Abstract
The topology of polytopic membrane proteins is determined by topogenic sequences in the protein, protein-translocon interactions, and interactions during folding within the protein and between the protein and the lipid environment. Orientation of transmembrane domains is dependent on membrane phospholipid composition during initial assembly as well as on changes in lipid composition postassembly. The membrane translocation potential of negative amino acids working in opposition to the positive-inside rule is largely dampened by the normal presence of phosphatidylethanolamine, thus explaining the dominance of positive residues as retention signals. Phosphatidylethanolamine provides the appropriate charge density that permits the membrane surface to maintain a charge balance between membrane translocation and retention signals and also allows the presence of negative residues in the cytoplasmic face of proteins for other purposes.
Keywords: phosphatidylethanolamine, protein topology, positive-inside rule, lactose permease, lipochaperones
INTRODUCTION
Membrane proteins account for 30% of all open reading frames in sequenced genomes, and they fulfill a wide range of central functions, such as solute transport, energy production, membrane and protein biogenesis, intracellular signaling, and cell-cell communication. More than 60% of all drug targets are membrane proteins, making them prime therapeutic targets. The majority of integral membrane proteins are composed of hydrophobic α-helical transmembrane domains (TMs) that zigzag across the membrane and are connected by hydrophilic loops alternately exposed to opposing compartments separated by the membrane lipid bilayer (Figure 1). A fundamental architectural feature of membrane proteins is topological organization, i.e., the number of TMs and their orientation with respect to the plane of the lipid bilayer. The α-helical organization of TMs positions the predominantly hydrophobic amino acid side chains in direct contact with the membrane lipid bilayer interior and allows the hydrogen bonds of the polypeptide backbone to be satisfied within the α-helical backbone. Thus, the formation of a folded α-helix is thermodynamically favored for TMs of integral membrane proteins and is the predominant motif among all integral membrane proteins. A less common motif, not considered here, is where hydrogen bonding is satisfied between polypeptide backbones arranged in an antiparellel β-pleated sheet resulting in a barrel structure with hydrophilic side chains facing an aqueous interior pore and hydrophobic amino acids facing a hydrophobic exterior of the barrel.
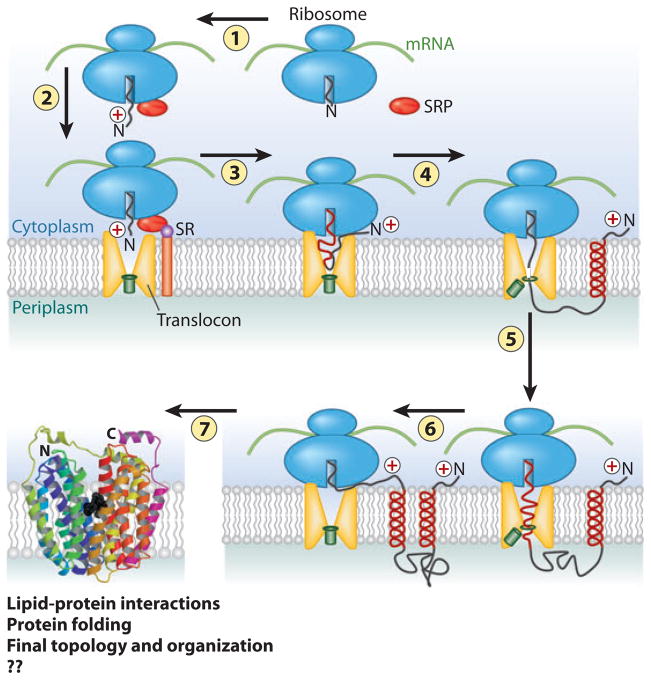
Figure 1.
Overview of membrane protein synthesis, membrane insertion, and assembly. 1. As the N-terminal domain of a nascent polypeptide chain emerges from the ribosome, it complexes with the signal recognition particle (SRP). 2. The resulting complex is delivered to the membrane in association with the translocon through binding with the SRP receptor (SR). 3. Translation proceeds with threading of the nascent chain into the translocon (SecYEG in prokaryotes and Sec61 in eukaryotes) pore, which is sealed by the plug (green rectangle). YidC (not shown) in bacteria plays a dual role with or independent of the translocon in the integration and assembly of a subset inner membrane proteins. 4. Translation proceeds with the synthesis of the first transmembrane domain (TM), which exits laterally as an α-helix into the lipid bilayer with retention of the positively charged N-terminal domain (according to the positive-inside rule) on the cytoplasmic side of the membrane. 5. The plug opens to allow the next extramembrane domain to cross the membrane and exit into the periplasm. 6. The next TM enters the translocon channel followed by lateral exit into the membrane and cytoplasmic orientation of the next extramembrane domain. Steps 4 through 6 are repeated until synthesis is complete. 7. As TMs enter the membrane, lipid-protein interactions, short-range protein-protein interactions, and long-range helix packing now govern topological and folding events [in a coordinated but undefined (??) manner], resulting in a final compact native structure. In eukaryotes, the periplasm corresponds to the lumen of the endoplasmic reticulum. Figure was adapted and extensively modified from Reference 29. Folded protein is lactose permease (LacY) of Escherichia coli from Reference 79 and reprinted with permission from American Association for the Advancement of Science.
Topological organization is well established, or readily determinable, for most membrane proteins, but the process of membrane protein topogenesis and the factors that influence topogenesis are less well defined. In this review, the overall process of synthesis, insertion, and folding of membrane proteins is briefly covered as background for a more detailed review of co- and postinsertion events with an emphasis on the role of lipids in topogenesis. Multiple modes of protein insertion into the membrane exist. This review focuses on proteins that utilize the translocon machinery where a role for membrane lipids is well establihsed.
Overview of Membrane Protein Assembly
Polytopic membrane protein biogenesis requires the coordination of several events: recognition and targeting of the nascent chain to the membrane localized translocon and accompanying machinery; integration and orientation of TMs coupled with coordinated folding of extramembrane domains; helical packing within the lipid bilayer; and proper formation of a lipid-embedded, tertiary structure (Figure 1). A fundamental architectural principle of the structure of polytopic membrane proteins is membrane protein topology, i.e., the number of TM segments and their orientation relative to the membrane bilayer. The vast majority of prokaryotic and eukaryotic integral membrane proteins are cotranslationally inserted into and oriented within the membrane in a signal recognition particle- and translocon-dependent manner (1, 2). However, there are numerous examples of rearrangements that take place in a posttranslational manner while the nascent protein is still engaged with the ribosome and the translocon (3, 4). Hydrophobic segments or flanking hydrophilic domains first enter the translocon and then in a sequential manner laterally partition into the membrane bilayer (5) or extend into the interfacial regions bordering the membrane, respectively, governed largely by thermodynamic considerations (6, 16).
Membrane protein topogenesis, which leads to a predictable final organization for many but not all membrane proteins (4, 7), is directed by a combination of several factors. Polytopic membrane protein topology is determined by an interplay between the topogenic signals residing within the protein sequence (8–10), the interaction of the protein with the translocon machinery (11, 12), internal protein interactions, and protein-lipid interactions during final folding into a compact membrane protein (13–15). During protein topogenesis, these factors influence final topological organization simultaneously or sequentially (3), resulting in most cases in a unique topology.
An initial topological decision appears to be made at the translocon, which provides the permissive environment required for concurrent insertion of TMs into the membrane and for the positioning of hydrophilic flanking regions on either side of the membrane according to the positive-inside and/or charge difference rules (11, 16, 17) summarized below. While in the translocon, large protein segments composed of TMs with flanking extramembrane sequences (18, 19) can explore possible topologies favored by the different topogenic signals until protein synthesis is completed (12) or until further reorientation is terminated by unknown mechanisms within a limited time window (8, 20). TMs may adopt an initial topology according to the positive-inside rule by direct charge interactions between the translocon (11) and the protein. However, the contribution of the translocon to making a topological decision is limited by time (8, 12), size of the newly synthesized protein (18, 19), and the effective size of the translocation pore, which is still a matter of debate (21, 22). Therefore, irrespective of the number of TMs accommodated by the translocon pore, final protein topology and organization after exit from the translocon must follow thermodynamically driven routes dictated by direct interaction of the TMs and associated extramembrane domains both within the protein and with the lipid bilayer (6, 16).
In eukaryotic cells, this dynamic reorientation process can be blocked by glycosylation (23) that proceeds in the endoplasmic reticulum simultaneously with insertion, thus providing accuracy of orientation and irreversibility after insertion. The delay in the translocation of positively charged extramembrane domains relative to negatively charged domains might contribute to the preferential glycosylation of the latter (8). However, prokaryotes lack this initial ability to fix topology and instead may “proofread” the topology of proteins in their membranes well after exit of TMs from the translocon (3, 4, 13, 24).
It is generally accepted that the topology of most polytopic membrane proteins is established cotranslationally during membrane insertion and, once established, is maintained during subsequent steps of biogenesis, cellular trafficking, and function. According to the simplest model for membrane protein insertion, the most N-terminal TM defines its own orientation as well as the orientations of all subsequent TMs, which simply insert in alternating orientations into the bilayer (25). However, for E. coli lactose permease (LacY) and maltose transporter, deletion of individual TMs (26, 27) or perturbation of the orientation of the N-terminal segment of the tetracycline/H+ antiporter (28) did not affect the topology of downstream TMs, leaving open the possibility of nonsequential insertion mechanisms and postinsertion reorientation of TMs. Owing to their hydrophobic properties, TMs passively partition into the lipid bilayer with flanking charged residues positioned near the aqueous-membrane interface (14, 29). Therefore, the physical properties of the lipid bilayer can have an affect on membrane protein organization in a retrograde manner (3, 4, 13, 24) as discussed below.
Topogenic Signals and Topological Rules
Membrane protein topogenesis is governed by a set of topogenic signals encoded in the amino acid sequence (Figure 2), which are decoded by processes not completely understood. In combination, these determinants are sufficient to achieve a unique final orientation for any TM, but the role that factors, such as the translocon and the membrane lipid environment, play in decoding these topogenic signals has only recently been made evident.
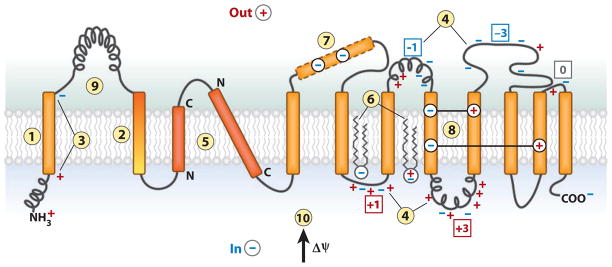
Figure 2.
The features of polytopic membrane proteins and biological membranes that determine folding and final transmembrane domain (TM) topology. TM integration is driven by (1) the hydrophobicity of TMs, and initial orientation is influenced by (2) the hydrophobic gradient (highest outward) along a TM, (3) the charge difference or (4) the positive-inside rule, (5) preferential orientation of highly hydrophobic TMs with the C terminus out if short and the N terminus out if long. After exit from the translocon, (6) final topology is influenced by lipid headgroup-protein charge interactions. During protein folding, interactions between TMs can stabilize (7) a domain containing charged residues as (8) a TM by compensating salt bridges. (9) A rapidly folding extramembrane domain can prevent reorientation of TMs during late folding events. (10) The positive outward membrane potential favors translocation of net negative domains and retention of net positive domains.
Overall hydrophobicity of individual TMs is the primary energetically favorable driving force for membrane integration (30). Orientation correlates best with total hydrophobicity rather than length (31) but is strongly affected by the hydrophobicity gradient within a TM (32). The most hydrophobic terminus is preferentially translocated across the membrane. Translocation of the N terminus is favored by long hydrophobic sequences and translocation of the C terminus by short hydrophobic sequences (33).
In many cases, the hydrophobicity of a TM is sufficiently strong to drive the translocation of flanking segments. However, according to the biological free-energy scale for predicting insertion into the bilayer, the total hydrophobicity is not the sole determinant of membrane insertion; residue position also has an important influence on insertion efficiency and probability (16, 29). Tryptophan and tyrosine in contrast to phenylalanine and leucine are statistically over represented near both TM ends, and the former two strongly reduce membrane insertion when placed centrally where they appear to pull the helix out of the membrane. TMs, containing charged amino acids or domains with insufficient hydrophobicity, are able to insert synergistically into the membrane by specific interactions with other TMs (34–36). Although simple hydrophobicity is the predominant factor determining the insertion efficiency, analysis of whole genomic data by the TM tendency scale revealed an overlap of TMs and soluble sequences in the “semihydrophobic” range (37). This raises the possibility that a significant number of proteins have sequences that lie close to an equilibrium between insertion and exclusion that can result in switching between TM and non-TM states depending on environmental and physiological conditions.
The number of integral membrane proteins containing TMs with unusually low hydrophobicity is quite limited. The presence of two charged residues within a TM domain generally requires salt bridge neutralization through interaction with other TMs containing the opposite charges (38). Non-orthodox or “leave-one-out” topologies or mixed topologies with frustrated hydrophobic domains facing the solvent have been observed for bacterial (39) and eukaryotic proteins (40–42). Deletion, duplication, or changes in protein sequence, resulting in a change from an even to odd number of TMs, generally result in inversion of protein topology (43). The 13-TM Pseudomonas aeruginosa ChrA protein is composed of two homologous 6-TM bundles, which are separated by a hydrophobic TM (TMVII) and are in opposite orientation with respect to each other. However, the homologous two 6-TM bundles of the 12-TM Cupriavidus metallidurans ChrA are arranged in the same orientation with respect to each other (44). Sequence comparison between ChrA from P. aeruginosa and from C. metallidurans shows that TMVII of the former has no charged amino acids, whereas the corresponding C. metallidurans sequence (a 20-residue extramembrane domain) is polar and includes two positively charged residues. This suggests that both ChrA proteins arose from the same intragenic duplication of an ancestral 6-TM protein, followed by the insertion of an intervening sequence whose hydrophobicity determined the relative orientation of the duplicated domains. Another topologically important feature of polytopic membrane proteins is the folding state of the extramembrane hydrophilic domains (45). Translocation of extramembrane domains occurs in the unfolded state. Therefore, rapid folding of hydrophilic domains in the cytoplasm may prevent an export of a domain, and tight folding of a domain on the trans side of the membrane may ensure the location of that domain.
The Positive-Inside Rule
The final TM topology of polytopic membrane proteins is dictated by the encoding sequence, which has coevolved with the translocation-insertion machinery and the unique lipid environment of each organism. Gross protein topology appears to be primarily determined by charged residues flanking the hydrophobic core and, in most cases, can be described by the positive-inside rule (46), which derives from the fact that positively charged residues are four times more abundant on the cytoplasmic side of membrane proteins as compared to the trans side (47). Moving even a single positively charged residue from the cytoplasmic side to the opposite side of a TM results in inversion of topology (9). Mutations that violate the positive-inside rule can prevent insertion of a TM to maintain the positive-inside constraint (39). In most cases, the precise position of the charge is unimportant, the contribution of charges is additive, and the density of charge within the extramembrane domain does matter (48). However, the positive-inside rule is not absolute because cytoplasmic domains with a net negative charge are found (49, 50).
The initial topological decision appears to be made by the translocon, which provides the permissive hydrophilic environment required for translocation of the flanking regions to the extramembrane space according to the positive-inside and/or charge difference rule (20, 29). In cells with a positive outward membrane electrochemical gradient (51), the gradient may actively promote translocation of negative residues (52, 53) and impede translocation of positive residues (54), especially when the hydrophobicity of an associated TM is low (55). Although the positive-inside rule discounts the importance of negatively charged residues, the topological effect of positively charged residues can be attenuated (56) or even overridden by negatively charged residues in a position-specific manner for bacterial (48) and eukaryotic membrane proteins (57). Negatively charged residues appear to be topologically active only if they are present in high numbers (9), flank a marginally hydrophobic TM (55), or lie within a window of six residues from the end of a highly hydrophobic TM (10). However, several negative residues are required to translocate a cytoplasmic domain with even a single positive residue (9). The fact that the orientation of membrane proteins can be reversed either by the addition or removal of a single positively charged residue or by introduction of negatively charged residues situated close to the ends of TMs raises the question of the relative topological power of the charged residues. Although eukaryotic membrane proteins display a positive charge bias for cytoplasmic domains, the net electrical charge difference, or balance between the two flanking segments (more positive on the cytosolic side), rather than the presence of positively charged residues per se has been shown statistically and experimentally to correlate with protein orientation (32, 58).
Why do positively charged residues appear more potent topological determinants than negatively charged residues? What is the balance between negatively and positively charged residues that determines final topological organization? Are there cellular factors that act in concert with protein sequence to establish topology? As described below, the membrane lipid composition is also a determinant in defining topology, and alterations in lipid composition can also affect topology, demonstrating again that membrane protein topogenic signals have evolved within the context of a given membrane lipid composition.
UNCOVERING A SPECIFIC ROLE FOR LIPIDS IN TOPOGENESIS
A primary role of phospholipids is to define the permeability barrier of cells and organelles by forming a bilayer. The lipid bilayer serves as the matrix and support for a vast array of proteins involved in diverse and essential cellular processes (59, 60). The large diversity in lipid structures allows for a broad spectrum of chemical and physical properties within the membrane bilayer and provides a complex solvent with a diverse array of hydrophilic and hydrophobic surfaces and positive and negative charges that affects the folding and orientation of membrane proteins. Therefore, manipulation of membrane lipid composition might be expected to result in pleiotropic or lethal effects, which has made defining the function of lipids in molecular terms in cellular processes a challenging undertaking. Although the physical and chemical properties of individual lipids and lipid mixtures have been extensively studied, it is not clear how to relate this information to the in vivo state. Lipids have no inherent catalytic activity, so biochemical studies have relied on the secondary effects of lipids on in vitro reconstituted processes. Given these obstacles to the study of specialized roles for individual lipids, most studies of membrane protein topogenesis and assembly treat the lipid bilayer as a passive amphipathic solvent for membrane proteins with little consideration of how more specific lipid-protein interactions might play a role in final protein organization.
Although the translocon provides a transient folding environment that enables nascent TMs to sample alternate conformations prior to adapting an intermediate or final transmembrane orientation, the folding pathways of TMs after clearance of the translocon must follow thermodynamically driven routes involving direct interaction of the TMs and associated extramembrane domains with the surrounding lipid bilayer (14, 29). TMs passively partition into the bilayer on the basis of their affinity for the hydrophobic lipid core of the membrane while flanking aromatic and charged residues are positioned near or within the aqueous-membrane interface (14). Therefore, early TM interactions and folding events are impacted by the physical properties of the surrounding lipids, which further decode the topogenic signals within nascent chain sequences to influence TM orientation and final folding events. Although the topology of membrane proteins is dictated primarily by the encoded amino acid sequence, topogenesis of the membrane proteins LacY (13, 61, 62), phenylalanine permease (PheP) (15), and γ-aminobutyrate permease (GabP) (63) of E. coli are also governed by the membrane lipid composition.
Uncovering a specific role for membrane lipid composition during topogenesis was made possible by the development of strains of E. coli in which membrane phospholipid composition can be controlled and varied systematically over a broad range while maintaining cell viability. E. coli membranes contain approximately 70% phosphatidylethanolamine (PE), 20% phosphatidylglycerol (PG), and 5% cardiolipin (CL) (64); the first is zwitterionic and therefore has no net charge, and the latter two carry a net negative charge (Figure 3). The first evidence that lipid-protein interactions play a role in topogenesis (65) came from studies of a mutant of E. coli in which the anionic phospho-lipid content (PG plus CL) could be reduced from wild-type levels in a systematic manner, thus reducing the net negative charge density of the membrane surface. Expression of the pgsA gene (encodes the committed step to PG and CL biosynthesis) (Figure 4) in this mutant was placed under the regulation of the inducible lac-OP (lactose operon promoter). By varying both the positive charge density of the cytoplasmic domain of a chimeric bitopic membrane protein and the anionic phospholipid content of the membrane, it was established that cytoplasmic retention was proportional to both the number of positive charges within the cytoplasmic extramembrane domain and the membrane content of anionic phospholipids, thus suggesting that lipid-protein charge interactions are determinants of protein topology.
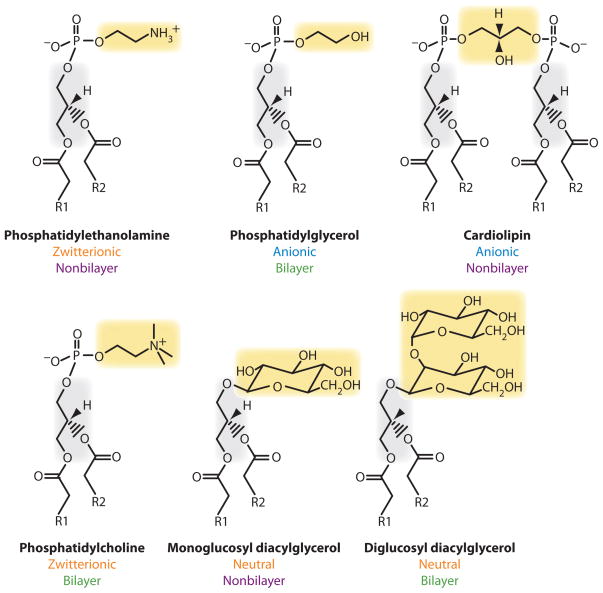
Figure 3.
Structure and physical chemical properties of lipids. Stick diagram of the carbon backbone of lipids with R substituting for the long hydrocarbon portion of fatty acids. Phospholipids found normally in E. coli are in the top row. Foreign lipids that have been introduced into E. coli mutants lacking phosphatidylethanolamine are in the bottom row. The charge nature of the lipid headgroups is noted. Depending on fatty acid composition, solvent, and temperature, those lipids that assume nonbilayer organization in solution are indicated in contrast to those lipids that only form bilayer structures.
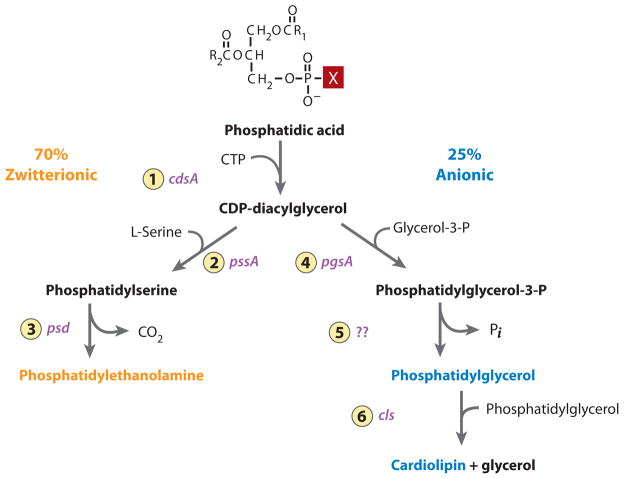
Figure 4.
Pathway for synthesis of the major phospholipids of E. coli. The following enzymes with their respective genes indicated are 1. CDP-diacylglycerol synthase; 2. phosphatidylserine synthase; 3. phosphatidylserine decarboxylase; 4. phosphatidylglycerol-3-P synthase; 5. phosphatidylglycerol-3-P phosphatase; 6. cardiolipin synthase. The X in phosphatidic acid is an OH and is in the position that changes depending on the downstream pathways. The remaining 5% of total phospholipids are primarily the minor lipids (black).
However, the most compelling evidence for lipids as determinants of membrane protein topology comes from either null (66) or engineered inducible mutants (13, 67) of the pssA gene (which encodes the committed step to PE synthesis) (Figure 4) of E. coli that result in either the complete lack of PE (and almost exclusively includes PG and CL) or the ability to control the level of PE in vivo, respectively. In wild-type E. coli, insertion and folding of membrane proteins occur in the presence of an abundance of PE, so lipid-assisted and -unassisted folding pathways cannot be distinguished. However, a role for phospholipids in establishing proper topology was discovered using a mutant of E. coli lacking PE. As genetic manipulation of the protein sequence established and verified the positive-inside rule, genetic manipulation of membrane lipid composition revealed rules relating lipid composition to topological determination.
Lipid-Assisted Folding
LacY is a 12-TM-spanning polytopic protein (Figure 5a) of the inner membrane of E. coli and is responsible for the active uphill energy-dependent and facilitated downhill energy-independent transport of lactose across the cytoplasmic membrane (68). Early studies (69) on reconstitution of LacY into proteoliposomes indicated an important role for lipid composition in protein function. Liposomes composed of E. coli lipids supported uphill and downhill transport, but liposomes that lacked PE and containing only PG, CL, and/or phosphatidylcholine (PC, a zwitterionic phospholipid similar to PE) (Figure 3) only supported downhill transport, suggesting a possible requirement for a specific lipid to support full function of LacY. Although such in vitro studies are very useful tools in analyzing biological function, in vivo verification is always required to eliminate unknown artifacts introduced by in vitro systems. Comparing LacY function in PE-lacking cells with wild-type cells not only verified a specific requirement for PE in supporting uphill transport (70) but also provided a means for a detailed analysis of the role of lipid composition on insertion, organization, and function of membrane proteins. The obvious possibility of defects in the bioenergetics of PE-lacking cells was ruled out (70) and, given the direct correlation with in vitro studies, suggested a direct role for PE.
Figure 5.
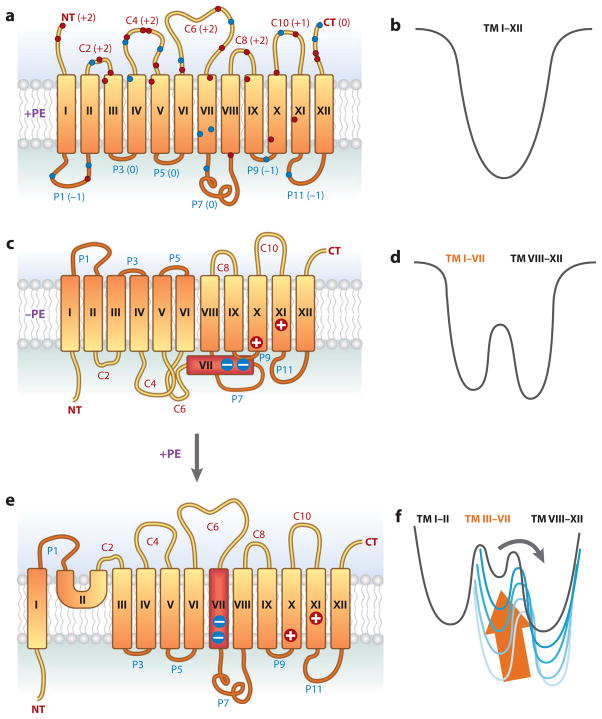
Topological organization of LacY as a function of membrane lipid composition. (a) The topology of LacY initially assembled in E. coli with wild-type phospholipid composition is shown. Rectangles define the transmembrane domains (TMs) (79), oriented with the cytoplasm above the figure. TMs (Roman numerals), extramembrane domains [P for periplasmic and C for cytoplasmic as oriented in phosphatidylethanolamine (PE)-containing cells], N terminus (NT), and C terminus (CT) are indicated. The net charge of each extramembrane domain is indicated next to the domain name. The approximate locations of negatively charged (blue) and positively charged (red ) residues are indicated. (b) A funnel-shaped energy landscape that defines the folding pathway of a membrane protein such as LacY in wild-type cells is depicted. The horizontal axis ( funnel circumference) represents the conformational space occupied by protein as it folds, and the vertical axis represents the internal free energy of a given polypeptide conformation. As TMs I–XII fold, they move down the funnel to the lowest energy state as defined by the final organization of native LacY. (c) The topology of LacY assembled in E. coli cells lacking PE is shown. TMs I–VI are inverted with respect to TMs VIII–XII, which still exhibit native topology. TM VII (red ) is exposed to the periplasm, resulting in breakage of the salt bridges with TM X and TM XI. (d ) A new energy landscape defines the energetics of folding in PE-lacking cells of the two halves of LacY with the N-terminal bundle forming a new energy minimum separated by a large activation energy, which disconnects it from the native state. (e) The topological organization of LacY after synthesis of PE in cells after assembly of LacY. All of LacY assumes a native topological organization except the domain defined by NT-TM I-P1-TM II. ( f ) The presence of PE raises the energy minimum (arrow) of the inverted topology of TM III–VII, resulting in low activation energy for a return to the native topology. A large energy barrier remains for TM I flipping with a new miniloop structure (TM II) allowing all domains to exist at their respective energy minima.
The molecular basis for the lack of full LacY function initially appeared to be a subtle alteration in structure, which was determined by use of a conformation-specific monoclonal antibody directed at the extramembrane domain P7 (Figure 5a) exposed to the periplasmic side of the membrane (71). The P7 domain was readily recognized in spheroplasts (the exposed outer surface of the inner membrane) containing LacY in PE-containing but not PE-lacking cells. The monoclonal antibody also recognized LacY from the former but not the latter cells after denaturation by sodium dodecyl sulfate (SDS) and Western blotting analysis. A monoclonal conformation-specific antibody directed against a discontinuous epitope composed of domains C8 and C10 facing the cytoplasm recognized LacY from both sources. Thus, LacY is “denatured” in vivo with respect to the P7 domain by assembly in a non-native lipid environment. Perturbation of the P7 domain had previously been linked to loss of uphill transport function (72). Therefore, PE appeared to be required during initial assembly of LacY to establish the proper conformation of the P7 domain, and the conformation of this domain was retained after partial denaturation with SDS, followed by removal during electroblotting. However, more extensive denaturation by SDS in the presence of urea resulted in loss of recognition of the P7 domain. Therefore, once information was imparted during folding of LacY in vivo in the presence of PE, this lipid was no longer required for maintenance of proper protein conformation, and LacY assembled in PE-containing membranes retains sufficient “conformational memory” to reform the native structure of the epitope after removal of SDS.
The misfolding of LacY attributed to assembly in PE-lacking cells was corrected by employing the Eastern-Western blotting technique in which LacY was exposed to hydrated phospholipids on a solid support during renaturation from SDS in the standard Western blotting procedure, followed by probing with a conformation-specific monoclonal antibody (71, 73, 74). This technique allows renaturation of proteins in the presence of lipids as the SDS is removed during electroblotting. Using this procedure, LacY from PE-lacking cells regained recognition by the monoclonal antibody directed at domain P7 if PE, mono-or dimethyl-PE, or phosphatidylserine was present on the solid support during the Eastern-Western procedure but not with PG or CL or PC. The restoration of monoclonal antibody binding specifically by aminophospholipids indicates that PE plays a positive role as a folding determinant, rather than acting to exclude a negative determinant in the form of anionic phospholipids PG and CL. Finally, when LacY from PE-deficient membranes was solubilized in the presence of PE-containing liposomes before SDS-PAGE, no restoration of monoclonal antibody binding to domain P7 was observed, indicating that formation of the epitope is dependent on removal of SDS in the presence of PE rather than simply exposure of the denatured protein to PE. Furthermore, there was additional specificity within PE species in that the wild-type chirality at the sn-2 position of the glycerol backbone of PE was required as were the PE species, which were bilayer forming (containing saturated fatty acids) as opposed to nonbilayer forming (containing unsaturated fatty acids). Therefore, the minimum requirement for proper refolding of the P7 domain is a zwitterionic bilayer-forming phospholipid, with an ionizable amine and wild-type chirality.
This level of specificity extends the role of lipids well beyond that of a passive amphipathic solvent for membrane proteins. Traditionally, molecular chaperones have been a class of proteins that bind transiently to substrate proteins to promote their proper folding by interacting noncovalently with nonnative folding intermediates and not with either the native or totally unfolded protein. Retention of a properly folded domain P7 after subjecting native LacY to Western blotting was found not to be caused by residual PE (73), but rather, once information was imparted during folding of LacY in vivo in the presence of PE, this lipid was no longer required for maintenance of proper conformation of the protein, which is consistent with the minimal definition of a molecular chaperone.
Although refolding of denatured or partially denatured full-length protein provides very important information, refolding of the protein starts from a complete polypeptide chain, whereas the de novo folding of a nascent peptide in vivo begins during translation. Therefore, the postassembly influence of PE on structure was also extended to a more biological system (73). When LacY was synthesized in vitro in the presence of E. coli membranes isolated from either wild-type or PE-lacking cells, LacY inserted into the membranes and folded sufficiently to be detected by the monoclonal antibody directed against the C8–C10 epitope. However, the monoclonal antibody directed against the P7 domain recognized LacY from wild-type membranes but not from PE-lacking membranes unless PE was resupplied biosynthetically after insertion of in vitro-translated LacY. Therefore, PE is not required for a good insertion yield or initial folding of LacY but is required in a postinsertion folding step to assure the proper conformation of P7. Thus, PE appears to facilitate in vitro refolding and in vivo folding into a fully native conformation by interacting with late folding and nonnative intermediates, as do most conventional protein molecular chaperones. On the basis of these results, lipids can function as lipochaperones that specifically mediate the folding of proteins, thereby extending the definition of chaperones to other biomolecules in addition to proteins (71, 75).
Lipids as Determinants of Membrane Protein Topology
What are the structural alterations in LacY that result in the misfolding of domain P7 and the loss of uphill transport function in the absence of PE? The availability of strains of E. coli in which steady-state PE content can be regulated (13, 76) coupled with methods, such as the substituted cysteine accessibility method as applied to TMs (SCAM™), to determine the orientation of TMs with respect to the plane of the membrane bilayer (77) were used to analyze LacY organization in the membrane as a function of lipid composition. In SCAM™, single-cysteine replacements are made in cytoplasmic or periplasmic loops or TMs in an otherwise cysteineless LacY, so that their susceptibility to a membrane-impermeable sulfhydryl reagent in either intact cells (periplasmic location only) or cells in which the cell membrane has been disrupted (all extramembrane cysteines) can be probed; lack of reaction can be due to either residency in a TM or local effects on an extramembrane cysteine. Analysis of LacY assembled in PE-containing cells by SCAM™ (Figure 5a) showed the expected topological organization (13) as previously defined (78) and later verified by X-ray crystallography (79). However, analysis of LacY assembled in PE-lacking cells revealed a dramatic misassembly of LacY (13) with the N-terminal six-TM helical bundle (TM I–VI) and adjacent extramembrane domains completely inverted with respect to the plane of the bilayer and the C-terminal five-TM helical bundle (TM VIII–XII) (Figure 5c). Domain P7 remained exposed to the periplasm but TM VII (colored red) appears to now be exposed to the periplasm (67) and acts as a molecular hinge along with domain C6, which is now periplasmic, to allow the N- and C-terminal bundles to respond differentially to the membrane lipid composition. The periplasmic exposure of TM VII is also consistent with its low hydrophobicity owing to two aspartate residues that are normally in salt bridges with positive amino acids in TM IX and TM X. Increasing the hydrophobicity of TM VII by electrostatic neutralization through a D240I mutation within this hinge prevented TM VII from being exposed to the periplasm in PE-deficient cells and simultaneously blocked inversion of the N-terminal bundle. These results are consistent with the antibody studies because the primary disruption in the TM helical packing of LacY occurs in TM VII adjacent to P7, and the C8-C10 epitope remains intact. These results demonstrate that the rules governing TM orientation are dependent on both the protein sequence and the lipid composition.
Postassembly Transmembrane Domain Flipping
Even more dramatic is that the aberrant topological organization of LacY is nearly completely reversible postassembly in vivo (13, 67). By placing the pssA gene under the control of an inducible promoter, cells grown in the absence of an inducer have less than 3% PE, whereas addition of an inducer brings PE levels back to normal. This made possible the growth of cells induced for LacY synthesis in the absence of significant levels of PE, followed by removal of the inducer for LacY synthesis and addition of an inducer for PE synthesis. Therefore, the effect of PE on preformed and assembled LacY could be assessed to extend the observation in the in vitro experiments outlined above, which involved only the postassembly refolding of the P7 domain (73). Postassembly synthesis of PE resulted in the regain of uphill transport function by LacY, near-native topology of LacY with the return of TMs II–VI (and their associated extramembrane domains) to their native orientation (67), and reinsertion of TM VII across the membrane (Figure 5c,e). The N terminus and TM I do not revert. TM II formed a miniloop, which does not traverse the membrane, to act as a bridge between native and aberrant topologies. This result clearly demonstrated that simply changing the lipid composition of the membrane could reverse topology of multiple TMs.
Whether other proteins acting as molecular chaperones are involved in reorganization is not known, but clearly this is a lipid-driven process. Therefore, it is entirely possible that reorientation is spontaneous and driven by simple thermodynamic considerations rather than being under kinetic control of a chaperone catalyst (13, 24). Furthermore, the topogenic influence of lipids appears to be largely independent of protein folding history or other protein factors since topological organization and transport function of LacY reconstituted into protein-free liposomes are determined solely by the final phospholipid composition independent of the lipid composition of the cells from which LacY was derived (61). Because LacY is functional in downhill transport and is stable in PE-lacking membranes, it must be folded into a compact structure. The simplest interpretation of the reorganization process is that introduction of PE into the membrane destabilizes the folded state of the protein, resulting in reorientation of most of the N-terminal helical bundle in order for the protein to assume its new minimum energy state (Figure 5b,d,f ).
Such postassembly reorganization has dramatic implications for a number of biological processes. First, the dogma that TMs are not prone to reorientation postinsertion is not supported. Current dogma assumes that the initial topology of a protein in the endoplasmic reticulum membrane accurately reflects the topology of the protein elsewhere in the cell. However, because large topological reorientations involving multiple TMs are possible, protein trafficking between organelles of different membrane lipid compositions or transient changes in local lipid composition have the potential for inducing large topological changes that could result in activation, inactivation, degradation, or changes in function of a protein.
Why can some proteins or protein domains undergo large TM movement but others do not? What is necessary and sufficient for TM reorientation within a membrane in response to changes in membrane lipid composition? The differential response of the N-terminal bundle to the lipid environment may be due to the intrinsic structural flexibility provided by the C6-TM VII-P7 hinge region. Without a flexible hinge region, as demonstrated by increasing the hydrophobicity of TM VII, LacY assembled in PE-lacking membranes either would be forced to assume an overall fold that balances the opposing thermodynamic minimum of each domain with the unfavorable aqueous exposure of a hydrophobic domain or would not fold into a compact structure and would be degraded. The former may be the case for many essential proteins in PE-lacking cells, which would explain the viability of this mutant. Therefore, long-range interactions can dominant over local effects, which are generally the basis for the failure of in silico predictions of the structure of some membrane proteins.
How universal is the requirement for PE? Thus far, a number of secondary transport proteins have been demonstrated to require PE for proper topology and/or function, making this requirement of broader significance. Both PheP (15) and GabP (63) show an inversion of only the N-terminal two-TM hairpin in PE-lacking cells with nearly complete loss of uphill transport function. Only PheP was investigated for reorientation and regain of activity by postassembly synthesis of PE, which did occur. The uphill transport of proline (PutP), lysine (LysP), tryptophan (AroP and/or TrpP), and melibiose (MelB), presumably owing to misassembly of their respective permeases, is also defective in PE-lacking cells (M. Bogdanov & W. Dowhan, unpublished data). However, if this phenomenon is broad based, why are PE-lacking cells still viable? Clearly no essential function is completely defective, but then neither is lactose transport completely defective. Not all proteins, even those with lipid-dependent topological motifs, may be affected by changes in lipid composition if other structural features, such as a functional hinge region, are not present. Lipid composition and the positive-inside rule are only singular determinants of topological organization, and final topology results from an integration of local and long-range interactions.
It is very difficult to predict how and when such long-range intramolecular interactions might overcome early folding events to influence final topology, but there is precedence for such effects. The glycosylation of the C-terminal tail of a hepatitis C virus protein NS4B before the N-terminal tail suggests that the N-terminal tail of NS4B is translocated across the membrane well after exit from the translocon and after synthesis of the C-terminal tail (80). A similar posttranslational rearrangement was observed in aquaporin 1 (81). The protein initially adopts a four-TM-spanning topology with the extramembrane loop 2 mislocalized to the cytosolic face of the endoplasmic reticulum membrane. To acquire its mature topology, TMs 2, 3, and 4 and two interhelix loops are reoriented during late stages of biogenesis. Therefore, the synthesis of downstream topogenic determinants can rearrange the topology of preexisting TMs. The E. coli SecG protein shows an unusual property of inverting its orientation in the membrane, which is tightly coupled to its function and linked with the insertion-deinsertion cycle of SecA (82). The C terminus of E. coli TatA is accessible from the periplasm in the resting state but is accessible from the cytoplasm in the functional state (83). Colicin Ia translocates spontaneously and reversibly a 68-amino acid hydrophilic segment across a lipid bilayer in conjunction with membrane depolarization (84). The topology of chimeric model membrane proteins can be manipulated by a ligand in vivo (85).
Unassisted post-insertional rearrangement or reorientation of membrane proteins is not unprecedented. Polar domains of up to 85 amino acid residues appended to the C terminus of a tail-anchored protein and in an unfolded state are efficientlty translocated across a protein-free lipid bilayer (86) dependent on the lipid composition (87). Colicin Ia translocates spontaneously and reversibly a large internal hydrophilic domain (at least 68 amino acids long and containing 15 basic and 8 acidic residues) completely across a lipid bilayer in conjunction with the formation of an ion-conducting channel using unknown mechanisms to overcome the apparent unfavorable free energy of transferring charged residues from the aqueous phase into and through the bilayer (84).
Therefore, according to this new topology switch paradigm, it is possible that specific regions of a membrane protein can undergo reversible TM reorganizations in vivo, dictated not only by phospholipids but also by components of the translocation machinery, ligands, and substrates as well as by other cellular events, such as membrane depolarization.
Properties of Lipids that Affect Topology and Function
What specific structural, chemical, and phase-forming features of lipid molecules are important for proper topogenesis (Figure 3)? E. coli is not only viable in the face of large changes in native lipid composition but tolerates introduction of foreign lipids, many of which improve the growth and functional properties of mutants lacking native lipids. This affords the possibility of broadening possible lipid compositions and studying the properties of lipids that support native topology of lipid-sensitive domains. The effect on topology of LacY has been studied for the following diacylglycerol-containing foreign lipids with different physical and chemical properties (Figure 3): Monoglucosyl diacylglycerol (GlcDAG) and diglucosyl diacylglycerol (GlcGlcDAG) headgroups are neutral, carry no charge, and are capable of hydrogen bonding; PC and PE are zwitterionic and carry no net charge but only the headgroup of PE is capable of hydrogen bonding. PC and GlcGlcDAG tend to be bilayer-forming lipids, and GlcDAG and PE tend to be nonbilayer-forming lipids.
Introduction of the gene from Acholeplasma laidlawii encoding the GlcDAG synthase partially corrects the cell division defect of PE-lacking cells and reduces the dependence on divalent cations for viability (88). PE-lacking cells expressing this gene contain about 30–40 mole % GlcDAG with a proportional reduction of PG and CL levels. Remarkably, the overall topology of LacY is normal in these cells (62). In addition, LacY displays both uphill and downhill transport function. Introduction of the genes from A. laidlawii that synthesize GlcDAG and GlcGlcDAG results in about 30–40 mole % GlcGlcDAG with less than 1% GlcDAG (89). In this membrane environment, LacY topology is normal, but LacY only displays downhill transport activity. Finally, introduction into E. coli of the gene from Legionella pneumophila that confers the ability to synthesize PC results in about 70 mole % PC with the remainder being PG plus CL (M. Bogdanov, P. Heacock, & W. Dowhan, unpublished data). In agreement with in vitro reconstitution experiments in liposomes containing PC in place of PE (61), LacY also displayed wild-type topology in cells containing PC in place of PE. However, the replacement of PE by PC also supported uphill transport function, which was not the case in liposomes with the same substitution (61, 69). This latter result emphasizes the need to verify in vitro results with in vivo observations before a definitive conclusion can be made about a role for a particular lipid.
Since zwitterionic PE and PC and the uncharged glycolipids GlcDAG and GlcGlcDAG support native topology, the dilution of the negative charge character of the membrane surface is a more important topological determinant than the positive charge within the headgroup of PE or PC or the phase-forming properties of the lipids. In general, the same conclusions can be made with respect to the dependence of LacY function on lipid composition (now that PC also supports full activity in vivo) except it is not clear why GlcGlcDAG does not support uphill transport. It is tempting to speculate that during the course of evolution, both proteins and lipids coevolved in the context of the lipid environment of membrane systems in which both are mutually dependent on each other. Although it is surprising that E. coli tolerates such changes in membrane lipid composition, these results emphasize the importance of the similar collective charge properties of lipids in supporting cell functions rather than of a strict structural requirement, as observed for proteins. Therefore, the property of the membrane bilayer that governs topological orientation of polytopic membrane proteins is the balance between anionic lipids and those with no net charge as evidenced by the shared capacity of lipids with net zero charge to promote proper polytopic membrane topogenesis.
The Charge Balance Hypothesis
The effects of different lipids on topology indicates that topology of the N-terminal bundle of LacY is sensitive to the negative charge density of the membrane surface, suggesting that charge interactions between the extramembrane domains of the protein and the lipids is a determinant of topology. Therefore, are there topogenic sequences within membrane proteins that make their topology sensitive to membrane lipid composition?
All periplasmic domains of LacY carry either a net neutral or net negative charge, and all cytoplasmic domains strictly follow the positive-inside rule and carry a net positive charge (Figure 5a). However, the N-terminal cytoplasmic domains are significantly enriched in negatively charged residues relative to those of the C-terminal domain. Furthermore, these negatively charged residues lie within six residues of the end of the TM helices; this location of negatively charged residues has been shown to destabilize cytoplasmic retention of monotopic model membrane proteins (10). If the topological inversion of the N-terminal bundle in PE-lacking cells is due to the presence of negative residues in the cytoplasmic surface, why do these negative residues reduce the retention potential of positive residues only in the absence of PE? Inversion in PE-lacking cells also appears to be in conflict with an expected increase in retention potential afforded by an increase in anionic phospholipid content interacting with positively charged amino acids (65). To investigate whether the presence of mixed proximal topogenic signals, i.e., both negative and positive residues, is the basis for topological sensitivity to the lipid environment and to examine the effect of lipids on the relative potency of opposing topogenic signals, charged residues within the cytoplasmic domains of the N-terminal bundle were altered, and their topology was studied as a function of lipid composition (67).
Changing one acidic amino acid to its corresponding amide in any one of domains C2, C4, and C6 (Figure 5a) resulted in native topology for LacY when expressed in PE-lacking cells (67). Adding a positively charged amino acid to domain C2 also resulted in native topology, but elimination of both a negative and positive charge (no net change in charge) in C2 did not prevent inversion. Therefore, increasing the net charge by plus one of the cytoplasmic surface of the N-terminal bundle prevented inversion in PE-lacking cells in a position-independent manner. If net charge of the cytoplasmic surface is the primary determinant, then increasing the negative charge density should induce inversion in PE-containing cells. This proved to be the case, but inversion of the N-terminal bundle only occurred after making the net negative charge minus two in each of the three cytoplasmic domains (C2–C6), i.e., a change of net charge from plus six to minus six for the whole cytoplasmic surface of the bundle. The N-terminal bundle appears to respond to topological signals as a single domain rather than as individual TMs, and the net charge of the domain surface in a position-independent manner determines the sensitivity of the whole domain to the lipid environment. Remarkably, the effects of net charge were the same whether these resulted from the lipid or the protein, thus supporting charge interaction in a complementary manner between the cytoplasmic domains and the bilayer surface as a determinant of topological organization.
The results from changes in the amino acid sequence further support a lack of translocation machinery involvement in final topological decisions. Neutralizing a single acidic amino acid in C6, which is six TMs from the N terminus, prevented topology inversion of LacY in PE-lacking cells. In addition, the hydrophobicity of TM VII also determined the final topology of the N-terminal bundle. These results, coupled with the behavior of the N-terminal bundle as an independent folding unit (90), are most consistent with final topology being determined after all of the TMs comprising the bundle exit the translocon and begin to fold, which also requires interaction of TM VII with the C-terminal bundle through salt bridges. Although the translocon may impart initial TM orientation, final topology is governed by short-range and long-range interactions within the protein and interactions between the protein and the lipid environment that occur after exit from the translocon.
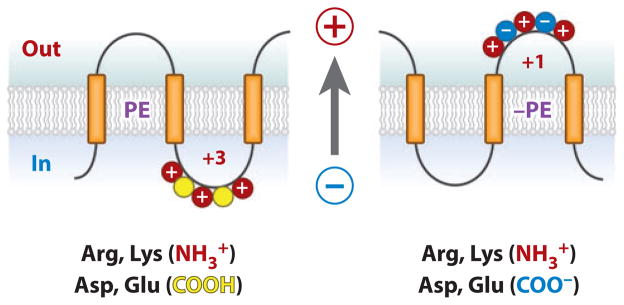
These results reveal a new rule governing topological organization and suggest an important physiological role for neutral lipids, e.g., PE, in cell function. The presence of PE dampens the translocation potential of acidic residues and explains the dominance of positive residues over negative residues as topogenic signals. Whatever the precise mechanism, PE clearly attenuates the topological effects of acidic residues, thereby providing a means for inclusion of acidic residues in the cytoplasmic face of membrane proteins for functional or structural reasons without affecting topology. Therefore, lipids and proteins have coevolved to follow a set of interdependent rules governing topogenesis that also support function.
The above basis for PE function does not provide directionality because PE resides on both sides of the membrane bilayer. However, the proton electrochemical potential directed positive outward was demonstrated as the driving force for translocation of negatively charged domains and the retention of positively charged domains (52–54). Therefore, in PE-containing cells, the net charge contributed by positive residues is dominant, resulting in retention of domains with a mixture of positive and negative residues (Figure 6). However, in the absence of PE, the charge contribution of negative residues becomes significant, favoring translocation of domains with a mixture of charges. Reintroduction of PE postassembly would reverse the charge dominance and establish a new minimum energy state for LacY, thereby driving reorganization (Figure 5f ).
Figure 6.
Phosphatidylethanolamine (PE) and the positive-inside rule. (left) A cytoplasmic domain is shown containing a mixture of negative and positive amino acids. PE is shown to suppress or neutralize the presence of negative residues ( yellow), which increases the effective positive charge potential, thus favoring retention on the cytoplasmic side of the membrane. (right) In the absence of PE, negative residues exert their full potential and result in translocation of a domain that exhibits a lower effective net positive charge. The proton motive force (arrow) positive outward determines domain directional movement depending on the domain effective charge. Postassembly addition of PE changes the effective net charge of these domains and favors reorientation.
The ease of “flipping” LacY postassembly suggests that the activation energy for this transition in vivo is low, which is further supported by the high flexibility and water accessibility of the N-terminal six TMs (91, 92). Therefore, the –11 kcal of translocation driving force provided by the proton motive force (53) may be sufficient to allow spontaneous flipping without an additional catalyst. Future studies are needed to determine if flipping is spontaneous or catalyzed by a protein system that may be involved in general proofreading of aberrant protein structures.
GENERALITY OF LIPID-PROTEIN INTERACTIONS AS TOPOLOGICAL DETERMINANTS
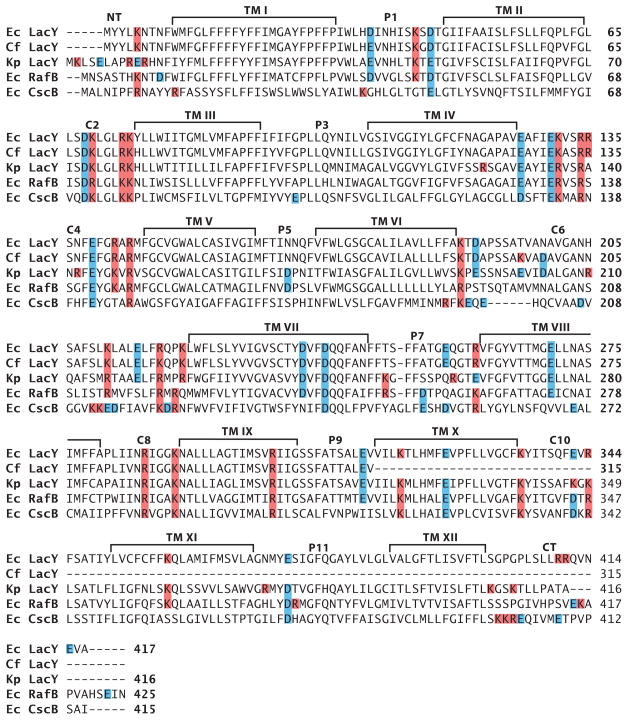
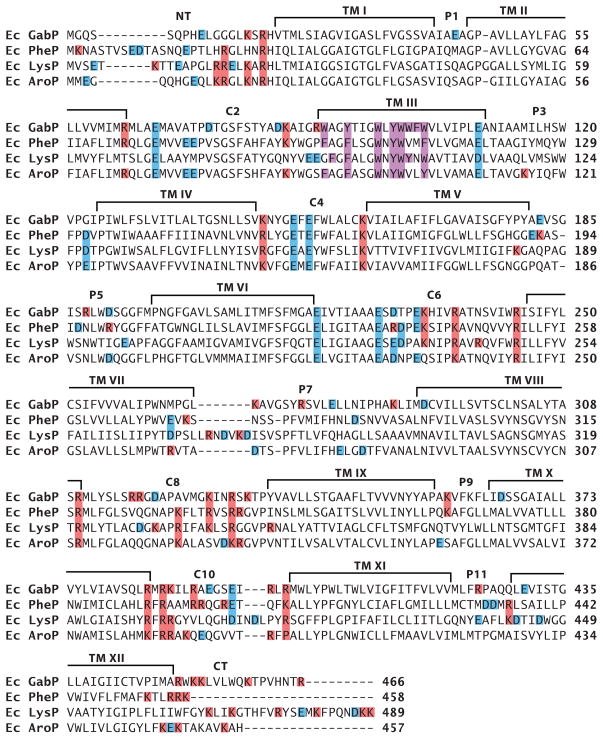
On the basis of the current properties of lipid-sensitive domains, some predictions can be made about domains within other proteins that might require PE or other net neutral lipids to display native topology. A primary feature would be the presence of cytoplasmic domains containing a mixture of negatively and positively charged residues flanking a TM. Such conflicted domains must be free of restrictions preventing different orientations relative to other parts of the protein. The second feature would be a molecular hinge region allowing a differential response to the lipid environment of flanking domains. The former property is relatively easy to predict, whereas the latter property is more difficult because it is a function of more global and long-range interactions within a protein. In the case of LacY, the low hydrophobicity of TM VII provides this flexible molecular hinge. C10 and the CT domains of LacY also contain potentially conflicting charges, but the lack of conflicting charges in C8 may stabilize the whole C-terminal domain in the proper orientation in the absence of PE. Inspection of the charge distribution of cytoplasmic domains of sugar permeases closely related to LacY (67) predicts that the topology of all, with the possible exception of sucrose permease (CscB protein of E. coli ), would be affected by the absence of PE (Figure 7). The sucrose permease has only one acidic residue in its TM VII, which might prevent this domain from behaving as a reversible hinge similar to the LacY D240I derivative. Such molecular hinges cannot yet be predicted. In the case of amino acid permeases closely related to PheP and GabP (Figure 8), the putative conflicted domains requiring PE for native orientation are NT and C2, with TM III supplying the molecular hinge via a possible miniloop structure. Decreasing the net negative charge of either the NT or C2 domain resulted in a mixed topology (60% in the native orientation) for the N-terminal hairpin of PheP expressed in the absence of PE (M. Bogdanov, P. Heacock, & W. Dowhan, unpublished data). Failure to completely prevent misorientation may be due to the high negative charge of C2. In the amino acid permeases, the TM III hinge domain is hydrophobic but is highly enriched in interface-prone aromatic residues, which may be why this domain can form an aberrant hinge structure. C4 and C6 of this group of proteins are also conflicted yet lie on the wrong side of the hinge domain and may be constricted by the downstream cytoplasmic domains carrying a net positive charge. Therefore, predictions of topological dependence on lipid composition are currently very difficult to make with any certainty, but with current information, putative domains can be identified at least within related proteins and subjected to experimental evaluation to build a database similar to those supporting the positive-inside rule.
Figure 7.
Distribution of charged amino acids in homologous sugar permeases. Sequence alignments are shown for E. coli (Ec) lactose (LacY), raffinose (RafB), and sucrose (CscB) permeases and for Klebsiella pneumoniae (Kp) and Citrobacter freundii (Cf ) lactose permeases. Positively or negatively charged amino acids are colored red or blue, respectively.
Figure 8.
Distribution of charged amino acids in homologous amino acid permeases. Sequence alignments shown for E. coli γ-aminobutyrate permease (GabP), phenylalanine (PheP), lysine (LysP), and aromatic (AroP) permeases. Positively or negatively charged amino acids are colored with red or blue, respectively, and aromatic residues in TM III are colored purple.
SUMMARY POINTS.
Lipids appear to facilitate membrane protein refolding in vitro and folding in vivo into a fully native conformation by transient interaction with late folding and nonnative intermediates in a manner similar to most protein molecular chaperones.
TM orientation may be initially influenced by the translocation machinery, but final topology is determined outside the translocon by long-range interactions within the protein and short-range interactions between the protein and the lipids as independently folding domains assume their final structure.
Topological organization of a membrane protein, once established, is not static but is dynamic in response to changes in lipid environment.
Lipids and proteins have coevolved to follow a set of interdependent rules governing topological organization of membrane proteins.
A protein’s sequence determines its topology, but its sequence is written for a specific membrane lipid environment.
Lipid-protein interactions affect the potency of charged residues as topological signals. The balance of net charge owing to the presence of opposing topogenic signals (discrete opposite charges) can either represent a retention or driving force, which either supports or overrides the positive-inside rule depending on presence of neutral lipids (e.g, PE) and on the magnitude of negative charge density of the membrane surface.
The translocation potential of negative amino acids working in opposition to the positive-inside rule is largely dampened by the presence of PE, thus enhancing the effect of the positive-inside rule and explaining the dominance of positive residues as retention signals even in the presence of excess negative residues.
An important physiological role for PE is to allow the presence of negatively charged amino acids in the cytoplasmic domains of membrane proteins to support structure and function without affecting topological organization governed by the positive-inside rule.
FUTURE ISSUES.
How general is lipid-dependent topogenesis and can more predictive rules be generated?
Can membrane proteins adopt multiple topology states within one biological membrane and are there fluctuations among these states with time and environmental condition?
Is there functional significance associated with lipid-sensitive protein domains?
Is postassembly topological inversion spontaneous or is it catalyzed by a topological proofreading process?
Is topological switching a function of bidirectional lipid composition changes? Are there levels of PE where multiple protein orientations exist?
Do topological differences originate cotranslationally during membrane insertion or are they induced by changes in lipid composition as eukaryotic proteins move through different organelles to their final destination?
Are postassembly lipid-dependent topological changes biologically important control mechanisms?
Do lipid-dependent topological disorders exist? Diseases in which proper protein folding or topological organization is not attained due to either demonstrated or proposed involvement of a lipid (93) are becoming more evident. Therefore, how lipids influence the folding, assembly, and function of proteins will be useful in developing novel therapeutic approaches for treatment of conformational and topological protein disorders.
Acknowledgments
This review was supported in part by a National Institutes of Health grant to W.D. (GM20478) and by the John S. Dunn Research Foundation.
- Integral membrane protein
a protein with at least one domain that completely spans the membrane bilayer
- TM
transmembrane domain
- Topogenesis
the process that establishes membrane protein topology
- Membrane protein topology
orientation of transmembrane domains and flanking extramembrane domains with respect to the plane of the membrane lipid bilayer
- Translocon
a protein complex responsible for translocating soluble proteins across the membrane and integrating membrane proteins into the membrane
- Topogenic signals
amino acid sequences that predetermine the final topology of membrane proteins
- LacY
lactose permease
- Topological determinants
cellular factors and processes that decode topogenic signals to establish final topological organization of integral membrane proteins
- PheP
phenylalanine permease
- GabP
γ-aminobutyrate permease
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- CL
cardiolipin
- PC
phosphatidylcholine
- SCAM™
a substituted cysteine accessibility method, as applied to transmembrane domains, used to establish the topological organization of membrane proteins
- GlcDAG
monoglucosyl diacylglycerol
- GlcGlcDAG
diglucosyl diacylglycerol
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
Contributor Information
William Dowhan, Email: William.Dowhan@uth.tmc.edu.
Mikhail Bogdanov, Email: Mikhail.V.Bogdanov@uth.tmc.edu.
LITERATURE CITED
- 1.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–67. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–69. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 3.Pitonzo D, Skach WR. Molecular mechanisms of aquaporin biogenesis by the endoplasmic reticulum Sec61 translocon. Biochim Biophys Acta. 2006;1758:976–88. doi: 10.1016/j.bbamem.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 4.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–18. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 5.Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat Struct Mol Biol. 2005;12:870–88. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- 6.Hessa T, White SH, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 7.Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–40. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 8.Monné M, Hessa T, Thissen L, von Heijne G. Competition between neighboring topogenic signals during membrane protein insertion into the ER. FEBS J. 2005;272:28–36. doi: 10.1111/j.1742-4658.2004.04394.x. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson I, von Heijne G. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell. 1990;62:1135–41. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- 10.Rutz C, Rosenthal W, Schulein R. A single negatively charged residue affects the orientation of a membrane protein in the inner membrane of Escherichia coli only when it is located adjacent to a transmembrane domain. J Biol Chem. 1999;274:33757–63. doi: 10.1074/jbc.274.47.33757. [DOI] [PubMed] [Google Scholar]
- 11.Goder V, Junne T, Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell. 2004;15:1470–78. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goder V, Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–53. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–16. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White SH, von Heijne G. Do protein-lipid interactions determine the recognition of transmembrane helices at the ER translocon? Biochem Soc Trans. 2005;33:1012–15. doi: 10.1042/BST20051012. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J Biol Chem. 2003;278:50128–35. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 16.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–81. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 17.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–30. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 18.Ismail N, Crawshaw SG, Cross BC, Haagsma AC, High S. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem J. 2008;411:495–506. doi: 10.1042/BJ20071597. [DOI] [PubMed] [Google Scholar]
- 19.Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J Cell Biol. 2007;179:1441–52. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–22. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 21.van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 22.Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40–60 Å during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–44. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 23.Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol. 1999;147:257–66. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie KR. Folding and stability of α-helical integral membrane proteins. Chem Rev. 2006;106:1931–77. doi: 10.1021/cr0404388. [DOI] [PubMed] [Google Scholar]
- 25.Rapoport TA. Extensions of the signal hypothesis–sequential insertion model versus amphipathic tunnel hypothesis. FEBS Lett. 1985;187:1–10. doi: 10.1016/0014-5793(85)81202-3. [DOI] [PubMed] [Google Scholar]
- 26.Bibi E, Stearns SM, Kaback HR. The N-terminal 22 amino acid residues in the lactose permease of Escherichia coli are not obligatory for membrane insertion or transport activity. Proc Natl Acad Sci USA. 1992;89:3180–84. doi: 10.1073/pnas.89.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrmann M, Beckwith J. Proper insertion of a complex membrane protein in the absence of its amino-terminal export signal. J Biol Chem. 1991;266:16530–33. [PubMed] [Google Scholar]
- 28.Guo D, Liu J, Motlagh A, Jewell J, Miller KW. Efficient insertion of odd-numbered transmembrane segments of the tetracycline resistance protein requires even-numbered segments. J Biol Chem. 1996;271:30829–34. doi: 10.1074/jbc.271.48.30829. [DOI] [PubMed] [Google Scholar]
- 29.White SH, von Heijne G. How translocons select transmembrane helices. Annu Rev Biophys. 2008;37:23–42. doi: 10.1146/annurev.biophys.37.032807.125904. [DOI] [PubMed] [Google Scholar]
- 30.Lee E, Manoil C. Mutations eliminating the protein export function of a membrane-spanning sequence. J Biol Chem. 1994;269:28822–28. [PubMed] [Google Scholar]
- 31.Wahlberg JM, Spiess M. Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. J Cell Biol. 1997;137:555–62. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harley CA, Holt JA, Turner R, Tipper DJ. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J Biol Chem. 1998;273:24963–71. doi: 10.1074/jbc.273.38.24963. [DOI] [PubMed] [Google Scholar]
- 33.Sakaguchi M, Tomiyoshi R, Kuroiwa T, Mihara K, Omura T. Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc Natl Acad Sci USA. 1992;89:16–19. doi: 10.1073/pnas.89.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao G, Cheng S, Whitley P, von Heijne G, Kuhn A, Dalbey RE. Synergistic insertion of two hydrophobic regions drives Sec-independent membrane protein assembly. J Biol Chem. 1994;269:26898–903. [PubMed] [Google Scholar]
- 35.Carveth K, Buck T, Anthony V, Skach WR. Cooperativity and flexibility of cystic fibrosis trans-membrane conductance regulator transmembrane segments participate in membrane localization of a charged residue. J Biol Chem. 2002;277:39507–14. doi: 10.1074/jbc.M205759200. [DOI] [PubMed] [Google Scholar]
- 36.Heinrich SU, Rapoport TA. Cooperation of transmembrane segments during the integration of a double-spanning protein into the ER membrane. EMBO J. 2003;22:3654–63. doi: 10.1093/emboj/cdg346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao G, London E. An amino acid “transmembrane tendency” scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: relationship to biological hydrophobicity. Protein Sci. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engelman DM, Chen Y, Chin CN, Curran AR, Dixon AM, et al. Membrane protein folding: beyond the two stage model. FEBS Lett. 2003;555:122–25. doi: 10.1016/s0014-5793(03)01106-2. [DOI] [PubMed] [Google Scholar]
- 39.Gafvelin G, von Heijne G. Topological “frustration” in multispanning E. coli inner membrane proteins. Cell. 1994;77:401–12. doi: 10.1016/0092-8674(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 40.Kanki T, Sakaguchi M, Kitamura A, Sato T, Mihara K, Hamasaki N. The tenth membrane region of band 3 is initially exposed to the luminal side of the endoplasmic reticulum and then integrated into a partially folded band 3 intermediate. Biochemistry. 2002;41:13973–81. doi: 10.1021/bi026619q. [DOI] [PubMed] [Google Scholar]
- 41.Sato M, Mueckler M. A conserved amino acid motif (R-X-G-R-R) in the Glut1 glucose transporter is an important determinant of membrane topology. J Biol Chem. 1999;274:24721–25. doi: 10.1074/jbc.274.35.24721. [DOI] [PubMed] [Google Scholar]
- 42.Tector M, Hartl FU. An unstable transmembrane segment in the cystic fibrosis transmembrane conductance regulator. EMBO J. 1999;18:6290–98. doi: 10.1093/emboj/18.22.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saier MH., Jr Tracing pathways of transport protein evolution. Mol Microbiol. 2003;48:1145–56. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 44.Jimenez-Mejia R, Campos-Garcia J, Cervantes C. Membrane topology of the chromate transporter ChrA of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2006;262:178–84. doi: 10.1111/j.1574-6968.2006.00386.x. [DOI] [PubMed] [Google Scholar]
- 45.Granseth E, von Heijne G, Elofsson A. A study of the membrane-water interface region of membrane proteins. J Mol Biol. 2005;346:377–85. doi: 10.1016/j.jmb.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson J, Persson B, von Heijne G. Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins. 2005;60:606–16. doi: 10.1002/prot.20583. [DOI] [PubMed] [Google Scholar]
- 47.von Heijne G. Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature. 1989;341:456–58. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 48.Andersson H, Bakker E, von Heijne G. Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. J Biol Chem. 1992;267:1491–95. [PubMed] [Google Scholar]
- 49.Allard JD, Bertrand KP. Membrane topology of the pBR322 tetracycline resistance protein. TetA-PhoA gene fusions and implications for the mechanism of TetA membrane insertion. J Biol Chem. 1992;267:17809–19. [PubMed] [Google Scholar]
- 50.Pi J, Chow H, Pittard AJ. Study of second-site suppression in the pheP gene for the phenylalanine transporter of Escherichia coli. J Bacteriol. 2002;184:5842–47. doi: 10.1128/JB.184.21.5842-5847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishtalik LI, Cramer WA. On the physical basis for the cis-positive rule describing protein orientation in biological membranes. FEBS Lett. 1995;369:140–43. doi: 10.1016/0014-5793(95)00756-y. [DOI] [PubMed] [Google Scholar]
- 52.Andersson H, von Heijne G. Membrane protein topology: effects of ΔμH+ on the translocation of charged residues explain the ‘positive inside’ rule. EMBO J. 1994;13:2267–72. doi: 10.1002/j.1460-2075.1994.tb06508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao G, Kuhn A, Dalbey RE. The translocation of negatively charged residues across the membrane is driven by the electrochemical potential: evidence for an electrophoresis-like membrane transfer mechanism. EMBO J. 1995;14:866–75. doi: 10.1002/j.1460-2075.1995.tb07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao G, Dalbey RE. Translocation of N-terminal tails across the plasma membrane. EMBO J. 1994;13:4662–69. doi: 10.1002/j.1460-2075.1994.tb06789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delgado-Partin VM, Dalbey RE. The proton motive force, acting on acidic residues, promotes translocation of amino-terminal domains of membrane proteins when the hydrophobicity of the translocation signal is low. J Biol Chem. 1998;273:9927–34. doi: 10.1074/jbc.273.16.9927. [DOI] [PubMed] [Google Scholar]
- 56.Andersson H, von Heijne G. Position-specific Asp-Lys pairing can affect signal sequence function and membrane protein topology. J Biol Chem. 1993;268:21389–93. [PubMed] [Google Scholar]
- 57.Parks GD, Lamb RA. Role of NH2-terminal positively charged residues in establishing membrane protein topology. J Biol Chem. 1993;268:19101–9. [PubMed] [Google Scholar]
- 58.Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–90. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dowhan W. Molecular basis for membrane phospholipid diversity: Why are there so many phospholipids. Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 60.Dowhan W, Bogdanov M, Mileykovskaya E. Functional roles of lipids in membranes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 5 San Diego, CA: Elsevier; 2008. pp. 1–37. [Google Scholar]
- 61.Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–81. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyl diacyl-glycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J Biol Chem. 2006;281:19172–78. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology. Phosphatidylethanolamine is required for the proper topological organization of the γ-aminobutyric acid permease (GabP) of Escherichia coli. J Biol Chem. 2005;280:26032–38. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 64.Raetz CR, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem. 1990;265:1235–38. [PubMed] [Google Scholar]
- 65.van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. Anionic phospholipids are determinants of membrane protein topology. EMBO J. 1997;16:4261–66. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeChavigny A, Heacock PN, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–32. [PubMed] [Google Scholar]
- 67.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: protein-lipid charge interactions are a determinant of final membrane protein topology. J Cell Biol. 2008;182:925–35. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abramson J, Iwata S, Kaback HR. Lactose permease as a paradigm for membrane transport proteins. Mol Membr Biol. 2004;21:227–36. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 69.Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J Biol Chem. 1984;259:10150–58. [PubMed] [Google Scholar]
- 70.Bogdanov M, Dowhan W. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J Biol Chem. 1995;270:732–39. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- 71.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J Biol Chem. 1996;271:11615–18. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 72.Sun J, Wu J, Carrasco N, Kaback HR. Identification of the epitope for monoclonal antibody 4B1 which uncouples lactose and proton translocation in the lactose permease of Escherichia coli. Biochemistry. 1996;35:990–98. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 73.Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J. 1998;17:5255–64. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein: minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–45. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 75.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J Biol Chem. 1999;274:36827–30. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 76.Dowhan W, Mileykovskaya E, Bogdanov M. Diversity and versatility of lipid-protein interactions revealed by molecular genetic approaches. Biochim Biophys Acta. 2004;1666:19–39. doi: 10.1016/j.bbamem.2004.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM™): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–71. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calamia J, Manoil C. lac permease of Escherichia coli: topology and sequence elements promoting membrane insertion. Proc Natl Acad Sci USA. 1990;87:4937–41. doi: 10.1073/pnas.87.13.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–15. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 80.Lundin M, Monné M, Widell A, von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428–38. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y, Turnbull IR, Bragin A, Carveth K, Verkman AS, Skach WR. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2973–85. doi: 10.1091/mbc.11.9.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sugai R, Takemae K, Tokuda H, Nishiyama KI. Topology inversion of SecG is essential for cytosolic SecA-dependent stimulation of protein translocation. J Biol Chem. 2007;282:29540–48. doi: 10.1074/jbc.M704716200. [DOI] [PubMed] [Google Scholar]
- 83.Gouffi K, Gerard F, Santini CL, Wu LF. Dual topology of the Escherichia coli TatA protein. J Biol Chem. 2004;279:11608–15. doi: 10.1074/jbc.M313187200. [DOI] [PubMed] [Google Scholar]
- 84.Slatin SL, Nardi A, Jakes KS, Baty D, Duche D. Translocation of a functional protein by a voltage-dependent ion channel. Proc Natl Acad Sci USA. 2002;99:1286–91. doi: 10.1073/pnas.022480199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ikeda M, Kida Y, Ikushiro S, Sakaguchi M. Manipulation of membrane protein topology on the endoplasmic reticulum by a specific ligand in living cells. J Biochem. 2005;138:631–37. doi: 10.1093/jb/mvi157. [DOI] [PubMed] [Google Scholar]
- 86.Brambillasca S, Yabal M, Borgese N. Unassisted translocation of large polypeptide domains across phospholipid bilayers. J Cell Biol. 2006;175:767–77. doi: 10.1083/jcb.200608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brambillasca S, Yabal M, Soffientini P, Stefanovic S, Makarow M, et al. Transmembrane topogenesis of a tail-anchored protein is modulated by membrane lipid composition. EMBO J. 2005;24:2533–42. doi: 10.1038/sj.emboj.7600730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, et al. Monoglucosyl diacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem. 2004;279:10484–93. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 89.Wikström M, Kelly A, Georgiev A, Eriksson H, Rosén-Klement M, et al. Lipid-engineered Escherichia coli membranes reveal critical lipid head-group size for protein function. J Biol Chem. 2009;284:954–65. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagamori S, Vazquez-Ibar JL, Weinglass AB, Kaback HR. In vitro synthesis of lactose permease to probe the mechanism of membrane insertion and folding. J Biol Chem. 2003;278:14820–26. doi: 10.1074/jbc.M300332200. [DOI] [PubMed] [Google Scholar]
- 91.Bennett M, D’Rozario R, Sansom MS, Yeagle PL. Asymmetric stability among the transmembrane helices of lactose permease. Biochemistry. 2006;45:8088–95. doi: 10.1021/bi060355g. [DOI] [PubMed] [Google Scholar]
- 92.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–33. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]