Abstract
STAT3 is a ubiquitous transcription factor that is indispensable during early embryogenesis. To study the functions of STAT3 postnatally, we generated conditional STAT3-deficient mice. To that end, STAT3lox/lox mice were crossed with mice expressing Cre under the control of rat insulin II gene promoter (RIP-Cre mice). Immunohistochemical and Western blot analyses showed that STAT3 is deleted from β cells in the islets of Langerhans. Genomic DNA PCR revealed that STAT3 deletion also occurred in the hypothalamus. Hypothalamic Cre expression was further confirmed by crossing RIP-Cre/STAT3lox/lox mice with the ROSA26 Cre reporter strain and staining for lacZ activity. Double immunohistochemical staining confirmed that deletion of STAT3 occurred in leptin receptor (OB-Rb isoform)-positive neurons. RIP-Cre/STAT3lox/lox mice are mildly hyperglycemic and hyperinsulinemic at the time of weaning, become hyperphagic immediately after weaning, and exhibit impaired glucose tolerance. Body weight, body fat, and mRNA and protein levels of leptin are all significantly increased in RIP-Cre/STAT3lox/lox mice. Administration of recombinant leptin by intracerebroventricular infusion failed to cause complete loss of body fat in RIP-Cre/STAT3lox/lox mice. Transplantation of wild-type islets into RIP-Cre/STAT3lox/lox mice also failed to decrease adiposity or to correct other abnormalities in these mice. These data thus suggest that loss of STAT3 in the hypothalamus caused by RIP-Cre action likely interferes with normal body weight homeostasis and glucose metabolism.
Signal transducers and activators of transcription (STAT) proteins are a family of latent cytoplasmic transcription factors that are produced in many cell types and that are activated by tyrosine phosphorylation and dimerization in response to a wide variety of extracellular ligands, such as cytokines and growth factors (12, 36). One member of this family, STAT3, is expressed ubiquitously and is transiently activated by a large number of ligands, including epidermal growth factor, platelet-derived growth factor, interleukin 6 (IL-6), ciliary neurotrophic factor (CNTF), oncostatin M, leukemia inhibitory factor, leptin, growth hormone, and prolactin, as well as a number of oncogenic receptor and nonreceptor (Src-like) tyrosine kinases (12). While gene disruption approaches have been used extensively to define the functions of members of the STAT family of transcription factors (18), the knockout of STAT3 results in early embryonic lethality (42). At the cellular level, STAT3 is required in order to maintain the pluripotency of embryonic stem cells, as demonstrated by the reduced ability of cells to undergo undifferentiated clonal growth when the level of STAT3 is reduced (33).
The early embryonic lethality of STAT3 knockout mice prevents any type of physiological study (42). To overcome this limitation, many laboratories have employed tissue-specific conditional gene targeting to study STAT3 function in adult mice (2, 3, 24, 39). These efforts have led to the elucidation of the roles played by STAT3 in various aspects of cytokine and growth factor signaling in different tissues and cell types. For instance, in T cells, STAT3 functions to transduce the antiapoptotic function of IL-6 independently from that of Bcl-2 (41); in macrophages and neutrophils, STAT3 is required to suppress the overshooting of inflammatory stimulus-induced proinflammatory response (40); in keratinocytes, loss of STAT3 results in compromised wound healing (19, 34, 35); in the mammary gland, loss of STAT3 causes delayed mammary gland involution after weaning (9); in the liver, STAT3 is required to mediate the ability of both IL-6- and lipopolysaccharide-induced acute-phase gene expressions (4); in sensory neurons, loss of STAT3 is associated with their enhanced death, which is normally prevented by neurotrophic factors such as CNTF and leukemia inhibitory factor (5); motor neuron survival also requires a higher dose of CNTF for survival in the absence of STAT3 (37). While these studies have shed light on the functions of STAT3 in these tissues and cell types, the roles of STAT3 in other biological processes remain to be determined. In particular, STAT3 is specifically activated by leptin in vivo and in vitro (8, 25, 43), but whether and/or how STAT3 mediates leptin's biological activity has not been determined.
Previous studies have suggested that STAT3 may play a very important role in the regulation of mammalian body weight and energy homeostasis. Leptin, the adipocyte-specific hormone with a potent weight reducing effect in vivo, activates STAT3 in the hypothalamus and several other tissues, including pancreatic β cells (30, 43). Mice deficient for leptin (Lepob) or resistant to the actions of leptin (Leprdb) are severely obese and hyperphagic, and they develop diabetes at around 6 weeks of age. These phenotypes are caused by loss of leptin signaling (10, 23, 46). While STAT3 is a known downstream component of leptin action, its contribution to these aspects of energy homeostasis has yet to be determined.
To address the function of STAT3 in pancreatic β cells, which are direct and indirect targets of many cytokines, including leptin (16), we generated mice with selective deletion of the STAT3 gene in vivo by using the Cre-Lox technology. To generate RIP-Cre/STAT3lox/lox mice (32, 41), mice containing LoxP-flanked STAT3 alleles (STAT3lox/lox mice) were crossed to Cre mice that express Cre under the control of rat insulin II gene promoter (RIP-Cre). STAT3 deletion was confirmed in pancreatic β cells by immunohistochemical analysis and Western blotting. Genomic DNA PCR and ROSA26 Cre reporter analysis revealed that STAT3 deletion also occurred in the hypothalamus. Surprisingly, RIP-Cre/STAT3lox/lox mice developed mild hyperglycemia and hyperinsulinemia immediately after weaning and exhibited impaired glucose tolerance. They also developed hyperphagia and obesity, having increased leptin mRNA and protein levels. The effectiveness of centrally administered leptin to reduce adipose tissue mass in RIP-Cre/STAT3lox/lox mice was impaired. Likewise, transplantation of wild-type islets into RIP-Cre/STAT3lox/lox mice failed to correct the obesity phenotype of RIP-Cre/STAT3lox/lox mice. These results demonstrate that while RIP-Cre action causes STAT3 deletion in both β cells and the hypothalamus, loss of STAT3 from the hypothalamus interferes with normal body weight homeostasis and glucose metabolism.
MATERIALS AND METHODS
Mice and genotyping.
STAT3lox/lox mice and RIP-Cre transgenic mice have been described previously as having been maintained on a mixed (C57BL/6 × 129/SV) genetic background (32, 41). RIP-Cre mice were crossed with STAT3lox/lox mice for two generations in order to obtain RIP-Cre/STAT3lox/lox mice. First, RIP-Cre/STAT3+/+ mice were mated with STAT3lox/lox mice to generate RIP-Cre/STAT3lox/+ mice, which make up about half of the progenies produced from such crosses. Second, RIP-Cre/STAT3lox/+ mice were mated again with STAT3lox/lox mice to generate RIP-Cre/STAT3lox/lox mice; this crossing served to delete both copies of the STAT3 alleles in RIP-Cre-expressing cells. All experiments were performed by using progenies of STAT3lox/lox mice crossed with RIP-Cre/STAT3lox/lox mice. The genotypes of the progenies are either wild type (STAT3lox/lox) or knockout (RIP-Cre/STAT3lox/lox) at an observed Mendelian ratio of 1/1 (for example, from 51 mice in four litters, 26 mice were RIP-Cre/STAT3lox/lox, while the remaining 25 were STAT3lox/lox). All mice were housed in specific-pathogen-free barrier facilities, maintained on a 12-h light-dark cycle, and fed a standard rodent chow diet (Teklad, Madison, Wis.). Genotyping was performed by PCR amplification of tail DNA from each mouse at 2 weeks of age. Primers specific for Cre recombinase amplify a fragment of 0.8 kb, and primers that flank one of the two LoxP sites of STAT3 genomic DNA will amplify genomic DNA of 0.3 kb. The primer sequences are: Cre forward primer (CL326), 5′-CTCTGGCCATCTGCTGATCCA-3′ and reverse primer (CL196), 5′CTAAGTGCCTTCTTCTACACCTG3′; and LoxP forward primer (CL270), 5′-CCTGAAGACCAAGTTCATCTGT-3′ and reverse primer (CL271), 5′-CACACAAGCCATCAAACTCTGG-3′.
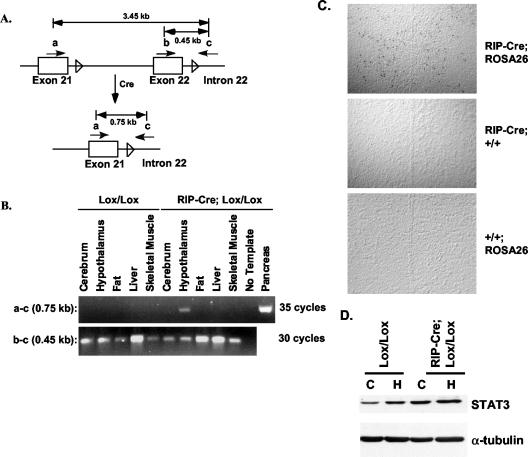
To detect Cre-mediated recombination at the genomic DNA level by PCR, primers were designed that are located on exon 21 (primer a, CL381), exon 22 (primer b, CL384), and intron 22 (primer c, CL271) (see Fig. 2A). The primer pair a and CL271 will give rise to PCR products of 3.45 or 0.75 kb before and after Cre-mediated STAT3 deletion, respectively. Primer pair b and CL271 amplifies a product of 0.45 kb and is used as a control for the quality of the template. The primer sequences were primer a, 5′-GATCCAGTCTGTAGAGCCATACACCAAG-3′, and primer b, 5′-CGCCGTCTGGGAGAGGCAGGAGGATTC-3′.
FIG. 2.
Assessment of STAT3 deletion in extrapancreatic cells of RIP-Cre/STAT3lox/lox mice. (A) Schematic illustration of genomic DNA PCR strategy to detect Cre recombinase-mediated deletion of STAT3 genomic DNA. Genomic DNA structure of exons 21 and 22 of the STAT3 gene is shown, with the LoxP sequences indicated by an empty arrowhead. Cre action removes exon 22 and other sequences between the two LoxP sites. (B) Survey of tissues from STAT3lox/lox (Lox/Lox) and RIP-Cre/STAT3lox/lox mice (RIP-Cre; Lox/Lox) to detect Cre activity outside of pancreatic β cells. Shown is the result of PCR product amplified from genomic DNA of indicated tissues from 3-month-old mice and run on agarose gel. Deletion of STAT3 is detected in the pancreas and hypothalamus of RIP-Cre/STAT3lox/lox mice (RIP-Cre; Lox/Lox) but not in any other tissues, nor was deletion detected in any tissue from STAT3lox/lox (Lox/Lox) mice. Note that 35 cycles of PCR are required to detect STAT3 deletion from the hypothalamus. (C) lacZ staining of brain sections from RIP-Cre; ROSA26, RIP-Cre; +/+, or +/+; ROSA26 mice. lacZ staining was found throughout the hypothalamus in RIP-Cre; ROSA26 mice but not in RIP-Cre; +/+ or +/+; ROSA26 mice. The midline in each panel shows the third ventricle. (D) Western blotting to determine STAT3 protein levels in the hypothalamus (H) and cerebrum (C) of STAT3lox/lox (Lox/Lox) mice and RIP-Cre/STAT3lox/lox (RIP-Cre; Lox/Lox) mice. Tubulin staining was performed on the same filter to show equal loading of proteins in all lanes. No decrease of STAT3 protein levels was detected from the hypothalamus of RIP-Cre/STAT3lox/lox mice.
In all cases, PCR conditions were 94°C for 1 min, followed by 30 to 35 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min. A final extension step of 7 min at 68°C was performed to ensure complete synthesis of all annealed products.
All in vivo procedures were performed in accordance with the policies of the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center.
Dual-energy X-ray absorptiometry (DEXA) and nuclear magnetic resonance (NMR) measurement of body fat.
Body fat content in mice was measured by two methods. During the initial phases of the project, the DEXA method was used. Briefly, mice were anesthetized with a cocktail containing ketamine, xylazine, and acepromazine. A stock solution was prepared by mixing 5 ml of ketamine (100 mg/ml), 2.5 ml of xylazine (20 mg/ml), 1.0 ml of acepromazine (10 mg/ml), and 1.5 ml of H2O to a final volume of 10 ml which was then diluted 1:1 with H2O before use. Each mouse received 0.1 ml of cocktail per 20 g of body weight intraperitoneally. The percentage of body fat was determined by scanning using a Lunar PIXImus densitometer (GE Medical Systems Lunar, Madison, Wis.). The instrument was first calibrated with an aluminum-lucite phantom before anesthetized mice were placed on the imaging tray, in accordance with the manufacturer's instructions. Results shown in Fig. 6 and 7 were conducted with a Bruker Minispec mq7.5 NMR analyzer (Bruker Optics, The Woodlands, Tex.). The accuracy and precision of both instruments were cross-calibrated by measuring the same groups of mice with different adiposity.
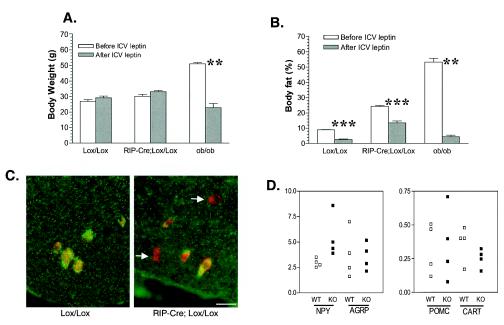
FIG. 6.
RIP-Cre/STAT3lox/lox mice are resistant to central leptin action on adiposity. STAT3lox/lox mice (Lox/Lox), RIP-Cre/STAT3lox/lox mice (RIP-Cre/Lox/Lox), or ob/ob mice were infused with recombinant leptin into the third ventricle. Body weight (A) and body fat levels (B) before and after a 4-week leptin infusion are shown. ICV-administered leptin caused a reduction of body weight by more than half in ob/ob mice, and the percentage of body fat decreased to less than 5%, as determined by NMR. ICV-administered leptin has no appreciable effect on the body weight of 2-month-old STAT3lox/lox mice or RIP-Cre/STAT3lox/lox mice, but it does reduce the body fat of STAT3lox/lox mice to visually undetectable levels (data not shown), with a reading of less than 4% as determined by NMR. In contrast, RIP-Cre/STAT3lox/lox mice after 4 weeks of leptin infusion still have 13% body fat as determined by NMR. There were six mice per group for STAT3lox/lox mice or RIP-Cre/STAT3lox/lox mice; there were two ob/ob mice. **, P < 0.01; ***, P < 0.0001. (C) Deletion of STAT3 from OB-Rb-positive neurons. Sections were stained with a rabbit polyclonal antibody against OB-Rb (red) and a monoclonal antibody against STAT3 (green) and then superimposed. Arrows point to OB-Rb neurons (red) where STAT3 protein is absent. (D) Real-time PCR analysis of expression levels of NPY, AGRP, POMC, and CART. Expression levels are normalized to cyclophilin. There were four mice each for STAT3lox/lox mice (WT) and RIP-Cre/STAT3lox/lox mice (KO). Mice were 2 months old at the time of sacrifice.
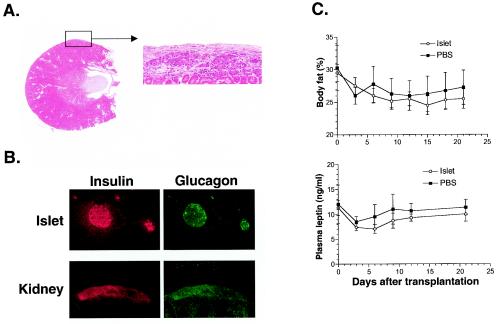
FIG. 7.
Transplantation of islets from control mice failed to correct the phenotypes of RIP-Cre/STAT3lox/lox mice. Transplanted islets were examined postmortem by hematoxylin and eosin staining (A) and immunohistochemical analysis (B). Intact islets were found beneath the kidney capsule at the site of transplantation and stained positively for both insulin and glucagon (panel B). However, neither body fat nor plasma leptin levels were reduced in RIP-Cre/STAT3lox/lox recipients (C). Islet functionality was confirmed by transplantation into C57BL/6-Ins2Akita/+ mice and STZ-treated STAT3lox/lox mice (data not shown). Arrow from the boxed area in panel A shows the same area at a higher magnification.
Assessment of RIP-Cre-mediated recombination by immunohistochemical analysis and Western blotting.
Pancreases were dissected from anesthetized STAT3lox/lox and RIP-Cre/STAT3lox/lox mice, and the specimens were immediately fixed in Bouin's solution. The organs were embedded in paraffin and sectioned onto glass slides, which were then deparaffinized sequentially in a graded series of xylene, ethanol, and water. Slides were washed in phosphate-buffered saline (PBS), blocked with 1% bovine serum albumin, and incubated with primary antibody solution in a humid chamber. Slides were washed in PBS, and secondary antibody was applied in the dark, washed, and coverslipped for microscopy. Primary antibodies used were goat anti-STAT3 from Santa Cruz Biotechnology (catalog no. sc482) at a 1:20 dilution and guinea pig anti-human insulin and glucagon from Linco Research (St. Louis, Mo.), also used at a 1:20 dilution. Secondary antibodies were from Molecular Probes, Inc. (Eugene, Oreg.) and included goat anti-rabbit immunoglobulin G (IgG) conjugated to Alexa Fluor 488 (green) and goat anti-guinea pig IgG conjugated to Alexa Fluor 594 (red). Imaging was performed with a Zeiss LSM 510 confocal microscope. To determine the amount of STAT3 protein in islets, 25 islets each from RIP-Cre/STAT3lox/lox mice or from control STAT3lox/lox littermates were lysed directly in sample buffer and run on an SDS 8%-polyacrylamide gel. Antibodies against tubulin and SHP-2 were purchased from Sigma (St. Louis, Mo.) or generated as described earlier (25).
To assess Cre recombinase activity in the hypothalamus, ROSA26 Cre reporter mice, which carry a Cre-activatable lacZ transgene, were crossed with RIP-Cre/STAT3lox/lox mice (38). Progenies from such crosses are 25% double positive for the RIP-Cre and ROSA26 allele. Two-week-old RIP-Cre-containing progenies with or without the ROSA26 allele were used to detect the site of RIP-Cre expression by lacZ staining. Briefly, mice were deeply anesthetized and perfused with 0.2% glutaraldehyde in PBS. Whole mouse brain was rapidly removed, put into a mold, and kept in place by immersing into optimum cutting temperature compound and frozen on dry ice. Seven-micrometer sections were made and stained for the presence of lacZ activity with the β-galactosidase staining kit (Roche, Indianapolis, Ind.). In parallel, hypothalamus and cerebrum were snap-frozen in liquid nitrogen, and total protein extracts were prepared to determine the level of STAT3 proteins in each sample by Western blot analysis.
Immunohistochemical staining for OB-Rb and STAT3 in hypothalamic slices.
Mouse brains for immunostaining of long-form leptin receptor (OB-Rb) and STAT-3 were harvested and cryoembedded in tissue freezing medium (Triangle Biomedical Sciences, Durham, N.C.) following gravity-forced transcardial perfusion with Ca2+-free Tyrode's solution and formalin-picric acid fixative (30 ml each; 25-cm pressure column) (14). For β-galactosidase staining, mouse brains from Rip-Cre × Rosa26 and non-Cre × Rosa26 crosses were harvested and cryoembedded in tissue freezing medium without fixation. Both sets of cryoembedments were rapidly frozen by partial immersion in liquid nitrogen-supercooled 2-methyl-butane. Eight-micrometer cryosections were cut from prepared blocks and thaw mounted on silanated microscope slides (Fisher Scientific, Pittsburgh, Pa.). Embedments and mounted slides were prepared according to standard histologic practices and stored at −80°C between preparation, sectioning, and staining. Rabbit anti-OB-Rb used for immunohistochemistry was raised in-house against a glutathione S-transferase fusion protein fused to the cytoplasmic domain of mouse OB-Rb. Antibody was affinity purified by using the same antigen. Mouse anti-STAT-3 (clone 84) was obtained from Transduction Laboratories (Lexington, Ky.).
Staining was carried out according to previously described immunofluorescence methods (6, 11). Briefly, sections were hydrated and blocked against nonspecific secondary binding by utilizing mouse-on-mouse blocking kit components prior to subjecting the sections to 1-h incubation in either OB-Rb/STAT-3 primary antibody cocktail in PBS or PBS substitution controls. Unbound primary antibody was washed from slides with PBS, and OB-Rb/STAT-3 was detected with biotinylated anti-mouse IgG and Cy-3 anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, Pa.). Streptavidin-fluorescein was used to detect bound anti-mouse, followed by final PBS washes and coverslipping with Vectashield fluorescence mounting medium. Except where noted, all immunochemical reagents were obtained from Vector Laboratories (Burlingame, Calif.).
Review and photography of histologic preparations were carried out on a Zeiss Axioplan 2i photomicroscope equipped with epifluorescence, differential interference-contrast illumination, and computer-controlled motorization. Photomicrography was achieved by using this microscope and an Axiocam monochromatic charge-coupled device camera with a CRI RGB color filter. Single z-plane images (β-galactosidase) and multiple z-plane image stacks (OB-Rb/STAT-3) were captured by using Openlab 3.0.3 acquisition, analysis, and deconvolution software (Improvision, Inc., Boston, Mass.). Image stacks of OB-Rb/STAT-3 immunostaining, comprising 32 250-nm-interval images, were deconvolved to fluorophore-separate, confocal representations and subsequently pseudocolored and overlaid with Adobe Photoshop version 5.5.
Taqman PCR.
Total RNA from the hypothalamus of RIP-Cre/STAT3lox/lox mice and STAT3lox/lox mice were isolated with TRIzol reagent. RNA samples were treated with DNA-free from Ambion (Austin, Tex.) and reverse transcribed to first-strand cDNA with the reverse transcription reagent at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min. To quantitate mRNA levels encoding neuropeptide Y (NPY), pro-opiomelanocortin (POMC), agouti-related protein (AGRP), and cocaine- and amphetamine-regulated transcript (CART), real-time PCR was performed. The primer sequences were NPY, CTACTCCGCTCTGCGACACT (forward), AGTGTCTCAGGGCTGGATCTC (reverse); POMC, GAGGCCACTGAACATCTTTGTC (forward), GCAGAGGCAAACAAGATTGG (reverse); AGRP, CTTTGGCGGAGGTGCTAGA (forward), GGACTCGTGCAGCCTTACACA (reverse); and CART, CGAGAAGAAGTACGGCCAAGTC (forward), CCGATCCTGGCCCCTTT (reverse). The reaction was performed on a 96-well plate and detected by an ABI Prism 7000 sequence detection system. The relative levels of expression of each neuropeptide were compared between RIP-Cre/STAT3lox/lox mice and STAT3lox/lox mice after normalization with cyclophilin.
Islet isolation and insulin secretion assays.
Islets were isolated from RIP-Cre/STAT3lox/lox mice and control littermates by centrifugation through a Ficoll gradient. Briefly, pancreases were harvested and transferred to a 15-ml conical tube containing 5 ml of cold Liberase enzyme solution (Roche) and were incubated for ∼20 to 25 min in a 37°C water bath. The sample was shaken vigorously for 10 or 30 s and transferred into a new 15-ml conical tube and spun; supernatant was siphoned off, to which 40 ml of cold quenching buffer was added, and the sample was vortexed. The sample was then mixed with 25% Ficoll, and 23, 21, and 13% concentrations of Ficoll were sequentially added to the top. Following spinning at ∼450 to 500 × g (1,800 revolutions/min; SORVALLRT7 PLUS) for 20 min at 4°C, islets form a band at levels of Ficoll between 23 and 21%. Islets were collected and incubated with culture media for insulin secretion assays.
Plasma preparation and analysis.
Blood samples were collected from the tail vein into Eppendorf tubes coated with EDTA. Plasma was prepared by low-speed centrifugation (5,000 × g for 5 min) and used for measurement of glucose, insulin, and leptin. Plasma glucose was measured by Sigma Diagnostics glucose (Trinder) reagent. Insulin and leptin levels in mice plasma were measured with a rat insulin radioimmunoassay kit and rat leptin radioimmunoassay kit from Linco Research, respectively.
Intracerebroventricular infusion of leptin.
Mice were anesthetized with tribromoethanol at 0.4 mg/g of body weight. Intracerebroventricular (ICV) infusion of leptin has been described previously (13). Briefly, the calvarium was exposed under anesthesia, and a 0.7-mm hole was drilled upon the bregma. A 28-gauge cannula (brain infusion kit II; Alza) was implanted into the third ventricle according to the following coordinates: midline, −0.3 anterior pituitary, 3 mm ventral (zero-point bregma). The cannula was secured to the skull and attached with tubing to an osmotic pump (Alza) placed in the dorsal subcutaneous space of the animal. The pump delivers human leptin (Sigma) at 8 ng/h for 28 days. Mice were housed in single cages with ad libitum access to food and water.
Glucose tolerance test.
Mice at 2 months of age were fasted overnight with free access to water. Glucose was injected into the peritoneal cavity at 2 g/kg of body weight. Blood samples were collected immediately before and at 15, 30, 60, 120, 180, and 240 min after glucose injection. Blood glucose levels were determined as described above.
Northern blot analysis of adipose tissue gene expression.
Total RNA was isolated from the white adipose tissue of both control (STAT3lox/lox) and knockout (RIP-Cre/STAT3lox/lox) mice as described previously (17). RNA was resolved on formaldehyde-agarose gel, transferred to a nylon membrane, and hybridized with a random primer-labeled cDNA probe.
Islet transplantation.
Islet transplantation was performed by following previously published procedures (27). Briefly, 400 islets were isolated from C57BL/6J mice (The Jackson Laboratory) and implanted beneath the renal capsule of three recipient groups. Group A consisted of 10-week-old C57BL/6J-Ins2Akita males (n = 2) used as positive control, and group B comprised 12-week-old RIP-Cre/STAT3lox/lox mice (n = 6), which were used to test if wild-type islets can rescue the phenotype in these mice. A third group, group C, included age-matched RIP-Cre/STAT3lox/lox mice (n = 4) that were injected with PBS instead of islets. Because RIP-Cre/STAT3lox/lox mice are on a mixed C57BL/6 × 129/SV background, an additional group, group D, consisting of STAT3lox/lox littermates of RIP-Cre/STAT3lox/lox mice, was treated with streptozotocin (STZ) and used as islet recipients to monitor the survival and functionality of islets under such mixed background. Before transplantation, mice were anesthetized with the same cocktail as described above. Four hundred islets or sterile PBS in a volume of 50 μl was carefully deposited under the kidney capsule of recipients by the use of a small catheter (PE50 tubing; Becton Dickinson Company). The graft function was followed by serial blood glucose measurements of diabetic C57BL/6J-Ins2Akita mice by using the 510-A glucose kit (Sigma). Mice had free access to water and were given a standard diet. Plasma glucose, leptin, and insulin levels were determined twice weekly posttransplantation. Twenty-five days following transplantation, all mice were sacrificed, and kidneys were removed for histological examination and immunohistochemical analysis.
Statistical analysis.
Results were analyzed using GraphPad Prism software. Results are expressed as means ± standard errors of the means. Differences were considered significant when P values were <0.05.
RESULTS
Generation of RIP-Cre/STAT3lox/lox mice and confirmation of STAT3 deletion from β cells.
In earlier studies, RIP-Cre mice were found to be identical to wild-type mice in terms of body weight homeostasis and glucose metabolism (21). To simplify the analysis, we used progenies of crosses between STAT3lox/lox (control) and RIP-Cre/STAT3lox/lox (knockout) mice throughout the study. Both genotypes of mice were born at the expected 1:1 Mendelian frequency, suggesting that there is no embryonic lethality due to STAT3 deletion from RIP-Cre-mediated recombination.
To assess the efficiency of Cre-mediated STAT3 deletion in pancreatic β cells in RIP-Cre/STAT3lox/lox mice, the localization and quantity of STAT3 protein were determined by immunohistochemical analysis and Western blotting. To distinguish the different cell types in the islet and to localize STAT3 within the same islet, we costained islets with primary antibodies against insulin, glucagon, and STAT3. Secondary antibodies were conjugated to Alexa Fluor dyes that recognize insulin (green; data not shown), STAT3 (green), or glucagon (red). In control STAT3lox/lox mice, STAT3 expression was detected throughout the islets (Fig. 1A). In contrast, STAT3 staining was absent in cells that are positive for insulin in islets of RIP-Cre/STAT3lox/lox mice but remained in glucagon-positive cells in the periphery, demonstrating that STAT3 has been efficiently and selectively deleted from the β cells in RIP-Cre/STAT3lox/lox mice. When islet extracts were blotted for STAT3 protein, signal from RIP-Cre/STAT3lox/lox mice was barely detectable. This is congruent with the results of immunohistochemical analysis (Fig. 1B). The remaining STAT3 signal was most likely from non-β cells in the islets of Langerhans, such as cells that produce glucagon, somatostatin, and pancreatic polypeptide. When the same filter was blotted for SH2 domain-containing protein tyrosine phosphatase 2 or α-tubulin, equivalent signal intensity was observed in all lanes, demonstrating that samples were loaded evenly.
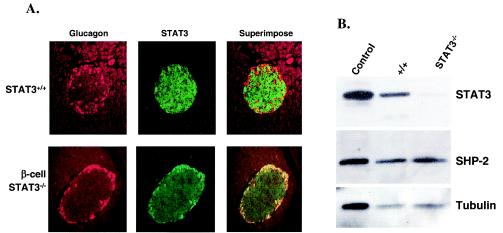
FIG. 1.
Assessment of STAT3 deletion in pancreatic β cells of RIP-Cre/STAT3lox/lox mice. (A) Fluorescence immunohistochemical analysis for STAT3 on serial pancreatic sections of control mice (STAT3+/+) and RIP-Cre/STAT3lox/lox mice (β-cell STAT3−/−). Pancreata from 3-month-old control or RIP-Cre/STAT3lox/lox mice were used to prepare sections for immunohistochemical analysis. Sections were stained with an antibody against glucagon (Glucagon), followed by detection with Alexa Fluor 594-conjugated goat anti-guinea pig IgG. The same sections were also stained with an antibody against STAT3 (STAT3), followed by detection with Alexa Fluor 488-conjugated goat anti-rabbit IgG. Images for STAT3 and glucagon were superimposed (Superimpose) to reveal costaining of STAT3 and glucagon. Residual staining of STAT3 in the islet of RIP-Cre/STAT3lox/lox mice does not overlap with insulin staining. (B) Western blotting of islet extracts from STAT3lox/lox mice (+/+) and RIP-Cre/STAT3lox/lox mice (STAT3−/−). Ten micrograms of islet extract was loaded on each lane and sequentially blotted for STAT3, SHP-2, and α-tubulin. The control lane contains 10 μg of protein from lysate of 293 cells. Islet extract from RIP-Cre/STAT3lox/lox mice showed barely detectable STAT3 signal, which arose from non-β cells in the islets.
Activity of RIP-Cre transgene outside pancreatic β cells.
The RIP-Cre transgenic mouse line used in the present study has been used in other instances to delete several other genes from the pancreatic β cells, such as those encoding glucokinase, the insulin receptor, and the receptor for insulinlike growth factor 1 (21, 22, 32). In all cases, the RIP-Cre-mediated gene knockout was clearly demonstrated to have occurred in β cells. However, because several recent studies have shown that the RIP-driven transgene also expresses outside islets (44, 45), we determined whether RIP-Cre activity could be found outside β cells in RIP-Cre/STAT3lox/lox mice by using STAT3lox/lox mice as a negative control group. This is a necessary control because of the two common pitfalls of conditional-knockout experiments: that gene deletion in a given tissue is rarely complete and that the tissue specificity of a promoter is always relative. If deletion of a gene occurs outside of the tissue intended, then the possibility that such deletion may contribute to the observed phenotype needs to be considered.
To determine the extraislet expression of the RIP-Cre transgene and whether it causes STAT3 deletion at these sites during mouse ontogeny, we performed genomic DNA PCR, Western blotting, Cre reporter staining, and immunohistochemical analysis (Fig. 2 and data not shown). As illustrated in Fig. 2A, primers used for PCR analyses were designed to surround both LoxP sequences on STAT3 genomic DNA (primers a and c) so that amplification is easily achieved only when RIP-Cre-mediated recombination has occurred (3.45 kb without recombination versus 0.75 kb after recombination). By preparing genomic DNA from the cerebrum, hypothalamus, fat, liver, skeletal muscle, and pancreas, STAT3 deletion from the pancreas of RIP-Cre/STAT3lox/lox mice was readily detected, as expected. Unexpectedly, STAT3 deletion was also detectable in the hypothalamus but not in any other tissues examined (Fig. 2B).
To further confirm that STAT3 deletion in the hypothalamus has occurred, we performed Cre reporter staining (Fig. 2C) and Western blotting of hypothalamic extracts for STAT3 protein (Fig. 2D). Immunohistochemical analysis of brain slices (sectioned through the hypothalamus) for STAT3 did not reveal distinguishable differences between STAT3lox/lox and RIP-Cre/STAT3lox/lox mice (data not shown). Western blotting of hypothalamic extract from STAT3lox/lox and RIP-Cre/STAT3lox/lox mice (Fig. 2D) also revealed no distinguishable differences. However, when lacZ staining was performed on whole-brain sections to detect Cre activity in RIP-Cre/ROSA26 mice, diffuse staining was observed throughout the hypothalamus but not in other brain regions, a finding that was in agreement with the genomic DNA PCR result (Fig. 2B). These results suggest that the phenotype of RIP-Cre/STAT3lox/lox mice may arise from STAT3 deletion from both the β cells and the hypothalamus.
RIP-Cre/STAT3lox/lox mice exhibit mild hyperglycemia, hyperinsulinemia, and impaired glucose disposal.
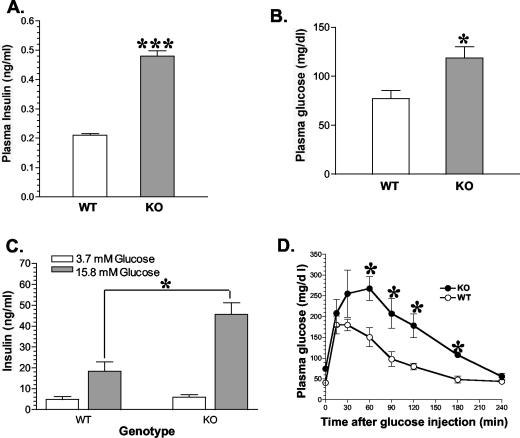
Earlier studies in other laboratories have demonstrated that STAT3 DNA binding activity in β cells can be activated by leptin and other cytokines (29, 30). To determine the consequences of RIP-Cre-mediated STAT3 deletion in vivo on β-cell function, we first measured plasma immunoreactive insulin and glucose levels. Figure 3A shows that in newly weaned mice, the fasting plasma insulin level more than doubled in RIP-Cre/STAT3lox/lox mice compared with that of control STAT3lox/lox littermates (0.48 ± 0.02 versus 0.21 ± 0.01 ng/ml, P = 0.0001). Fasting-state plasma glucose levels were also elevated in RIP-Cre/STAT3lox/lox mice (118 ± 11 versus 77 ± 8 mg/dl, P = 0.01) (Fig. 3B).
FIG. 3.
Mild fasting-state hyperinsulinemia, hyperglycemia, increased insulin secretion, and impaired glucose tolerance in RIP-Cre/STAT3lox/lox mice. Fasting-state levels of plasma insulin (A) and glucose (B) were both increased in RIP-Cre/STAT3lox/lox mice. Insulin secretion of isolated islets from RIP-Cre/STAT3lox/lox mice was also significantly increased at stimulating glucose concentration (15.8 mM) but not under basal glucose concentration (3.7 mM) (C). RIP-Cre/STAT3lox/lox mice exhibited reduced glucose tolerance (D), as revealed by delayed glucose disposal at the indicated time points. WT, wild-type STAT3lox/lox mice; KO, RIP-Cre/STAT3lox/lox mice. *, P < 0.05; ***, P < 0.0001.
To determine if there were changes in the size and/or number of islets between RIP-Cre/STAT3lox/lox mice and control littermates, we assayed the insulin content of islets as well as their insulin secretion profiles upon glucose stimulation. Isolated islets from RIP-Cre/STAT3lox/lox mice showed enhanced insulin release at stimulating glucose concentrations (15.8 mM) but not under basal glucose concentrations (3.5 mM) (Fig. 3C). While the number of islets obtained per mouse was similar, islets from RIP-Cre/STAT3lox/lox mice appeared to be larger than those from control littermates. Insulin content was also increased in RIP-Cre/STAT3lox/lox mice (data not shown). Taken together, these results suggest that in the absence of STAT3 caused by RIP-Cre-mediated recombination, both insulin synthesis and glucose-stimulated secretion are increased in vivo.
Fasting-state hyperglycemia and hyperinsulinemia levels in RIP-Cre/STAT3lox/lox mice suggested that these mice might be insulin resistant. To test this possibility further, we performed the glucose tolerance test on 1-month-old fasted mice. Compared with littermate controls, RIP-Cre/STAT3lox/lox mice exhibited delayed clearance of glucose at all time points after glucose injection (Fig. 3D), demonstrating that RIP-Cre-mediated conditional STAT3 deletion causes impaired glucose disposal and insulin resistance.
RIP-Cre/STAT3lox/lox mice are hyperphagic and develop early-onset obesity.
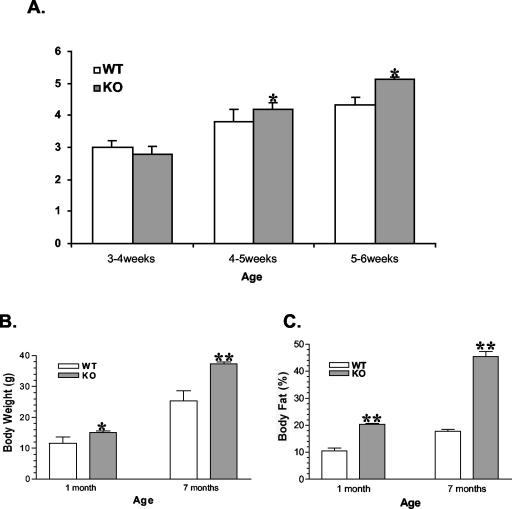
The evidence of a hypothalamic deletion of STAT3 led us to study body weight regulation. Although the body weights of both genotypes were not significantly different at weaning, increased adiposity was already evident before weaning, as assessed by mouse NMR analyzer measurement (data not shown). Immediately after mice were weaned, food intake of the RIP-Cre/STAT3lox/lox mice was higher than that of STAT3lox/lox littermates (Fig. 4A). Increased food intake persists well into adulthood (data not shown). Correspondingly, at 1 and 7 months of age, body weight of RIP-Cre/STAT3lox/lox mice was 30 and 35% higher than that of the STAT3lox/lox controls (15.06 ± 0.60 g versus 11.54 ± 0.80 g [P = 0.0055] at 1 month; 37.38 ± 0.60 g versus 25.33 ± 1.00 g [P < 0.0001] at 7 months) (Fig. 4B), respectively.
FIG. 4.
Increased food intake (A; units are grams per day per mouse), body weight (B), and adiposity (C) in RIP-Cre/STAT3lox/lox mice. Male RIP-Cre/STAT3lox/lox mice and control littermates were housed in single cages with free access to food and water. Food intake and body weight were measured daily. Body weight (B) and body fat (C) of mice at 1 and 7 months are shown. In all cases, the number of mice ranged from six to eight. WT, wild-type STAT3lox/lox mice; KO, RIP-Cre/STAT3lox/lox mice. *, P < 0.05; **, P < 0.01. Similar results were observed in female mice.
To determine if increased body weight was due to an increase in the weight of all organs in the mouse or to increased adiposity, we measured the percentage of body fat of STAT3lox/lox and RIP-Cre/STAT3lox/lox mice. These parameters were obtained by the use of a PIXImus mouse densitometer (Lunar Corporation) that measured body composition by using DEXA (31). We determined the percentage of body fat of mice at different ages. Figure 4C shows that at 1 and 7 months of age, the fat content levels of RIP-Cre/STAT3lox/lox mice were also statistically significantly higher than those of the STAT3lox/lox controls at both time points. At 1 month, the percentage of body fat was 10.54% ± 0.32% for STAT3lox/lox mice and 20.38% ± 0.34% for RIP-Cre/STAT3lox/lox mice, respectively; at 7 months of age, STAT3lox/lox mice had 17.78% ± 0.20% of body fat, while RIP-Cre/STAT3lox/lox mice contained 45.44% ± 1.90% of body fat—levels which were significantly higher than those of STAT3lox/lox mice (P < 0.001%). Both values were, however, lower than those of the Leprdb mice, which are deficient in leptin signaling (20).
RIP-Cre/STAT3lox/lox mice show increased leptin expression.
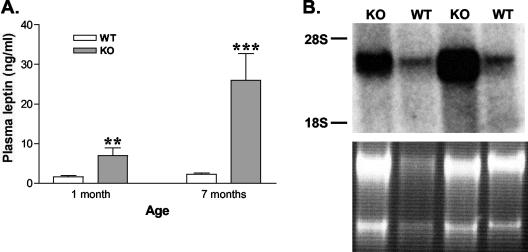
The increased body weight and adiposity are indications that are reminiscent of the syndromes associated with defective leptin signaling, such as those shown in Lepob and Leprdb mice. We compared plasma leptin levels as well as leptin gene expression in the adipose tissue of STAT3lox/lox mice and RIP-Cre/STAT3lox/lox mice. At 1 month of age, the plasma leptin level of RIP-Cre/STAT3lox/lox mice is more than fourfold higher than that in control littermates (6.9 ± 2.0 versus 1.6. ± 0.4 ng/ml). At 7 months of age, the difference was more pronounced, increasing to 26.0 ± 6.8 ng/ml compared with that of controls, 2.3 ± 0.3 ng/ml. This increase was mediated, at least in part, by increased leptin expression: when total RNA from the adipose tissue was blotted for leptin, significantly increased mRNA levels were detected in RIP-Cre/STAT3lox/lox mice (Fig. 5B). While both body weight and percentage of body fat are lower in RIP-Cre/STAT3lox/lox mice than in Leprdb mice, leptin mRNA levels in the adipose tissue and circulating leptin protein levels are comparable (26).
FIG. 5.
Increased circulating leptin (A) and mRNA (B) levels in RIP-Cre/STAT3lox/lox mice. Plasma was obtained from mice at 1 or 7 months of age. Circulating leptin levels were measured with a radioimmunoassay kit. There were five to eight mice in each group. Elevated plasma leptin is observed in RIP-Cre/STAT3lox/lox mice (KO) at both time points (panel A). Panel B shows a Northern blot for leptin mRNA from the white adipose tissue of wild-type littermates (WT) or RIP-Cre/STAT3lox/lox mice (KO). RNA loading is shown in the bottom by ethidium bromide staining. **, P < 0.01; ***, P < 0.0001.
RIP-Cre/STAT3lox/lox mice are partially resistant to central leptin action on adiposity.
Increases of food intake, body weight, adiposity, and plasma leptin levels were all suggestive of a defect in central leptin action. To address this eventuality, we tested whether centrally administered leptin could reduce body fat content in RIP-Cre/STAT3lox/lox mice. When given via ICV infusion into Lepob mice or wild-type mice, leptin induced a complete disappearance of body fat. Both male and female RIP-Cre/STAT3lox/lox mice at 2 months of age as well as their STAT3lox/lox littermates were tested for their ability to respond to centrally administered leptin through ICV infusion. Lepob mice were included as a control. As shown in Fig. 6A, leptin infusion into Lepob mice results in a decrease of body weight by more than half, similar to previous results reported by other groups (15). The percentage of body fat of treated Lepob mice was also decreased from 53 to 4%, as measured by NMR (Fig. 6B). The 4% remaining body fat in Lepob mice after leptin treatment may reflect residual body fat in nonadipose tissues or baseline reading of the instrument, since dissection of sacrificed mice at the end of 4-week ICV leptin treatment did not reveal any visible fat depot (data not shown). Similar loss of body fat was also observed in STAT3lox/lox mice, as assessed by both NMR measurement and dissection (Fig. 6B and data not shown), though body weight remained essentially unchanged. However, ICV leptin treatment of RIP-Cre/STAT3lox/lox mice failed to achieve the complete loss of body fat (Fig. 6B). In agreement with the NMR measurement, sacrificed RIP-Cre/STAT3lox/lox mice showed an abundance of adipose tissue in the abdominal cavity (data not shown). These results demonstrate that centrally administered leptin is only partially active in RIP-Cre/STAT3lox/lox mice.
Because STAT3 is a downstream effector of leptin (43), we determined whether STAT3 deletion had occurred in OB-Rb-positive neurons. Double immunostaining of hypothalamic slices with antibodies against OB-Rb and STAT3 revealed that STAT3 is deleted in a subset of OB-Rb-positive neurons (Fig. 6C). We also performed quantitative real-time PCR analysis for brain neuropeptides that are known to be involved in leptin-regulated feeding behavior (Fig. 6D). Consistent with the phenotype of RIP-Cre/STAT3lox/lox mice, expression levels of NPY showed a trend toward being increased in RIP-Cre/STAT3lox/lox mice, while those of other neuropeptides were not significantly altered.
Increased leptin mRNA transcription and protein synthesis observed in RIP-Cre/STAT3lox/lox mice might be a feedback mechanism in response to the impairment of central leptin action. Litter size or nursing of the young is similar to that found in STAT3lox/lox mice (data not shown). Even though central leptin action with regard to adiposity is impaired, further studies are required to elucidate the mechanism underlying the obesity phenotype in RIP-Cre/STAT3lox/lox mice, since other factors, such as CNTF, also cause weight reduction through activation of the STAT3 pathway (28).
Transplantation of islets from control mice fails to correct the phenotypes of RIP-Cre/STAT3lox/lox mice.
To address whether the obesity and insulin resistance phenotypes in RIP-Cre/STAT3lox/lox mice were due to the loss of STAT3 function in pancreatic β cells or in hypothalamic neurons, we performed an islet transplantation experiment on 12-week-old RIP-Cre/STAT3lox/lox mice. Islets from C57BL/6J mice were isolated and transplanted under the kidney capsule of recipients, per published protocols (27). Because RIP-Cre/STAT3lox/lox mice are on a mixed C57BL/6 × 129/SV background, STAT3lox/lox littermates of RIP-Cre/STAT3lox/lox mice were treated with STZ and used as islet recipients to monitor survival and functionality of islets under such mixed background. C57BL/6-Ins2Akita/+ mice, a new mouse model of insulin-dependent diabetes mellitus, were also transplanted as a positive control (27). RIP-Cre/STAT3lox/lox mice were implanted with 400 islets or were treated with PBS. Food intake, body weight, body fat, and levels of glucose, insulin, and leptin in plasma were measured weekly for 3 weeks before all mice were sacrificed for histological examination. While blood glucose levels remained stable and at a normal range for C57BL/6-Ins2Akita/+ and STZ-treated STAT3lox/lox littermates, no statistically significant correction was detected in RIP-Cre/STAT3lox/lox islet recipients compared with those receiving PBS (data not shown). Fasting-state plasma glucose levels (islet group, 73.88 ± 5.55 mg/dl; PBS group, 92.31 ± 12.86 mg/dl) and insulin levels (islet group, 0.488 ± 0.055 ng/ml; PBS group, 0.559 ± 0.084 ng/ml) remained similar between RIP-Cre/STAT3lox/lox mice that received islets or PBS, as did food intake (islet group, 8.55 ± 0.85 g/mouse/3 days; PBS group, 9.35 ± 0.30 g/mouse/3 days) and body weight (islet group, 27.68 ± 0.86 g; PBS group, 27.50 ± 0.32 g). Hematoxylin and eosin staining as well as immunohistochemical analysis of kidney sections at the implantation site demonstrated that implanted islets were well granulated and positively stained for both insulin and glucagon (Fig. 7A and B). However, body fat, plasma leptin (Fig. 7C), and other hormonal and metabolic profiles showed no statistically significant improvement over sham-operated PBS controls. Even though both body fat and plasma leptin levels tended to decrease compared to those of controls, the absolute values are still much higher than those for control littermates (Fig. 6B and 5A). Taken together, these results suggest that transplantation of wild-type islets fails to reverse the obese and insulin-resistant phenotype of RIP-Cre/STAT3lox/lox mice.
DISCUSSION
By performing conditional gene targeting with the Cre-Lox technology, we demonstrate that the transcription factor STAT3 plays crucial roles in the control of food intake, body weight, insulin sensitivity, and leptin sensitivity. RIP-Cre/STAT3lox/lox mice exhibit increased adiposity before weaning and increased food intake immediately after weaning, when they are also mildly hyperglycemic and hyperinsulinemic compared to STAT3lox/lox littermates. Adult RIP-Cre/STAT3lox/lox mice are obese and hyperleptinemic, showing impaired glucose tolerance and impaired response to central leptin action upon reduction of body fat.
This study demonstrates that RIP-Cre caused quantitative STAT3 deletion in β cells and in a subset of neurons in the hypothalamus. Because RIP-Cre/STAT3lox/lox mice are partially resistant to central leptin infusion, and since transplantation of wild-type islets into RIP-Cre/STAT3lox/lox mice failed to reverse their phenotypes, STAT3 deletion in the hypothalamus is likely responsible for the obese and insulin-resistant phenotype present in our mouse model. Interestingly, Western blotting of hypothalamic extracts did not reveal a decrease of STAT3 protein levels in RIP-Cre/STAT3lox/lox mice, suggesting that STAT3 action in a subset of cells is critical to the maintenance of normal body weight and glucose homeostasis.
Our initial intent was to determine the function of STAT3 in β cells by the selective deletion of STAT3 in this cell type. Earlier studies in other laboratories have demonstrated that STAT3 DNA binding activity in β cells can be activated by leptin and other cytokines (29, 30). Since the target genes of STAT3 in this cell type are not very well defined, deletion of STAT3 from this site was performed in the present study in order to elucidate the functional significance of STAT3 activation in β cells. This goal was achieved by our demonstration that STAT3 protein is indeed diminished in islets of RIP-Cre/STAT3lox/lox mice by independent criteria, i.e., immunocytochemical analysis for STAT3 in pancreas sections and Western blotting of islet extracts. Islets from RIP-Cre/STAT3lox/lox mice harbor altered glucose-stimulated insulin secretion as well as increased triglyceride content (Fig. 3C and data not shown).
The unexpected obesity and central leptin resistance in RIP-Cre/STAT3lox/lox mice led us to further examine the potential involvement of STAT3 deletion in extraislet sites that may have accounted for the observed phenotype. Genomic DNA PCR performed on tissues of RIP-Cre/STAT3lox/lox mice and β-galactosidase staining of progenies between RIP-Cre mice and the reporter strain Rosa 26 mice both revealed that STAT3 deletion had also occurred in the hypothalamus but in no other tissues. The elevated leptin mRNA and protein levels in RIP-Cre/STAT3lox/lox mice are consistent with a defect in central leptin action, a finding confirmed by the impairment of ICV-delivered leptin to completely reduce body fat (Fig. 6). That the leptin pathway of STAT3 activation, but not those of other growth factors or cytokines, is impaired in RIP-Cre/STAT3lox/lox mice is further supported by the recent finding that mice defective for STAT3 binding to the leptin receptor are also obese (7).
We performed islet transplantation experiments in an attempt to rescue the obese and other phenotypes of RIP-Cre/STAT3lox/lox mice and to test whether STAT3 deletion from this site alone is responsible for the phenotypes that we observed. There are precedents that defects in certain pathways at one site can affect actions of insulin and other hormones at different sites. One example is the muscle and liver insulin resistance in mice with selective GLUT-4 deletion from the adipose tissue (1). However, in the present study, RIP-Cre/STAT3lox/lox recipients of wild-type islets did not show improvement compared to sham-operated controls in all parameters measured, supporting the notion that STAT3 deletion in the islets may not be the cause of the obese and insulin-resistant phenotype of RIP-Cre/STAT3lox/lox mice. However, if deletion of STAT3 in the islets early in life prior to islet transplantation caused irreversible hormonal and metabolic changes in RIP-Cre/STAT3lox/lox mice, rescue by wild-type islets would not be possible at a later stage. Because the increased adiposity phenotype of RIP-Cre/STAT3lox/lox mice occurs before weaning, as determined by NMR analyzer measurement (data not shown), this possibility cannot be excluded. Alternatively, STAT3 deletion in other cryptic sites in the body due to leaky Cre expression could also contribute to the phenotype observed.
In summary, we have demonstrated, by using conditional STAT3 deletion with RIP-Cre mice, that STAT3 is essential in the regulation of food intake, body weight, and metabolism and that STAT3 deletion in the pancreas may not be involved in the phenotypic abnormalities of RIP-Cre/STAT3lox/lox mice. Our findings thus indicate that STAT3 activity in hypothalamic neurons might be a useful target for future study in the pharmacological management of obesity, diabetes, and related metabolic disorders.
Acknowledgments
We thank Kiyoshi Takeda and Shizuo Akira (Osaka University, Japan) for STAT3lox/lox mice, Mark Magnuson (Vanderbilt University) for RIP-Cre mice, Bob Hammer (UT Southwestern) for the ROSA26 mice and advice on LacZ staining, Edward H. Leiter (The Jackson Laboratory) for advice on islet transplantation, Joyce Repa for advice on real-time PCR and use of ABI Prism 7000 sequence detection system, and Alexia Thomas for performing DEXA analysis. We also thank Roger Unger, Chris Newgard (Duke University Medical Center), Jeff Friedman (Rockefeller University), Tom Südhof, and Andrew Zinn for many discussions and/or a critical review of the manuscript.
This work was supported by grants to O.K.O. from the Cain Foundation and the National Institutes of Health (grant number HD01463) and to C.L. from JDRF (grant number 5-2002-619) and the National Institutes of Health (grant number DK60137). C.L. is also the recipient of a career development award from the American Diabetes Association.
REFERENCES
- 1.Abel, E. D., O. Peroni, J. K. Kim, Y. B. Kim, O. Boss, E. Hadro, T. Minnemann, G. I. Shulman, and B. B. Kahn. 2001. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 409:729-733. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S. 1999. Functional roles of STAT family proteins: lessons from knockout mice. Stem Cells 17:138-146. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S. 2000. Roles of STAT3 defined by tissue-specific gene targeting. Oncogene 19:2607-2611. [DOI] [PubMed] [Google Scholar]
- 4.Alonzi, T., D. Maritano, B. Gorgoni, G. Rizzuto, C. Libert, and V. Poli. 2001. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene activation in the liver. Mol. Cell. Biol. 21:1621-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonzi, T., G. Middleton, S. Wyatt, V. Buchman, U. A. Betz, W. Muller, P. Musiani, V. Poli, and A. M. Davies. 2001. Role of STAT3 and PI 3-kinase/Akt in mediating the survival actions of cytokines on sensory neurons. Mol. Cell. Neurosci. 18:270-282. [DOI] [PubMed] [Google Scholar]
- 6.Antos, C. L., T. A. McKinsey, N. Frey, W. Kutschke, J. McAnally, J. M. Shelton, J. A. Richardson, J. A. Hill, and E. N. Olson. 2002. Activated glycogen synthase-3β suppresses cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. USA 99:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bates, S. H., W. H. Stearns, T. A. Dundon, M. Schubert, A. W. Tso, Y. Wang, A. S. Banks, H. J. Lavery, A. K. Haq, E. Maratos-Flier, B. G. Neel, M. W. Schwartz, and M. G. Myers, Jr. 2003. STAT3 signalling is required for leptin regulation of energy balance but not reproduction. Nature 421:856-859. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, L. R., T. J. Farruggella, A. Symes, M. L. Karow, G. D. Yancopoulos, and N. Stahl. 1998. Enhancing leptin response by preventing SH2-containing phosphatase 2 interaction with Ob receptor. Proc. Natl. Acad. Sci. USA 95:6061-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman, R. S., P. C. Lourenco, E. Tonner, D. J. Flint, S. Selbert, K. Takeda, S. Akira, A. R. Clarke, and C. J. Watson. 1999. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev. 13:2604-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H., O. Charlat, L. A. Tartaglia, E. A. Woolf, X. Weng, S. J. Ellis, N. D. Lakey, J. Culpepper, K. J. Moore, R. E. Breitbart, G. M. Duyk, R. I. Tepper, and J. P. Morgenstern. 1996. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell 84:491-495. [DOI] [PubMed] [Google Scholar]
- 11.Cianga, P., C. Medesan, J. A. Richardson, V. Ghetie, and E. S. Ward. 1999. Identification and function of neonatal Fc receptor in mammary gland of lactating mice. Eur. J. Immunol. 29:2515-2523. [DOI] [PubMed] [Google Scholar]
- 12.Dandoy-Dron, F., J. M. Itier, E. Monthioux, D. Bucchini, and J. Jami. 1995. Tissue-specific expression of the rat insulin 1 gene in vivo requires both the enhancer and promoter regions. Differentiation 58:291-295. [DOI] [PubMed] [Google Scholar]
- 13.Darnell, J. E. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 14.Ducy, P., M. Amling, S. Takeda, M. Priemel, A. F. Schilling, F. T. Beil, J. Shen, C. Vinson, J. M. Rueger, and G. Karsenty. 2000. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell 100:197-207. [DOI] [PubMed] [Google Scholar]
- 15.Hakansson-Ovesjo, M. L., M. Collin, and B. Meister. 2000. Down-regulated STAT3 messenger ribonucleic acid and STAT3 protein in the hypothalamic arcuate nucleus of the obese leptin-deficient (ob/ob) mouse. Endocrinology 141:3946-3955. [DOI] [PubMed] [Google Scholar]
- 16.Halaas, J. L., K. S. Gajiwala, M. Maffei, S. L. Cohen, B. T. Chait, D. Rabinowitz, R. L. Lallone, S. K. Burley, and J. M. Friedman. 1995. Weight-reducing effects of the plasma protein encoded by the obese gene. Science 269:543-546. [DOI] [PubMed] [Google Scholar]
- 17.Hara, M., X. Wang, T. Kawamura, V. P. Bindokas, R. F. Dizon, S. Y. Alcoser, M. A. Magnuson, and G. I. Bell. 2003. Transgenic mice with green fluorescent protein-labeled pancreatic β cells. Am. J. Physiol. Endocrinol. Metab. 284:E177-E183. [DOI] [PubMed] [Google Scholar]
- 18.Huang, L., and C. Li. 2000. Leptin: a multifunctional hormone. Cell Res. 10:81-92. [DOI] [PubMed] [Google Scholar]
- 19.Huang, L., Z. Wang, and C. Li. 2001. Modulation of circulating leptin levels by its soluble receptor. J. Biol. Chem. 276:6343-6349. [DOI] [PubMed] [Google Scholar]
- 20.Ihle, J. N. 2001. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 13:211-217. [DOI] [PubMed] [Google Scholar]
- 21.Kira, M., S. Sano, S. Takagi, K. Yoshikawa, J. Takeda, and S. Itami. 2002. Stat3 deficiency in keratinocytes leads to compromised cell migration through hyperphosphorylation of p130Cas. J. Biol. Chem. 277:12931-12936. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi, K., T. M. Forte, S. Taniguchi, B. Y. Ishida, K. Oka, and L. Chan. 2000. The db/db mouse, a model for diabetic dyslipidemia: molecular characterization and effects of Western diet feeding. Metabolism 49:22-31. [DOI] [PubMed] [Google Scholar]
- 23.Kulkarni, R. N., J. C. Bruning, J. N. Winnay, C. Postic, M. A. Magnuson, and C. R. Kahn. 1999. Tissue-specific knockout of the insulin receptor in pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96:329-339. [DOI] [PubMed] [Google Scholar]
- 24.Kulkarni, R. N., M. Holzenberger, D. Q. Shih, U. Ozcan, M. Stoffel, M. A. Magnuson, and C. R. Kahn. 2002. β-Cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter β-cell mass. Nat. Genet. 31:111-115. [DOI] [PubMed] [Google Scholar]
- 25.Lee, G. H., R. Proenca, J. M. Montez, K. M. Carroll, J. G. Darvishzadeh, J. I. Lee, and J. M. Friedman. 1996. Abnormal splicing of the leptin receptor in diabetic mice. Nature 379:632-635. [DOI] [PubMed] [Google Scholar]
- 26.Levy, D. E., and C.-K. Lee. 2002. What does Stat3 do? J. Clin. Investig. 109:1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, C., and J. M. Friedman. 1999. Leptin receptor activation of SH2 domain containing protein tyrosine phosphatase 2 modulates Ob receptor signal transduction. Proc. Natl. Acad. Sci. USA 96:9677-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maffei, M., J. Halaas, E. Ravussin, R. E. Pratley, G. H. Lee, Y. Zhang, H. Fei, S. Kim, R. Lallone, S. Ranganathan, P. A. Kern, and J. M. Friedman. 1995. Leptin levels in human and rodent: measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat. Med. 1:1155-1161. [DOI] [PubMed] [Google Scholar]
- 29.Mathews, C. E., S. H. Langley, and E. H. Leiter. 2002. New mouse model to study islet transplantation in insulin-dependent diabetes mellitus. Transplantation 73:1333-1336. [DOI] [PubMed] [Google Scholar]
- 30.Mattson, M. P. 2001. Lose weight STAT: CNTF tops leptin. Trends Neurosci. 24:313-314. [DOI] [PubMed] [Google Scholar]
- 31.Morton, N. M., R. P. de Groot, M. A. Cawthorne, and V. Emilsson. 1999. Interleukin-1β activates a short STAT-3 isoform in clonal insulin-secreting cells. FEBS Lett. 442:57-60. [DOI] [PubMed] [Google Scholar]
- 32.Morton, N. M., V. Emilsson, P. de Groot, A. L. Pallett, and M. A. Cawthorne. 1999. Leptin signalling in pancreatic islets and clonal insulin-secreting cells. J. Mol. Endocrinol. 22:173-184. [DOI] [PubMed] [Google Scholar]
- 33.Nagy, T. R., and A. L. Clair. 2000. Precision and accuracy of dual-energy X-ray absorptiometry for determining in vivo body composition of mice. Obes. Res. 8:392-398. [DOI] [PubMed] [Google Scholar]
- 34.Postic, C., M. Shiota, K. D. Niswender, T. L. Jetton, Y. Chen, J. M. Moates, K. D. Shelton, J. Lindner, A. D. Cherrington, and M. A. Magnuson. 1999. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β-cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274:305-315. [DOI] [PubMed] [Google Scholar]
- 35.Raz, R., C. K. Lee, L. A. Cannizzaro, P. d'Eustachio, and D. E. Levy. 1999. Essential role of STAT3 for embryonic stem cell pluripotency. Proc. Natl. Acad. Sci. USA 96:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sano, S., S. Itami, K. Takeda, M. Tarutani, Y. Yamaguchi, H. Miura, K. Yoshikawa, S. Akira, and J. Takeda. 1999. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18:4657-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sano, S., Y. Takahama, T. Sugawara, H. Kosaka, S. Itami, K. Yoshikawa, J. Miyazaki, W. van Ewijk, and J. Takeda. 2001. Stat3 in thymic epithelial cells is essential for postnatal maintenance of thymic architecture and thymocyte survival. Immunity 15:261-273. [DOI] [PubMed] [Google Scholar]
- 38.Schindler, C. W. 2002. Series introduction: JAK-STAT signaling in human disease. J. Clin. Investig. 109:1133-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schweizer, U., J. Gunnersen, C. Karch, S. Wiese, B. Holtmann, K. Takeda, S. Akira, and M. Sendtner. 2002. Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J. Cell Biol. 156:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 41.Takeda, K., and S. Akira. 2001. Multi-functional roles of Stat3 revealed by conditional gene targeting. Arch. Immunol. Ther. Exp. 49:279-283. [PubMed] [Google Scholar]
- 42.Takeda, K., B. E. Clausen, T. Kaisho, T. Tsujimura, N. Terada, I. Forster, and S. Akira. 1999. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10:39-49. [DOI] [PubMed] [Google Scholar]
- 43.Takeda, K., T. Kaisho, N. Yoshida, J. Takeda, T. Kishimoto, and S. Akira. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: generation and characterization of T cell-specific Stat3-deficient mice. J. Immunol. 161:4652-4660. [PubMed] [Google Scholar]
- 44.Takeda, K., K. Noguchi, W. Shi, T. Tanaka, M. Matsumoto, N. Yoshida, T. Kishimoto, and S. Akira. 1997. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 94:3801-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaisse, C., J. L. Halaas, C. M. Horvath, J. E. Darnell, Jr., M. Stoffel, and J. M. Friedman. 1996. Leptin activation of Stat3 in the hypothalamus of wild-type and ob/ob mice but not db/db mice. Nat. Genet. 14:95-97. [DOI] [PubMed] [Google Scholar]
- 46.Vasavada, R. C., A. Garcia-Ocana, W. S. Zawalich, R. L. Sorenson, P. Dann, M. Syed, L. Ogren, F. Talamantes, and A. F. Stewart. 2000. Targeted expression of placental lactogen in the β cells of transgenic mice results in β-cell proliferation, islet mass augmentation, and hypoglycemia. J. Biol. Chem. 275:15399-15406. [DOI] [PubMed] [Google Scholar]