Abstract
Purpose
Malignant brain tumors are characterized by extensive infiltration into the normal brain tissue. Tumor migration is a complicated process which results from the interplay of a number of mechanisms, and the extent to which anatomic structure determined the migration pattern has not been extensively addressed. In the present study, we labeled C6 glioma cells with iron oxide nanoparticles and monitored the fate of the cells in vivo with magnetic resonance imaging (MRI).
Procedures
C6 glioma cells were labeled with ferumoxide–poly-L-lysine complexes and their migration in the brains of rats tracked by T2-weighted MRI. The same amount of iron-laden cells were implanted into the caudate nucleus (CN) and at the vicinity of anterior commissure (AC), respectively, and MRI was performed during the course of 20-day monitoring period to track tumor growth and migration.
Results
A clear tendency of tumor migration along the white matter fiber tracts was observed in the AC group, which is consistent with the previous reports; by contrast, tumor expanded to but remained confined within the boundary of right hemisphere in the CN group.
Conclusion
We successfully demonstrated the ability of MRI to investigate the impact of anatomical structure on the glioma migration pathway in vivo.
Keywords: Superparamagnetic iron oxide (SPIO) nanoparticles, Cell trafficking, Glioma, Magnetic resonance imaging (MRI), Tumor invasion and migration
Introduction
Tumor cell invasion and migration is closely associated with cancer malignancy. Understanding the underlying mechanisms is a topic of considerable significance in cancer research. In general, tumor cells are highly adaptable, adopting various invasion mechanisms in response to changes in their environment. Taking brain tumor as an example, previous studies have found that diffuse astrocytomas, especially glioblastoma multiforme, invade the brain preferentially along the white matter fiber tracts [1, 2]. However, most previous studies on tracking glioma migration were conducted with histological approaches such as by fast blue and Dil staining on biopsy samples [3]. Due to the limited amount of tissue sample available and the heterogeneous nature of tumor, the staining results may be unrepresentative and misleading. Successful cell trafficking with lac-Z transfected human glioma cells in rat brain has been reported [2]. This strategy too, however, requires ex vivo analysis, not to mention the need to euthanize a large number of animals to achieve a statistically significant temporal profile [4]. Therefore, although substantial progress has been made in vitro and ex vivo, it is still poorly understood at the in vivo level which factors are implicated in the invasion and migration of glioma [5].
Recent advances in molecular imaging have made it possible to track cell migration in vivo in a non-invasive and real-time manner; and along this line, a variety of cell-labeling techniques have been developed. In preclinical studies, optical reporter genes such as luciferases [6–9] and fluorescent proteins [10, 11] have been adopted for genetic cell labeling. While proving useful, the application of these methodologies is limited by the inherent low resolution and poor depth of penetration of optical imaging. Also, efforts have been made towards developing approaches of labeling cells with highly sensitive PET and SPECT techniques [12–14]. These may be limited by problems in labeling cells with radionuclides, including efflux of the radionuclides, short half lives of radionuclides, and relatively low spatial resolution of PET and SPECT.
Magnetic resonance imaging (MRI), on the other hand, has emerged as a powerful tool in studying cell migration with excellent spatial and temporal resolution. In addition, MRI is used in both preclinical and clinical settings, potentially providing a role as a bridge between clinical and preclinical studies. As for cell labeling, exogenous compounds with magnetic properties, such as iron oxide nanoparticles, are widely utilized [15–18]. Cells are labeled in vitro via phagocytosis, and in vivo, the contrast agents induce strong signal reduction on T2-weighted maps, resulting in negative contrast that can facilitate the tracking of the transplanted cells.
In the current study, we used poly-L-lysine-conjugated superparamagnetic iron oxide particles (PLL-SPIO) to label C6 glioma cells. We then intracerebrally injected 1×106 cells into the caudate nucleus (CN) or at the vicinity of the anterior commissure (AC) of Sprague–Dawley rats and tracked by MRI migration of the implanted cells during tumor growth. The purpose of this study was to demonstrate to what extent anatomical structure will affect the path of glioma migration.
Materials and Methods
Cell Culture of C6 Glioma Cells and Labeling of Glioma Cells with Iron Oxide Nanoparticles
The PLL-SPIO nanoparticles were generously provided by Dr. Ning Gu (Laboratory of Molecular and Biomolecular Electronics, Southeast University, Nanjing, China). The details of the particle synthesis and labeling have been previously reported [19]. C6 rat glioma cells were grown as a monolayer in 75-cm2 sterile plastic flasks in F12K media (Gibco) with 10% FBS. Cells were cultured in an incubator at 37°C in an atmosphere of 95% air and 5% CO2 until confluent.
The nanoparticles were added to the culture media to arrive at a final concentration of 25 μg Fe per milliliter, and the labeling was carried on for 24 h. To determine the SPIO labeling efficiency, cells were stained with Prussian blue, and the percentage of the stained cells was counted under a microscope (Leica, Germany). The cell viability was measured at 24, 48, and 72 h after SPIO labeling via 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays and was compared with that of the unlabeled cells (Fig. 1a). The C6 cells were seeded at 4×103/well into 96-well plates, and Fe2O3-PLL was added the next day to reach a final iron concentration of 25 μg Fe per milliliter. The incubation was stopped at 24, 48, and 72 h, and 20 μL of MTT solution (5 mg/mL, Fluka Co., Switzerland) was added into each well and incubated for 4 h at 37°C under 5% CO2 atmosphere. After the supernatant was discarded, 150 μL of dimethyl sulfoxide (Shanghai Bioengineering Co., Shanghai, China) was added into each well, and the plate was gently shaken for 10 min. The absorption at 570 nm was measured by a spectrophotometer (Sepctra MAX 250, Molecular Devices Corp, Sunnyvale, CA, USA).
Fig. 1.
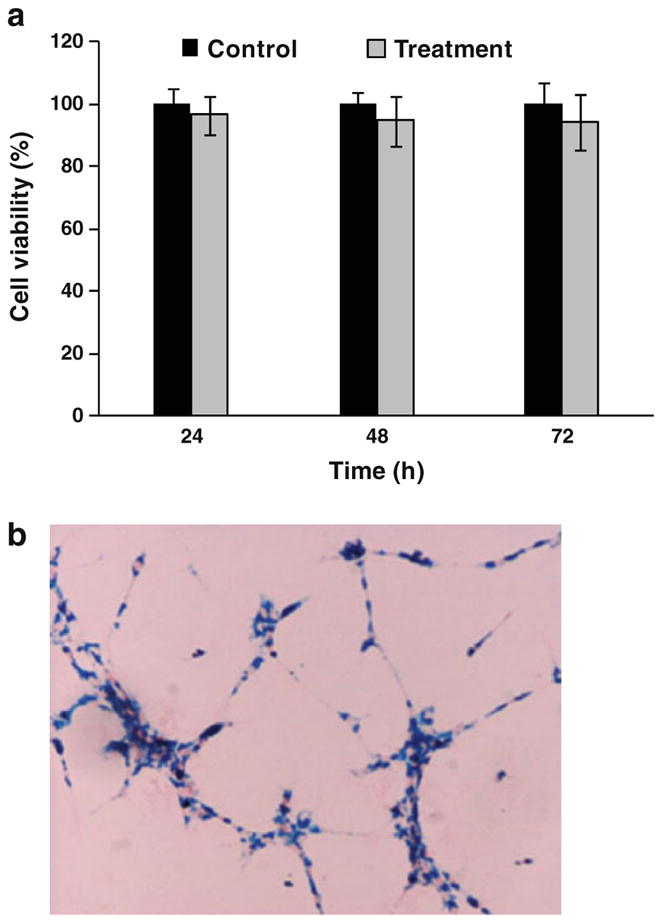
a MTT assay of C6 glioma cells incubated with PLL-SPIOs at 25 μg Fe per milliliter for 24, 48, and 72 h. b Prussian blue staining of C6 glioma cells after incubation with PLL-SPIOs at 25 μg Fe per milliliter for 24 h.
After cell labeling, the cells were detached from the flask surface with 0.05% trypsin-EDTA solution and were thoroughly washed three times with phosphate-buffered saline (PBS). The cells were resuspended in PBS at a concentration of 1×105 cell/μL for transplantation.
Animal Model Preparation
All the animal studies were approved by the ethics committee of Medical College of Nanjing University and were conducted in compliance with the local guidelines for animal care. Adult male Sprague–Dawley rats weighing 220–220 g were group-housed in a temperature-controlled room with access to chow and water ad libitum under 12 h light–dark conditions. The rats (n=8) were anesthetized with 10% chloral hydrate (4 mL/kg i.p.) before surgery. A small skin incision was made over the right hemisphere and a high-speed drill was used to create a 0.5-mm-in-diameter burr hole through the skull. Subsequently, the rats were divided into two groups, and cells in PBS were infused at the following coordinates in relation to bregma using a stereotaxic apparatus (Stoelting company): (1) the caudate nucleus (CN): anterior–posterior, 10 mm; lateral, 25 mm; ventral, −45 mm; and (2) the vicinity of anterior commissure (AC): anterior–posterior, 16 mm; lateral, 25 mm; ventral, −60 mm. For each rat, a total amount of 10 μL (1×106 C6 cells) was transplanted using a Hamilton syringe fitted with a 30 G 0.5-inch needle. Two minutes after the injection, the needle was slowly withdrawn, and the burr hole was filled with bone wax to minimize the extracerebral extension of the tumor tissue. The skin was sutured together, and the rats were allowed to recover.
In Vivo MRI Trafficking of the Iron-Loaded Glioma Cells
The two groups of rats were subjected to the MRI scans on day 3, 6, 12, 16, and 20 after tumor cell implantation on a 7.0 T MR scanner (Biospin, Bruker, Germany). For MRI examination, rats were anesthetized with 10% chloral hydrate (4 mL/kg i.p.) and were maintained at 37°C inside the imaging chamber using a heated circulating water blanket. Multi-slice axial and coronal images were acquired using a standard spin echo sequence with the following parameters: TR/TE=3,500/36, FOV=40×40 mm, matrix=386× 512, slice thickness=0.4 mm, and slice separation=0.65 mm.
Histology
After completion of the final scan, the animals were euthanized and were transcardially perfused with saline followed by 4% paraformaldehyde (PFA). Brains were collected and fixed for 24 h in 3% PFA, followed by cryopreservation in 3% PFA/sucrose for another 48 h. Subsequently, the brains were cut into 40 μm sections. Iron deposits were detected using Prussian blue staining on adjacent brain sections. In brief, slices were incubated in 2% potassium ferrocyanide (Perl’s reagent; Sigma) with 2% HCl for 20 min, and were then washed and counterstained with erythrosin. The Fe content was visualized as blue dots after the staining (Fig. 1b).
Results
In Vitro C6 Cell Labeling with PLL-SPIOs
The effect of PLL-SPIOs on C6 proliferation was evaluated with an MTT assay. At a concentration of 25 μg/mL, no significant difference in viability was observed between the labeled cells and the control (P=0.061), 48 h (P=0.064), and 72 h (P=0.189) after labeling with PLL-SPIOs (Fig. 1a). Prussian blue staining was conducted on C6 cells after 24 h incubation. As displayed in Fig. 1b, the loaded Fe, visualized as blue spots, was found in the cytoplasm of over 90% of the cells.
In Vivo Tracking of the Implanted C6 Glioma Cells
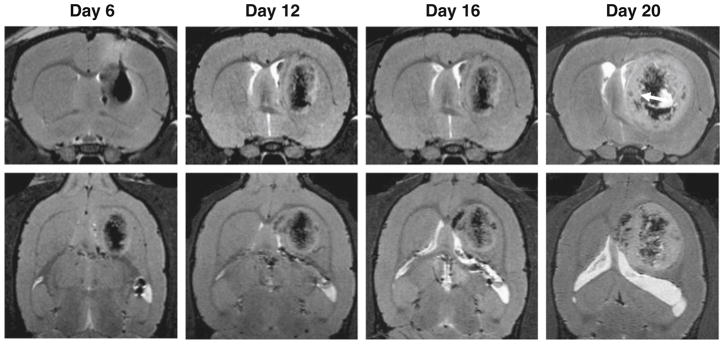
Eight rats were divided evenly into two groups, and each rat was implanted with 1×106 iron-laden C6 cells at the right caudate nucleus (CN) (n=4) or the vicinity of anterior commissure (AC) (n=4). A series of MRI scans were performed after the C6 cell implantation. The first MRI scan on day 3 found areas of strong hypointensities at the right caudate nucleus and the vicinity of anterior commissure for the CN group and the AC group, respectively. The locations of the “dark” areas were well correlated with the designated injection sites and were believed to be due to the presence of Fe-laden cells (Fig. 2). In the CN group, the implanted cells, aside from limited diffusion, maintained their position. On day 12 and 16, we observed a radical cell diffusion pattern from the original injection place and, concomitantly, a decrease in the hypointensity, which we attributed to the dilution of iron through cell proliferation (Fig. 3). Notably, the hypointensities were found expanded to but remained confined within the boundary of right hemisphere on day 20.
Fig. 2.
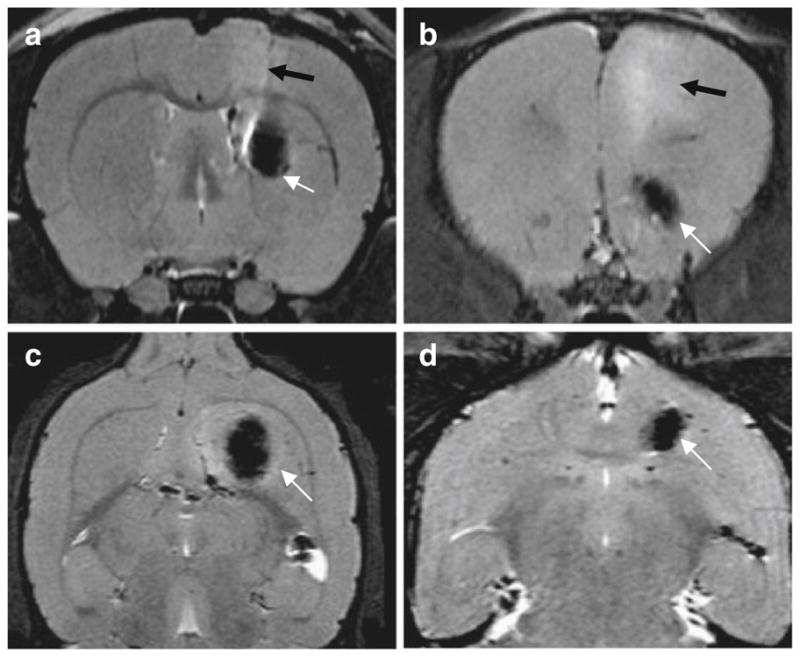
Axial (a, b) and coronal (c, d) magnetic resonance images of a rat brain transplanted with SPIO-labeled cells. The images were taken on day 3 after implantation in the right caudate nucleus (a, c) and at vicinity of anterior commissure (b, d). SPIO-labeled cells causes strong hypointensities around the injection sites (white arrows). Some surgically related edema was observed as diffuse hyperintense areas in the axial T2-weighted images (a, b) (black arrows).
Fig. 3.
High-resolution MR brain images taken on day 6, 12, 16, and 20 after right caudate nucleus (CN) cell implantation (upper panel: coronal; lower panel: transaxial). The tumor continued to grow but was remained in the caudate nucleus from day 12 to day 20. The hypointense signals were diffuse and diluted in tumor mass gradually. Tumor encapsulation could be observed at day 20 post tumor inoculation (arrow).
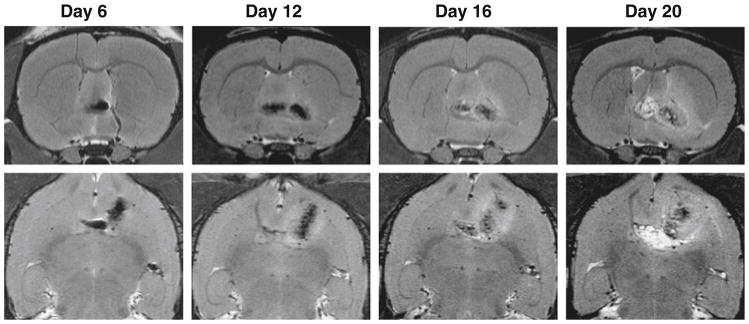
On the other hand, clear cell migration along the anterior commissure was observed in the AC group on day 6 (Fig. 4) which continued throughout the rest of the observation period. The hypointense signals, originally observed in the vicinity of anterior commissure of the right hemisphere, trespassed into the left hemisphere by day 20.
Fig. 4.
High-resolution MR brain images taken on day 6, 12, 16, and 20 after implanting cells in the vicinity of right anterior commissure (AC) (upper panel: coronal; lower panel: transaxial). On day 6 after transplantation, hypointense signals from SPIO-labeled C6 glioma cells were found extending along the anterior commissure (white arrow). On days 12, 16, and 20, tumor size increased rapidly, but tumor cells were not extended along the anterior commissure. Tumor encapsulation could be observed on day 20 (arrow).
Histology Studies
After the day 20 imaging, the rats were euthanized, and Prussian blue staining was performed on the tissue samples. For the AC group, positive Prussian blue staining was found in the right hemisphere as well as across the anterior commissure in the left hemisphere (Fig. 5). For the CN group, positive Prussian blue staining was found exclusively within the right caudate nucleus, along with the observation of brown plaques that were indicative of hemosiderin (Fig. 6).
Fig. 5.
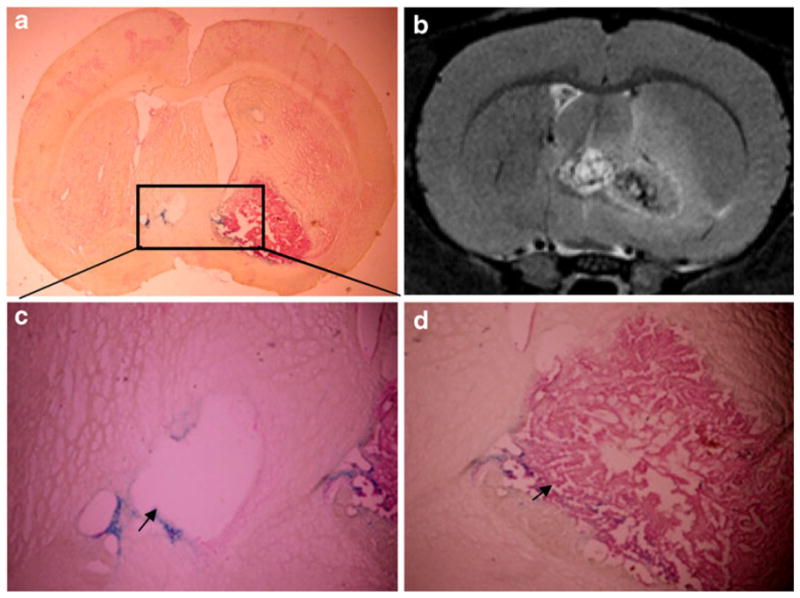
Ex vivo histological studies on brain sample (CN group) taken after the day 20 scan. The tumor was easily recognized in the whole-brain histology sections (a), which correlated well with the corresponding MRI observation (b). A magnification (×20) of the region of tumor encapsulation which crosses the anterior commissure into the left hemisphere (arrow) (c). The blue dots represent nanoparticles of tumor cells. Magnified images (×20) of the boundary of the tumor showing SPIO-labeled tumor cells (arrow) (d).
Fig. 6.
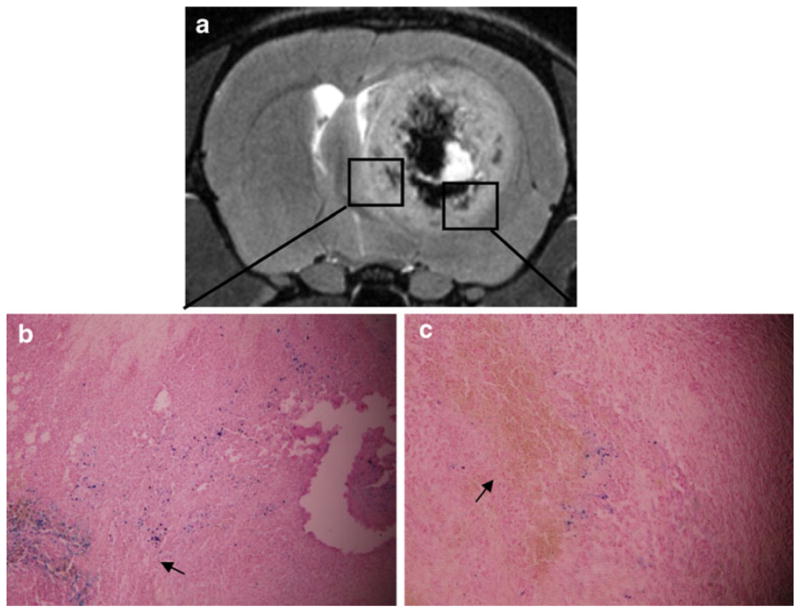
Ex vivo histological studies on the AC group taken after the day 20 scan (a) indicate that the cells do not migrate along the nerve tracks. Magnified (×20) images from different regions of the tumor (b, c). Blue dots represent nanoparticles in tumor cells (arrow) (b). Brown plaques indicate hemosiderin (arrow) (c).
Discussion
Previous studies have revealed that gliomas are capable of spreading diffusely through the brain, preferentially infiltrating along white matter tracts [1, 2]. Although the molecular mechanism of tumor cell invasion is not well understood, it is believed to be implicated with the constitutive extracellular ligands that are expressed along the dissemination pathway [20]. Meanwhile, matrix metalloproteases, which remodel the extracellular matrix, may also play a role and create a more permissive environment in favor of tumor invasion [21]. However, it has not been studied previously whether the tendency of tumor cells migrating along the fiber tracts of the white matter can be enhanced by specific anatomical structures in the brain.
In the current study, SPIO-labeled C6 cells were implanted into the caudate nucleus (CN) and at the vicinity of anterior commissure (AC). We then used high-resolution MRI to monitor tumor cell migration. On day 6, other than radical expansion, we observed no migration of cells in the CN group; by contrast, clear cell migration along the anterior commissure was found in the AC group. The pattern of cell distribution is similar to that previously report by Pederson et al. [2], who found that glioma cells migrate over the corpus callosum and around blood vessels on day 3 after tumor spheroids implantation. Indeed, from day 12 of our study, C6 cells were found across anterior commissure into the left hemisphere in the AC group, but not in the CN group. In the CN group, despite a rapid growth rate, the tumor was found restricted in the right hemisphere. Such differences clearly show that the tumor invasion and migration pattern is affected by its location.
The fibers of the anterior commissure connect the olfactory tracts and the piriform, prepiriform, and amygdaloid bodies. Lower animals typically have a larger olfactory tracts and large anterior commissure than humans [5]. Unlike the peripheral nervous system (PNS), most central nervous system (CNS) axons are myelinated. Andersson et al. found that tumor cells preferentially migrate on myelinated fiber tracts rather than on unmyelinated fiber tracts or on gray matter [3]. This gives a terrain advantage to the AC group and helps to explain the cellular migration observed in the AC group but not in the CN group.
In this study, we used non-invasive MRI to investigate the influence of anatomical structure on glioma migration pathway. Compared with the traditional histopathological methods that are highly invasive, MRI technology, in combination with efficient iron oxide nanoparticle labeling techniques, offers an alternative strategy that allows longitudinal trafficking of implants with a high spatial resolution and a much improved animal attrition rate. However, just like any another method, such techniques have their own limitations. One disadvantage is that the SPIO, as non-genetic materials, will be degraded and divided among the daughter cells through the tumor growth [22]. This dilution attenuates signal with time, limiting the effective observation period to a maximum of 2–3 weeks [22]. Moreover, confounding factors other than particle susceptibility may contribute to the signal attenuation, which cannot be distinguished by MRI. Among these factors, hemorrhage (tumor necrosis) might be the most common, as was observed in our study. It suggests that, in many cases, histologic analysis may be indispensable in validating the imaging results [23].
We are aware of several limitations in our work. First, the present study is based on a C6 glioma model, which exhibits rapid tumor growth. It is possible that other tumor types, especially those with less invasiveness, may demonstrate different migration patterns under the same conditions. It is of our interest to evaluate in future studies those potential discrepancies and reveal the underlying mechanisms of tumor invasion and migration. Second, in contrast with nuclear medicine techniques such as PET and SPECT, MRI is known for being of low sensitivity. It is therefore interesting to label cells with more than one tag and use multimodal methods to improve the data quality and reduce the risk of data misinterpretation. Last but not least, the implanted cells might die during the observation period. This causes a release of the imbedded SPIO, which might remain in the intercellular space or be phagocytized by macrophages. In either case, the associated hypointensities cease to report the whereabouts of the implanted cells. Careful histologic assays are thus needed to confirm or correct the observation made by the MRI.
Conclusion
We have shown in this study that C6 glioma cells can be labeled effectively and efficiently by PLL-SPIOs with negligible influence on cell viability or proliferation rates. We then implanted labeled C6 cells into CN and at the vicinity of AC of rat brain and used MRI to track the migration of the cells over time. A tendency of tumor cells migrating along the white matter fiber tracts was observed. Meanwhile, difference in migration pattern was found between the CN group and the AC group, suggesting the impact of anatomical structure on the tumor migration pathway.
Acknowledgments
F Zhang, Y He, and G Lu are partially supported by 973 grants (2006, CB705707, China) and National Natural Science Foundation (30930028, China). F Zhang, J Xie, G Liu, and X Chen are supported by the Intramural Research Program (IRP) at NIBIB, NIH.
Footnotes
Conflict of Interest Disclosure. The authors declare that they have no conflict of interest.
References
- 1.Belien AT, Paganetti PA, Schwab ME. Membrane-type 1 matrix metalloprotease (MT1-MMP) enables invasive migration of glioma cells in central nervous system white matter. J Cell Biol. 1999;144:373–384. doi: 10.1083/jcb.144.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen PH, Edvardsen K, Garcia-Cabrera I, Mahesparan R, Thorsen J, Mathisen B, Rosenblum ML, Bjerkvig R. Migratory patterns of lac-Z transfected human glioma cells in the rat brain. Int J Cancer. 1995;62:767–771. doi: 10.1002/ijc.2910620620. [DOI] [PubMed] [Google Scholar]
- 3.Andersson C, Tytell M, Brunso-Bechtold J. Transplantation of cultured type 1 astrocyte cell suspensions into young, adult and aged rat cortex: cell migration and survival. Int J Dev Neurosci. 1993;11:555–568. doi: 10.1016/0736-5748(93)90045-f. [DOI] [PubMed] [Google Scholar]
- 4.Blasberg RG. Molecular imaging and cancer. Mol Cancer Ther. 2003;2:335–343. [PubMed] [Google Scholar]
- 5.Go Y, Chintala SK, Oka K, Gokaslan Z, Sawaya R, Rao JS. Invasive pattern of lac-Z-transfected human glioblastoma cells in nude mice brain. Cancer Lett. 1996;110:225–231. doi: 10.1016/s0304-3835(96)04515-6. [DOI] [PubMed] [Google Scholar]
- 6.Greer LF, 3rd, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence. 2002;17:43–74. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 7.Cao F, Lin S, Xie X, Ray P, Patel M, Zhang X, Drukker M, Dylla SJ, Connolly AJ, Chen X, et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113:1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Cao F, De A, Cao Y, Contag C, Gambhir SS, Wu JC, Chen X. Trafficking mesenchymal stem cell engraftment and differentiation in tumor-bearing mice by bioluminescence imaging. Stem Cells. 2009;27:1548–1558. doi: 10.1002/stem.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Chen X. Imaging mesenchymal stem cell migration and the implications for stem cell-based cancer therapies. Future Oncol. 2008;4:623–628. doi: 10.2217/14796694.4.5.623. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Hazelrigg T. Implications for bcd mRNA localization from spatial distribution of exu protein in Drosophila oogenesis. Nature. 1994;369:400–403. doi: 10.1038/369400a0. [DOI] [PubMed] [Google Scholar]
- 11.Yang M, Baranov E, Jiang P, Sun FX, Li XM, Li L, Hasegawa S, Bouvet M, Al-Tuwaijri M, Chishima T, et al. Whole-body optical imaging of green fluorescent protein-expressing tumors and metastases. Proc Natl Acad Sci USA. 2000;97:1206–1211. doi: 10.1073/pnas.97.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adonai N, Nguyen KN, Walsh J, Iyer M, Toyokuni T, Phelps ME, McCarthy T, McCarthy DW, Gambhir SS. Ex vivo cell labeling with 64Cu-pyruvaldehyde-bis(N4-methylthiosemicarbazone) for imaging cell trafficking in mice with positron-emission tomography. Proc Natl Acad Sci USA. 2002;99:3030–3035. doi: 10.1073/pnas.052709599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li ZB, Chen K, Wu Z, Wang H, Niu G, Chen X. 64Cu-labeled PEGylated polyethylenimine for cell trafficking and tumor imaging. Mol Imaging Biol. 2009;11:415–423. doi: 10.1007/s11307-009-0228-x. [DOI] [PubMed] [Google Scholar]
- 14.Kraitchman DL, Tatsumi M, Gilson WD, Ishimori T, Kedziorek D, Walczak P, Segars WP, Chen HH, Fritzges D, Izbudak I, et al. Dynamic imaging of allogeneic mesenchymal stem cells trafficking to myocardial infarction. Circulation. 2005;112:1451–1461. doi: 10.1161/CIRCULATIONAHA.105.537480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolte IS, Gungor S, Erber R, Plaxina E, Scharf J, Misselwitz B, Gerigk L, Przybilla H, Groden C, Brockmann MA. In vitro labeling of glioma cells with gadofluorine M enhances T1 visibility without affecting glioma cell growth or motility. Magn Reson Med. 2008;59:1014–1020. doi: 10.1002/mrm.21503. [DOI] [PubMed] [Google Scholar]
- 16.Valable S, Barbier EL, Bernaudin M, Roussel S, Segebarth C, Petit E, Remy C. In vivo MRI tracking of exogenous monocytes/macrophages targeting brain tumors in a rat model of glioma. Neuroimage. 2007;37(Suppl 1):S47–S58. doi: 10.1016/j.neuroimage.2007.05.041. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Jiang Q, Jiang F, Ding G, Zhang R, Wang L, Zhang L, Robin AM, Katakowski M, Chopp M. In vivo magnetic resonance imaging tracks adult neural progenitor cell targeting of brain tumor. Neuroimage. 2004;23:281–287. doi: 10.1016/j.neuroimage.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Xie J, Wang J, Niu G, Huang J, Chen K, Li X, Chen X. Human serum albumin coated iron oxide nanoparticles for efficient cell labeling. Chem Commun. 2010;46:433–435. doi: 10.1039/b917195a. Camb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju S, Teng G, Zhang Y, Ma M, Chen F, Ni Y. In vitro labeling and MRI of mesenchymal stem cells from human umbilical cord blood. Magn Reson Imaging. 2006;24:611–617. doi: 10.1016/j.mri.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Demuth T, Berens ME. Molecular mechanisms of glioma cell migration and invasion. J Neurooncol. 2004;70:217–228. doi: 10.1007/s11060-004-2751-6. [DOI] [PubMed] [Google Scholar]
- 21.Uhm JH, Dooley NP, Villemure JG, Yong VW. Mechanisms of glioma invasion: role of matrix-metalloproteinases. Can J Neurol Sci. 1997;24:3–15. doi: 10.1017/s0317167100021028. [DOI] [PubMed] [Google Scholar]
- 22.Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed. 2004;17:484–499. doi: 10.1002/nbm.924. [DOI] [PubMed] [Google Scholar]
- 23.Stroh A, Zimmer C, Werner N, Gertz K, Weir K, Kronenberg G, Steinbrink J, Mueller S, Sieland K, Dirnagl U, et al. Tracking of systemically administered mononuclear cells in the ischemic brain by high-field magnetic resonance imaging. Neuroimage. 2006;33:886–897. doi: 10.1016/j.neuroimage.2006.07.009. [DOI] [PubMed] [Google Scholar]