Abstract
The transcription factor Snail has been described as a direct repressor of E-cadherin expression during development and carcinogenesis; however, the specific mechanisms involved in this process remain largely unknown. Here we show that mammalian Snail requires histone deacetylase (HDAC) activity to repress E-cadherin promoter and that treatment with trichostatin A (TSA) is sufficient to block the repressor effect of Snail. Moreover, overexpression of Snail is correlated with deacetylation of histones H3 and H4 at the E-cadherin promoter, and TSA treatment in Snail-expressing cells reverses the acetylation status of histones. Additionally, we demonstrate that Snail interacts in vivo with the E-cadherin promoter and recruits HDAC activity. Most importantly, we demonstrate an interaction between Snail, histone deacetylase 1 (HDAC1) and HDAC2, and the corepressor mSin3A. This interaction is dependent on the SNAG domain of Snail, indicating that the Snail transcription factor mediates the repression by recruitment of chromatin-modifying activities, forming a multimolecular complex to repress E-cadherin expression. Our results establish a direct causal relationship between Snail-dependent repression of E-cadherin and the modification of chromatin at its promoter.
The regulation of E-cadherin expression is a controlled process that requires strict spatiotemporal tuning during natural processes such as development, organogenesis, and tissue formation. However, the regulation of E-cadherin also plays an essential role in pathological processes such as tumor progression. The loss of expression or function of the E-cadherin cell-to-cell adhesion molecule has emerged as an important event for the local invasion of epithelial tumor cells, leading to the consideration of E-cadherin as an invasion suppressor gene (7, 8, 14, 49).
The molecular mechanisms involved in E-cadherin downregulation during physiological and pathological processes have started to be uncovered in recent years. Several mechanisms have been implicated in the regulation of E-cadherin expression during tumor progression, including genetic, epigenetic, and transcriptional changes. While genetic alterations of the E-cadherin loci have been found only infrequently in tumors, particularly, in lobular breast carcinomas and diffuse gastric carcinomas (6, 7, 21, 44), the majority of carcinomas with downregulated E-cadherin maintain an intact E-cadherin locus. Epigenetic processes involving hypermethylation of the E-cadherin promoter and/or transcriptional alterations have emerged as the main mechanisms responsible for E-cadherin downregulation in most carcinomas (13, 14, 23, 42). Several transcriptional repressors of E-cadherin have been recently identified, including the zinc finger factors Snail (5, 11) and Slug (10, 22), the two-handed zinc factors ZEB1(δEF1) and ZEB2 (SIP-1) (15, 20), and the bHLH factor E12/E47 (40). Factors belonging to the Snail family are in fact involved in E-cadherin repression and in epithelial to mesenchymal transitions (EMTs) when they are overexpressed in epithelial cell lines (5, 10, 11) as well as in embryonic development (reviewed in reference 37), and it has been proposed that these factors act as inducers of the invasion process (9, 11). The generation of mice lacking Snail has firmly established the role of this factor in EMT and as an E-cadherin gene repressor, as the null Snail embryos die at gastrulation and fail to undergo a complete EMT process, forming an altered mesodermal layer which maintains the expression of E-cadherin (12). Despite all the above information, the molecular mechanisms involved in the repression by factors of the Snail family are still poorly understood (37, 50). A previous work established that human Slug, a Snail family member, is a transcriptional repressor with an N-terminal 32-amino-acid repression domain and postulated the possible involvement of histone deacetylation in the repression mechanism (26).
Chromatin remodeling and histone modifications have emerged as the main mechanisms of the control of gene expression. Hyperacetylation of histones H3 and H4 is generally associated with transcriptionally active chromatin (47), while the chromatin of inactive regions is enriched in deacetylated histones H3 and H4. The acetylation status of histones at specific DNA regulatory sequences depends on the recruitment of histone acetyltransferases or histone deacetylase (HDAC) activities, usually as part of large multiprotein complexes of coactivators or corepressors, respectively. Several corepressor complexes have been identified to date (such as the SIN3, Mi-2/NuRD, and CoREST complexes) with the ability to interact with several transcriptional repressors (1, 27). Interestingly, during the past 5 years, the connection between DNA methylation and histone deacetylation in the silencing of genes has been established, and the mechanisms involve the participation of proteins belonging to the family of methyl-CpG binding domain proteins and HDACs (4). Moreover, other histone modifications, such as histone methylation, appear to be associated with gene regulation (32), thus suggesting the participation of different histone and DNA modifying activities in multiprotein complex regulators.
To gain further understanding of the mechanisms implicated in E-cadherin repression by Snail, we have investigated the involvement of HDACs and other potential corepressors. We report here that the endogenous E-cadherin promoter of Snail-expressing cells is enriched in deacetylated histones H3 and H4 and dimethylated H3 at K9 and that Snail-mediated repression is abolished by treatment with trichostatin A (TSA). Snail interacts directly with the endogenous E-cadherin promoter, as demonstrated by chromatin immunoprecipitation (ChIP) assays, and recruits HDAC activity. Moreover, in vivo and in vitro interactions of Snail with histone deacetylase 1 (HDAC1) and HDAC2 and the corepressor mSin3A have been detected. These interactions depend on the SNAG N-terminal domain of Snail and are required for an efficient repression of the E-cadherin promoter, which supports the idea that Snail mediates the repression of E-cadherin by the recruitment of a corepressor complex containing HDAC1 and HDAC2 (HDAC1/2) and Sin3A.
MATERIALS AND METHODS
Generation of plasmids, expression vectors, and stable cell lines.
All the Snail constructions were generated by using the full-length Snail cDNA as template (11). Snail mutants were generated by PCR with the following primers (restriction sites are indicated in bold): for ΔSNAG, 5′-TTCAAGCTTATGAAGCCGTCCGAC-3′ (direct) and 5′-GCCGAATTCCCCGGACAAGGC-3′ (reverse); and for ΔNt, 5′-GCCAAGGGATCCCAGATGCGGAAG-3′ (direct) and 5′-GCCGAATTCCCCGGACAAGGC-3′ (reverse). The PCR products were cloned in plasmids pcDNA3 or pGEX (Amersham). Snail-hemagglutinin (HA) construction was generated by mutation of the stop codon by PCR with the primers (restriction sites are indicated in bold) 5′-TCTGCGAATTCATGCCGCGCTCCT-3′ (direct) and 5′-CTCGAGGGCGCGAGGGCCTCCGGA-3′ (reverse) and cloning in vector pcDNA3-HA (Promega). Green fluorescent protein (GFP)-Snail construction was generated by PCR with the same direct primer that was used for Snail-HA and the reverse primer 5′-ATCCCGGGCGCGAGGGCCTCCGGA-3′, with cloning in vector pEGFP-C1 (Clontech). MDCK-GFP and MDCK-GFP-Snail cell lines were generated by stable transfection with 3 μg of plasmids pEGFP-C1 and pEGFP-Snail, respectively, and selection with 400 μg of G418/ml for 3 to 4 weeks. The generation of MDCK-CMV and MDCK-Snail cell lines has been previously described (11).
Cell culture and treatments.
MDCK-II, MDCK-CMV, MDCK-Snail, MDCK-GFP, MDCK-GFP-Snail, human embryonic kidney (HEK) 293T, mouse keratinocytes Pam212, and spindle CarC and CarB cells were grown in Dulbecco's modified Eagle's medium, and mouse keratinocytes MCA3D and PDV cells were grown in Ham's F-12 medium, supplemented with 10% fetal bovine serum, 10 mM glutamine (Gibco BRL), 100 μg of ampicillin/ml, and 32 μg of gentamicin/ml (Sigma Chemical Co). Cells were grown at 37°C in a humidified CO2 atmosphere. The origin, tumorigenic properties, and E-cadherin expression levels of the mouse cell lines have been previously described (11, 35). TSA (Sigma) was dissolved in ethanol and added to the culture medium at 300 nM. A corresponding volume of ethanol was added to control untreated cells.
E-cadherin promoter analysis.
The mouse E-cadherin promoter sequences (positions −178 to +92) cloned into vector pGL2 (Invitrogen) that was fused to a firefly luciferase reporter gene (10) was used to determine the activity of the E-cadherin promoter as previously described (39). Except where indicated, TSA treatments were performed for 24 h after transfection. Cotransfections were carried out in the presence of the indicated amounts of pcDNA3-Snail constructs, or 100 ng of pcDNA3-HDAC1 and pSC2-mSin3A (a gift of R. N. Eisenmam, Fred Hutchinson Cancer Research Center, Seattle, Wash.). Luciferase and Renilla activities were measured with the dual-luciferase reporter assay kit (Promega) and normalized to the wild-type promoter activity detected in mock-transfected (pcDNA3) or untreated cells.
Transient transfections, immunoprecipitations, and pull-down assays.
A total of 6 × 105 HEK 293T cells grown in P60 dishes were transiently transfected in a P60 dish with Lipofectamine reagent (Invitrogen) and 2 μg of each of the following plasmids: pcDNA3, pcDNA3-Snail-HA, pSC2-mSin3A-myc, and/or pcDNA3-HDAC1-Flag, pcDNA3-HDAC2-Flag, pcDNA3-HDAC3-Flag (provided by E. Seto, Moffit Cancer Center, Tampa, Fla.). For immunoprecipitation, cells were lysed 36 h after transfection in 500 μl of IPH buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.2 mM phenylmethylsulfonyl fluoride) at 4°C for 30 min and then centrifuged at 16,060 × g for 15 min to remove cell debris. The cleared lysate was then subjected to immunoprecipitation with the indicated antibodies overnight at 4°C. Protein A- or G-Sepharose beads (Sigma) were added, and incubation continued for 2 h at 4°C. Precipitates were washed four times with IPH buffer (1 ml) and then resuspended in 1× Laemmli buffer. Proteins were separated on a sodium dodecyl sulfate-7.5 to 10% polyacrylamide gel and blotted onto Immobilon-P (Millipore Co.) membranes. Endogenous mSin3A was immunoprecipitated or detected by Western blotting with rabbit anti-mSin3A (dilution, 1:200) (AK-11; SantaCruz Biotechnology). Blots were also incubated (at the dilutions shown) with rat anti-HA (1:400) (Roche), rabbit anti-HDAC2 and anti-HDAC3 (both, 1:500) (Abcam), mouse anti-Flag (1:3,000), and rabbit anti-myc (1:500) (Sigma) antibodies. The secondary antibodies used (at the dilutions shown) were goat anti-rabbit conjugated to horseradish peroxidase (HRP) (1:4,000) (Nordic), sheep anti-mouse-HRP (1:1,000) (Amersham), or goat anti-rat-HRP (1:10,000) (Nordic).
For pull-down assays, HEK 293T cells were transiently transfected with the indicated plasmids and lysed in IPH buffer as above. Extracts were then incubated with the glutathione S-transferase (GST)-fusion proteins and the bound fraction was purified through glutathione-Sepharose beads (Amersham). Bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting to the indicated antibodies.
Immunofluorescence.
For immunofluorescence, cells grown on coverslips and transfected as above were fixed in methanol (at −20°C) for 1 min and stained (at the dilutions shown) for rat anti-HA (1:100), rabbit anti-mSin3A (1:50), anti-HDAC1, anti-HDAC2, anti-HDAC3 (all anti-HDACs, 1:300), mouse anti-Flag (1:300), or rabbit anti-myc (1:100). The secondary antibodies used were anti-rat Alexa 594 (1:1,000), anti-mouse Alexa 647 (1:200) (Molecular Probes), and anti-rabbit fluorescein isothiocyanate (1:500) (Jackson). The cells were mounted on Mowiol and the preparations were visualized with a Leica confocal TCS SP2 microscope.
HDAC activity.
HEK 293T cells were transiently transfected with GFP or GFP-Snail plasmids, and immunoprecipitates were obtained in IPH buffer as described above but with anti-GFP antibodies (Clontech). The immunoprecipitated beads were used to assay the associated HDAC activity. The beads were incubated with 10 μl of [H3]acetate-labeled histones (1.8 nCi/μg) in 100 μl of activity buffer (25 mM Tris-HCl [pH 7.5], 10% glycerol, 1 mM EDTA, 50 mM NaCl) for 2 h at 37°C. The reaction was stopped by the addition of 50 μl of 1 N HCl-0.4 M acetate and the released [H3]acetate was extracted with 600 μl of ethyl acetate. After centrifugation, a 450-μl aliquot of the organic phase was counted in 3 ml of scintillation cocktail.
ChIP assays.
To investigate the modification status of histones at the E-cadherin promoter, standard ChIP assays were performed as previously described (17) in cells with or without TSA (300 nM) treatment. Cells were cross-linked with formaldehyde prior to DNA sonication. Immunoprecipitations of the cross-linked chromatin were carried out with commercial antibodies (anti-acetyl-histone H3, anti-acetyl-histone H4, anti-dimethyl-K4 histone H3, and anti-dimethyl-K9 histone H3) (Upstate Biotech). A similar protocol was used for analysis of the binding of GFP-Snail to the E-cadherin promoter but with anti-GFP antibodies (Clontech). To investigate the association of the HDACs with the E-cadherin promoter, an extra cross-linking step was introduced in the assay. In this case, prior to formaldehyde cross-linking, cells were treated with 10 mM dimethyl adipimidate, a protein-to-protein cross-linking agent, and 0.25% dimethyl sulfoxide in phosphate-buffered saline for 45 min (31). Anti-HDAC1, anti-HDAC2, and anti-HDAC3 antibodies were from Abcam. In all cases, chromatin was sheared to an average length of 0.25 to 1 kb for this analysis. PCR amplification was performed in 25 μl with specific primers for each of the analyzed promoters. The sensitivity of PCR amplification was evaluated on serial dilutions of total DNA collected after sonication (input fraction). A ∼250-bp fragment of the mouse E-cadherin promoter was amplified with the primers 5′-TAGGAAGCTGGGAAG-3′ (direct) and 5′-TGCGGTCGGGCAGGG-3′ (reverse); a ∼150-bp fragment of canine E-cadherin promoter (15) was amplified with the primers 5′-CCCGCCGCAGGTGCAGCCGCAGC-3′ (direct) and 5′-GAGGCGGCGCGAGGCCGGCAG-3′ (reverse). In both cases PCR was carried out according to the following program: 32 cycles at 94°C for 40 s, 65 to 68°C for 40 s, and 72°C for 40 s. The amplified DNA was separated on 2% agarose gel and visualized with ethidium bromide.
Reverse transcription (RT)-PCR analysis of E-cadherin expression.
Expression of E-cadherin after TSA treatment was performed by RT-PCR. Total RNA was extracted with the RNeasy kit (QIAGEN) from cells treated for 24 h with 300 nM TSA or the vehicle. RT-PCR of E-cadherin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts were performed as previously described (10, 40), with Taq-DyNAzyme polymerase (Fynnzyme) and appropriate primers for the amplification of mouse and canine cDNAs.
RESULTS
Snail requires HDAC activity to repress E-cadherin promoter.
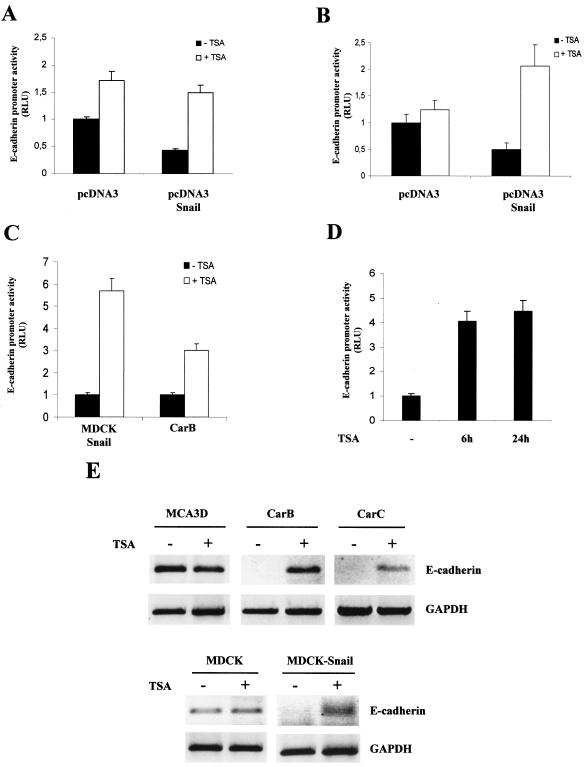
It has been suggested that histone deacetylation could be one of the mechanisms by which other members of the Snail family, such as human Slug, repress transcription (26). We decided to investigate whether Snail could be mediating the repression of E-cadherin by the recruitment of HDACs. First, we analyzed the effects of TSA, which specifically inhibits HDACs (56) in the Snail-mediated repression of E-cadherin. To this end, we studied the repressor effect of Snail on a reporter gene system in which the mouse E-cadherin proximal promoter was fused upstream of the luciferase cDNA (10) in several epithelial cell lines, such as MDCK and the mouse keratinocyte cell line MCA3D (Fig. 1). The transfection of Snail (50 ng) in MDCK cells reduced the activity of the E-cadherin promoter to 40%, as previously observed (10, 11). This repressive effect was totally eliminated by treatment with TSA (Fig. 1A). The elimination of the Snail-repressing activity by TSA was also observed in the MCA3D cell line (Fig. 1B), in which the reduction of the E-cadherin promoter activity by Snail transfection was also fully relieved by TSA treatment. These results strongly suggest that HDAC activity is required for efficient Snail repression of the E-cadherin promoter. TSA treatment induced a moderate increase (less than twofold) in the activity of the E-cadherin promoter in MDCK cells transfected with control vector pcDNA3 (Fig. 1A and B), which suggests a TSA effect independent of Snail in this cell line. However, RT-PCR analysis showed no increase in E-cadherin transcripts after TSA treatment in either MCDK or MCA3D cells (Fig. 1E).
FIG. 1.
TSA inhibits the Snail-mediated repression of the E-cadherin promoter. (A and B) E-cadherin promoter activity was analyzed in epithelial MCDK (A) and epidermal keratinocyte MCA3D (B) cells in the presence of plasmids pcDNA3 control and pcDNA-Snail. Where indicated, cells were treated with TSA (300 nM; white bars) or with ethanol (black bars) for 24 h after transfection. (C and D) The activity of the E-cadherin promoter was analyzed in MDCK-Snail (C and D) and spindle carcinoma CarB (C) cells. Cells were treated with TSA (300 nM; white bars) or with ethanol (black bars) for 24 h (C) or at the indicated time points (D) after transfection. Luciferase and Renilla activities were determined 24 h after transfection; the promoter activity is represented as the relative luciferuse units (RLU) for control untreated cells. Results represent the averages ± standard deviations of at least two independent experiments performed in duplicate. − (D), cells treated with ethanol for 24 h after transfection. (E) RT-PCR analysis of E-cadherin levels. The indicated cell lines were treated for 24 h with TSA (300 nM) (lanes marked with plus sign) or with vehicle (lanes marked with a minus sign); total RNA was extracted and subjected to RT-PCR analysis with specific E-cadherin primers. The levels of GAPDH were analyzed as a control of the amount of cDNA.
We next decided to analyze the activity of the E-cadherin promoter in the presence or absence of TSA in E-cadherin-negative cells in which Snail determines E-cadherin repression. We used the stably transfected MDCK-Snail cell line to assay the activity of the E-cadherin promoter after TSA treatment. The low basal activity of the E-cadherin promoter in this cell line (10, 11) could be upregulated up to sixfold after 24 h of TSA treatment (Fig. 1C). Similar levels of derepression were obtained after only 6 h of treatment with TSA (Fig. 1D), indicating that HDAC activity is required for Snail repression. Similar effects were observed after TSA treatment in the E-cadherin-deficient mouse spindle carcinoma cell line CarB expressing endogenous Snail (11). As previously reported (16, 42), we detected very low E-cadherin promoter activity in CarB cells, but the treatment with TSA upregulates the activity by threefold (Fig. 1C), indicating that the transcription factor(s) implicated in the repression of E-cadherin in this cell line also requires HDAC activity to function effectively. In agreement with these observations, RT-PCR analysis showed the reexpression of E-cadherin transcripts after TSA treatment in MDCK-Snail and spindle CarB cells (Fig. 1E). Reexpression of E-cadherin after TSA treatment could also be detected in CarC cells, an additional spindle cell line deficient in E-cadherin and with endogenous Snail expression (reference 35 and data not shown).
Snail expression is correlated with E-cadherin promoter deacetylation.
The initial studies suggested that HDACs participate in the Snail-mediated repression of E-cadherin. Should the TSA treatment directly affect E-cadherin gene expression levels, one would expect to observe changes in the histone acetylation status at its promoter. Therefore, our next goal was to check whether Snail expression correlated with histone deacetylation at the E-cadherin promoter. To address this point, we first analyzed the acetylation status of histones H3 and H4 at the E-cadherin promoter, comparing MDCK and MDCK-Snail cell lines by means of ChIP assays.
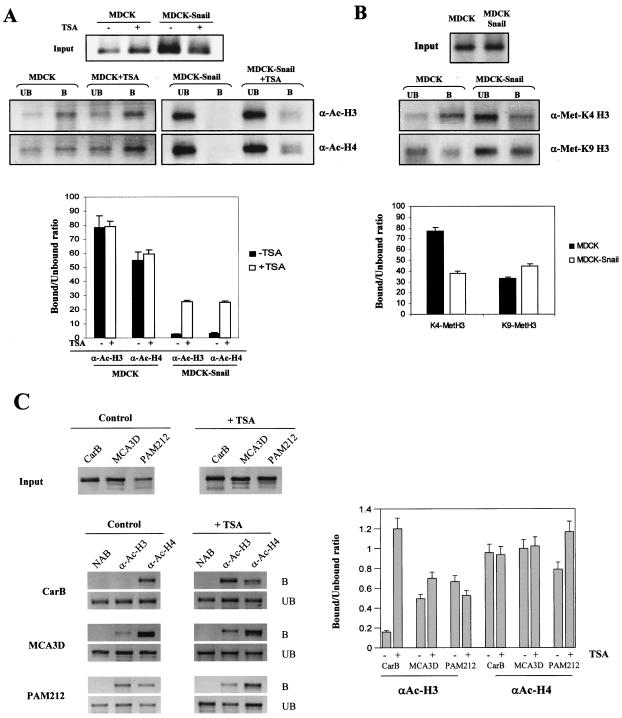
After the formaldehyde cross-linking of MDCK-CMV and MDCK-Snail cells, ChIP assays were performed with antibodies against acetylated histones H3 and H4. The precipitated DNA was subjected to PCR with specific primers for the endogenous dog E-cadherin proximal promoter region (15). The analysis revealed that Snail expression is associated with a strong decrease in the levels of acetylated histones H3 and H4 at the E-cadherin promoter (Fig. 2A). Moreover, TSA treatment led to a strong increase in histone acetylation at the E-cadherin promoter in MDCK-Snail cells, while it did not significantly change the histone acetylation status of MDCK control cells (Fig. 2A).
FIG. 2.
Histone acetylation and methylation analysis at the endogenous E-cadherin promoter in Snail-expressing and Snail-deficient cells. (A and B) ChIP analysis of the modification status of histones H3 and H4 at the endogenous E-cadherin promoter in MDCK-CMV (MDCK) and MDCK-Snail cells, with anti-acetyl-histone H3 (α-Ac-H3) and anti-acetyl-histone H4 (α-Ac-H4) and anti-dimethyl-K4 histone H3 (α-Met-K4-H3) and anti-dimethyl-K9 histone H3 (α-Met-K9-H3) antibodies. Where indicated, cells were treated with TSA (300 nM) for 24 h before formaldehyde cross-linking. The amplified dog E-cadherin promoter sequences in the input and the immunoprecitated bound and unbound fractions are shown in the upper panels. (C) ChIP assays of the histone H3 and H4 acetylation status at the endogenous E-cadherin promoter in mouse keratinocyte MCA3D, Pam212, and spindle CarB cells, with anti-acetyl-histone H3 (α-Ac-H3) and anti-acetyl-histone H4 (α-Ac-H4) antibodies. Where indicated, cells were treated with TSA (300 nM) for 24 h before formaldehyde cross-linking. The amplified mouse E-cadherin promoter sequences in the input (upper panels) and bound and unbound (lower panels) fractions are shown. Results from controls with no antibody (NAB), in which no amplification occurs, are also included for each cell line. Quantification of the amplified sequences in the immunoprecipitated fractions (represented as the ratio of bound to unbound fractions) with each antibody and corresponding cells lines and treatments is shown in the lower (A and B) and right (C) panels. Results represent the averages ± standard deviations of at least two experiments. B, bound; UB, unbound.
During recent years it has been proposed that the patterns of different histone modifications constitute a sort of code that is read by different factors to bring about distinct downstream events (47). Active genes are rich in acetylated histones H3 and H4. Also, K4 of histone H3 has been found to be methylated in active euchromatic regions (46). On the other hand, the histone deacetylation and methylation of K9 of histone H3 have been associated with gene silencing (32). In order to determine whether the histone modification pattern of the E-cadherin promoter was also affected by stable transfection of Snail, we performed ChIP assays with antibodies against dimethyl-K4 histone H3 and dimethyl-K9 histone H3 amplified with primers for the E-cadherin promoter. ChIP assays with anti-dimethyl-K4 histone H3 indicated that K4 of H3 is methylated in a higher proportion in MDCK control cells than in MDCK-Snail cells (Fig. 2B, upper panel), as expected from their E-cadherin expression patterns. As shown in Fig. 2B (lower panel), ChIP assays with antibodies against dimethyl-K9 histone H3 indicated that methylation of K9 of histone H3 at the E-cadherin promoter occurs at higher levels in MDCK-Snail than in MDCK-CMV cells, which is compatible with the silenced state of the gene in this cell line.
To further confirm that Snail expression is correlated with the deacetylation status of the E-cadherin promoter, we performed ChIP assays in several keratinocyte cell lines. We used the keratinocyte cell lines MCA3D and Pam212 characterized by high E-cadherin expression and no Snail expression and the spindle carcinoma CarB cell line that expresses Snail and does not express E-cadherin (references 11 and 35 and data not shown). The analysis revealed that acetylated histone H3 at the mouse E-cadherin promoter was almost undetectable in CarB cells, in contrast to results for MCA3D and Pam212 cells, while similar levels of acetylated histone H4 were detected in all three cell lines (Fig. 2C, bound and unbound panels). Furthermore, treatment of CarB cells with TSA led to a strong increase in the immunoprecipitated fraction with anti-acetyl H3 antibodies, but it did not affect the level of acetylated histone H4 bound to the E-cadherin promoter (Fig. 2C), indicating that TSA does indeed affect the histone H3 acetylation status at the E-cadherin promoter in CarB cells.
Snail interacts in vivo with the E-cadherin promoter and recruits HDACs.
It has been described previously that Snail interacts with the E-Pal element of the mouse E-cadherin promoter to repress transcription (10, 11); however, the interaction at the chromatin level has not yet been demonstrated. To assess this point, and because of the lack of highly specific anti-Snail antibodies, we generated a fusion GFP-Snail construct and obtained stable transfectants from MDCK cells. Two independent MDCK-GFP-Snail clones and a control MCDK-GFP clone were used in ChIP assays with anti-GFP antibodies. As shown in Fig. 3A, this analysis confirmed that there is an interaction between the GFP-Snail protein and the endogenous E-cadherin promoter in MDCK-GFP-Snail cells (Fig. 3A, right lanes); this interaction is specific for the GFP-Snail protein as revealed by the absence of E-cadherin promoter sequences in the immunoprecipitate from control MDCK-GFP cells (Fig. 3A, left lane).
FIG. 3.
Snail interacts with the endogenous E-cadherin promoter and recruits HDAC activity. (A) MDCK-GFP and MDCK-GFP-Snail cells were analyzed by ChIP assays with anti-GFP (α-GFP) antibodies. Amplification of the endogenous dog E-cadherin promoter in the input and immunoprecipitated fractions of a control MDCK-GFP clone and two independently isolated MDCK-GFP-Snail clones is shown. (B) HDAC activity was determined in the α-GFP immunoprecipitated fractions obtained from HEK 293T cells transiently transfected with GFP (left lanes) and GFP-Snail (right lanes) vectors and either untreated (black bars) or treated with TSA (300 nM) (white bars) for 24 h.
Since all the previous results indicated that Snail-dependent repression of E-cadherin is affected by TSA treatment and resulted in E-cadherin promoter histone deacetylation, we wondered whether Snail could, in fact, be recruiting HDAC activity to achieve its repressor effect. To address this issue, an HDAC activity assay was performed in HEK 293T cells transiently transfected with GFP or GFP-Snail plasmids in which the GFP immunoprecipitates were incubated with 3H-labeled histones and the released [3H]acetate was measured. We observed a significantly higher level of HDAC activity in the GFP-Snail immunoprecipitated fraction compared to the level in the GFP negative control. The HDAC activity was inhibited by TSA treatment in the GFP-Snail immunoprecipitate (Fig. 3B, right columns), while it did not affect the basal HDAC activity in the control GFP immunoprecipitate (Fig. 3B, left columns).
Snail associates with HDAC1/2 and mSin3A.
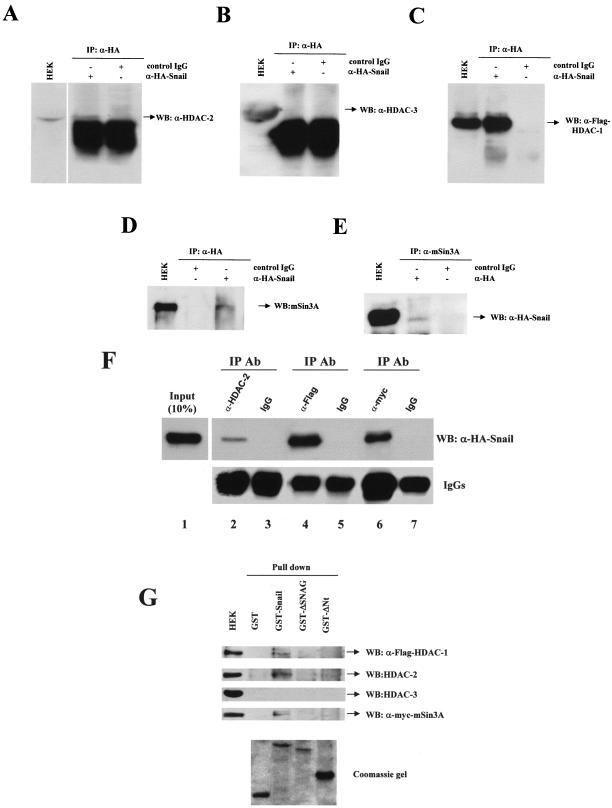
To get further insights into the model of Snail-mediated repression, we decided to analyze the association of specific HDACs with Snail. For this purpose, we analyzed the possibility that Snail could be interacting with the class I family of HDACs. To this end, transient transfections of an HA-tagged version of Snail were performed in HEK 293T cells, and coimmunoprecipitation with several class I HDAC proteins was detected. We first analyzed the interaction of Snail-HA with the endogenous HDAC2/3 proteins. As shown in Fig. 4A, the immunoprecipitate of Snail-HA associates with endogenous HDAC2, as detected by Western blotting. A similar analysis of the anti-HA immunoprecipitate with anti-HDAC3 antibodies did not show an interaction between Snail-HA and HDAC3 (Fig. 4B). We next sought to confirm the interaction of Snail with HDAC1; for this purpose, we coexpressed HDAC1-Flag with Snail-HA and analyzed the fraction immunoprecipitated with anti-HA. The analysis by Western blotting with anti-Flag antibodies showed that there is an interaction between Snail and HDAC1 (Fig. 4C).
FIG. 4.
Snail associates with HDAC1/2 and the mSin3A corepressor through the SNAG domain. (A to D) HEK 293T cells were transiently transfected with Snail-HA and HDAC1-Flag constructs. Cell extracts were immunoprecipitated with anti-HA antibodies (α-HA) and control IgG, as indicated, and analyzed by Western blotting with anti-HDAC2 (α-HDAC-2), anti-HDAC3 (α-HDCA-3), anti-Flag (α-Flag), and anti-mSin3A (α-mSin3A) antibodies. (E) HEK 293T cells transiently transfected as above were immunoprecipitated with anti-mSin3A antibodies and analyzed by Western blotting with anti-HA. Cell extracts (HEK) were analyzed in parallel in all panels. (F) HEK 293T cells transiently transfected withSnail-HA, HDAC1-Flag, and mSin3A-myc constructs were immunoprecipitated with the indicated antibodies and analyzed by Western blotting with anti-HA antibodies (upper panel). Detection of IgG heavy chain is shown in the lower panel as an internal control. Input (10%) of whole cell extract was also analyzed in parallel. (G) Cell extracts obtained from HEK 293T cells transiently transfected as above were incubated with the indicated GST fusion proteins. The bound fractions from the glutathione-Sepharose beads and the input cell extract were analyzed by Western blotting with the indicated antibodies (upper panel). Analysis of the different recombinant GST fusion proteins used is shown in the lower panel. WB, Western blotting; IP, immunoprecipitation; Ab, antibody.
HDAC1/2 are associated in at least three different multiprotein complexes named SIN3, Mi-2/NuRD, and CoREST (1, 27). To determine if Snail could be recruiting additional proteins, we analyzed the potential interaction of Snail with the corepressor mSin3A (a specific component of the SIN3 complex) in HEK 293T cells. Analysis of the immunoprecipitated fraction of Snail-HA showed the existence of an interaction between endogenous mSin3A and Snail-HA (Fig. 4D). This interaction was also confirmed by coimmunoprecipitation analysis with anti-mSin3A and Western blotting with anti-HA antibodies (Fig. 4E). The specificity of the Snail-HA interactions with HDAC1/2 and mSin3A were confirmed by the use of a control (unrelated) immunoglobulin G (IgG) (Fig. 4A to E). The ability of Snail-HA to interact in vivo in a complex with HDAC1/2 and exogenous mSin3A was also confirmed in reverse immunoprecipitation assays. Immunoprecipitation with specific antibodies against endogenous HDAC2, HDAC1-Flag, and mSin3A-myc, followed by Western blotting against Snail-HA, revealed the coimmunoprecipitation of Snail-HA with all three proteins (Fig. 4F, lanes 2, 4, and 6) and the specificity of these interactions, as shown with the IgG control immunoprecipitations (Fig. 4F, lanes 3, 5, and 7). In contrast, no interaction of Snail-HA with endogenous HDAC3 could be detected in the anti-HDAC3 immunoprecipitate (data not shown). Higher levels of Snail-HA were apparently immunoprecipitated by anti-Flag than by anti-myc antibodies (Fig. 4F; lanes 4 and 6), suggesting that in overexpressing cells Snail might interact with HDAC1 in additional complexes independent of mSin3A. Nevertheless, different affinities of the antibodies may also explain the above results, and this possibility could not be formally discarded at present.
The above evidence supports the existence of physical interactions between Snail and HDAC1/2 and the corepressor mSin3A, suggesting the formation of a multimolecular complex to repress E-cadherin expression. To analyze whether Snail interacts with the three proteins in a direct or indirect fashion and to determine the Snail domains involved in those interactions, GST pull-down assays were carried out on HEK 293T cells. As shown in Fig. 4G, endogenous HDAC2 and exogenous HDAC1-Flag and mSin3A-myc are able to interact with a full-length GST-Snail protein, while no interaction with endogenous or exogenous HDAC3 could be detected (Fig. 4G and data not shown). As expected, the C-terminal half of Snail carrying the zinc finger DNA-binding domain does not support the interaction with any of the three partners (Fig. 4G, GST-ΔNt), indicating that the N-terminal half of Snail is required for the interactions. Indeed, the N-terminal SNAG domain of Snail, previously involved in the repression function (5, 34), is required to establish the interaction with HDAC1/2 and the corepressor mSin3A, since its deletion is sufficient to eliminate interactions with the three proteins (Fig. 4G, GST-ΔSNAG). These results, therefore, indicate that the SNAG domain is essential for Snail recruitment of HDAC1/2 and the mSin3A corepressor.
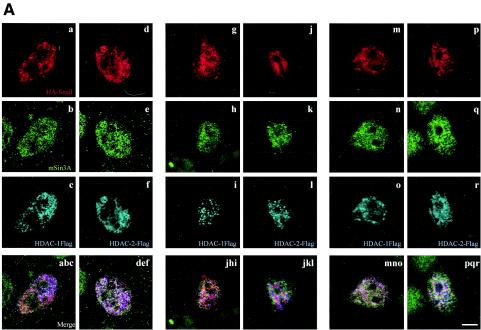
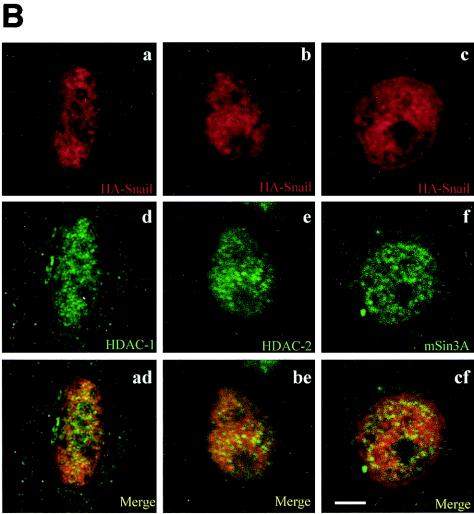
To further analyze the in vivo functionality of the complexes comprised of Snail and the HDCAs and mSin3, confocal immunofluorescence analyses of transiently transfected HEK 293T, MDCK, and CarB cells were performed. Nuclear colocalization of endogenous mSin3A with Snail-HA and HDAC1-Flag (Fig. 5A, left panels) or HDAC2-Flag (Fig. 5A, right panels) proteins could be detected in a punctuate pattern, indicative of defined nuclear substructures, and with apparent nucleolar exclusion in the three cell lines (Fig. 5A). To confirm this point we also performed costaining of Snail-HA with endogenous HDAC-1 (Fig. 5B, left panels), HDAC-2 (Fig. 5B, central panels), and mSin3A (Fig. 5B, right panels). Colocalization of the molecules could clearly be detected in punctuate nuclear substructures, which are indicative of active transcription sites in CarB, MDCK, and HEK 293T cells (Fig. 5B and data not shown). In agreement with the immunoprecipitation and pull-down assays, endogenous HDAC3 did not reveal a colocalization with Snail-HA (data not shown), thus indicating that the nuclear colocalization of Snail-HA with HDAC1/2 is specific.
FIG. 5.
Snail colocalizes with HDAC1/2 and mSin3A in the nucleus. (A) MDCK (a to f), CarB (g to l), and HEK 293T (m to r) cells were transiently transfected with Snail-HA and HDAC1-Flag (a to c, g to i, and m to o) or with Snail-HA and HDAC2-Flag (d to f, j to l, and p to r) constructs and costained with anti-HA (red), anti-mSin3A (green), and anti-Flag (cyan) antibodies. Note the nuclear colocalization of the proteins (white to pink) in the merged images of the corresponding three channels presented in the lower panels. (B) CarB cells were transiently transfected with the Snail-HA construct and costained with anti-HA (a to c) and anti-HDAC1 (d), anti-HDAC2 (e), or anti-mSin3A (f). Merged images of the corresponding two channels show the nuclear colocalization of Snail-HA with endogenous HDAC1 (ad), endogenous HDAC2 (be), and endogenous mSin3A (ef). Bar, 5 μm.
Taken together with the immunoprecipitation analyses, these results confirm the in vivo formation of a multiprotein complex involving the repressor Snail, the corepressor mSin3A, and HDAC1/2 in the nucleus.
HDAC1/2 interact with the E-cadherin promoter.
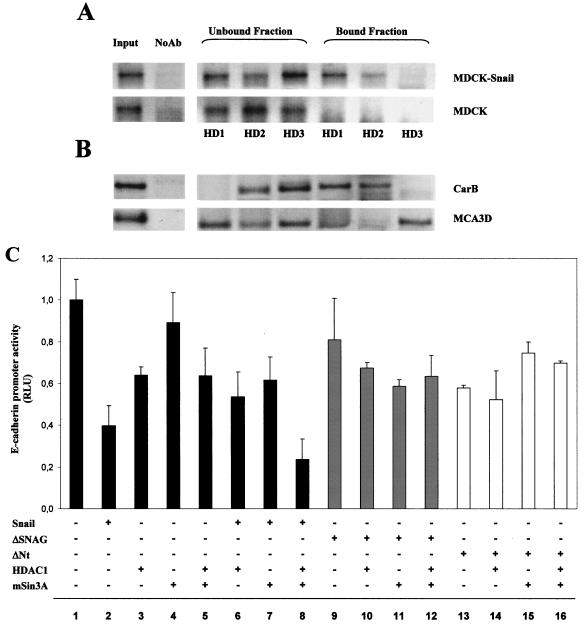
The previous results indicate that Snail directly interacts with the E-cadherin promoter and recruits HDAC activity. In addition, we have demonstrated the existence of a physical interaction between Snail and HDAC1/2 and the corepressor mSin3A. To get additional evidence for the functionality of these interactions in E-cadherin regulation, we performed ChIP assays with antibodies against HDAC1/2/3 to determine whether these proteins are indeed interacting with the E-cadherin promoter in Snail-expressing cells.
Interestingly, we found a specific interaction of HDAC1/2 with the E-cadherin promoter in MDCK-Snail cells (Fig. 6A, upper panel). In contrast, our results indicated a lack of association of HDAC3 with the E-cadherin promoter that showed an increased amount in the unbound fraction compared with results for HDAC1/2. In MDCK-CMV cells, these interactions between HDAC1/2 and the E-cadherin promoter were absent (Fig. 6A, lower panel). Similar ChIP assays were also performed in mouse MCA3D and CarB cells, selected because of their patterns of expressing E-cadherin and Snail (11). Spindle CarB cells (E-cadherin negative and Snail positive) showed a ChIP pattern similar to MDCK-Snail cells: the association of HDAC1/2, but not HDAC3, at the mouse E-cadherin promoter (Fig. 6B, upper panel), supporting the participation of HDAC1/2 in E-cadherin repression. In contrast, keratinocyte MCA3D cells (E-cadherin positive and Snail negative) showed a distinct ChIP pattern: the association of HDAC3 and, to a lesser extent, of HDAC1 and lack of interaction of HDAC2 at the E-cadherin promoter (Fig. 6B, lower panels). These results suggest that HDAC3 and/or HDAC1 could be mediating a basal E-cadherin repression in MCA3D cells, as indicated by the TSA effect on the promoter in this cell line (Fig. 1B), which might be recruited by other endogenous weaker repressors, such as Slug expressed in MCA3D cells (10, 11). Nevertheless, the lack of changes in the levels of E-cadherin transcripts in MCA3D cells after TSA treatment (Fig. 1E) support a transient TSA effect on the E-cadherin promoter in this cell line.
FIG. 6.
HDAC1/2 proteins are recruited at the endogenous E-cadherin promoter and cooperate with mSin3A in Snail-mediated repression of the promoter activity. (A and B) ChIP assays of HDAC1/2/3 at the endogenous E-cadherin promoter in MDCK-Snail (A, upper panel) and MDCK-CMV (A, lower panel) cells, and in spindle CarB (B, upper panel) and epidermal keratinocyte MCA3D (B, lower panel) cells. ChIP assays were performed as indicated in Materials and Methods with antibodies specific for HDAC1 (HD1), HDAC2 (HD2), and HDAC3 (HD3). The amplified sequences of the dog or mouse E-cadherin promoter detected in the input and in the immunoprecipitated bound and unbound fractions are shown. Results for controls with no antibody (NoAb), in which no amplification occurs, are also included for each cell line. (C) The activity of the E-cadherin promoter was analyzed in MDCK cells in the presence of the indicated Snail expression vectors (50 ng) and in the absence or presence of cotransfection with HDAC1 and/or mSin3A expression vectors (100 ng). Promoter activity was determined 24 h after transfection, as indicated in the legend of Fig. 1.
Cooperation of Snail, mSin3A, and HDAC1 to repress the E-cadherin promoter requires the SNAG domain of Snail.
To look into the functionality of the identified protein interactions on the E-cadherin promoter, we analyzed the effect of HDAC1 and the corepressor mSin3A in the absence or presence of Snail on the E-cadherin promoter activity in wild-type MDCK cells. To observe potential cooperation, pcDNA3-Snail was transfected under conditions (50 ng) that resulted in a partial repression (60%) of the promoter in this cell line (Fig. 1A and 6C, lane 2). Transfection of HDAC1 induced a modest repression (40%) of the E-cadherin promoter (Fig. 6C, lane 3); no effect was detected after transfection of mSin3A (Fig. 6C, lane 4) and cotransfection of both HDAC1 and mSin3A induced a repression level similar to that induced by HDAC1 alone (Fig. 6C, lane 5). Cotransfection of either HDAC1 or mSin3A with Snail resulted in a similar or even lower repression than that induced by Snail alone (Fig. 6C, lanes 6 and 7). However, a strong repression (80%) was observed after cotransfection of Snail with HDAC1 and mSin3A (Fig. 6C, lane 8), indicating that the three proteins have an additive effect in E-cadherin promoter repression. Similar results were obtained in HEK 293T cells (data not shown), supporting the idea that the model of cooperation between Snail, HDAC1, and mSin3A in E-cadherin repression operates in other epithelial cell lines.
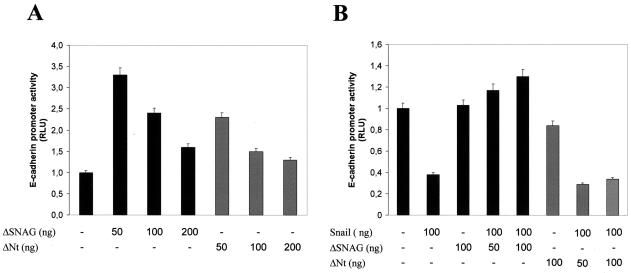
The interaction analyses, shown in Fig. 4G, indicated that the SNAG domain of Snail is required for the recruitment of HDACs and the mSin3A corepressor. To further establish the participation of the SNAG domain in the repressor function of Snail on the E-cadherin promoter, cotransfection assays were performed with two N-terminal Snail deletion mutants (ΔSNAG and ΔNt). Both Snail mutants were unable to efficiently repress E-cadherin promoter activity (Fig. 6C, lanes 9 and 13) and to cooperate with HDAC1 and mSin3A in the repression of the promoter in MDCK cells (Fig. 6C, lanes 10 to 12 and 14 to 16). As expected from these results, the ΔSNAG construct can act as a dominant negative of Snail in different cell systems. ΔSNAG derepresses the E-cadherin promoter activity in Snail-expressing cells, such as CarB and MDCK-Snail (Fig. 7A, and data not shown) to a larger extent than the ΔNt construct. In addition, ΔSNAG is able to fully overcome the E-cadherin promoter repression induced by transient transfection of Snail in keratinocyte PDV cells (Fig. 7B) deficient in Snail expression (11). These results, together with the interaction assays, strongly indicate that the SNAG domain is required for the repression function of Snail through the recruitment of HDAC1/2 and corepressor mSin3A.
FIG. 7.
The ΔSNAG construct acts as a dominant negative of Snail in E-cadherin promoter repression. (A) The activity of the E-cadherin promoter in Snail-expressing CarB cells was more efficiently derepressed by the ΔSNAG than by the ΔNt Snail mutant. (B) E-cadherin promoter activity in Snail-deficient PDV cells. The repression of the E-cadherin promoter induced by cotransfection of Snail was fully relieved by the ΔSNAG mutant but not affected by the ΔNt mutant. Promoter activity was determined 24 h after transfection, as indicated in the legend of Fig. 1.
DISCUSSION
Downregulation or loss of function of E-cadherin has been firmly implicated in the acquisition of invasive potential by carcinomas (8, 14, 23). A large body of evidence accumulated in recent years suggests that epigenetic mechanisms involving chromatin modification, hypermethylation of CpG islands, and/or transcriptional repression are the main mechanisms responsible for E-cadherin silencing in different types of tumors and cancer cell lines (13, 14, 42). Several transcriptional repressors of E-cadherin have been identified in the last 3 years. Among them, the transcription factor Snail has been described as a strong repressor of E-cadherin in different murine and human carcinoma and melanoma cell lines and tumors (5, 9, 11, 13, 41, 48, 55) as well as during Drosophila and mouse embryo development (12, 25, 37, 38). An additional member of the Snail family, Slug, has also been reported to act as an E-cadherin repressor (10, 22). Both factors, as well as additional identified repressors, E47 bHLH and ZEB2 (SIP-1), bind to specific E boxes of the proximal E-cadherin promoter (5, 10, 11, 15, 40). However, the affinity of Snail binding for specific E boxes is much higher than for the other repressors (10).
To date, the molecular mechanisms underlying E-cadherin repression by factors belonging to the Snail family have not been elucidated, although some previous works have provided insights into the function of the N-terminal region. The repressor effect of Snail is associated with a motif found in a short amino-terminal sequence (5, 34) called SNAG because it was first described as a repressor domain of the Gfi1 protein (19). In Drosophila embryos, the N-terminal region of Snail has been shown to recruit the corepressor CtBP (C-terminal binding protein) to exert its transcriptional repression (36). On the other hand, the repressor domain of human Slug has been ascribed to a 32-amino-acid N-terminal domain (containing the SNAG motif) and proposed to require association with HDACs at sites of active transcription (26). In contrast to Drosophila Snail and to vertebrate Slug factors, the mouse and human Snail proteins do not contain a CtBP binding domain (25). In fact, in vertebrate development there is no evidence for the interaction of Snail with corepressors, and the relevance of the SNAG motif in mouse or human Snail to support interactions with other proteins such as HDACs or associated corepressors has not been previously established (25).
In the present study, we report the identification of a functional association of mouse Snail (mSnail) and HDAC1/2 and the corepressor mSin3A that interact to repress E-cadherin expression through the SNAG motif of Snail. Our results represent the first evidence that the Snail protein can interact with a corepressor complex to recruit HDAC activity and repress E-cadherin expression by modification of the local structure of the chromatin.
Snail-mediated E-cadherin repression requires HDAC activity.
Repression of the E-cadherin promoter mediated by mSnail requires the recruitment of HDAC activity, as demonstrated by the derepression exerted by treatment with the HDAC inhibitor TSA. The derepression effect of TSA was detected after cotransfection of exogenous mSnail in E-cadherin-expressing cells and, more significantly, in Snail-expressing E-cadherin-deficient cells in which the TSA treatment increased the basal activity of the E-cadherin promoter up to three- to fourfold in spindle carcinoma CarB cells or up to six- to sevenfold in MDCK-Snail cells, leading to reexpression of the E-cadherin transcripts (Fig. 1). These results are, therefore, in agreement with previous observations indicating that the repression effect of the human Slug protein on artificial promoters requires HDACs (26). In addition, the ChIP assays performed here to analyze the acetylation status of histones H3 and H4 at the endogenous E-cadherin promoter have in fact detected very reduced levels of acetylated histones H3 and H4 in MDCK-Snail cells compared to levels in control MDCK cells. A similar ChIP analysis in spindle CarB cells, expressing endogenous Snail, showed very reduced levels of acetylated histone H3 compared with levels in MCA3D and Pam212 cells deficient in Snail expression (see Fig. 2). In agreement with the above observations, TSA treatment induced a significant increase in the level of acetylated histones H3 and H4 or of H3 associated with the endogenous E-cadherin promoter in Snail-expressing cells, further supporting the recruitment of HDACs to the proximal promoter. Indeed, association of HDAC activity with Snail has been demonstrated in the immunoprecipitates of GFP-Snail-expressing cells, in which ChIP assays with anti-GFP antibodies have additionally confirmed that GFP-Snail associates to the endogenous E-cadherin proximal promoter. All these results suggest that Snail represses the E-cadherin promoter by HDAC recruitment, indicating that the repression mechanism involves the modification of the local chromatin structure.
Snail associates with HDAC1/2 and mSin3A in a multimolecular complex.
HDACs are frequently recruited to specific DNA sites by association with corepressor molecules, which together with other associated proteins operate as multimolecular repressor complexes (27). The two major complexes containing class I HDACs are the NuRD and SIN3 complexes (1, 29). The SIN3 complex has been previously described as containing the Sin3A and Sin3B corepressors, class I HDAC1/2, and additional associated proteins. To date several transcriptional repressors have been shown to associate with the SIN3 complex to mediate repression of specific target genes (3, 24, 30, 51, 57). Interestingly, the methyl CpG binding protein, MeCP2, has also been found to be associated with the SIN3 complex (28, 52). Here, we present evidence for the in vitro and in vivo association of Snail with the mSin3A corepressor and class I HDAC1/2, indicating the Snail-mediated recruitment of a multimolecular complex to the E-cadherin promoter. The four proteins are colocalized to the cell nucleus, apparently in defined nuclear domains and with a nucleolar exclusion pattern (Fig. 5), supporting their joint association to transcription sites. In contrast, no association of Snail with the class I HDAC3 and no nuclear colocalization have been observed. In agreement with these observations, the HDAC1/2 proteins but not HDCA3 have been found associated to the endogenous E-cadherin promoter in Snail-expressing cells. Moreover, the cooperation of HDAC1 and mSin3A with Snail seems to be required for effective repression of the E-cadherin promoter. These results strongly indicate that Snail is associated in vivo with several components of the SIN3 repressor complex to mediate transcriptional repression of the E-cadherin promoter.
Based on the pull-down assays, the interaction of Snail with HDAC1/2 and mSin3A seems to be direct and depends on the presence of the SNAG transactivation domain (Fig. 4G). Significantly, the ΔSNAG mutant is unable to cooperate with HDAC1 and mSin3A in E-cadherin promoter repression, supporting the functional participation of the SNAG domain in the recruitment of the complex comprised of Sin3A and HDAC1/2. These results, apart from reinforcing the previously assigned repressor function of the SNAG domain (5, 34), provide for the first time a molecular mechanism for Snail repression of the E-cadherin promoter, which is linked to the recruitment of the Sin3A and HDAC1/2 components of the SIN3 complex. The participation of additionally associated components of the SIN3 complex in the Snail-mediated repression of E-cadherin remains an open question. However, and as suggested by the colocalization analysis (see Fig. 5), it is likely that the SIN3 complex recruited by Snail might represent a minor subfraction of the endogenous nuclear SIN3 complex specifically recruited to the E boxes of the E-cadherin promoter, as reported for other repressors that recruit particular SIN3 complexes to specific promoter sequences (30, 33, 43, 51, 53, 54, 57, 58).
Regulation of E-cadherin repression might involve several repressor complexes.
Our present results clearly indicated that mSnail recruits several components of the SIN3 repressor complex to downregulate E-cadherin expression in epithelial cell lines. Nevertheless, the participation of Snail in other corepressor complexes recruiting HDAC1/2 or containing additional histone- or chromatin-modifying activities, such as methyltransferases, cannot presently be discarded. In fact, our ChIP assays have shown a lower level of dimethyl-K4 of histone H3 and a higher level of dimethyl-K9 of histone H3 at the E-cadherin promoter in MDCK-Snail cells compared to levels in control MDCK cells (Fig. 2B), suggesting the participation of additional histone- and chromatin-modifying activities in Snail-mediated regulation. Interestingly, CarB and CarC cells present hypermethylation of the CpG islands of the proximal E-cadherin promoter, and their treatment with the DNA demethylating agent 5-aza-2′deoxycytidine leads to reexpression of E-cadherin in both cell lines (M. Fraga et al., unpublished data), suggesting that cooperation between methyl-CpG binding domain proteins and HDACs recruited by Snail might operate in these two cell lines, as demonstrated in other systems (4).
Besides Snail, additional repressors of E-cadherin have been identified in the last 2 years, among them the ZEB1 (δEF1) and ZEB2 (SIP-1) two-handed zinc binding factors (15, 20). Very recently, a new CtBP corepressor complex has been described, containing histone-modifying activities (HDAC1/2 and K9 methyltransferase) and ZEB1 (δEF1) and ZEB2 (SIP-1), with the ability to affect the histone modification status and the activity of the E-cadherin promoter in osteosarcoma cells (45). Taken together with our present data, these results indicate that at least two distinct corepressor complexes, SIN3 and CtBP, can be recruited by different repressors to the E-cadherin promoter to mediate its transcriptional silencing. Whether both repressor complexes can operate on the same cell system or whether they are targeted to the same regulatory sequences of the E-cadherin promoter remains to be established. In this regard, previous studies have determined that mSnail binds to two adjacent E boxes present in the E-Pal element (positions −90 to −70) of the mouse E-cadherin promoter with a much higher affinity than other repressors, such as E47 and mSlug (10). In contrast, Snail does not bind to a proximal −30 E box (10), to which the ZEB2 (SIP-1) factor binds in conjunction with the −79 E box (15). Therefore, it might be possible that the independent transcriptional repressors Snail, ZEB1, and ZEB2 can target distinct repressor complexes, such as the SIN3 and CtBP complexes, to specific regulatory sites of the E-cadherin promoter. However, the high binding affinity of Snail for the more distal E boxes and the absence of a CtBP binding site in mSnail (25) make it unlikely that the Snail-recruited SIN3 complex can compete with a CtBP complex for E-cadherin promoter repression in the same cellular context. Additional work is clearly needed to further clarify this important issue.
Understanding the mechanisms involved in the regulation of Snail and other E-cadherin repressors is an important issue to advance in our understanding of the tumor invasion process. Recently it was reported that the Mi-2/NuRD corepressor complex is involved in the transcriptional repression of Snail in an estrogen receptor-dependent fashion in breast cancer cells (18). This information suggests additional levels of regulation for the Snail-mediated repression of E-cadherin, involving the participation of at least two corepressor complexes. It is tempting to speculate that these corepressors might operate in a hierarchical fashion in some specific contexts, such as in estrogen-dependent tissues. In such scenarios, under a positive estrogen receptor signal an active Mi-2/NuRD complex will repress Snail and maintain E-cadherin expression, while its inactivation will lead to the expression of Snail, which can then recruit the SIN3 complex to repress E-cadherin expression. In the field of tumor invasion and metastasis, an important issue that remains to be solved in the near future is the possible modification of either corepressor complex by external growth factors signals, such as transforming growth factor β which operates as a tumor promoter at advanced stages of tumor progression (2) and was recently reported to act as an inducer of Snail expression (39).
Acknowledgments
We thank E. Seto and R. N. Eisenman for providing reagents and A. Montes for her excellent technical assistance. Special thanks go to D. Megias and M. Cortés-Canteli for helping us with the confocal immunofluorescence analysis and to P. de la Peña-Ingelmo and J. Manzano for their suggestions in PCR experiments.
A. Cano's laboratory is supported by Spanish Ministry of Science and Technology grant SAF2001-2819 and by grants from the Instituto de Salud Carlos III (01/1074 and 031C03/10). M. Esteller's laboratory is supported by Spanish Ministry of Science and Technology grant SAF2001-0059 and the International Rett Syndrome Association. H. Peinado is a predoctoral fellow of the Spanish Ministry of Education, Culture and Sports. E. Ballestar is funded by the Ramón y Cajal Programme of the Spanish Ministry of Science and Technology.
REFERENCES
- 1.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and R. Derynck. 2001. TGF-beta signaling in cancer—a double-edged sword. Trends Cell Biol. 11:S44-S51. [DOI] [PubMed] [Google Scholar]
- 3.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 4.Ballestar, E., and M. Esteller. 2002. The impact of chromatin in human cancer: linking DNA methylation to gene silencing. Carcinogenesis 23:1103-1109. [DOI] [PubMed] [Google Scholar]
- 5.Batlle, E., E. Sancho, C. Franci, D. Dominguez, M. Monfar, J. Baulida, and A. Garcia De Herreros. 2000. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell. Biol. 2:84-89. [DOI] [PubMed] [Google Scholar]
- 6.Becker, K. F., M. J. Atkinson, U. Reich, I. Becker, H. Nekarda, J. R. Siewert, and H. Hofler. 1994. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 54:3845-3852. [PubMed] [Google Scholar]
- 7.Berx, G., A. M. Cleton-Jansen, F. Nollet, W. J. de Leeuw, M. van de Vijver, C. Cornelisse, and F. van Roy. 1995. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 14:6107-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birchmeier, W., and J. Behrens. 1994. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta 1198:11-26. [DOI] [PubMed] [Google Scholar]
- 9.Blanco, M. J., G. Moreno-Bueno, D. Sarrio, A. Locascio, A. Cano, J. Palacios, and M. A. Nieto. 2002. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene 21:3241-3246. [DOI] [PubMed] [Google Scholar]
- 10.Bolos, V., H. Peinado, M. A. Perez-Moreno, M. F. Fraga, M. Esteller, and A. Cano. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116:499-511. [DOI] [PubMed] [Google Scholar]
- 11.Cano, A., M. A. Perez-Moreno, I. Rodrigo, A. Locascio, M. J. Blanco, M. G. del Barrio, F. Portillo, and M. A. Nieto. 2000. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell. Biol. 2:76-83. [DOI] [PubMed] [Google Scholar]
- 12.Carver, E. A., R. Jiang, Y. Lan, K. F. Oram, and T. Gridley. 2001. The mouse Snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol. Cell. Biol. 21:8184-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng, C. W., P. E. Wu, J. C. Yu, C. S. Huang, C. T. Yue, C. W. Wu, and C. Y. Shen. 2001. Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene. Oncogene 20:3814-3823. [DOI] [PubMed] [Google Scholar]
- 14.Christofori, G., and H. Semb. 1999. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem. Sci. 24:73-76. [DOI] [PubMed] [Google Scholar]
- 15.Comijn, J., G. Berx, P. Vermassen, K. Verschueren, L. van Grunsven, E. Bruyneel, M. Mareel, D. Huylebroeck, and F. van Roy. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell 7:1267-1278. [DOI] [PubMed] [Google Scholar]
- 16.Faraldo, M. L., I. Rodrigo, J. Behrens, W. Birchmeier, and A. Cano. 1997. Analysis of the E-cadherin and P-cadherin promoters in murine keratinocyte cell lines from different stages of mouse skin carcinogenesis. Mol. Carcinog. 20:33-47. [DOI] [PubMed] [Google Scholar]
- 17.Fournier, C., Y. Goto, E. Ballestar, K. Delaval, A. M. Hever, M. Esteller, and R. Feil. 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 21:6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita, N., D. L. Jaye, M. Kajita, C. Geigerman, C. S. Moreno, and P. A. Wade. 2003. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113:207-219. [DOI] [PubMed] [Google Scholar]
- 19.Grimes, H. L., T. O. Chan, P. A. Zweidler-McKay, B. Tong, and P. N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16:6263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grooteclaes, M. L., and S. M. Frisch. 2000. Evidence for a function of CtBP in epithelial gene regulation and anoikis. Oncogene 19:3823-3828. [DOI] [PubMed] [Google Scholar]
- 21.Guilford, P., J. Hopkins, J. Harraway, M. McLeod, N. McLeod, P. Harawira, H. Taite, R. Scoular, A. Miller, and A. E. Reeve. 1998. E-cadherin germline mutations in familial gastric cancer. Nature 392:402-405. [DOI] [PubMed] [Google Scholar]
- 22.Hajra, K. M., D. Y. Chen, and E. R. Fearon. 2002. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62:1613-1618. [PubMed] [Google Scholar]
- 23.Hajra, K. M., and E. R. Fearon. 2002. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer 34:255-268. [DOI] [PubMed] [Google Scholar]
- 24.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 25.Hemavathy, K., S. I. Ashraf, and Y. T. Ip. 2000. Snail/Slug family of repressors: slowly going into the fast lane of development and cancer. Gene 257:1-12. [DOI] [PubMed] [Google Scholar]
- 26.Hemavathy, K., S. C. Guru, J. Harris, J. D. Chen, and Y. T. Ip. 2000. Human Slug is a repressor that localizes to sites of active transcription. Mol. Cell. Biol. 20:5087-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jepsen, K., and M. G. Rosenfeld. 2002. Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115:689-698. [DOI] [PubMed] [Google Scholar]
- 28.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 29.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 30.Koipally, J., A. Renold, J. Kim, and K. Georgopoulos. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18:3090-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurdistani, S. K., D. Robyr, S. Tavazoie, and M. Grunstein. 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31:248-254. [DOI] [PubMed] [Google Scholar]
- 32.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 33.Li, J., Q. Lin, W. Wang, P. Wade, and J. Wong. 2002. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 16:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama, H., I. C. Scott, and J. C. Cross. 1998. The transition to endoreduplication in trophoblast giant cells is regulated by the mSNA zinc finger transcription factor. Dev. Biol. 199:150-163. [DOI] [PubMed] [Google Scholar]
- 35.Navarro, P., M. Gómez, A. Pizarro, C. Gamallo, M. Quintanilla, and A. Cano. 1991. A role for the E-cadherin cell-cell adhesion molecule in tumor progression of mouse epidermal carcinogenesis. J. Cell Biol. 115:517-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nibu, Y., H. Zhang, E. Bajor, S. Barolo, S. Small, and M. Levine. 1998. dCtBP mediates transcriptional repression by Knirps, Kruppel and Snail in the Drosophila embryo. EMBO J. 17:7009-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieto, M. A. 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell. Biol. 3:155-166. [DOI] [PubMed] [Google Scholar]
- 38.Oda, H., S. Tsukita, and M. Takeichi. 1998. Dynamic behavior of the cadherin-based cell-cell adhesion system during Drosophila gastrulation. Dev. Biol. 203:435-450. [DOI] [PubMed] [Google Scholar]
- 39.Peinado, H., M. Quintanilla, and A. Cano. 2003. Transforming growth factor beta 1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial-mesenchymal transitions. J. Biol. Chem. 278:21113-21123. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Moreno, M. A., A. Locascio, I. Rodrigo, G. Dhondt, F. Portillo, M. A. Nieto, and A. Cano. 2001. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 276:27424-27431. [DOI] [PubMed] [Google Scholar]
- 41.Poser, I., D. Dominguez, A. G. de Herreros, A. Varnai, R. Buettner, and A. K. Bosserhoff. 2001. Loss of E-cadherin expression in melanoma cells involves up-regulation of the transcriptional repressor Snail. J. Biol. Chem. 276:24661-24666. [DOI] [PubMed] [Google Scholar]
- 42.Rodrigo, I., A. C. Cato, and A. Cano. 1999. Regulation of E-cadherin gene expression during tumor progression: the role of a new Ets-binding site and the E-pal element. Exp. Cell Res. 248:358-371. [DOI] [PubMed] [Google Scholar]
- 43.Rundlett, S. E., A. A. Carmen, N. Suka, B. M. Turner, and M. Grunstein. 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392:831-835. [DOI] [PubMed] [Google Scholar]
- 44.Sarrio, D., G. Moreno-Bueno, D. Hardisson, C. Sanchez-Estevez, M. Guo, J. G. Herman, C. Gamallo, M. Esteller, and J. Palacios. 2003. Epigenetic and genetic alterations in APC and CDH1 genes in lobular breast cancer: relationships with abnormal E-cadherin and catenin expression and microsatellite instability. Int. J. Cancer 106:208-215. [DOI] [PubMed] [Google Scholar]
- 45.Shi, Y., J. Sawada, G. Sui, B. Affarel, J. R. Whetstine, F. Lan, H. Ogawa, M. P. Luke, and Y. Nakatani. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 46.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 48.Sugimachi, K., S. Tanaka, T. Kameyama, K. Taguchi, S. Aishima, M. Shimada, K. Sugimachi, and M. Tsuneyoshi. 2003. Transcriptional repressor Snail and progression of human hepatocellular carcinoma. Clin. Cancer Res. 9:2657-2664. [PubMed] [Google Scholar]
- 49.Takeichi, M. 1993. Cadherins in cancer: implications for invasion and metastasis. Curr. Opin. Cell Biol. 5:806-811. [DOI] [PubMed] [Google Scholar]
- 50.Thiery, J. P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2:442-454. [DOI] [PubMed] [Google Scholar]
- 51.Vietor, I., S. K. Vadivelu, N. Wick, R. Hoffman, M. Cotten, C. Seiser, I. Fialka, W. Wunderlich, A. Haase, G. Korinkova, G. Brosch, and L. A. Huber. 2002. TIS7 interacts with the mammalian SIN3 histone deacetylase complex in epithelial cells. EMBO J. 21:4621-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 53.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, S. H., E. Vickers, A. Brehm, T. Kouzarides, and A. D. Sharrocks. 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21:2802-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokoyama, K., N. Kamata, E. Hayashi, T. Hoteiya, N. Ueda, R. Fujimoto, and M. Nagayama. 2001. Reverse correlation of E-cadherin and snail expression in oral squamous cell carcinoma cells in vitro. Oral Oncol. 37:65-71. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida, M., S. Horinouchi, and T. Beppu. 1995. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17:423-430. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y., and M. L. Dufau. 2002. Silencing of transcription of the human luteinizing hormone receptor gene by histone deacetylase-mSin3A complex. J. Biol. Chem. 277:33431-33438. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, W., M. Foehr, J. B. Jaynes, and S. D. Hanes. 2001. Drosophila SAP18, a member of the Sin3/Rpd3 histone deacetylase complex, interacts with Bicoid and inhibits its activity. Dev. Genes Evol. 211:109-117. [DOI] [PubMed] [Google Scholar]