-cultured UH18301 and EZ55. The * in Panel B indicates that only 1 of 3 replicates survived to this point, leading to a very large standard deviation for cell counts.
(TIF)
Photoinactivation of purified catalase. Catalase was added to sterile Pro99 media at 1 U/mL and incubated in a Sunbox incubator under low light conditions (see Methods). Aliquots were removed immediately after sample preparation (circles), after 1 d (up triangles), and after 3 d (down triangles). 0.8 µM HOOH was added to these aliquots, and the change in HOOH was monitored for 6 h.
(TIF)
Alkaline phosphatase activity in EZ55-treated media. Alteromonas sp. EZ55 was added to Pro99 at 106 cells mL−1 and incubated in the dark for 24 h. Alkaline phosphatase activity was measured as described in the Methods either before (solid line) or after (dashed line) 0.2 µm filtration of this medium through low-protein-binding PVDF membranes. Readings were taken every 10 min for 24 h. Error bars are the standard deviation of three replicate well on a single 96-well plate.
(TIF)
Effects of chronic HOOH exposure on Prochlorococcus UH18301. UH18301 was inoculated into sterile, unamended seawater containing the indicated concentration of HEPES at 105 cells mL−1. All cultures had a constant 10 mM concentration of buffer; the difference was made up using TAPS. The control cultures for this experiment are plotted in Figure 1C. A) HOOH accumulated in proportion to the concentration of HEPES in the medium. Values are the zero-order rate constants for HOOH formation, in µM d−1, calculated over the first 3 d. Down triangles plot the same data shown by red circles in Figure 1C for comparison. B) Changes in cell concentration were observed over time.
(TIF)
Effects of HOOH exposure on Prochlorococcus ecotypes. Cultures representing 2 LL (MIT9313 and VOL3) and 2 HL (VOL1 and VOL7) ecotypes with (green circles) or without (red squares) EZ55 were exposed to the indicated [HOOH]. Cultures were inoculated at ecologically relevant concentrations (105 Prochlorococcus and 106 EZ55 cells mL−1) in Pro99 medium and grown under low light. Vertical red line represents the SMC HOOH concentration described in the text. LP−1, inverse lag proportion as described in the text and in the legend for Figure 2; *, no growth was detectable after 60 d in all 3 biological replicates.
(TIF)
Photophysiological parameters of Prochlorococcus ecotypes following exposure to SMC (0.8 µM) HOOH exposure. Fv/Fm and Fm(MT/ST) of representative strains of Prochlorococcus were measured after 24 h in sterile unamended seawater with 0.05 or 0.8 µM HOOH. Low light, maximum 40 µmol quanta m−2s−1; high light, maximum 250 µmol quanta m−2s−1. All error bars are the standard error of three biological replicates. *, axenic, HOOH-treated cultures are significantly different (t-test, df = 4, p<0.05) than both untreated axenic cultures and HOOH-treated cultures containing EZ55.
(TIF)
Acknowledgments
The authors would like to thank J. Chandler, E. Mann, K. Barbeau, B. Binder, L. Poorvin, A. Anderson, W. Cooper, and G. O'Toole for advice and technical support; the captains and crews of R/V Kilo Moana, R/V New Horizon, and R/V Cape Hatteras; S. Allman for assistance with flow cytometry; J. Dunlap for assistance with scanning electron microscopy; and S. Wilhelm and G. Sayler for access to equipment and facilities.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by NSF grants OCE0526072 to ERZ and OCE0550798 to ZIJ, and a Phycological Society of America grant-in-aid to JJM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sarthou G, Baker AR, Kramer J, Laan P, Laes A, et al. Influence of atmospheric inputs on the iron distribution in the subtropical Northeast Atlantic Ocean. Mar Chem. 2007;104:186–202. [Google Scholar]
- 2.Kirchman DL, editor. Hoboken, NJ: Wiley; 2008. Microbial Ecology of the Oceans. 2nd ed.620 [Google Scholar]
- 3.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:10. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fogg GE, Thake B. Madison, WI: University of Wisconsin Press; 1987. Algal Cultures and Phytoplankton Ecology.269 [Google Scholar]
- 7.Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH. Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci USA. 2006;103:8607–8612. doi: 10.1073/pnas.0600540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goericke R, Welschmeyer NA. The marine prochlorophyte Prochlorococcus contributes significantly to phytoplankton biomass and primary production in the Sargasso Sea. Deep-Sea Res I. 1993;40:2283–2294. [Google Scholar]
- 9.DiTullio GR, Geesey ME, Jones DR, Daly KL, Campbell L, et al. Phytoplankton assemblage structure and primary productivity along 170°W in the South Pacific Ocean. Mar Ecol Prog Ser. 2003;255:55–80. [Google Scholar]
- 10.Partensky F, Blanchot J, Lantoine F, Neveux J, Marie D. Vertical structure of picophytoplankton at different trophic sites of the subtropical northeastern Atlantic. Deep-Sea Res I. 1996;43:1191–1213. [Google Scholar]
- 11.Ras J, Claustre H, Uitz J. Spatial variability of phytoplankton pigment distributions in the subtropical South Pacific Ocean: Comparison between in situ and predicted data Biogeosciences. 2008;5:353–369. [Google Scholar]
- 12.Veldhuis MJW, Kraay GW. Phytoplankton in the subtropical Atlantic Ocean: towards a better assessment of biomass and composition. Deep-Sea Res I. 2004;51:507–530. [Google Scholar]
- 13.Ahlgren NA, Rocap G, Chisholm SW. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 14.West NJ, Scanlan DJ. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl Environ Microbiol. 1999;65:2585–2591. doi: 10.1128/aem.65.6.2585-2591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol. 2006;72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore LR, Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol Oceanogr. 1999;44:628–638. [Google Scholar]
- 17.Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, et al. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr. 2007;52:2205–2230. [Google Scholar]
- 18.Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, et al. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific Oceans. ISME J. 2010;4:1252–1264. doi: 10.1038/ismej.2010.60. [DOI] [PubMed] [Google Scholar]
- 19.Bouman HA, Ulloa O, Scanlan DJ, Zwirglmaier K, Li WKW, et al. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science. 2006;312:918–921. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- 20.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 21.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 22.Partensky F, Garczarek L. Prochlorococcus: advantages and limits of minimalism. Annu Rev Mar Sci. 2010;2:305–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- 23.Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:2515–2528. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper WJ, Zika RG, Petasne RG, Plane JMC. Photochemical formation of H2O2 in natural waters exposed to sunlight. Environ Sci Technol. 1988;22:1156–1160. doi: 10.1021/es00175a004. [DOI] [PubMed] [Google Scholar]
- 25.Gerringa LJA, Rijkenberg MJA, Timmermans KR, Buma AGJ. The influence of solar ultraviolet radiation on the photochemical production of H2O2 in the equatorial Atlantic Ocean. J Sea Res. 2004;51:3–10. [Google Scholar]
- 26.Hanson AK, Tindale NW, Abdel-Moati MAR. An equatorial Pacific rain event: influence on the distribution of iron and hydrogen peroxide in surface waters. Mar Chem. 2001;75:69–88. [Google Scholar]
- 27.Kieber RJ, Peake B, Willey JD, Jacobs B. Iron speciation and hydrogen peroxide concentrations in New Zealand rainwater. Atmos Environ. 2001;35:6041–6048. [Google Scholar]
- 28.Yuan J, Shiller AM. The variation of hydrogen peroxide in rainwater over the South and Central Atlantic Ocean. Atmos Environ. 2000;34:3973–3980. [Google Scholar]
- 29.Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Annu Rev Physiol. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- 30.Moffett JW, Zika RG. Reaction kinetics of hydrogen peroxide with copper and iron in seawater. Environ Sci Technol. 1987;21:804–810. doi: 10.1021/es00162a012. [DOI] [PubMed] [Google Scholar]
- 31.Drabkova M, Admiraal W, Marsalek B. Combined exposure to hydrogen peroxide and light – selective effects on cyanobacteria, green algae, and diatoms. Environ Sci Technol. 2007;41:309–314. doi: 10.1021/es060746i. [DOI] [PubMed] [Google Scholar]
- 32.Willey JD, Kieber RJ, Avery GB., Jr Effects of rainwater iron and hydrogen peroxide on iron speciation and phytoplankton growth in seawater near Bermuda. J Atmos Chem. 2004;47:209–222. [Google Scholar]
- 33.Dummermuth AL, Karsten U, Fisch KM, König GM, Wiencke C. Responses of marine macroalgae to hydrogen peroxide stress. J Exp Mar Biol Ecol. 2003;289:103–121. [Google Scholar]
- 34.Xenopoulos MA, Bird DF. Effect of acute exposure to hydrogen peroxide on the production of phytoplankton and bacterioplankton in a mesohumic lake. Photochem Photobiol. 1997;66:471–478. [Google Scholar]
- 35.Suggett DJ, Warner ME, Smith DJ, Davey P, Hennige S, et al. Photosynthesis and production of hydrogen peroxide by Symbiodinium (Pyrrhophyta) phylotypes with different thermal tolerances. J Phycol. 2008;44:948–956. doi: 10.1111/j.1529-8817.2008.00537.x. [DOI] [PubMed] [Google Scholar]
- 36.Samuilov VD, Bezryadnov DV, Gusev MV, Kitashov AV, Fedorenko TA. Hydrogen peroxide inhibits the growth of cyanobacteria. Biochemistry-Moscow. 1999;64:47–53. [PubMed] [Google Scholar]
- 37.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol. 2008;74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernroitner M, Zamocky M, Furtmuller PG, Peschek GA, Obinger C. Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J Exp Bot. 2009;60:423–440. doi: 10.1093/jxb/ern309. [DOI] [PubMed] [Google Scholar]
- 39.Hebrard M, Viala JPM, Meresse S, Barras F, Aussel L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009;191:4605–4614. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tytler EM, Wong T, Codd GA. Photoinactivation in vivo of superoxide dismutase and catalase in the cyanobacterium Microcystis aeruginosa. FEMS Microbiol Lett. 1984;23:239–242. [Google Scholar]
- 42.Lepe-Zuniga JL, Zigler JS, Jr, Gery I. Toxicity of light-exposed HEPES media. J Immunol Meth. 1987;103:145. doi: 10.1016/0022-1759(87)90253-5. [DOI] [PubMed] [Google Scholar]
- 43.Zigler JS, Jr, Lepe-Zuniga JL, Vistica B, Gery I. Analysis of the cytotoxic effects of light-exposed HEPES-containing culture medium. In Vitro Cell Dev Biol. 1985;21:282–287. doi: 10.1007/BF02620943. [DOI] [PubMed] [Google Scholar]
- 44.Zika RG, Saltzman ES, Cooper WJ. Hydrogen peroxide concentrations in the Peru upwelling area. Mar Chem. 1985;17:265–275. [Google Scholar]
- 45.Yuan J, Shiller AM. Distribution of hydrogen peroxide in the northwest Pacific Ocean. Geochem Geophys Geosyst. 2005;6:1–3. [Google Scholar]
- 46.Avery GBJ, Cooper WJ, Kieber RJ, Willey JD. Hydrogen peroxide at the Bermuda Atlantic Time Series Station: temporal variability of seawater hydrogen peroxide. Mar Chem. 2005;97:236–244. [Google Scholar]
- 47.Miller WL, Kester DR. Peroxide variations in the Sargasso Sea. Mar Chem. 1994;48:17–29. [Google Scholar]
- 48.Johnson KS, Willason SW, Wiesenburg DA, Lohrenz SE, Arnone RA. Hydrogen peroxide in the western Mediterranean Sea: a tracer for vertical advection. Deep-Sea Res I. 1989;36:241–254. [Google Scholar]
- 49.Yuan J, Shiller AM. The distribution of hydrogen peroxide in the southern and central Atlantic ocean. Deep-Sea Res I. 2001;48:2947–2970. [Google Scholar]
- 50.Draper WM, Crosby DG. The photochemical generation of hydrogen peroxide in natural waters. Arch Environ Contam Toxicol. 1983;12:121–126. [Google Scholar]
- 51.Cooper WJ, Zika RG. Photochemical formation of hydrogen peroxide in surface and ground waters exposed to sunlight. Science. 1983;220:711–712. doi: 10.1126/science.220.4598.711. [DOI] [PubMed] [Google Scholar]
- 52.Petasne RG, Zika RG. Hydrogen peroxide lifetimes in south Florida coastal and offshore waters. Mar Chem. 1997;56:215–225. [Google Scholar]
- 53.Cooper WJ, Zepp RG. Hydrogen peroxide decay in waters with suspended soils: evidence for biologically mediated processes. Can J Fish Aquat Sci. 1990;47:888–893. [Google Scholar]
- 54.Schreiber U, Krieger A. Two fundamentally different types of variable chlorophyll fluorescence in vivo. FEBS Lett. 1996;397:131–135. doi: 10.1016/s0014-5793(96)01176-3. [DOI] [PubMed] [Google Scholar]
- 55.Halliwell B, Gutteridge JMC. New York: Oxford University Press; 2007. Free Radicals In Biology and Medicine.851 [Google Scholar]
- 56.Ma M, Eaton JW. Multicellular oxidant defense in unicellular organisms. Proc Natl Acad Sci USA. 1992;89:7924–7928. doi: 10.1073/pnas.89.17.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fazzini RAB, Preto MJ, Quintas ACP, Bielecka A, Timmis KN, et al. Consortia modulation of the stress response: proteomic analysis of single strain versus mixed culture. Environ Microbiol. 2010;12:2449. doi: 10.1111/j.1462-2920.2010.02217.x. [DOI] [PubMed] [Google Scholar]
- 58.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66:637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubinin AV, Gerasimenko LM, Venetskaya SL, Gusev MV. Cyanobacterium Microcoleus chthonoplastes fails to grow in pure culture. Microbiology. 1992;61:41–46. [Google Scholar]
- 60.Paerl HW, Pinckney JL. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb Ecol. 1996;31:225–247. doi: 10.1007/BF00171569. [DOI] [PubMed] [Google Scholar]
- 61.Mouget JL, Dakhama A, Lavoie MC, de la Noije J. Algal growth enhancement by bacteria: is consumption of photosynthetic oxygen involved? FEMS Microbiol Ecol. 1995;18:35–44. [Google Scholar]
- 62.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 63.de Bashan LE, Antoun H, Bashan Y. Involvement of indole-3-acetic-acid produced by the growth-promoting bacterium Azospirillum spp. in promoting growth of Chlorella vulgaris. J Phycol. 2008;44:938–947. doi: 10.1111/j.1529-8817.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 64.D'Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, et al. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem Biol. 2010;17:254–264. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rippka R, Coursin T, Hess W, Lichtle C, Scanlan DJ, et al. Prochlorococcus marinus Chisholm et al. 1992 subsp pastoris subsp nov strain PCC 9511, the first axenic chlorophyll a(2)/b(2)-containing cyanobacterium (Oxyphotobacteria). Int J Syst Evol Microbiol. 2000;50:1833–1847. doi: 10.1099/00207713-50-5-1833. [DOI] [PubMed] [Google Scholar]
- 66.Moore LR, Coe A, Zinser ER, Saito MA, Sullivan MB, et al. Culturing the marine cyanobacterium Prochlorococcus. Limnol Oceanogr: Methods. 2007;5:353–362. [Google Scholar]
- 67.Zepp RG, Faust BC, Hoigne J. Hydroxyl radical formation in aqueous reactions (pH 3-8) of iron(II) with hydrogen peroxide: the photo-Fenton reaction. Environ Sci Technol. 2002;26:313–319. [Google Scholar]
- 68.Halsey TA, Vazquez-Torres A, Gravdahl DJ, Fang FC, Libby SJ. The ferritin-like dps protein Is required for Salmonella enterica serovar typhimurium oxidative stress resistance and virulence. Infect Immun. 2004;72:1155–1158. doi: 10.1128/IAI.72.2.1155-1158.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pena MMO, Bullerjahn GS. The dpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein - linkage of the dps and bacterioferritin protein families. J Biol Chem. 1995;270:22478–22482. doi: 10.1074/jbc.270.38.22478. [DOI] [PubMed] [Google Scholar]
- 70.Blankenship RE. London: Blackwell; 2002. Molecular Mechanisms of Photosynthesis.321 [Google Scholar]
- 71.Kojima K, Oshita M, Nanjo Y, Kasai K, Tozawa Y, et al. Oxidation of elongation factor G inhibits the synthesis of the D1 protein of photosystem II. Mol Microbiol. 2007;65:936–947. doi: 10.1111/j.1365-2958.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- 72.Bertilsson S, Berglund O, Pullin MJ, Chisholm SW. Release of dissolved organic matter by Prochlorococcus. Vie Et Milieu - Life Environ. 2005;55:225–231. [Google Scholar]
- 73.Choler P, Michalet R, Callaway RM. Facilitation and competition on gradients in alpine plant communities. Ecology. 2001;82:3295–3308. [Google Scholar]
- 74.Kranner I, Cram WJ, Zorn M, Wornik S, Yoshimura I, et al. Antioxidants and photoprotection in a lichen as compared with its isolated symbiotic partners. Proc Natl Acad Sci. 2005;USA102:3141–3146. doi: 10.1073/pnas.0407716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richier S, Furla P, Plantivaux A, Merle PL, Allemand D. Symbiosis-induced adaptation to oxidative stress. J Exper Biol. 2005;208:277–285. doi: 10.1242/jeb.01368. [DOI] [PubMed] [Google Scholar]
- 76.Bertness MD, Callaway R. Positive interactions in communities. Trends Ecol Evol. 1994;9:191–193. doi: 10.1016/0169-5347(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 77.Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 78.Chelikani P, Fita I, Loewen PC. Diversity of structures and properties among catalases. Cell Mol Life Sci. 2004;61:192–208. doi: 10.1007/s00018-003-3206-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 80.Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J Bacteriol. 2007;189:4485–4493. doi: 10.1128/JB.01948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vieira-Silva S, Touchon M, Rocha EPC. No evidence for elemental-based streamlining of prokaryotic genomes. Trends Ecol Evol. 2010;25:319–320. doi: 10.1016/j.tree.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 83.Marais GAB, Calteau A, Tenaillon O. Mutation rate and genome reduction in endosymbiotic and free-living bacteria. Genetica. 2008;134:205–210. doi: 10.1007/s10709-007-9226-6. [DOI] [PubMed] [Google Scholar]
- 84.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 85.Heck DE, Vetrano AM, Mariano TM, Laskin JD. UVB light stimulates production of reactive oxygen species - Unexpected role for catalase. J Biol Chem. 2003;278:22432–22436. doi: 10.1074/jbc.C300048200. [DOI] [PubMed] [Google Scholar]
- 86.King DW, Cooper WJ, Rusak SA, Peake BM, Kiddle JJ, et al. Flow injection analysis of H2O2 in natural waters using acridinium ester chemiluminescence: method development and optimization using a kinetic model. Anal Chem. 2007;79:4169–4176. doi: 10.1021/ac062228w. [DOI] [PubMed] [Google Scholar]
- 87.Moore LR, Ostrowski M, Scanlan DJ, Feren K, Sweetsir T. Ecotypic variation in phosphorus acquisition mechanisms within marine picocyanobacteria. Aquat Microb Ecol. 2005;39:257–269. [Google Scholar]
- 88.Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, et al. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS ONE. 2009;4:e5135. doi: 10.1371/journal.pone.0005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cavender-Bares KK, Frankel SL, Chisholm SW. A dual sheath flow cytometer for shipboard analyses of phytoplankton communities from oligotrophic seas. Limnol Oceanogr. 1998;43:1383–1388. [Google Scholar]
- 90.Eaton AD, Clesceri LS, Rice EW, Greenberg AE, Franson MAH, editors. American Public Health Association; 2005. Standard Methods for the Examination of Water and Wastewater. 21st ed. [Google Scholar]
- 91.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 92.Johnson ZI. Description and application of the background irradiance gradient-single turnover fluorometer (BIG-STf). Mar Ecol Progr Ser. 2004;283:73–80. [Google Scholar]
- 93.Marchetti A, Varela DE, Lance VP, Johnson Z, Palmucci M, et al. Iron and silicic acid effects on phytoplankton productivity, diversity, and chemical composition in the central equatorial Pacific Ocean. Limnol Oceanogr. 2010;55:11–29. [Google Scholar]
- 94.Bruyant F, Babin M, Genty B, Prasil O, Behrenfeld MJ, et al. Diel variations in the photosynthetic parameters of Prochlorococcus strain PCC 9511: Combined effects of light and cell cycle. Limnol Oceanogr. 2005;50:850–863. [Google Scholar]
- 95.Bertilsson S, Berglund O, Karl DM, Chisholm SW. Elemental composition of marine Prochlorococcus and Synechococcus: Implications for the ecological stoichiometry of the sea. Limnol Oceanogr. 2003;48:1721–1731. [Google Scholar]
- 96.Thompson AW. Cambridge, MA: Massachusetts Institute of Technology; 2009. Iron and Prochlorococcus. Ph.D. Thesis.251 [Google Scholar]
- 97.Neidhardt FC, Umbarger HE. Chemical composition of Escherichia coli. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 13–16. [Google Scholar]
- 98.Kubitschek HE, Friske JA. Determination of bacterial cell volume with the Coulter counter. J Bacteriol. 1986;168:1466–1467. doi: 10.1128/jb.168.3.1466-1467.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Measurement of HOOH using a microplate luminometer.
(DOC)
Strains used in this study.
(DOC)
Results of helper assays in liquid Pro99. Prochlorococcus was inoculated at 100 cells mL−1 in media with the indicated treatment.
(DOC)
Field sampling stations.
(DOC)
Estimated effects of katG loss from a hypothetical catalase positive ancestor of Prochlorococcus MED4 on cell quotas for selected nutrients.
(DOC)
HOOH removal by Prochlorococcus spp. and Alteromonas sp. EZ55. Prochlorococcus was added to autoclaved Pro99 media at the indicated initial inoculum (cells mL−1) either with or without 106 cells mL−1 EZ55. Changes in [HOOH] were followed using acridinium ester chemiluminescence (Methods S1).
(TIF)
Influence of Prochlorococcus UH18301 and Alteromonas EZ55 on Pro99 medium chemistry. A) EZ55 cell concentration; B) Prochlorococcus UH18301 cell concentration; C) pH; D) HCO3 − concentration; E) CO2 concentration, representing the sum of dissolved CO2 and H2CO3; F) dissolved O2 concentration; G) Total organic carbon in 0.22 µm-filtered media. Error bars are the standard error of three biological replicates. Black circles, sterile media; green triangles, axenic UH18301; red circles, axenic EZ55; blue squares, co-cultured UH18301 and EZ55. The * in Panel B indicates that only 1 of 3 replicates survived to this point, leading to a very large standard deviation for cell counts.
(TIF)
Photoinactivation of purified catalase. Catalase was added to sterile Pro99 media at 1 U/mL and incubated in a Sunbox incubator under low light conditions (see Methods). Aliquots were removed immediately after sample preparation (circles), after 1 d (up triangles), and after 3 d (down triangles). 0.8 µM HOOH was added to these aliquots, and the change in HOOH was monitored for 6 h.
(TIF)
Alkaline phosphatase activity in EZ55-treated media. Alteromonas sp. EZ55 was added to Pro99 at 106 cells mL−1 and incubated in the dark for 24 h. Alkaline phosphatase activity was measured as described in the Methods either before (solid line) or after (dashed line) 0.2 µm filtration of this medium through low-protein-binding PVDF membranes. Readings were taken every 10 min for 24 h. Error bars are the standard deviation of three replicate well on a single 96-well plate.
(TIF)
Effects of chronic HOOH exposure on Prochlorococcus UH18301. UH18301 was inoculated into sterile, unamended seawater containing the indicated concentration of HEPES at 105 cells mL−1. All cultures had a constant 10 mM concentration of buffer; the difference was made up using TAPS. The control cultures for this experiment are plotted in Figure 1C. A) HOOH accumulated in proportion to the concentration of HEPES in the medium. Values are the zero-order rate constants for HOOH formation, in µM d−1, calculated over the first 3 d. Down triangles plot the same data shown by red circles in Figure 1C for comparison. B) Changes in cell concentration were observed over time.
(TIF)
Effects of HOOH exposure on Prochlorococcus ecotypes. Cultures representing 2 LL (MIT9313 and VOL3) and 2 HL (VOL1 and VOL7) ecotypes with (green circles) or without (red squares) EZ55 were exposed to the indicated [HOOH]. Cultures were inoculated at ecologically relevant concentrations (105 Prochlorococcus and 106 EZ55 cells mL−1) in Pro99 medium and grown under low light. Vertical red line represents the SMC HOOH concentration described in the text. LP−1, inverse lag proportion as described in the text and in the legend for Figure 2; *, no growth was detectable after 60 d in all 3 biological replicates.
(TIF)
Photophysiological parameters of Prochlorococcus ecotypes following exposure to SMC (0.8 µM) HOOH exposure. Fv/Fm and Fm(MT/ST) of representative strains of Prochlorococcus were measured after 24 h in sterile unamended seawater with 0.05 or 0.8 µM HOOH. Low light, maximum 40 µmol quanta m−2s−1; high light, maximum 250 µmol quanta m−2s−1. All error bars are the standard error of three biological replicates. *, axenic, HOOH-treated cultures are significantly different (t-test, df = 4, p<0.05) than both untreated axenic cultures and HOOH-treated cultures containing EZ55.
(TIF)