SUMMARY
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic tumor resulting from the malignant transformation of immature T-cell progenitors. Originally associated with a dismal prognosis, the outcome of T-ALL patients has improved remarkably over the last two decades as a result of the introduction of intensified chemotherapy protocols. However, these treatments are associated with significant acute and long-term toxicities, and the treatment of patients presenting with primary resistant disease or thoserelapsing after a transient response remains challenging.
T-ALL is a genetically heterogeneous disease in which numerous chromosomal and genetic alterations cooperate to promote the aberrant proliferation and survival of leukemic lymphoblasts. However, the identification of activating mutations in the NOTCH1 gene in over 50% of T-ALL cases has come to define aberrant NOTCH signaling as a central player in this disease. Therefore, the NOTCH pathway represents an important potential therapeutic target. In this review, we will update our current understanding of the molecular basis of T-ALL, with a particular focus on the role of the NOTCH1 oncogene and the development of anti-NOTCH1 targeted therapies for the treatment of this disease.
Keywords: T-ALL, NOTCH, gamma-secretase inhibitors, targeted therapy, prognosis, leukemia
Introduction
Acute lymphoblastic leukemias (ALL) are characterized by the uncontrolled clonal proliferation of immature lymphoid cells which infiltrate the bone marrow. In T-cell acute lymphoblastic leukemias (T-ALL) the malignant clone is derived from T-cell progenitor cells and expresses immature immunophenotypic markers characteristic of the T-cell lineage. T-ALL represents about 15% of pediatric and 25% of adult ALLs and is typically associated with very high white cell counts, mediastinal masses with pleural effusions, and increased risk of leptomeningeal infiltration at diagnosis [1]. Although initially associated with a particularly bad prognosis, the introduction of intensified treatment protocols has improved the outcome of this disease and current therapies achieve five-year relapse-free survival rates of about 75% in children and 50% in adults [2–8].
T-cell transformation is a multistep oncogenic process in which multiple lesions involving different oncogenes and tumor suppressor genes cooperate to disrupt the normal circuitry that controls cell proliferation, differentiation and survival during T-cell development [9–12]. Most of what we know about the molecular basis of T-ALL has been learned from the study of recurrent cytogenetic alterations [9]. The most common genetic alteration in T-ALL is the presence of deletions in the CDKN2A tumor suppressor locus containing the P16/INK4A and the P14/ARF tumor suppressor genes, which control cell cycle progression and p53 mediated apoptosis, respectively [13]. In addition, over 50% of T-ALLs harbor activating mutations in the NOTCH signaling pathway making of NOTCH1 the most prominent T-ALL specific oncogene [14] and defining T-ALL as a disease primarily characterized by aberrant NOTCH1 activation [15, 16]. However, T-ALL is a heterogeneous disease in which different molecular groups, primarily defined by the expression of T-ALL transcription factor oncogenes, are associated with specific patterns of gene expression, a specific block in cell differentiation and distinct clinical characteristics [10–12]. Thus, T-ALL-associated chromosomal translocations typically result in the juxtaposition of a selective group of oncogenic transcription factors next to strong regulatory elements located in the vicinity of the T-cell receptor β(TCRB) gene in chromosome 7q34 or the T-cell receptor α-δ (TCRAD) locus in chromosome 14q11 [9, 17]. These T-ALL-specific transcription factor oncogenes include basic helix-loop-helix (bHLH) family members such as TAL1 [18–21], TAL2 [22], LYL1 [23] and BHLHB1 [24]; LIM-only domain (LMO) factors such as LMO1 and LMO2 [25–29]; TLX1/HOX11[30–33], TLX3/HOX11L2 [34], NKX2.5 [35, 36] and HOXA homeobox genes [11, 37]; MYC [38–42] and MYB [43] oncogenes; and TAN1, a truncated and constitutively activated form of the NOTCH1 receptor [44, 45]. In some cases, these factors can also be activated in the context of other non-TCR-associated chromosomal abnormalities. This is the case for small deletions activating TAL1 [46] and LMO2 [47]; duplications of the MYB oncogene [48, 49]; and the t(5;14)(q32;q11) translocation which activates the TLX3 oncogene in chromosome 5 by relocating it to the vicinity of the BCL11B locus in chromosome 14.
Additional molecular alterations present in T-ALL include transcription factor fusion oncogenes such as PICALM/MLLT10/CALM-AF10 [50–52], MLL-MLLT1/MLL-ENL [53, 54], SET/NUP214 [55], NUP98-RAP1GDS1 [56, 57]; activation of signaling factors driving proliferation such as LCK [58], CCND2 [59, 60], JAK1 [61], NUP214-ABL1 [62], EML1-ABL1 [17], and NRAS [63, 64]; and the loss of tumor suppressor genes in the RAS (NF1 [65]) and PI3K (PTEN [66]) signaling pathways. However, the catalog of genetic alterations involved in the pathogenesis of T-ALL is not yet complete as shown by the recent identification of loss-of-function mutations in WT1 [67], the PTPN2 phosphatase [68] and the PHF6 tumor suppressor gene [69].
NOTCH1 signaling pathway
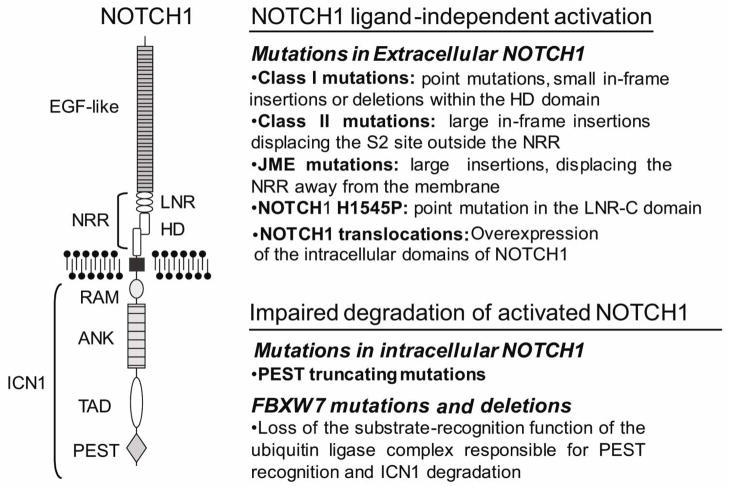
The NOTCH1 receptor is a class I transmembrane protein that functions as a ligand-activated transcription factor (Figure 1) [15]. Thus, NOTCH1 directly transduces information from extracellular signals into changes in gene expression in the nucleus. The main components of NOTCH1 signaling include: the Delta/Serrate/LAG-2 (DSL) family of ligands (Delta-like 1, 3, and 4 as well as Jagged 1 and 2); the NOTCH1 receptor (NOTCH1); the RBPJ/CSL (CBF1/Su(H)/LAG-1) DNA-binding protein; and the mastermind-like family of coactivators. In resting conditions, NOTCH1 sits in the membrane as a heterodimeric complex composed of an N-terminal extracellular subunit (NEC) and a C-terminal transmembrane and intracellular subunit (NTM). The NEC subunit interacts with Delta-like and Jagged ligands through 36 epidermal growth factor (EGF)-like repeat domains. In addition, it contains a negative regulatory region (NRR) composed of three Lin12/NOTCH repeats (LNRs). These LNR domains fold over and stabilize the heterodimerization domain (HD), which consists of the C-terminus of NEC and the N-terminus of NTM in close interaction, to prevent the spontaneous activation of the receptor in the absence of ligand. The NTM subunit contains a transmembrane sequence followed by a series of cytoplasmic domains, including a RAM domain, a series of ankyrin repeats, a transactivator domain, and several nuclear localization signals, which collectively function as a ligand activated transcription factor. The NTM also contains a C-terminal PEST (proline (P), glutamic acid (E), serine (S), and threonine (T) rich) domain, which is responsible for the proteosomal degradation of activated NOTCH1 in the nucleus [15].
Figure 1. NOTCH1 mutations in T-ALL.
Schematic representation of the NOTCH1 receptor structure and types of NOTCH1 mutations found in T-ALL. EGF-like, EGF-like repeats involved in ligand recognition. NRR, negative regulatory region. LNR, Lin12-NOTCH repeats. HD, heterodimerization domain. RAM, RAM domain. ANK, ankyrin repeats. TAD, transactivation domain. PEST, proline, glutamic acid, serine and threonine rich domain. ICN1, intracellular NOTCH1.
Under physiologic conditions the NOTCH1 receptor is activated via interaction with a Jagged or Delta-like ligand molecule. This ligand-receptor interaction induces a conformational change in the NRR regulatory region and triggers the cleavage of the HD domain by the ADAM10 and ADAM17 metalloproteases at the cell surface [70–74]. This first activation-associated cleavage, also known as S2, is then followed by a second proteolytic cleavage (S3) catalyzed by the γ-secretase complex in the transmembrane region of the receptor [70–72]. Thus, the γ-secretase complex releases the intracellular domain of NOTCH1 (ICN1) into the cytosol and allows its translocation into the nucleus, where it associates with the RBPJ/CSL DNA-binding protein, recruits members of the mastermind (MAML) family of coactivators and p300, and through these interactions, activates gene expression [15]. Finally, recruitment of the RNA polymerase II holoenzyme to the ICN1-RBPJ/CSL-MAML transcriptional complex triggers the phosphorylation of the PEST domain of NOTCH1 by cyclin-dependent kinase 8 and recruits the FBXW7-SCF ubiquitin ligase complex, which ultimately mediates the polyubiquitination and proteasomal degradation of the activated receptor in the nucleus [15].
NOTCH1 in T-cell development
The NOTCH signaling pathway is responsible for cell fate specification and tissue patterning in multiple cellular and tissue contexts during development. In the lymphoid system NOTCH signals provided by the thymic microenvironment are essential for the specification and development of T-cell progenitors [75, 76]. Consistent with this model, conditional inactivation of NOTCH1 results in a complete ablation of T-cell lymphopoiesis and differentiation accompanied by ectopic B-cell development in the thymus [77, 78]. Likewise, overexpression of an active, intracellular form of NOTCH1 in bone marrow progenitors results in ectopic pre-T cell development in the bone marrow [79].
Upon T-cell specification, thymocytes differentiate into αβ or γδ T-cell lineages, and while the development of γδ T-cells seems to be independent of NOTCH [80, 81], αβ T-cells require continuous NOTCH1 activation for their maturation to the DN3 stage of development [81]. During this process, several important factors required for T-cell development are transcriptionally controlled by NOTCH1, including the pre-T-cell receptor alpha (PTCRA) [82], the IL7 receptor alpha (IL7RA) [83] and MYC [84]. Both preTCR signaling and NOTCH activation are needed for growth and survival at the so called β-selection checkpoint [80], at which point NOTCH1 signaling is critically required to sustain cell metabolism via activation of the PI3K-AKT cascade [85].
Aberrant NOTCH1 activation in T-ALL
The first evidence of the role of NOTCH1 in the pathogenesis of T-ALL resulted from the cloning of TAN1, a truncated and constitutively active form of NOTCH1, at the breakpoint of the t(7;9)(q34;q34.3) chromosomal translocation present in about 1% of T-ALL cases [44]. In this translocation, the NOTCH1 locus in 9q34 is broken so that the derivative chromosome 9 retains the N-terminal domains of NOTCH1, including the NRR region, while the transmembrane and intracellular domains of the receptor are translocated to the derivative chromosome 7 where they are aberrantly expressed under the control of the TCRB regulatory sequences. Ultimately this rearrangement results in constitutive activation of NOTCH1 signaling due to the expression of high levels of NTM and/or ICN1 in T-cell precursors (Figure 1) [44, 45]. The pathogenic role of activated NOTCH1 in T-ALL was fully demonstrated when irradiated mice reconstituted with bone marrow progenitors expressing activated forms of NOTCH1 developed clonal hematopoietic tumors characterized as T-ALL [86]. In addition, T-cell tumors generated by insertional mutagenesis showed a high incidence of retroviral integrations resulting in constitutive activation of NOTCH1 [87]. However, it was only after the identification of activating mutations in NOTCH1 in over 50% of human T-ALL cases (Figure 1) [14] that the central role of NOTCH1 in the pathogenesis of this disease was fully appreciated.
Activating mutations in NOTCH1 typically result in the disruption of molecular locks responsible for preventing the spontaneous activation of the receptor at the membrane or mediating the termination of NOTCH1 signaling in the nucleus [14]. Thus, most mutations in the HD domain (exon 26 and exon 27), which are present in approximately 40% of human T-ALLs, destabilize the interaction between the N-terminal and C-terminal HD subunits and result in ligand-independent activation or ligand hypersensitivity (HD class 1 mutations) (Figure 1) [88]. A second mutational hotspot is located at the 3′ end of the gene, which encodes the C-terminal PEST domain [14]. PEST domain mutations are present in about 15% of T-ALL samples and are typically truncating and nonsense mutations, which result in deletion of the recognition sequence for proteasomal degradation of ICN1 by the FBXW7/SCF complex in the nucleus (Figure 1). In rare cases, NOTCH1 is activated as the result of in-frame insertions in the distal part of the HD domain (HD class 2 mutations) that result in the displacement and constitutive processing at the ADAM protease cleavage site [88]. Alternative mechanisms of NOTCH1 activation include juxtamembrane expansion NOTCH1 mutations (JME alleles), which consist of extracellular in frame insertions that displace the HD domain away from the membrane (Figure 1) [89] and the NOTCH1 H1545P mutation –located in the NOTCH1 LNR-C repeat– that disrupts the activity of the NRR and facilitates S2 processing of the HD domain (Figure 1) [90]. In addition to these mutations in the NOTCH1 gene, about 15% of T-ALLs harbor mutations in FBXW7, which typically involve key arginine residues responsible for the recognition of phosphorylation sites in the PEST domain of NOTCH1 [91–93]. These FBXW7 mutations impair the substrate recognition function of the FBXW7/SCF complex and impair the degradation of activated NOTCH1 (Figure 1) [91, 92]. In addition, the oncogenic effects of FBXW7 mutations may extend beyond the NOTCH1 signaling pathway as this ubiquitin ligase also mediates the proteosomal degradation of additional oncoproteins such as MYC, JUN, Cyclin E, Aurora-A and m-TOR [94–96]. Finally, about 20% of T-ALL patients harbor either dual mutations in the HD and PEST domains of NOTCH1 or both a NOTCH1 HD allele and a FBXW7 mutation. The combined effect of these mutations results in exceedingly high levels of NOTCH signaling as a result of NOTCH ligand-independent activation at the membrane plus impaired ICN1 degradation in the nucleus[14, 91].
An important point worth emphasizing here is that not all NOTCH mutations are functionally equivalent. Indeed each of the different types of NOTCH1 alleles described above has very different effects in its mechanism of action and its level of activation. NOTCH1 PEST mutations, when present alone, are typically weak alleles and are predicted to be functional only in the presence of NOTCH ligands [14]. NOTCH1 HD alleles result in variable levels of spontaneous NOTCH1 activation and, although some may induce ligand-independent activation of the receptor, others probably only confer ligand hypersensitivity [88]. In contrast, truncated NOTCH1 alleles resulting from the t(7;9) translocation, NOTCH1 insertion mutations (class 2 HD mutations and JME alleles), and double mutant alleles (NOTCH1 HD plus PEST or NOTCH1 HD plus FBXW7 mutations) result in remarkably high levels of NOTCH1 activation [14]. Consistently, each of these alleles and allele combinations has shown very different effects in its capacity to induce leukemia when expressed in mouse hematopoietic progenitor cells [97]. Specifically, weak NOTCH1 alleles failed to induce T-ALL by themselves although they accelerate T-cell transformation in hematopoietic progenitors expressing the k-ras oncogene [97]. Overall, strong NOTCH1 mutants may work as major drivers of the tumor phenotype, acting potentially probably as initiating events in T-ALL, while weaker alleles may function as secondary events that contribute to tumor progression.
Genes and pathways controlled by NOTCH1 in T-cell transformation
The identification of genes and pathways controlled by NOTCH in T-ALL has been the focus of extensive research over the last years. These studies have defined a prominent role for NOTCH1 as a central regulator, promoting leukemia cell growth by multiple direct and indirect mechanisms. Gene expression profiling of T-ALL cell lines and ChIP-on-chip analysis of NOTCH1 in T-ALL cells revealed a prominent role of oncogenic NOTCH1 as a direct transcriptional activator of multiple genes involved in anabolic cell growth and metabolism [98]. In addition, this study also identified the MYC oncogene as a prominent direct target gene regulated by NOTCH1 in human leukemias [98]. Notably, most of the genes controlled by NOTCH1 that regulate cell growth, proliferation and metabolism are also targets of MYC [98, 99]. The resulting NOTCH1-MYC feed-forward-loop transcriptional regulatory network reinforces the expression of genes implicated in anabolic pathways, ribosome biosynthesis, protein translation and nucleotide and amino acid metabolism downstream of NOTCH1 [84, 98]. Consistent with these observations, analysis of mouse tumor cells also revealed c-Myc as a prominent NOTCH1 target gene in T-cell transformation [84, 100].
In addition to its direct effect on anabolic genes and facilitating cell growth via upregulation of MYC, NOTCH1 facilitates the activation of the PI3K-AKT-mTOR signaling pathway, a critical regulator of cell growth and metabolism [66, 85]. The first indication of the key interaction between NOTCH and the PI3K pathway was provided in a seminal manuscript by Ciofani and coworkers who demonstrated that NOTCH signals regulate cell size, glucose uptake and glycolysis via activation of the PI3K-AKT signaling pathway during T-cell development [85]. More recently, phosphoproteomic analysis demonstrated a marked suppression of mTOR signaling in T-ALL cells upon inhibition of NOTCH signaling [101]. Overall, NOTCH1 seems to facilitate the activation of the PI3K-AKT-mTOR pathway at multiple levels. In T-cell progenitors and T-ALL lymphoblasts, , a transcriptional repressor directly downstream of NOTCH1 signaling, HES1, can downregulate the expression of PTEN, a critical negative regulator of the PI3K pathway [66]. In addition NOTCH1 can activate AKT via the LCK tyrosine kinase in T-cells [102] and MYC can rescue the inhibitory effects of blocking NOTCH1 on the m-TOR pathway [101]. Finally, numerous signaling molecules upstream of PI3K, including the interleukin 7 receptor alpha chain (IL7RA) [83] and the pre-T-cell receptor alpha (PTCRA) [82], are upregulated upon activation of NOTCH1 signaling in T-cell progenitors and in T-ALL lymphoblasts.
The transcriptional program activated by oncogenic NOTCH1 also has a direct effect on cell cycle progression. For instance, oncogenic NOTCH1 signaling promotes G1/S cell cycle progression in T-ALL [14, 45, 103, 104] [105]. These effects are mediated in part by transcriptional upregulation of CCND3, CDK4, and CDK6 [103]. Notably, CCND3 is a direct NOTCH1 target gene in T-ALL and is strictly required for NOTCH1-induced transformation [106]. Moreover, inhibition of NOTCH signaling in T-ALL is associated with upregulation of the cyclin-dependent kinase inhibitors CDKN2D (p19/INK4d) and CDKN1B (p27/Kip1) [98, 105]. Finally, in hematopietic progenitors, NOTCH1 can induce the transcription of the S phase kinase-associated protein 2 (SKP2), which mediates the proteasomal degradation of CDKN1B (p27/Kip1) and CDKN1A (p21/Cip1) [104].
Finally, NOTCH1 signaling can also regulate the survival of T-ALL cells via interaction with the NFκB. Specifically, activation of NOTCH signaling upregulates NFκB activity by increasing expression of IkB-kinase [107] and upregulating both the expression and the nuclear localization of NFκB [108]. The critical role of this interaction is demonstrated by the antileukemic effects of NFκB inhibition in T-ALL and the strict requirement of NFκB signaling for NOTCH-induced transformation [109].
NOTCH1 mutations and clinical prognosis in T-ALL
Since the identification of activating mutations in NOTCH1, a number of studies have addressed the prognostic significance of these alterations in T-ALL. Initially, a study reporting results from a cohort of 157 pediatric T-ALL patients treated with the ALL-BFM 2000 protocol found that NOTCH1 mutations were associated with increased prednisone sensitivity, lower levels of minimal residual disease and favorable long-term outcomes [110]. Similarly, analysis of 55 pediatric T-ALL and 14 T-cell lymphoblastic lymphoma patients treated in the Japan Association of Childhood Leukemia Study (JACLS) protocols ALL-97 and NHL-98 showed an improved outcome in patients harboring NOTCH1 and/or FBXW7 mutations [111]. In adult T-ALL, analysis of patients treated in the LALA-94 or GRAALL-2003 studies also identified NOTCH1 and/or FBXW7 mutations as favorable prognostic markers [112]. However, these results have not been fully validated in other series. Analysis of 72 pediatric T-ALL patients treated with the ALL-7, ALL-8 or ALL-9 protocols by the Dutch Childhood Oncology Group [113] and a study analyzing a cohort of 88 adult T-ALL patients treated according to the MRC UKALLXII/ECOGE2993 protocol [114] failed to detect a significant association between NOTCH1 and FBXW7 mutations and clinical outcome.
Notably, three timely reports have recently readdressed the association of NOTCH activation with outcome, clarifying some of the uncertainties raised by earlier studies. First, a retrospective study on the relevance of NOTCH1/FBXW7 mutations in pediatric T-ALL analyzed patients enrolled on Dutch DCOG ALL7/8 or ALL9 or the German COALL-97 protocols and combined mutation analysis of NOTCH1 and FBXW7 with direct measurement of activated NOTCH1 protein using reverse-phase protein microarrays [115]. This analysis confirmed that NOTCH1 and FBXW7 mutations are associated with increased intracellular NOTCH1 levels in clinical samples [115]. In this series, the presence of NOTCH1/FBXW7 mutations was associated with a good initial in vivo prednisone response. However, this improved response to therapy did not translate into a superior outcome [115]. Similarly, analysis of NOTCH1 and FBXW7 mutations in 134 children with T-ALL enrolled in EORTC-CLG trials showed that mutation-positive patients have a better response to prephase therapy and lower levels of minimal residual disease at the end of induction [116]. However, this improved therapeutic response once again, did not result in improved outcome [116]. Finally, and in contrast with the results of these reports, an extended analysis of the effects of NOTCH1 and FBXW7 mutations in patients treated on ALL-BFM protocols confirmed the overall favorable effect of activating NOTCH1 mutations in prognosis originally observed in the BFM2000 study [117]. This series included 151 cases from the original report of the ALL-BFM 2000 protocol [110] and extended this series by including 150 new cases. NOTCH1 and FBXW7 mutations in this cohort were associated with rapid early treatment response both in terms of prednisone sensitivity and as measured by minimal residual disease [117]. Notably, this improved therapeutic response resulted in improved outcome and decreased risk of relapse [117]. Overall the results of these studies show that activation of NOTCH pathway is associated with improved therapeutic response and high sensitivity to glucocorticoid therapy in T-ALL. However, the ultimate effect of these mutations in terms of clinical outcome seems to be therapy-dependent.
Targeted inhibition of NOTCH1 for the treatment of T-ALL
Perhaps the most exciting opportunity derived from the identification of NOTCH1 mutations in T-ALL is the possibility of developing anti-NOTCH1 targeted therapies in this disease. The γ-secretase complex, responsible for the proteolytic processing and activation of NOTCH signaling can be inhibited with small molecule inhibitors (GSIs) and has been the focus of extensive research by pharmaceutical companies because of its role in the pathogenesis of Alzheimer’s disease [118, 119]. These GSIs function as pan-NOTCH inhibitors blocking the activity of all 4 NOTCH receptors. Early studies on the activity of GSIs as an anti NOTCH-therapy for T-ALL showed that treatment of T-ALL cell lines with these drugs resulted in rapid clearance of activated NOTCH1 protein and effective downregulation of NOTCH1 target genes [14, 45, 84, 98, 101]. Most notably, NOTCH inhibition reduced growth and proliferation by inducing G1 cell cycle arrest and decreasing cell size [14, 45, 66, 101]. Following these encouraging results, the Dana-Farber Cancer Institute performed a phase I clinical trial testing MK-0752, an oral GSI developed by Merck for the treatment of Alzheimer’s disease, in T-ALL patients [120]. Six adults and two children with leukemia (seven with T-ALL and one with AML) where enrolled in this study and four of the seven T-ALL patients showed activating mutations in NOTCH1. Treatment duration ranged from 2 to 56 days, and one patient with T-ALL and a NOTCH1 mutation achieved a 45% reduction in a mediastinal mass after 28 days. However, this patient subsequently progressed, and no patient achieved an objective response before discontinuation because of disease progression or drug-related toxicity [120]. The most common dose-limiting toxicity was grade 3/4 diarrhea, revealing an unfavorable toxicity profile most probably related to inhibition of NOTCH signaling in the gut. The development of gastrointestinal toxicity in the context of GSI therapy was not completely unanticipated and has emerged as a significant obstacle for the clinical development of these drugs. NOTCH1 and NOTCH2 play an important role in the intestinal epithelium, where they are involved in the control of cell proliferation and differentiation, and as noted above, GSIs are pan-NOTCH inhibitors that cause a systemic block of all 4 NOTCH receptors. Genetic inhibition of NOTCH signaling in the gut using animal models via deletion of the Rbpjk gene [121] or in the context of double Notch1/Notch2 conditional knockouts [122] induces cell cycle arrest and differentiation to secretory cell lineages at the expense of the absorptive epithelium; a phenotype that is recapitulated upon pharmacologic inhibition of the Notch pathway with GSIs [121, 123]. Overall, these results strongly suggest that alternative strategies with an improved therapeutic window may be needed for the successful implementation of GSIs as anti-NOTCH therapies in T-ALL. In this regard, a recent report from Merck has shown that three days of >70% Notch inhibition with a GSI is sufficient to induce effective antileukemic responses in T-ALL xenograft models and is well-tolerated [124]. A similar intermittent dosing approach has shown to reduce the toxicity associated with PF-03084014, a GSI developed by Pfizer [125]. These results illustrate that secretory metaplasia induced by GSIs is time- and dose-dependent and can be avoided using intermittent dosing schemes. An alternative approach to improve the safety and efficacy of anti-NOTCH therapies in T-ALL may result from the combined used of GSIs with chemotherapy or other molecularly targeted drugs. The idea is to use GSIs at high doses for short periods of time to avoid the development of gastrointestinal toxicity while using drug combinations that increase their antileukemic efficacy. Combination therapies of GSIs with CDK inhibitors [105], drugs targeting NFκB signaling [91], or small molecule inhibitors of CK2 [125] and the PI3K-AKT-mTOR pathway [66] [101, 126] have been shown to increase the antileukemic effects of these pan-NOTCH inhibitors. In addition, prolonged exposure to GSIs may increase the response to glucocorticoid treatment [127], and inhibition of NOTCH signaling with a GSI can sensitize glucocorticoid-resistant T-ALL cell lines to glucocorticoid-induced apoptosis [128]. Importantly, in vivo testing of GSIs and glucocorticoids in combination in a mouse model of glucocorticoid resistant T-ALL showed that glucocorticoid treatment has a direct protective effect against GSI-induced intestinal toxicity in mice [128, 129]. These results have now been confirmed and extended in a report showing that glucocorticoids abrogate the gastrointestinal toxicity induced by the GSI PF-03084014 and that delayed administration of glucocorticoids does not impair their protective effect against GSI-induced gut toxicity [130]. Overall, these results strongly suggest that glucocorticoid treatment may enhance the antileukemic effects of GSIs, while at the same time amelliorating the intestinal toxicity typically associated with systemic inhibition of NOTCH signaling [128].
Modulators of clinical response to GSI in T-ALL
Despite the prominent role of NOTCH1 in the pathogenesis of T-ALL, inhibition of NOTCH1 signaling seems to have only modest antileukemic effects against human T-ALL cell lines. Thus, inhibition of NOTCH signaling with GSIs is effective only in a fraction of these tumors and induces primarily a cytostatic effect [14, 45, 98], although it can also result in the induction of apoptosis in some instances [45, 124, 130]. In contrast, Notch-induced mouse T-ALLs seem to be more sensitive to inhibition of NOTCH signaling [97] [126]. Comparative analysis of GSI-sensitive and GSI-resistant T-ALL cell lines showed that GSI treatment can effectively decrease the level of active NOTCH1 protein and the expression of NOTCH1 target genes in both sensitive and resistant tumors [66, 98]. These results demonstrate that GSI resistance in T-ALL cell lines is not mediated by defects in drug uptake or impaired inhibition of the γ-secretase complex and suggests that human T-cell leukemia cell lines may have accumulated additional mutations that sustain leukemic cell growth and bypass the effects of NOTCH1 inhibition. Detailed molecular analysis of GSI-sensitive and GSI-resistant T-ALL cell lines showed a striking correlation between PTEN mutational status and GSI sensitivity, as all GSI-sensitive tumors were PTEN wild type while each of the GSI-resistant lines analyzed showed mutational loss of this tumor suppressor gene [66]. However, analyses of mouse models of NOTCH1-induced leukemias and primary T-ALL samples in culture suggest that additional mutations may be required to confer full resistance to GSI therapy [131]. Notably, FBXW7 mutations, which upregulate the expression of MYC, JUN and Cyclin E in addition to contributing to increased ICN1 stability, are also more prevalent in GSI-resistant T-ALL cell lines [91, 92]. As clinical trials testing the safety and efficacy of GSI therapy in T-ALL progress, it will be important to analyze the effect of these genetic alterations in the response to anti-NOTCH1 therapies.
New and emerging anti-NOTCH therapies
The limitations of GSIs in the clinic suggest that alternative strategies may be needed for the therapeutic targeting of NOTCH1 in T-ALL. One possibility resides in the use of synthetic peptides to block the NOTCH transcriptional complex directly in the cell nucleus. This approach would confer direct NOTCH inhibition and may have a more rapid inhibitory effect on NOTCH signaling in the cell than GSIs. Following this approach, Moellering and coworkers have recently shown that SAHM1, a cell-permeable, stabilized alpha-helical peptide targeting the protein-protein interface responsible for the recruitment of MAML1 into the NOTCH-CSL transactivation complex, can effectively block NOTCH signaling and has potent, NOTCH-specific antileukemic effects both in human T-ALL cell lines and in a mouse model of NOTCH1-induced T-ALL [132].
Finally, given that NOTCH proteins are surface molecules, specific antibodies could provide selective blocking of NOTCH1, specifically, while preserving the activity of the other three NOTCH family members. An elegant study by Wu and coworkers at Genentech has demonstrated that highly specialized antibodies can block NOTCH1 signaling by binding to and stabilizing the LNR-HD complex [133], locking the receptor in an “off” conformation. Notably, this anti-NOTCH1 antibody blocked leukemic cell growth in pre-clinical models and inhibited angiogenesis [133]. Moreover, the anti-Notch1 antibody did not affect the activity of Notch2, which precluded the development of overt gastrointestinal toxicity [133]. In a related study, Aste-Amézaga and coworkers showed that anti-NOTCH1 antibodies can block ligand-independent signaling driven by Notch1 receptors with diverse class I HD point mutations, the most common type of mutation found in T-ALL [134].
Conclusions and future directions
Aberrant activation of the NOTCH signaling pathway plays a central role in T-ALL, a tumor in which multiple oncogenic and tumor suppressor pathways coordinately disrupt the regulatory programs controlling cell proliferation, differentiation and survival in normal developing thymocytes. The introduction of anti-NOTCH therapies in clinical trials may result in urgently needed improvements in therapy, particularly for patients with primary refractory and relapsed disease. Detailed correlative studies analyzing the genetic background of the tumors treated as well as elucidation of the mechanisms that mediate response to therapy and the genetic events implicated in resistance and relapse will be instrumental in defining the next steps towards achieving novel, more effective targeted treatments in this disease.
Footnotes
Conflict of interest statement
Conflict of interest statement: The Ferrando lab is partially funded by sponsored research projects funded by Merck and Pfizer on the preclinical testing of anti-NOTCH therapies in T-ALL.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350(15):1535–48. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AJ, Horowitz MM, Pollock BH, Zhang MJ, Bortin MM, Buchanan GR, et al. Bone marrow transplants from HLA-identical siblings as compared with chemotherapy for children with acute lymphoblastic leukemia in a second remission. N Engl J Med. 1994;331(19):1253–8. doi: 10.1056/NEJM199411103311902. [DOI] [PubMed] [Google Scholar]
- 3.Biggs JC, Horowitz MM, Gale RP, Ash RC, Atkinson K, Helbig W, et al. Bone marrow transplants may cure patients with acute leukemia never achieving remission with chemotherapy. Blood. 1992;80(4):1090–3. [PubMed] [Google Scholar]
- 4.Dopfer R, Henze G, Bender-Gotze C, Ebell W, Ehninger G, Friedrich W, et al. Allogeneic bone marrow transplantation for childhood acute lymphoblastic leukemia in second remission after intensive primary and relapse therapy according to the BFM- and CoALL-protocols: results of the German Cooperative Study. Blood. 1991;78(10):2780–4. [PubMed] [Google Scholar]
- 5.Forman SJ, Schmidt GM, Nademanee AP, Amylon MD, Chao NJ, Fahey JL, et al. Allogeneic bone marrow transplantation as therapy for primary induction failure for patients with acute leukemia. J Clin Oncol. 1991;9(9):1570–4. doi: 10.1200/JCO.1991.9.9.1570. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder H, Gustafsson G, Saarinen-Pihkala UM, Glomstein A, Jonmundsson G, Nysom K, et al. Allogeneic bone marrow transplantation in second remission of childhood acute lymphoblastic leukemia: a population-based case control study from the Nordic countries. Bone Marrow Transplant. 1999;23(6):555–60. doi: 10.1038/sj.bmt.1701617. [DOI] [PubMed] [Google Scholar]
- 7.Marks DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993) Blood. 2009;114(25):5136–45. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pui CH, Pei D, Sandlund JT, Ribeiro RC, Rubnitz JE, Raimondi SC, et al. Long-term results of St Jude Total Therapy Studies 11, 12, 13A, 13B, and 14 for childhood acute lymphoblastic leukemia. Leukemia. 24(2):371–82. doi: 10.1038/leu.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrando AA, Look AT. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastic leukemia. Semin Hematol. 2000;37(4):381–95. doi: 10.1016/s0037-1963(00)90018-0. [DOI] [PubMed] [Google Scholar]
- 10.Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1(1):75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 11.Soulier J, Clappier E, Cayuela JM, Regnault A, Garcia-Peydro M, Dombret H, et al. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL) Blood. 2005;106(1):274–86. doi: 10.1182/blood-2004-10-3900. [DOI] [PubMed] [Google Scholar]
- 12.Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102(1):262–8. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- 13.Hebert J, Cayuela JM, Berkeley J, Sigaux F. Candidate tumor-suppressor genes MTS1 (p16INK4A) and MTS2 (p15INK4B) display frequent homozygous deletions in primary cells from T- but not from B-cell lineage acute lymphoblastic leukemias. Blood. 1994;84(12):4038–44. [PubMed] [Google Scholar]
- 14.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 15.Aster JC, Pear WS, Blacklow SC. Notch Signaling in Leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology Am Soc Hematol Educ Program. 2009:353–61. doi: 10.1182/asheducation-2009.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Keersmaecker K, Graux C, Odero MD, Mentens N, Somers R, Maertens J, et al. Fusion of EML1 to ABL1 in T-cell acute lymphoblastic leukemia with cryptic t(9;14)(q34;q32) Blood. 2005;105(12):4849–52. doi: 10.1182/blood-2004-12-4897. [DOI] [PubMed] [Google Scholar]
- 18.Finger LR, Kagan J, Christopher G, Kurtzberg J, Hershfield MS, Nowell PC, et al. Involvement of the TCL5 gene on human chromosome 1 in T-cell leukemia and melanoma. Proc Natl Acad Sci U S A. 1989;86(13):5039–43. doi: 10.1073/pnas.86.13.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begley CG, Aplan PD, Denning SM, Haynes BF, Waldmann TA, Kirsch IR. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc Natl Acad Sci U S A. 1989;86(24):10128–32. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Q, Cheng JT, Tasi LH, Schneider N, Buchanan G, Carroll A, et al. The tal gene undergoes chromosome translocation in T cell leukemia and potentially encodes a helix-loop-helix protein. Embo J. 1990;9(2):415–24. doi: 10.1002/j.1460-2075.1990.tb08126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernard O, Guglielmi P, Jonveaux P, Cherif D, Gisselbrecht S, Mauchauffe M, et al. Two distinct mechanisms for the SCL gene activation in the t(1;14) translocation of T-cell leukemias. Genes Chromosomes Cancer. 1990;1(3):194–208. doi: 10.1002/gcc.2870010303. [DOI] [PubMed] [Google Scholar]
- 22.Xia Y, Brown L, Yang CY, Tsan JT, Siciliano MJ, Espinosa R, III, et al. TAL2, a helix-loop-helix gene activated by the (7;9)(q34;q32) translocation in human T-cell leukemia. Proc Natl Acad Sci U S A. 1991;88(24):11416–20. doi: 10.1073/pnas.88.24.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellentin JD, Smith SD, Cleary ML. lyl-1, a novel gene altered by chromosomal translocation in T cell leukemia, codes for a protein with a helix-loop-helix DNA binding motif. Cell. 1989;58(1):77–83. doi: 10.1016/0092-8674(89)90404-2. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Jani-Sait SN, Escalon EA, Carroll AJ, de Jong PJ, Kirsch IR, et al. The t(14;21)(q11.2;q22) chromosomal translocation associated with T-cell acute lymphoblastic leukemia activates the BHLHB1 gene. Proc Natl Acad Sci U S A. 2000;97(7):3497–502. doi: 10.1073/pnas.97.7.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGuire EA, Hockett RD, Pollock KM, Bartholdi MF, O’Brien SJ, Korsmeyer SJ. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989;9(5):2124–32. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenberg JM, Boehm T, Sofroniew MV, Keynes RJ, Barton SC, Norris ML, et al. Segmental and developmental regulation of a presumptive T-cell oncogene in the central nervous system. Nature. 1990;344(6262):158–60. doi: 10.1038/344158a0. [DOI] [PubMed] [Google Scholar]
- 27.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci U S A. 1991;88(10):4367–71. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Royer-Pokora B, Loos U, Ludwig WD. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11) Oncogene. 1991;6(10):1887–93. [PubMed] [Google Scholar]
- 29.Warren AJ, Colledge WH, Carlton MB, Evans MJ, Smith AJ, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78(1):45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 30.Dube ID, Kamel-Reid S, Yuan CC, Lu M, Wu X, Corpus G, et al. A novel human homeobox gene lies at the chromosome 10 breakpoint in lymphoid neoplasias with chromosomal translocation t(10;14) Blood. 1991;78(11):2996–3003. [PubMed] [Google Scholar]
- 31.Hatano M, Roberts CW, Minden M, Crist WM, Korsmeyer SJ. Deregulation of a homeobox gene, HOX11, by the t(10;14) in T cell leukemia. Science. 1991;253(5015):79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- 32.Lu M, Gong ZY, Shen WF, Ho AD. The tcl-3 proto-oncogene altered by chromosomal translocation in T-cell leukemia codes for a homeobox protein. Embo J. 1991;10(10):2905–10. doi: 10.1002/j.1460-2075.1991.tb07840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy MA, Gonzalez-Sarmiento R, Kees UR, Lampert F, Dear N, Boehm T, et al. HOX11, a homeobox-containing T-cell oncogene on human chromosome 10q24. Proc Natl Acad Sci U S A. 1991;88(20):8900–4. doi: 10.1073/pnas.88.20.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen-Hagge TE, Schafer M, Kiyoi H, Morris SW, Whitlock JA, Koch P, et al. Disruption of the RanBP17/Hox11L2 region by recombination with the TCRdelta locus in acute lymphoblastic leukemias with t(5;14)(q34;q11) Leukemia. 2002;16(11):2205–12. doi: 10.1038/sj.leu.2402671. [DOI] [PubMed] [Google Scholar]
- 35.Nagel S, Kaufmann M, Drexler HG, MacLeod RA. The cardiac homeobox gene NKX2–5 is deregulated by juxtaposition with BCL11B in pediatric T-ALL cell lines via a novel t(5;14)(q35.1;q32.2) Cancer Res. 2003;63(17):5329–34. [PubMed] [Google Scholar]
- 36.Przybylski GK, Dik WA, Grabarczyk P, Wanzeck J, Chudobska P, Jankowski K, et al. The effect of a novel recombination between the homeobox gene NKX2-5 and the TRD locus in T-cell acute lymphoblastic leukemia on activation of the NKX2-5 gene. Haematologica. 2006;91(3):317–21. [PubMed] [Google Scholar]
- 37.Speleman F, Cauwelier B, Dastugue N, Cools J, Verhasselt B, Poppe B, et al. A new recurrent inversion, inv(7)(p15q34), leads to transcriptional activation of HOXA10 and HOXA11 in a subset of T-cell acute lymphoblastic leukemias. Leukemia. 2005;19(3):358–66. doi: 10.1038/sj.leu.2403657. [DOI] [PubMed] [Google Scholar]
- 38.Shima EA, Le Beau MM, McKeithan TW, Minowada J, Showe LC, Mak TW, et al. Gene encoding the alpha chain of the T-cell receptor is moved immediately downstream of c-myc in a chromosomal 8;14 translocation in a cell line from a human T-cell leukemia. Proc Natl Acad Sci U S A. 1986;83(10):3439–43. doi: 10.1073/pnas.83.10.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erikson J, Finger L, Sun L, ar-Rushdi A, Nishikura K, Minowada J, et al. Deregulation of c-myc by translocation of the alpha-locus of the T-cell receptor in T-cell leukemias. Science. 1986;232(4752):884–6. doi: 10.1126/science.3486470. [DOI] [PubMed] [Google Scholar]
- 40.Urashima M, Iyori H, Fujisawa K, Hoshi Y, Akatsuka J, Maekawa K. Establishment and characteristics of a T-cell acute lymphoblastic leukemia cell line, JK-T1, with a chromosomal translocation between 8q24 and 14q13. Cancer Genet Cytogenet. 1992;64(1):86–90. doi: 10.1016/0165-4608(92)90329-7. [DOI] [PubMed] [Google Scholar]
- 41.Inaba T, Murakami S, Oku N, Itoh K, Ura Y, Nakanishi S, et al. Translocation between chromosomes 8q24 and 14q11 in T-cell acute lymphoblastic leukemia. Cancer Genet Cytogenet. 1990;49(1):69–74. doi: 10.1016/0165-4608(90)90165-7. [DOI] [PubMed] [Google Scholar]
- 42.Shima-Rich EA, Harden AM, McKeithan TW, Rowley JD, Diaz MO. Molecular analysis of the t(8;14)(q24;q11) chromosomal breakpoint junctions in the T-cell leukemia line MOLT-16. Genes Chromosomes Cancer. 1997;20(4):363–71. doi: 10.1002/(sici)1098-2264(199712)20:4<363::aid-gcc7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 43.Clappier E, Cuccuini W, Kalota A, Crinquette A, Cayuela JM, Dik WA, et al. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 2007;110(4):1251–61. doi: 10.1182/blood-2006-12-064683. [DOI] [PubMed] [Google Scholar]
- 44.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66(4):649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 45.Palomero T, Barnes KC, Real PJ, Bender JL, Sulis ML, Murty VV, et al. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20(7):1279–87. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- 46.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by “illegitimate” V-(D)-J recombinase activity. Science. 1990;250(4986):1426–9. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 47.Van Vlierberghe P, van Grotel M, Beverloo HB, Lee C, Helgason T, Buijs-Gladdines J, et al. The cryptic chromosomal deletion del(11)(p12p13) as a new activation mechanism of LMO2 in pediatric T-cell acute lymphoblastic leukemia. Blood. 2006;108(10):3520–9. doi: 10.1182/blood-2006-04-019927. [DOI] [PubMed] [Google Scholar]
- 48.Lahortiga I, De Keersmaecker K, Van Vlierberghe P, Graux C, Cauwelier B, Lambert F, et al. Duplication of the MYB oncogene in T cell acute lymphoblastic leukemia. Nat Genet. 2007;39(5):593–5. doi: 10.1038/ng2025. [DOI] [PubMed] [Google Scholar]
- 49.O’Neil J, Tchinda J, Gutierrez A, Moreau L, Maser RS, Wong KK, et al. Alu elements mediate MYB gene tandem duplication in human T-ALL. J Exp Med. 2007;204(13):3059–66. doi: 10.1084/jem.20071637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996;93(10):4804–9. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlson KM, Vignon C, Bohlander S, Martinez-Climent JA, Le Beau MM, Rowley JD. Identification and molecular characterization of CALM/AF10fusion products in T cell acute lymphoblastic leukemia and acute myeloid leukemia. Leukemia. 2000;14(1):100–4. doi: 10.1038/sj.leu.2401629. [DOI] [PubMed] [Google Scholar]
- 52.Asnafi V, Radford-Weiss I, Dastugue N, Bayle C, Leboeuf D, Charrin C, et al. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood. 2003;102(3):1000–6. doi: 10.1182/blood-2002-09-2913. [DOI] [PubMed] [Google Scholar]
- 53.Chervinsky DS, Sait SN, Nowak NJ, Shows TB, Aplan PD. Complex MLL rearrangement in a patient with T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 1995;14(1):76–84. doi: 10.1002/gcc.2870140114. [DOI] [PubMed] [Google Scholar]
- 54.Rubnitz JE, Behm FG, Curcio-Brint AM, Pinheiro RP, Carroll AJ, Raimondi SC, et al. Molecular analysis of t(11;19) breakpoints in childhood acute leukemias. Blood. 1996;87(11):4804–8. [PubMed] [Google Scholar]
- 55.Van Vlierberghe P, van Grotel M, Tchinda J, Lee C, Beverloo HB, van der Spek PJ, et al. The recurrent SET-NUP214 fusion as a new HOXA activation mechanism in pediatric T-cell acute lymphoblastic leukemia. Blood. 2008;111(9):4668–80. doi: 10.1182/blood-2007-09-111872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussey DJ, Nicola M, Moore S, Peters GB, Dobrovic A. The (4;11)(q21;p15) translocation fuses the NUP98 and RAP1GDS1 genes and is recurrent in T-cell acute lymphocytic leukemia. Blood. 1999;94(6):2072–9. [PubMed] [Google Scholar]
- 57.Mecucci C, La Starza R, Negrini M, Sabbioni S, Crescenzi B, Leoni P, et al. t(4;11)(q21;p15) translocation involving NUP98 and RAP1GDS1 genes: characterization of a new subset of T acute lymphoblastic leukaemia. Br J Haematol. 2000;109(4):788–93. doi: 10.1046/j.1365-2141.2000.02106.x. [DOI] [PubMed] [Google Scholar]
- 58.Tycko B, Smith SD, Sklar J. Chromosomal translocations joining LCK and TCRB loci in human T cell leukemia. J Exp Med. 1991;174(4):867–73. doi: 10.1084/jem.174.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clappier E, Cuccuini W, Cayuela JM, Vecchione D, Baruchel A, Dombret H, et al. Cyclin D2 dysregulation by chromosomal translocations to TCR loci in T-cell acute lymphoblastic leukemias. Leukemia. 2006;20(1):82–6. doi: 10.1038/sj.leu.2404008. [DOI] [PubMed] [Google Scholar]
- 60.Karrman K, Andersson A, Bjorgvinsdottir H, Strombeck B, Lassen C, Olofsson T, et al. Deregulation of cyclin D2 by juxtaposition with T-cell receptor alpha/delta locus in t(12;14)(p13;q11)-positive childhood T-cell acute lymphoblastic leukemia. Eur J Haematol. 2006;77(1):27–34. doi: 10.1111/j.0902-4441.2006.t01-1-EJH2553.x. [DOI] [PubMed] [Google Scholar]
- 61.Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205(4):751–8. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36(10):1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 63.Bar-Eli M, Ahuja H, Foti A, Cline MJ. N-RAS mutations in T-cell acute lymphocytic leukaemia: analysis by direct sequencing detects a novel mutation. Br J Haematol. 1989;72(1):36–9. doi: 10.1111/j.1365-2141.1989.tb07648.x. [DOI] [PubMed] [Google Scholar]
- 64.Kawamura M, Ohnishi H, Guo SX, Sheng XM, Minegishi M, Hanada R, et al. Alterations of the p53, p21, p16, p15 and RAS genes in childhood T-cell acute lymphoblastic leukemia. Leuk Res. 1999;23(2):115–26. doi: 10.1016/s0145-2126(98)00146-5. [DOI] [PubMed] [Google Scholar]
- 65.Balgobind BV, Van Vlierberghe P, van den Ouweland AM, Beverloo HB, Terlouw-Kromosoeto JN, van Wering ER, et al. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111(8):4322–8. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- 66.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–10. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tosello V, Mansour MR, Barnes K, Paganin M, Sulis ML, Jenkinson S, et al. WT1 mutations in T-ALL. Blood. 2009;114(5):1038–45. doi: 10.1182/blood-2008-12-192039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 42(6):530–5. doi: 10.1038/ng.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Vlierberghe P, Palomero T, Khiabanian H, Van der Meulen J, Castillo M, Van Roy N, et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 42(4):338–42. doi: 10.1038/ng.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–16. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 71.Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228(2):151–65. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 72.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 73.van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284(45):31018–27. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009;29(21):5679–95. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nat Immunol. 2007;8(5):451–6. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 76.Hozumi K, Mailhos C, Negishi N, Hirano K, Yahata T, Ando K, et al. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008;205(11):2507–13. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–45. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 78.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–58. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 79.Pui JC, Allman D, Xu L, DeRocco S, Karnell FG, Bakkour S, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11(3):299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 80.Ciofani M, Zuniga-Pflucker JC. A survival guide to early T cell development. Immunol Res. 2006;34(2):117–32. doi: 10.1385/IR:34:2:117. [DOI] [PubMed] [Google Scholar]
- 81.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta Lineage Thymocytes. Immunity. 2002;16(6):869–79. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 82.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16(3):295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gonzalez-Garcia S, Garcia-Peydro M, Martin-Gayo E, Ballestar E, Esteller M, Bornstein R, et al. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7R{alpha} gene expression in early human thymopoiesis and leukemia. J Exp Med. 2009;206(4):779–91. doi: 10.1084/jem.20081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20(15):2096–109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6(9):881–8. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 86.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183(5):2283–91. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Girard L, Hanna Z, Beaulieu N, Hoemann CD, Simard C, Kozak CA, et al. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10(15):1930–44. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 88.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26(12):4642–51. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sulis ML, Williams O, Palomero T, Tosello V, Pallikuppam S, Real PJ, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112(3):733–40. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113(18):4381–90. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204(8):1825–35. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O’Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204(8):1813–24. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67(19):9006–12. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 94.Minella AC, Clurman BE. Mechanisms of tumor suppression by the SCF(Fbw7) Cell Cycle. 2005;4(10):1356–9. doi: 10.4161/cc.4.10.2058. [DOI] [PubMed] [Google Scholar]
- 95.Fujii Y, Yada M, Nishiyama M, Kamura T, Takahashi H, Tsunematsu R, et al. Fbxw7 contributes to tumor suppression by targeting multiple proteins for ubiquitin-dependent degradation. Cancer Sci. 2006;97(8):729–36. doi: 10.1111/j.1349-7006.2006.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321(5895):1499–502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118(9):3181–94. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103(48):18261–6. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Margolin AA, Palomero T, Sumazin P, Califano A, Ferrando AA, Stolovitzky G. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc Natl Acad Sci U S A. 2009;106(1):244–9. doi: 10.1073/pnas.0806445106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma VM, Calvo JA, Draheim KM, Cunningham LA, Hermance N, Beverly L, et al. Notch1 contributes to mouse T-cell leukemia by directly inducing the expression of c-myc. Mol Cell Biol. 2006;26(21):8022–31. doi: 10.1128/MCB.01091-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110(1):278–86. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sade H, Krishna S, Sarin A. The anti-apoptotic effect of Notch-1 requires p56lck-dependent, Akt/PKB-mediated signaling in T cells. J Biol Chem. 2004;279(4):2937–44. doi: 10.1074/jbc.M309924200. [DOI] [PubMed] [Google Scholar]
- 103.Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113(8):1689–98. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dohda T, Maljukova A, Liu L, Heyman M, Grander D, Brodin D, et al. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res. 2007;313(14):3141–52. doi: 10.1016/j.yexcr.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 105.Rao SS, O’Neil J, Liberator CD, Hardwick JS, Dai X, Zhang T, et al. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009;69(7):3060–8. doi: 10.1158/0008-5472.CAN-08-4295. [DOI] [PubMed] [Google Scholar]
- 106.Sicinska E, Aifantis I, Le Cam L, Swat W, Borowski C, Yu Q, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4(6):451–61. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 107.Song LL, Peng Y, Yun J, Rizzo P, Chaturvedi V, Weijzen S, et al. Notch-1 associates with IKKalpha and regulates IKK activity in cervical cancer cells. Oncogene. 2008;27(44):5833–44. doi: 10.1038/onc.2008.190. [DOI] [PubMed] [Google Scholar]
- 108.Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, et al. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006;25(1):129–38. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13(1):70–7. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 110.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108(4):1151–7. doi: 10.1182/blood-2005-12-4956. [DOI] [PubMed] [Google Scholar]
- 111.Park MJ, Taki T, Oda M, Watanabe T, Yumura-Yagi K, Kobayashi R, et al. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J Haematol. 2009;145(2):198–206. doi: 10.1111/j.1365-2141.2009.07607.x. [DOI] [PubMed] [Google Scholar]
- 112.Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113(17):3918–24. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- 113.van Grotel M, Meijerink JP, Beverloo HB, Langerak AW, Buys-Gladdines JG, Schneider P, et al. The outcome of molecular-cytogenetic subgroups in pediatric T-cell acute lymphoblastic leukemia: a retrospective study of patients treated according to DCOG or COALL protocols. Haematologica. 2006;91(9):1212–21. [PubMed] [Google Scholar]
- 114.Mansour MR, Sulis ML, Duke V, Foroni L, Jenkinson S, Koo K, et al. Prognostic implications of NOTCH1 and FBXW7 mutations in adults with T-cell acute lymphoblastic leukemia treated on the MRC UKALLXII/ECOG E2993 protocol. J Clin Oncol. 2009;27(26):4352–6. doi: 10.1200/JCO.2009.22.0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zuurbier L, Homminga IC, te Winkel V, Buijs-Gladdines ML, Kooi J, Smits C, Sonneveld WK, Veerman E, Kamps AJP, Horstmann WA, Petricoin M, Pieters EF, Meijerink R, JPP NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell Acute Lymphoblastic Leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010 doi: 10.1038/leu.2010.204. in press. [DOI] [PubMed] [Google Scholar]
- 116.Clappier EC, Grardel S, Girard N, Suarez S, Brunie L, Kaltenbach G, Yakouben S, Mazingue K, Robert F, Boutard A, Plantaz P, Rohrlich D, van Vlierberghe P, Preudhomme P, Otten C, Speleman J, Dastugue F, Suciu N, Benoit S, Bertrand Y, Cave YH. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment but not on outcome in children with T-cell acute lymphoblastic (T-ALL) leukemia treated on EORTC trials 58881 and 58951. Leukemia. 2010 doi: 10.1038/leu.2010.205. in press. [DOI] [PubMed] [Google Scholar]
- 117.Kox KZ, Stanulla M, Leibe M, Schrappe S, Ludwig M, Koehler WD, Tolle R, Bandapalli G, Breit OR, Muckenthaler S, Kulozik MUAE. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010 doi: 10.1038/leu.2010.203. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Evin G, Sernee MF, Masters CL. Inhibition of gamma-secretase as a therapeutic intervention for Alzheimer’s disease: prospects, limitations and strategies. CNS Drugs. 2006;20(5):351–72. doi: 10.2165/00023210-200620050-00002. [DOI] [PubMed] [Google Scholar]
- 119.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–97. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 120.Deangelo D, Stone R, Silverman L, Stock W, Attar E, Fearen I, et al. A phase I clinical trial of the notch inhibitor MK-0752 in patients with T-cell acute lymphoblastic leukemia/lymphoma (T-ALL) and other leukemias. Journal of Clinical Oncology; 2006 ASCO Annual Meeting Proceedings Part I; 2006. p. 6585. [Google Scholar]
- 121.van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–63. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 122.Riccio O, van Gijn ME, Bezdek AC, Pellegrinet L, van Es JH, Zimber-Strobl U, et al. Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27(Kip1) and p57(Kip2) EMBO Rep. 2008;9(4):377–83. doi: 10.1038/embor.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Milano J, McKay J, Dagenais C, Foster-Brown L, Pognan F, Gadient R, et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82(1):341–58. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- 124.Tammam J, Ware C, Efferson C, O’Neil J, Rao S, Qu X, et al. Down-regulation of the Notch pathway mediated by a gamma-secretase inhibitor induces anti-tumour effects in mouse models of T-cell leukaemia. Br J Pharmacol. 2009;158(5):1183–95. doi: 10.1111/j.1476-5381.2009.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 17(4):400–11. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113(24):6172–81. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.De Keersmaecker K, Lahortiga I, Mentens N, Folens C, Van Neste L, Bekaert S, et al. In vitro validation of gamma-secretase inhibitors alone or in combination with other anti-cancer drugs for the treatment of T-cell acute lymphoblastic leukemia. Haematologica. 2008;93(4):533–42. doi: 10.3324/haematol.11894. [DOI] [PubMed] [Google Scholar]
- 128.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15(1):50–8. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Real PJ, Ferrando AA. NOTCH inhibition and glucocorticoid therapy in T-cell acute lymphoblastic leukemia. Leukemia. 2009;23(8):1374–7. doi: 10.1038/leu.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wei P, Walls M, Qiu M, Ding R, Denlinger RH, Wong A, et al. Evaluation of selective gamma-secretase inhibitor PF-03084014 for its antitumor efficacy and gastrointestinal safety to guide optimal clinical trial design. Mol Cancer Ther. 9(6):1618–28. doi: 10.1158/1535-7163.MCT-10-0034. [DOI] [PubMed] [Google Scholar]
- 131.Medyouf H, Gao X, Armstrong F, Gusscott S, Liu Q, Gedman AL, et al. Acute T-cell leukemias remain dependent on Notch signaling despite PTEN and INK4A/ARF loss. Blood. 115(6):1175–84. doi: 10.1182/blood-2009-04-214718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–8. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 464(7291):1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 134.Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 5(2):e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]