Abstract
Cyclophilin A acts as protein folding chaperones and intracellular transports in many cellular processes. Previous studies have shown that cyclophilin A can interact with HIV-1 (human immunodeficiency virus type 1) gag protein and enhance viral infectivity. Many cyclophilin A inhibitors such as cyclosporin A can inhibit HIV-1 replication in vitro. Here, we report a structure-based identification of novel non-peptidic cyclophilin A inhibitors as anti-HIV lead compounds. Following a computer-aided virtual screening and subsequent surface plasmon resonance (SPR) analysis, 12 low molecular weight cyclophilin A ligands were selected for further evaluation of their in vitro inhibition of peptidyl-prolyl cis-trans isomerase (PPIase) activity of cyclophilin A and HIV-1 replication. Five of these compounds (FD5, FD8, FD9, FD10 and FD12) exhibited inhibition against both PPIase activity and HIV-1 infection. These active compounds will be used as leads for structure and activity relationship (SAR) and optimization studies in order to design more effective anti-HIV-1 therapeutics, and as probes for investigating the effect of cyclophilins on HIV-1 replication.
Keywords: Cyclophilin A, Cyclosporin A, Human immunodeficiency virus, Peptidyl prolyl cis-trans isomerase
1. Introduction
Cyclophilin A is a member of the cyclophilin protein family which is abundantly and ubiquitously expressed in a wide range of tissue types and organisms (Handschumacher et al., 1984; Liu et al., 1990). Cyclophilins possess the peptidyl-prolyl cis-trans isomerase (PPIase) activity and act as molecular chaperones to assist protein folding, assembly, and transportation processes (Bukrinsky, 2002). Like other cyclophilins, cyclophilin A has high binding affinity to cyclosporin A, a widely used immunosuppressive drug. Cyclosporin A can bind to the catalytic site of cyclophilin A, directly blocking cis-trans isomerization of cyclophilin A (Takahashi et al., 1989). In T cells, cyclosporin A can recruit cyclophilin A to form a protein complex which can bind to calcineurin and inhibit its phosphatase activity, resulting in the suppression of T cell activation (Zydowsky et al., 1992; Liu et al., 1991).
Cyclophilin A is involved in the replication process of the human immunodeficiency virus type 1 (HIV-1) (Braaten et al., 1996; Braaten and Luban, 2001). By directly binding to the capsid protein (CA) domain of HIV-1 Gag precursor polyprotein (Gamble et al., 1996), cyclophilin A can be packed into the viral capsid of HIV-1 (Franke et al., 1994; Thali et al., 1994). Viral infectivity can be weakened by cyclosporin A and its nonimmunosuppressive analogs (Sokolskaja et al., 2004; Steinkasserer et al., 1995) or by mutations in the CA domain which destruct the binding of cyclophilin A (Braaten et al., 1997; Dorfman et al., 1997).
The main aim of this study is to identify small molecule cyclophilin A ligands that can inhibit PPIase activity of cyclophilin A and HIV-1 replication. These compounds will be used as leads for rational design of novel anti-HIV-1 drugs without immunosuppressive activity.
2. Materials and methods
2.1. Cells and viruses
MT-2 cells and the HIV-1IIIB isolate were obtained from the NIH AIDS Research and Reference Reagent Program.
2.2. Proteins and compounds
Recombinant human cyclophilin A protein, the substrate N-succinyl-Ala-Ala-Pro-Phe-p- nitroanlilide (Suc-AAPF-pNA), α-chymotrypsin, and 2,2,2-trifluorethanal (TFE) were purchased from Sigma (St Louis, MO). The small organic compounds selected after virtual-screening were purchased from SPECS.
2.3. Computer-aided virtual screening
For the pharmaceutical value of cyclophilin members, we have carried out projects to screening for inhibitors of two cyclophilin members, cyclophilin A and cyclophilin J. Here, we report the virual screening and subsequent biochemical evaluation for cyclophilin A ligands. The DOCK program suite (DesJarlais et al., 1988; Shoichet et al., 1992) was employed to screen a small molecule database of SPECS (http://www.specs.net) for 80,000 commercially available compounds, and the methods have been described in great detail (Ring et al., 1993; Shoichet et al., 1993; Debnath et al., 1999). The 3D coordinates of the small molecules were generated by the Molecular Workbench software (Concord Consortium, Inc., Concord, MA) (link: http://mw.concord.org/modeler/index.html). Based on the X-ray structure of the cyclophilin A/cyclosporin A complex (PDB code: 1CWA), all residues surrounding cyclosporin A (Cutoff distance: 6.0 Å radius) were selected to generate a cavity. The molecules were then docked into the cavity, and the quality of the ligand binding was evaluated by a force-field scoring function. Five thousand top-scoring compounds were further evaluated by the FlexX program (BioSolveIT GmbH, Sankt Augustin, Germany) (link: http://www.biosolveit.de/FlexX/) and co-evaluated by using the CScore program (Tripos, Inc., St. Louis, MO). Molecules with FlexX energy scores from −25 to −40 or with X-score energy scores from 4 to 5 were visually analyzed.
2.4. Surface plasmon resonance (SPR) analysis
The binding affinity of the selected compounds to cyclophilin A was measured by SPR with a Biacore 3000 instrument (Biacore AB Corporation, Uppsala, Sweden) as previously described (Guo et al., 2005; Thurmond et al., 2001). Briefly, recombinant human cyclophilin A protein (10 µM) was coupled to a carboxylmethylated dextran surface (CM5 chip from Biacore, Inc., Piscataway, NJ) in a buffer containing 10 mM sodium acetate (pH 4.0) using standard amine coupling chemistry following the manufacturer’s instructions. Flow cell 1 was used as the control surface and flow cell 2 contained 9000 resonance units (RU) of cyclophilin A protein (1 RU corresponds to 1 pg of protein per mm2). All compounds were stocked in 100% dimethylsulfoxide at 10 mM and diluted at graded concentrations (0.3 to 10 µM) with HBS-EP buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, and 0.005% surfactant P20 at pH 7.4) containing 1% dimethylsulfoxide. The temperature of the instrument was set to 20°C, and the flow rate was set to 40 µl/min. Each compound (50 µl) was injected sequentially. NaOH (50 mM) was used to regenerate the surface. The 1:1 Langmuir binding model was used to determine the equilibrium dissociation constant (KD). For fast interactions, the Steady state model was used to determine the KD values. All experiments were carried out in triplicate.
2.5. Assay for inhibition of PPIase activity of cyclophilin A
Inhibition of each compound on PPIase activity of cyclophilin A was determined in an 8°C cold room based on a published method with some modifications (Kofron et al., 1991). Suc-AAPF-pNA was dissolved in TFE containing 480 mM of LiCl to a concentration of 3 mM. Then, α-chymotrypsin was dissolved in 1 mM HCl to a concentration of 42 mg/ml. Each test compound was diluted in 94 µl of assay buffer (50 mM HEPES, 100 mM NaCl; pH 8.0 at 0°C), and then mixed with 2 µl of cyclophilin A solution (5 µM). After pre-equilibrating for 3 h on ice, 2 µl of chymotrypsin solution and 2 µl of peptide substrate were added to the assay mixture. Absorbance at the wavelength of 390 nm was recorded on a Jasco V-550 spectrophotometer (Jasco, Inc., Easton, MD) for 20 s. Three independent experiments were performed for each test compound and the respective half-maximal inhibitory concentration (IC50) was calculated with OriginPro 7.5 software (OriginLab, Northampton, MA).
2.6. Detection of inhibitory activity of compounds on HIV-1 replication
The inhibitory activity of small molecule compounds on HIV-1 replication was determined using a method described by Sokolskaja et al. (2004) with modifications. In brief, MT-2 cells (5 × 105/well) were infected by HIV-1IIIB at 0.01 MOI (multiplicity of infection) in 24-wells cell culture plates at 37 °C for 1 h, followed by washing with PBS. The compounds stocked in 100% dimethylsulfoxide (10 mM) and diluted with RPMI 1640 medium containing 10% fetal bovine serum were added to the cells at the final concentration of 10 µM. After culture at 37 °C for 48 h, cells were fixed with 1% paraformadehyde-PBS and assayed for intracellular p24 by using KC57, a FITC-conjugated anti-p24 antibody (Beckman Coulter, Inc.) following the manufacturer’s procedure. Flow cytometry was performed on a Becton Dickinson FACSCalibur (Becton, Dickinson, and Company, San Jose, CA). The percent inhibition of HIV-1 replication was calculated with the following formula: [1 − (E − N)/(P − N)] × 100. E represents the number of p24+ cells in the presence of a compound, and P and N represent the positive control (no compound was added) and the negative control (neither virus nor compound was added), respectively.
2.7. Detection of in vitro cytotoxicity
The in vitro cytotoxicity of MT-2 cells for each compound was determined in 96-well plates using sodium3-[1-(phenylamino)-carbony]-3,4-tetrazolium-bis (4-methoxy-6-nitro)benzene-sulfonic acid hydrate (XTT) dye to measure cell viability (Debnath et al., 1999). Five percent Triton X-100 (10 µl) was added to the wells corresponding to positive controls (P), and 10 µl of medium was added to wells corresponding to negative controls (N). The cytotoxicity was calculated using the following formula: % cytotoxicity = [(E − N)/(P − N)] × 100, where E represents experimental data in the presence of a compound. The concentration corresponding to 50% cytotoxicity (CC50) for MT-2 cells was calculated using OriginPro 7.5 software.
3. Results
3.1. Identification of small molecule cyclophilin A ligands using computer-aided virtual screening and SPR analysis
To identify the inhibitory molecules of cyclophilin A, a virtual screening approach was conducted using commercially available compound libraries. Several crystal structures of cyclophilin A complexed with cyclosporin A, cyclosporin analogues, and various peptides available in the Protein Data Bank (http://www.rcsb.org/pdb/), were analyzed for potential cyclophilin ligands. Cyclophilin A contains an 8-stranded anti-parallel β-barrel structure, and a relatively hydrophobic groove formed by 11–13 residues via hydrogen bonds, polar and hydrophobic contacts. Cyclosporin A was found to bind to this groove (Mikol et al., 1993). The most important residues of the active site (Arg55, Phe60, Phe113, and His126) have been identified by site-directed mutagenesis (Liu et al., 1990; Zydowsky et al., 1992). We screened a database of 80,000 small organic molecules for compounds that can dock into the catalytic site of human cyclophilin A and selected 77 compounds with the best docking scores. These compounds were purchased SPECS and analyzed by SPR for their binding to cyclophilin A.
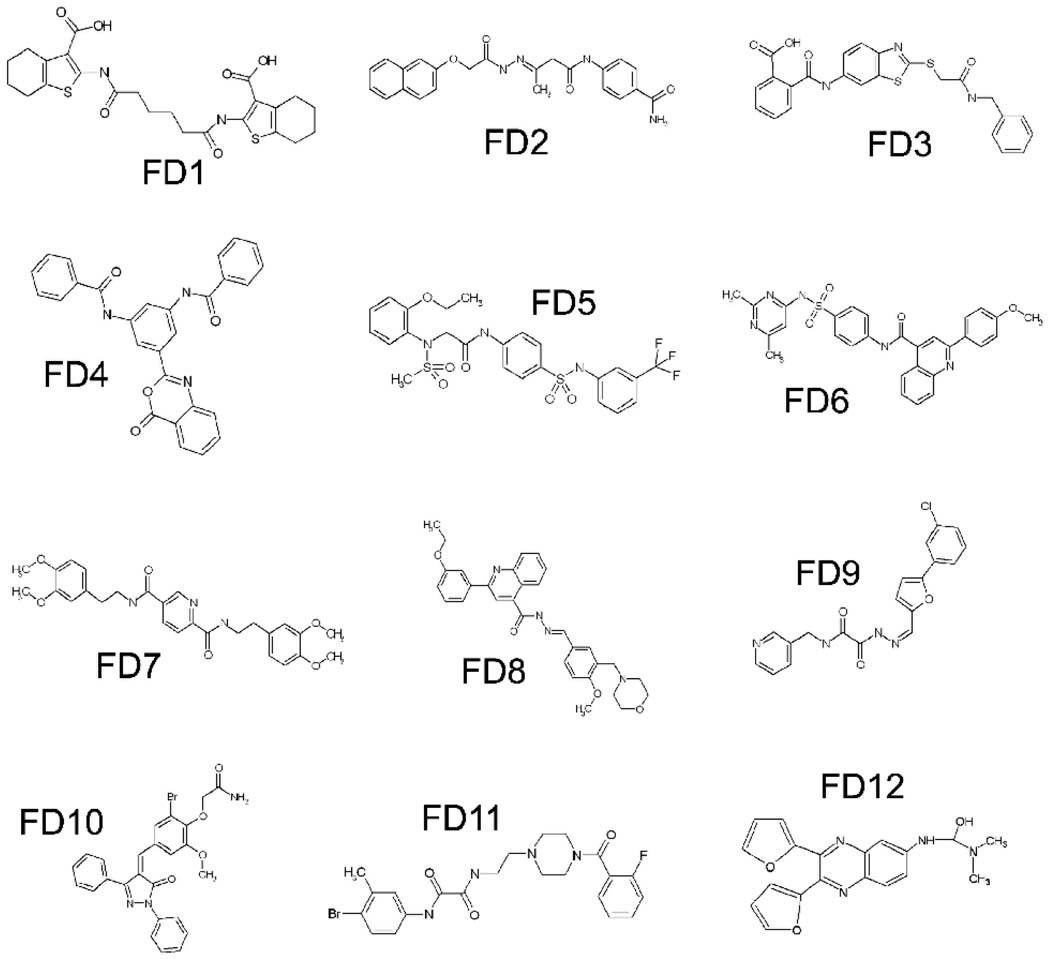
Among the 77 compounds selected, 12 compounds (their structures are shown in Fig. 1) were found to bind to cyclophilin A in a concentration-dependent manner with KD values ranging from 8 × 10−5 to 3 × 10−6 M (Table 1). Cyclosporin A, as a positive control, showed a KD value of binding to cyclophilin A at 2.18 ×10−7 M.
Fig. 1.
Chemical Structures of 12 cyclophilin A ligands selected after virtual screening and SPR analysis.
Table 1.
Affinity of FD1-FD12 and cyclosporin A binding to cyclophilin A, and their IC50 values for inhibition of PPIase activity of cyclophilin A and HIV-1 replication.
| Compound | Binding to cyclophilinA, KD (M) |
IC50 for inhibition of PPIase activity (µM) |
% Inhibition of HIV-1 replication |
CC50 (µM) |
|---|---|---|---|---|
| FD1 | 7.67±0.12×10−5 | 70.2±3.04 | 26.7 | >100 |
| FD2 | 6.78±0.08×10−5 | 72.8±1.12 | 23.0 | >100 |
| FD3 | 7.33±0.26×10−5 | 65.7±2.55 | 29.9 | >100 |
| FD4 | 2.11±0.15×10−5 | 56.2±0.97 | 22.5 | >100 |
| FD5 | 6.61±0.09×10−6 | 14.45±0.67 | 57.2 | 14.46±0.59 |
| FD6 | 9.19±1.03×10−6 | 21.65±2.20 | 18.2 | 41.81±2.52 |
| FD7 | 3.61±0.25×10−5 | 43.75±1.15 | 33.7 | 73.75±0.67 |
| FD8 | 8.42±0.13×10−6 | 18.67±1.74 | 62.6 | 40.91±0.59 |
| FD9 | 1.03±0.16×10−5 | 34.86±0.76 | 77.5 | 54.24±4.96 |
| FD10 | 5.95±0.24×10−6 | 6.98±1.39 | 64.2 | 65.66±4.77 |
| FD11 | 4.72±0.27×10−5 | 53.4±1.63 | 44.9 | 71.46±4.13 |
| FD12 | 3.19±0.22×10−6 | 3.47±0.92 | 65.2 | 69.73±0.75 |
| Cyclosporin A | 2.18±0.36×10−7 | 0.45±0.38 | 98.9 | 6.26±0.28 |
3.2. Identification of compounds with inhibitory activity against PPIase activity and HIV-1 replication
Since cyclophilin A belongs to cyclophilin protein family, the inhibitory activity of the compounds on PPIase could be determined by a standard spectrophotometric method. During the assay, the rate constants for the cis–trans conversion were evaluated by fitting the data to the integrated first-order rate equation through nonlinear least-square analysis. The inhibition results are shown in Table 1. The compounds FD5, FD6, FD7, FD8 and FD 9 showed IC50 values below 50 µM; FD10 and FD12 showed IC50 values below 10 µM. As the positive control, cyclosporin A showed an IC50 value of 0.45 µM against the PPIase activity of cyclophilin A.
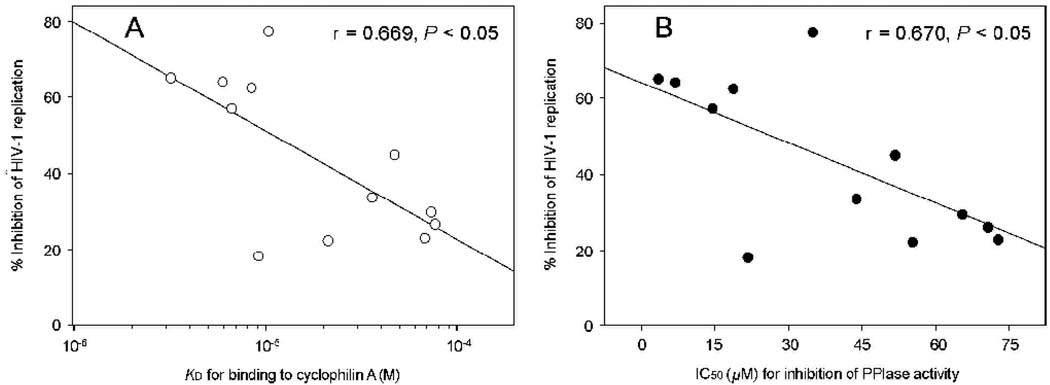
Cyclosporin A (positive control) and the 12 compounds that bound to cyclophilin A were evaluated for their effects on HIV-1IIIB p24 expression in MT-2 cells by flow cytometry and in vitro cytotoxicity by XTT assay. In the absence of inhibitors, 26.9% of the cells were HIV-1 p24-positive, which is higher than the percentage of the Jurket cells infected by the wild-type HIV-1NL4-3 (3% to 21%) (Sokolskaja et al., 2004). As shown in Table 1, cyclosporin A at 10 µM almost completely blocked HIV-1 replication, in agreement with the previous report (Sokolskaja et al., 2004). Five of the 12 compounds tested (FD5, FD8, FD9, FD10, FD12) exhibited more than 50% inhibition of HIV-1 infection at 10 µM. Cyclosporin A and FD5 showed higher cytotoxicity (CC50 < 15 µM) than other compounds tested (CC50 > 40 µM). Interestingly, the anti-HIV-1 activity of these 12 compounds is closely correlated with their cyclophilin A-binding affinity (r = 0.669, P < 0.05) and with their anti-PPIase activity (r = 0.670, P < 0.05), suggesting that these compounds block HIV-1 replication by targeting cyclophilin A (Fig. 2).
Fig. 2.
The anti-HIV-1 activity of the compounds is correlated with their binding affinity to cyclophilin A (A) and with their anti-PPIase activity (B).
4. Discussion
It has been known that cyclosporin A binds to the catalytic site of cyslophilin A and blocks its PPIase activity, resulting in inhibition of HIV-1 replication (Steinkasserer et al., 1995; Braaten et al., 1996; Braaten and Luban, 2001; Sokolskaja et al., 2004). However, cyclosporin A may not be suitable for use as an anti-HIV drug to treat HIV infection/AIDS due to its immunosuppressive property and high cytotoxicity. Therefore, it is essential to identify small-molecule anti-HIV compounds targeting cyclophilin A without immunosuppressive activity and yet low cytotoxicity. Efforts to find novel inhibitors of cyclophilin A have been made for a lot of pharmaceutical usage (Li et al. 2006a,b,c). Wu et al. (2003) identified a set of nonpeptidic cyclophilin ligands with neuroprotective/ neurotrophic activity in vitro. Guichou et al. (2006) have designed a number of inhibitors of human cyclophilin A and demonstrated that one of the inhibitors is effective in blocking HIV-1 replication.
In the present study, we carried out a computer-aided virtual screening of a database consisting of 80,000 small-molecule compounds based on the X-ray crystal structure of cyclophilin A/cyclosporin A complex. Seventy-seven compounds with high docking scores were purchased from SPECS. Their activity of binding to cyclophilin A was determined by SPR. Twelve of them with high binding affinity (< 8 × 10−5 M) were selected for evaluation of their in vitro inhibition on PPIase activity of cyclophilin A and HIV-1 replication. Five compounds (FD5, FD8, FD9, FD10 and FD12) with both anti-PPIase and anti-HIV-1 activity were identified (Table 1). Notably, the anti-HIV-1 activity of these compounds is well correlated with their cyclophilin A-binding affinity and anti-PPIase activity (Fig. 2). These results suggest that these compounds block HIV-1 replication by binding to cyclophilin A and inhibiting its PPIase activity. These compounds will be used as leads for structure and activity relationship (SAR) and optimization studies in order to design more effective and less toxic anti-HIV-1 therapeutics and as probes for determining the effect of cyclophilin on HIV-1 replication.
Acknowledgments
We would like to thank Dr. Susanne Heck of the Flow Cytometry Laboratory of the New York Blood Center for her technical assistance in flow cytometry and Ms. Veronica L. Kuhlemann at the Viral Immunology Laboratory of the New York Blood Center for editorial assistance. This work was supported by 863 High Technology Program of China (Grant No. 2005BA711A01) and Shanghai Basic Research Project from the Shanghai Science and Technology Commission (Grant No. 03DZ14086) to LY and the NIH grant of the United States (AI46221) to SJ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Braaten D, Franke EK, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten D, Ansari H, Luban J. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 1997;71:2107–2113. doi: 10.1128/jvi.71.3.2107-2113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten D, Luban J. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 2001;20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI. Cyclophilins: unexpected messengers in intercellular communications. Trends Immunol. 2002;23:323–325. doi: 10.1016/s1471-4906(02)02237-8. [DOI] [PubMed] [Google Scholar]
- Debnath AK, Radigan L, Jiang S. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 1999;42:3203–3209. doi: 10.1021/jm990154t. [DOI] [PubMed] [Google Scholar]
- DesJarlais RL, Sheridan RP, Seibel GL, Dixon JS, Kuntz ID, Venkataraghavan R. Using shape complementarity as an initial screen in designing ligands for a receptor binding site of known three-dimensional structure. J. Med. Chem. 1988;31:722–729. doi: 10.1021/jm00399a006. [DOI] [PubMed] [Google Scholar]
- Dorfman T, Weimann A, Borsetti A, Walsh CT, Gottlinger HG. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J. Virol. 1997;71:7110–7113. doi: 10.1128/jvi.71.9.7110-7113.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- Gamble TR, Vajdos FF, Yoo S, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- Guichou JF, Viaud J, Mettling C, Subra G, Lin YL, Chavanieu A. Structure-based design, synthesis, and biological evaluation of novel inhibitors of human cyclophilin A. J. Med. Chem. 2006;49:900–910. doi: 10.1021/jm050716a. [DOI] [PubMed] [Google Scholar]
- Guo HX, Wang F, Yu KQ, Chen J, Bai DL, Chen KX, Shen X, Jiang HL. Novel cyclophilin D inhibitors derived from quinoxaline exhibit highly inhibitory activity against rat mitochondrial swelling and Ca2+ uptake/ release. Acta Pharmacol. Sin. 2005;26:1201–1211. doi: 10.1111/j.1745-7254.2005.00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science. 1984;226:544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- Kofron JL, Kuzmic P, Kishore V, Colon-Bonilla E, Rich DH. Determination of kinetic constants for peptidyl prolyl cis-trans isomerases by an improved spectrophotometric assay. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang J, Chen J, Luo X, Zhu W, Shen J, Liu H, Shen X, Jiang H. Strategy for discovering chemical inhibitors of human cyclophilin a: focused library design, virtual screening, chemical synthesis and bioassay. J. Comb. Chem. 2006a;8 doi: 10.1021/cc0501561. 326-237. [DOI] [PubMed] [Google Scholar]
- Li J, Chen J, Gui C, Zhang L, Qin Y, Xu Q, Zhang J, Liu H, Shen X, Jiang H. Discovering novel chemical inhibitors of human cyclophilin A: virtual screening, synthesis, and bioassay. Bioorg. Med. Chem. 2006b;14:2209–2224. doi: 10.1016/j.bmc.2005.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen J, Zhang L, Wang F, Gui C, Zhang L, Qin Y, Xu Q, Liu H, Nan F, Shen J, Bai D, Chen K, Shen X, Jiang H. One novel quinoxaline derivative as a potent human cyclophilin A inhibitor shows highly inhibitory activity against mouse spleen cell proliferation. Bioorg. Med. Chem. 2006c;14:5527–5534. doi: 10.1016/j.bmc.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Albers MW, Chen CM, Schreiber SL, Walsh CT. Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc. Natl. Acad. Sci. USA. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Farmer JD, Jr, Lane WS, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell. 1991;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Mikol V, Kallen J, Pflugl G, Walkinshaw MD. X-ray structure of a monomeric cyclophilin A-cyclosporin A crystal complex at 2.1 A resolution. J. Mol. Biol. 1993;234:1119–1130. doi: 10.1006/jmbi.1993.1664. [DOI] [PubMed] [Google Scholar]
- Ring CS, Sun E, McKerrow JH, Lee GK, Rosenthal PJ, Kuntz ID, Cohen FE. Structure-based inhibitor design by using protein models for the development of antiparasitic agents. Proc. Natl. Acad. Sci. USA. 1993;90:3583–3587. doi: 10.1073/pnas.90.8.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK, Bodian DL, Kuntz ID. Molecular docking using shape descriptors. J. Comput. Chem. 1992;13:380–397. [Google Scholar]
- Shoichet BK, Stroud RM, Santi DV, Kuntz ID, Perry KM. Structure-based discovery of inhibitors of thymidylate synthase. Science. 1993;259:1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- Sokolskaja E, Sayah DM, Luban J. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 2004;78:12800–12808. doi: 10.1128/JVI.78.23.12800-12808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkasserer A, Harrison R, Billich A, Hammerschmid F, Werner G, Wolff B, Peichl P, Palfi G, Schnitzel W, Mlynar E, Rosenwirth B. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus type 1 (HIV-1): interference with early and late events in HIV-1 replication. J. Virol. 1995;69:814–824. doi: 10.1128/jvi.69.2.814-824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Hayano T, Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- Thali M, Bukovsky A, Kondo E, Rosenwirth B, Walsh CT, Sodroski J, Gottlinger HG. Functional association of cyclophilin A with HIV-1 virions. Nature. 1994;372:363–365. doi: 10.1038/372363a0. [DOI] [PubMed] [Google Scholar]
- Thurmond RL, Wadsworth SA, Schafer PH, Zivin RA, Siekierka JJ. Kinetics of small molecule inhibitor binding to p38 kinase. Eur. J. Biochem. 2001;268:5747–5754. doi: 10.1046/j.0014-2956.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- Wu YQ, Belyakov S, Choi C, Limburg D, Thomas IVBE, Vaal M, Wei L, Wilkinson DE, Holmes A, Fuller M, McCormick J, Connolly M, Moeller T, Steiner J, Hamilton GS. Synthesis and biological evaluation of non-peptidic cyclophilin ligands. J. Med. Chem. 2003;46:1112–1115. doi: 10.1021/jm020409u. [DOI] [PubMed] [Google Scholar]
- Zydowsky LD, Etzkorn FA, Chang HY, Ferguson SB, Stolz LA, Ho SI, Walsh CT. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1992;1:1092–1099. doi: 10.1002/pro.5560010903. [DOI] [PMC free article] [PubMed] [Google Scholar]