Abstract
Although mitochondria are essential organelles for long-term survival of eukaryotic cells, recent discoveries in biochemistry and genetics have advanced our understanding of the requirements for mitochondria in cell death. Much of what we understand about cell death is based on the identification of conserved cell death genes in Drosophila melanogaster and Caenorhabditis elegans. However, the role of mitochondria in cell death in these models has been much less clear. Considering the active role that mitochondria play in apoptosis in mammalian cells, the mitochondrial contribution to cell death in non-mammalian systems has been an area of active investigation. In this article, we review the current research on this topic in three non-mammalian models, C. elegans, Drosophila and Saccharomyces cerevisiae. In addition, we discuss how non-mammalian models have provided important insight into the mechanisms of human disease as they relate to the mitochondrial pathway of cell death. The unique perspective derived from each of these model systems provides a more complete understanding of mitochondria in programmed cell death.
Keywords: Apoptosis, Mitochondria, C. elegans, Drosophila, Yeast
I. Introduction
Apoptosis and other forms of programmed cell death have been investigated for the past century using genetically tractable non-mammalian eukaryotes such as Drosophila melanogaster and Caenorhabditis elegans. These studies have facilitated the characterization of a large number of conserved genes and signaling networks. Most of these networks are at least partially conserved in mammals, including those that regulate programmed cell death. Apoptosis is by far the best studied of the cell death mechanisms. Caspases, the critical effector proteases of apoptosis, were first identified in a screen for C. elegans mutants lacking developmental programmed cell death [1]. Sequence similarity between the worm CED-3 gene and a mammalian protein (ICE, Caspase-1) assigned protease function to this gene [2]. Similar to the orthologous CED-9, Bcl-2 was shown to promote cell survival [3]. The inhibitor of apoptosis (IAP) proteins were originally identified as cell death inhibitors in baculoviruses that infect insect cells, and were later found to be essential regulators of apoptosis in Drosophila [4, 5] and mammals [6, 7]. The central proapoptotic regulators in Drosophila, Reaper, Hid and Grim, were found to antagonize IAP-mediated caspase inhibition [8–12]. In addition to the fundamental contribution of non-mammalian models to apoptotic cell death, genetic screens for mutants affecting autophagy, primarily in S. cerevisiae, have identified and partially characterized most of genes involved in this process [13]. Only recently have the autophagy pathways become avidly studied in mammals [14] or shown to contribute to cell death [15]. Studies in multiple organisms have provided valuable insight into cell death mechanisms that can only come from viewing a problem from multiple perspectives. However there appears to be a dichotomy regarding the role of mitochondria as a central component of the apoptotic machinery, between mammals and other eukaryotes [16], and even between different non-mammalian eukaryotes [17].
In mammalian systems, mitochondria are intimately involved in the intrinsic cell death pathway. The pro-apoptotic Bcl-2 members BAX and BAK, BID, and others, initiate the mitochondrial cell death pathway by permeabilizing the mitochondrial outer membrane. Pro-apoptotic factors released from the mitochondria include Cytochrome c (Cyt-c), which binds to the adaptor protein Apaf-1 causing activation of Caspase-9 and downstream caspases. In addition to the release of Cyt-c, there are additional mitochondrial alterations that may contribute to cell death in mammals. These include disruption of electron transport, oxidative phosphorylation, and adenosine triphosphate (ATP) production, release of other proteins that trigger caspase activation, and changes of cellular reduction-oxidation (redox) potential [18]. Additionally, these organelles undergo significant ultrastructural changes during apoptosis. Considering the active role that mitochondria play in apoptosis in mammalian cells, they have become the prime suspects in the search for additional apoptotic roles non-mammalian eukaryotes. As mammalian cells have been found to die by potentially multiple mechanisms other than apoptosis, there is a growing need to explore the cell death mechanisms of multiple model organisms to develop a complete understanding of genetically controlled cell suicide. In this review, we discuss the current research on the role of mitochondria in programmed cell death in three non-mammalian models, C. elegans, Drosophila and yeast. Understanding the similarities and differences in these models will help identify the conserved functions of mitochondria in cell death.
II. Cell death pathways in non-mammalian models: Mitochondria as a docking site
In C. elegans, most cell death events are under the control of the central apoptotic pathway composed of the BH3-only protein EGL-1, the Bcl-2-like protein CED-9, the Apaf-1-like protein CED-4 and the caspase CED-3 [19]. In healthy cells, CED-9 sequesters CED-4 to mitochondrial membranes [20–23]. In cells destined to die, binding of EGL-1 to CED-9 disrupts its interaction with CED-4 [21, 24, 25]. Released CED-4 forms an octamer, translocates to perinuclear membranes, and activates the processing of CED-3, thereby initiating CED-3-dependent apoptosis [21, 26–29] (Figure 1B).
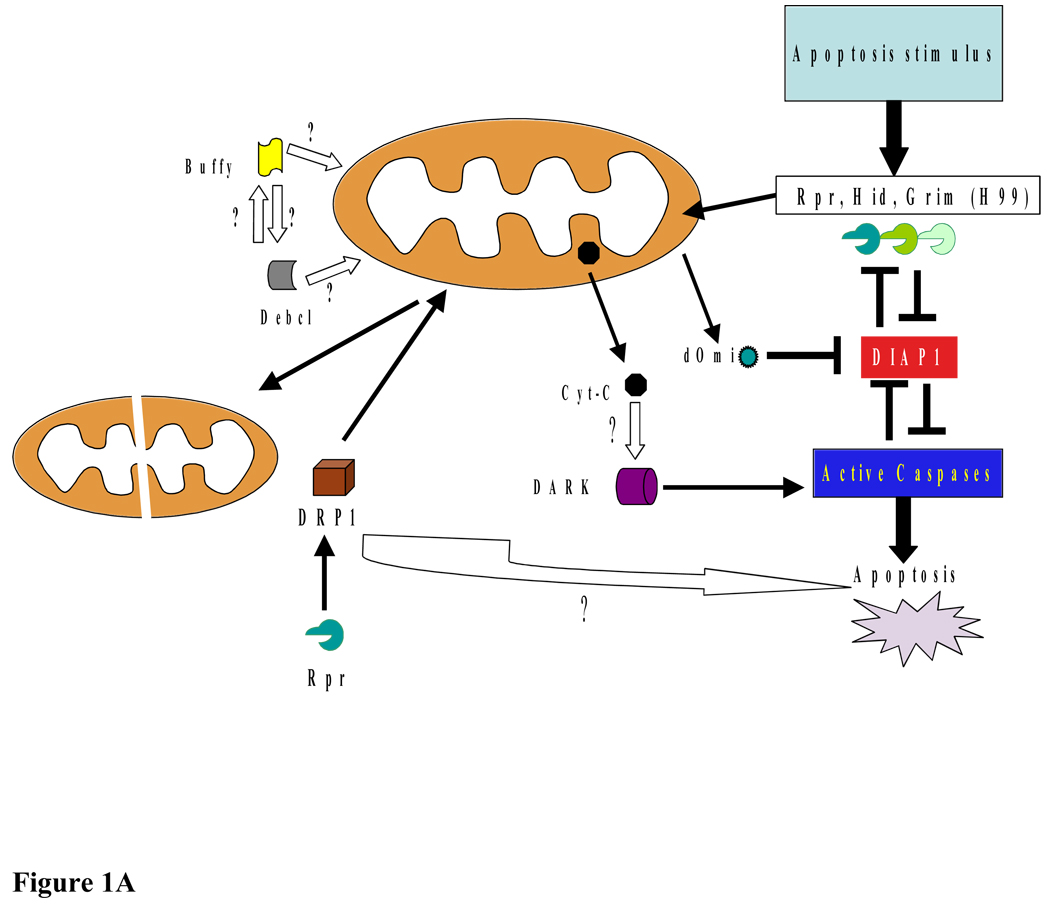
Figure 1.
A: Mitochondria and apoptosis in Drosophila. Cell death in Drosophila is activated by Rpr, Hid, Grim, and Skl, which bind to DIAP1 and inhibit DIAP1’s caspase inhibitory function. The role of cytochrome c in Drosophila caspase activation is not well characterized, however, a role for the mitochondria in the apoptotic process is suggested by the several findings, including Rpr, Grim, and Hid localization to the mitochondria. On initiation of apoptosis, Rpr and Hid are transcribed and rapidly localize to mitochondria, resulting in changes in mitochondrial ultrastructure and Cyt-c release. Drp 1-dependent mitochondrial disruption appears to be required for Drosophila apoptosis. Upon apoptosis induction, dOmi is released into the cytosol, where it binds DIAP1 causing caspase activation. The role of Drosophila Bcl-2-like proteins Buffy and Debcl in fly apoptosis remains unclear. Additionally, despite a requirement of Dark for caspase activation, the role of Drosophila apoptosome in caspase activation is not well understood.
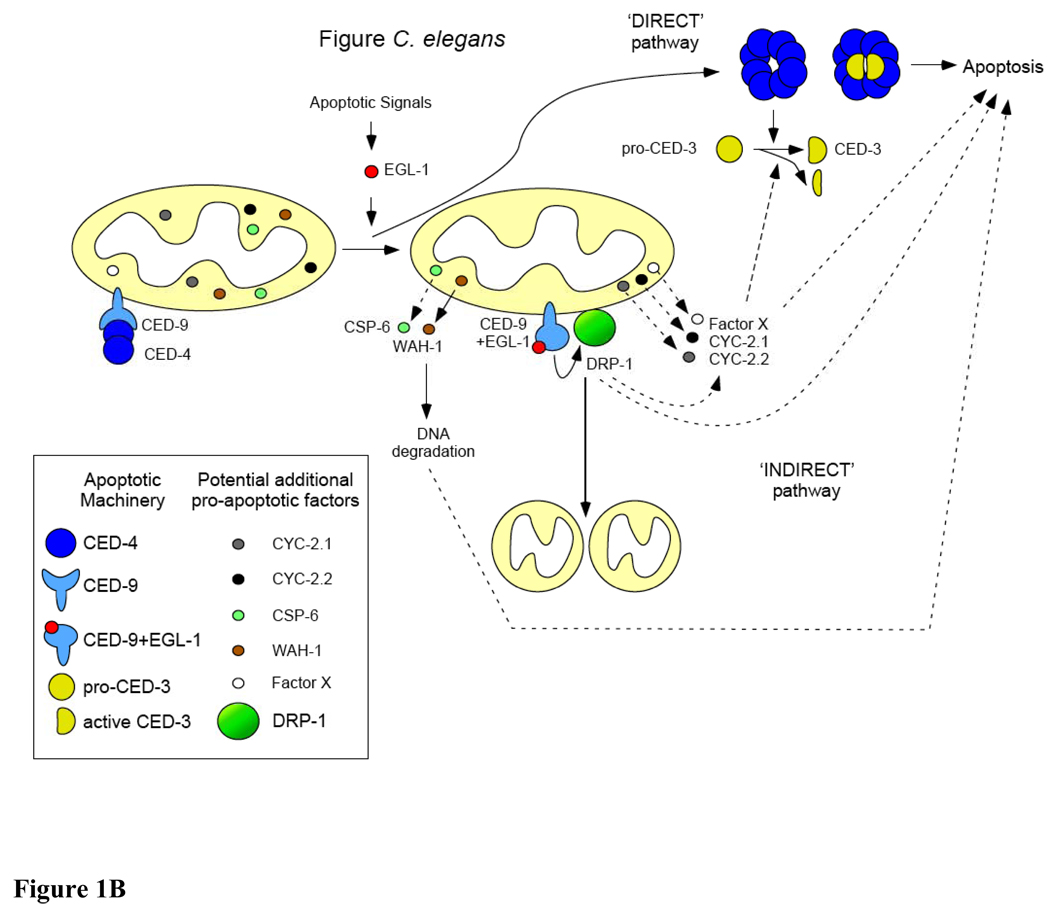
B: Mitochondria and apoptosis in C. elegans. Two apoptotic pathways are at work in C. elegans. During apoptosis, the pro-apoptotic BH3-only protein EGL-1 binds to the anti-apoptotic Bcl-2-like protein CED-9, thereby releasing the pro-apoptotic Apaf-1 like protein CED-4. Released CED-4 oligomerizes and promotes the processing of pro-caspase CED-3. CED-4 and active CED-3 form a holoenzyme that triggers apoptosis (‘DIRECT’ pathway). In a parallel pathway, when bound to EGL-1, CED-9 adopts a pro-apoptotic conformation and activates the dynamin-related protein DRP1. Active DRP1 promotes mitochondrial fragmentation and apoptosis by enhancing CED-3 processing or by acting in parallel to CED-3. The pro-apoptotic role of DRP1 could involve the release of potential pro-apoptotic factors (such as Cyt-c (CYC-2.1 or CYC-2.2) or an unknown factor X) but seems to be unrelated to its role in mitochondrial fission (‘INDIRECT’ pathway). Later during apoptosis, the endonuclease CSP-6 and the AIF homolog WAH-1 are released from mitochondria to degrade nuclear DNA, thereby participating in the dismantlement of the cell.
In addition to this ‘direct’ pathway of apoptosis induction, several lines of evidence suggest the existence of an ‘indirect’ pathway involving additional pro-apoptotic factors [30]. First, the inhibition of cell death seen in a weak ced-3 loss-of-function mutation, is enhanced by the loss of ced-9 function [31]. Hence, CED-9 has both anti-apoptotic and proapoptotic functions. In healthy cells CED-9 sequesters CED-4, but in dying cells CED-9 also has a pro-apoptotic function, as well as non-apoptotic functions related to mitochondrial dynamics (see section V). Second, some of the CED-4 protein localizes to the perinuclear membranes at permissive temperature in a ced-9 temperature-sensitive mutant (ced-9 (n1653ts)) without causing apoptosis [21]. Even though it cannot be excluded that the level of perinuclear CED-4 in this mutant is below a pro-apoptotic threshold, this experiment suggests that release of CED-4 from mitochondria is not sufficient to initiate apoptosis. This would leave a room for an CED-4 activation step potentially analogous to Cyt-c activation of mammalian Apaf-1 leading to caspase activation (See section IV).
In Drosophila, the pro-apoptotic regulators Reaper (Rpr), Hid, Grim and Sickle (known collectively as RGH proteins) regulate apoptosis [8, 32–34]. These factors induce apoptosis by binding to the Drosophila Inhibitor of Apoptosis Protein 1 (DIAP1), thereby inhibiting DIAP1 function, and this is followed by the activation of the effector caspases [16]. DIAP1 is the key anti-apoptotic molecule in Drosophila that ensures cell survival by inhibiting caspase activity through direct binding and (and/or) by ubiquitylation of caspases and other pro-apoptotic proteins [35]. Dronc, a CED-3/Caspase-9-like apical caspase is required for most developmental apoptosis [36]. Dronc associates with the Apaf-1/CED-4-like protein Dark, and becomes activated, in turn activating the effector caspases [37–39]. Current models suggest that Dronc activation is constitutive in most cells [17] (Figure 1A).
Yeast are a more recent addition to the list of model organisms used to study programmed cell death, and the topic of yeast cell death remains at the fringes in both the cell death field and the yeast genetics field. One of the hurdles to overcome in the study of yeast cell death was the long-standing assumption that single-cell species have no need of cell suicide pathways, and therefore lack evolutionary pressures to develop such mechanisms. However, this notion has been challenged and more recently overturned, in part due to revision of evolutionary theories regarding group selection. The yeast cell death field was further challenged by the difficult task of establishing experimental models to compellingly demonstrate that yeast have a purpose in dying in the absence of multicellular development. Nevertheless, several intriguing approaches have provided important insight, including evidence that yeast cell death plays a role in responding to viruses [40, 41]. In addition, the relationship between young and old yeast cells in a colony, which presumably reflects physiologically relevant events in the environment, have revealed that the death of old cells in a colony is essential to sustain younger cells, and some of the genes and signaling molecules required for this process have been identified [42, 43]. Analogous studies in liquid cultures have suggested that programmed cell suicide by a large portion of the population ensures long-term survival of the species, as cultures of cell death-resistant strains apparently consume the dwindling nutrient supply and all cells ultimately die [44].
Definitions of programmed cell death and the cell death nomenclature applied to yeast cell death further contribute to the controversies. For example, apoptosis has been applied to mean programmed cell death in general, but can strictly refer to caspase-dependent cell death [45, 46]. The existence of cell death pathways in yeast that resemble in part mammalian cell death pathways is supported by the fact that yeast encode several homologues of mammalian cell death regulators, but they lack many others. Yeast appear to lack the conserved caspase-activating apoptosome found in mammals, flies and worms, as yeast lack both an Apaf-1-like factor and a true caspase. However, yeast do encode a metacaspase Mca1/Yca1 that is related to plant metacaspases and appears to contribute importantly to yeast cell death induced by a multiple (but not all) stimuli based on genetic studies [47]. It is unclear if this yeast metacaspase, which lacks the Asp-cleaving activity characteristic of caspases, might have additional caspase-like functions. Thus far, the closest analogy is the identification of a conserved protein in humans (p100/SND1) and plants (TSN/Tudor staphylococcal nuclease) that is a substrate of human caspase-3 and of a plant metacaspase, respectively, during cell death [48]. Yeast also lack sequence similarity to Bcl-2 family proteins, although the existence of structural or functional equivalents in yeast cannot be ruled out. For example, several Bcl-2-like viral proteins that lack amino acid sequence similarity to Bcl-2 have been identified by 3-dimensional structure determination (e.g. 9 different ORFs in the vaccine strain of vaccinia virus) [49]. Although yeast lack recognizable homologues of the Drosophila IAP inhibitors Reaper, Grim and Hid, they do encode a protein containing an N-terminal BIR domain analogous to IAP proteins. Yeast Bir1 has been reported to protect yeast from cell death, but the detailed mechanisms have not been delineated [50]. Despite the prominent differences between the molecular machinery of yeast cell death compared to other species, mounting evidence supports the existence of gene-dependent death mechanisms in yeast. New evidence for the existence of potentially many non-apoptotic cell death mechanisms in mammals increases the likelihood that some of these pathways are represented in yeast. For example, autophagy has been extensively studied in yeast, though little it known about the role of autophagy in yeast cell death. Yeast as a model system to study programmed cell death is still catching up to flies and worms, but these are early days.
In both flies and worms, there is strong evidence for a role for mitochondria as a docking site for proteins involved in cell death. In C. elegans, the CED-9/CED-4 complex is localized to mitochondrial membranes in healthy cells, and disruption of this complex by EGL-1 results in cell death [20–25]. In Drosophila, Rpr, Grim, and Hid have been found to localize to mitochondria upon expression in Drosophila S2 cells, and this localization is important for mitochondrial permeabilization and efficient apoptosis activation [51–54]. The GH3 domain, an amphipathic helix conserved between Grim, Rpr and Skl, is required for mitochondrial localization, possibly through associate with Hid, which has a hydrophobic C-terminal mitochondrial targeting sequence [55]. In the absence of the Rpr GH3 domain, the IAP inhibitory activity of Rpr is preserved, but killing activity, and IAP degradation are inhibited [52, 53]. One possible interpretation of these data is that Rpr brings DIAP1 to a degradation complex localized on the mitochondria [53, 56]. Therefore, the GH3 domain appears to be critical for both Rpr mitochondrial localization and IAP destabilization. This indicates that mitochondrial localization maybe important for Rpr killing by DIAP1 destabilization, and not just DIAP1 inhibition [53]. In yeast, the pro-death function of Dnm1 (Drp1) appears to occur at mitochondria [57], and yeast AIF has been reported to translocate from mitochondria to the nucleus during death [58], but the subcellular localization of most cell death regulators in yeast are not yet known.
III. The role of mitochondrial permeabilization
Mitochondrial outer membrane permeabilization (MOMP) has been proposed as a “point of no return” of the mitochondrial apoptotic cascade. In mammalian cells it occurs in response to various apoptotic stimuli that cause the release of Cyt-c and other pro-apoptotic proteins from the intermembrane space of mitochondria into the cytoplasm [59]. It is a rapid concerted process and affects most, if not all, mitochondria and can occur in response to various stimuli and in different cell types with or without caspase activity [60]. Unlike in C. elegans, Bcl-2-family proteins in mammalian systems do not directly regulate Apaf-1 but instead regulate (positively and negatively) the earlier steps leading to release of Cyt-c from the mitochondria [61, 62].
The role of the Drosophila Bcl-2 proteins in mitochondrial permeabilization and cell death has not been yet fully characterized. In Drosophila, there are two homologs of the Bcl-2/CED-9 family proteins, Debcl/dBorg-1/dRob-1 and Buffy/dBorg-2 [63–65]. Both Debcl and Buffy share the BH1, BH2, BH3 and C-terminal transmembrane domains of the Bcl-2 family of proteins [66, 67]. Debcl has been shown to localize to mitochondria, while Buffy localizes to the endoplasmic reticulum [67]. Overexpression and knockdown studies suggest that Debcl has a pro-apoptotic function [63, 68, 69] while Buffy is antiapoptotic [70, 71]. However, a recent study showed that loss of Buffy can cause microchaete glial cell survival and blocks Grim induced cell death in the eye, suggesting a pro-apoptotic role for Buffy [72]. Moreover, Debcl has been suggested to protect against poly-glutamine induced death in the nervous system, which is antagonized by Buffy expression [73]. This situation in flies is potentially analogous to the mammalian nervous system where pro-apoptotic mammalian Bcl-2 proteins can exhibit potent anti-death activity [74, 75]. Debcl appears to have a limited role in Drosophila developmental apoptosis. However, a recent study showed that Debcl can be effectively recruited for killing by mammalian pro-apoptotic Bcl-2 gene family members [68]. Debcl mutations appear to reduce irradiation-induced cell death in the embryo [76], but they do not affect irradiation-induced cell death in the wing, despite increased morphological defects [68]. Gene disruption studies suggest that Debcl is required to inhibit the antiapoptotic function of Buffy [76]. Surprisingly, loss of Debcl does not appear to cause any defects in mitochondrial organization in living cells or in mitochondrial fragmentation in dying cells. In contrast, Buffy expression was shown to protect cells from mitochondrial changes and apoptosis caused by loss of function of Drosophila PTEN-induced kinase (Pink1) [71]. In summary, controversies persist regarding the specific functions of fly Bcl-2-like proteins (Buffy and Debcl) in cell death. Additional functional studies in vitro and in vivo are required to characterize the importance of these proteins in Drosophila apoptosis.
Although further studies may demonstrate a role for the Drosophila Bcl-2 proteins in regulating mitochondrial changes in apoptosis, it is also possible that other proteins such as Rpr, Grim, and Hid have taken over this function in flies. Drosophila mitochondria are rapidly permeablized when Rpr or Hid are expressed, both in cultured S2 cells and in vivo [51]. Mitochondrial permeabilization was also seen during DNA damage-induced apoptosis in fly embryos. This permeabilization requires RHG gene function. These data indicate that Rpr and Hid are both necessary and sufficient for mitochondrial permeabilization in Drosophila. Interestingly the GH3 domain of Rpr and Grim appears to have a pro-apoptotic activity that is independent of IAP inhibition and Caspase activation, suggesting that this domain might play a role in mitochondrial permeabilization and cell death [52, 77].
Little is known about mitochondrial permeabilization during C. elegans apoptosis. The only protein shown to be released from mitochondria in apoptotic cells is the AIF homolog, WAH-1 [78]. However, as discussed below, this release seems to be a late apoptotic event. In addition, WAN-1, one of four adenine nucleotide translocators (ANT) in C. elegans, has been shown to have pro-apoptotic activity [79]. In mammals, ANT is thought to participate in the formation of the Permeability Transition Pore (PTP) in the inner-mitochondrial membrane, although this is still under debate. The opening of the PTP during apoptosis has been proposed to cause mitochondrial swelling, rupture of the outer-mitochondrial membrane and release of pro-apoptotic factors localized in the inter membrane space [80]. Similarly, the mechanism by which WAN-1 exerts its pro-apoptotic activity in C. elegans remains unclear. Surprisingly, WAN-1 has been shown, by co-immunoprecipitation, to interact with CED-9 and CED-4 and these interactions have been shown to be disrupted in the presence of EGL-1 [79]. However, it is unclear how CED-9 and CED-4, which are both localized on the surface of mitochondria, can interact with an integral protein of the inner-mitochondrial membrane. In addition, there is no evidence that WAN-1 participates in the permeabilization of C. elegans mitochondria during apoptosis. Therefore, the role of WAN-1 in C. elegans apoptosis remains unclear.
In yeast, Cyt-c release from mitochondria during cell death has been observed and suggested to promote cell death, but a causal role is not firmly established [81, 82]. Given the absence of BAX-related proteins in yeast, the mechanism of Cyt-c release in yeast is unclear. The conserved mitochondrial protein AAC/ANT (ADP/ATP carrier) has been reported to be required for Cyt-c release in yeast and cell death induced by acetic acid but not H2O2 [83]. Release of yeast Pep4, homologue of mammalian Cathepsin D, from vacuoles during yeast cell death may play a role as well [84]. While intriguing, these events and their role in executing the yeast cell death program have not been fully investigated.
IV. Release of pro-apoptotic factors from mitochondria
In mammals, the release of Cyt-c from intermembrane space of the mitochondria into the cytoplasm occurs in response to a variety of pro-apoptotic stimuli and is regulated by the Bcl-2 family proteins [18, 61, 62]. Formation of the mammalian apoptosome, and subsequent caspase activation, requires Cyt c binding to Apaf-1 [85]. Several key mitochondrial changes appear to be required for complete Cyt-c release including changes in mitochondrial ultrastructure, mitochondrial permeability, and membrane potential [86]. In addition to Cyt-c, proteins released from mitochondria during mammalian apoptosis include: Apoptosis Inducing Factor (AIF), ARTS, SMAC/Diablo and Omi/HTRA2 [87–89].
In recent years several lines of evidence have supported a role for mitochondrial proteins in Drosophila apoptosis. Caspases in Drosophila S2 cell extracts were shown to be activated by mitochondria from apoptotic S2 cells, suggesting that mitochondrial factors contribute to caspase activation [90]. By contrast, other studies, using cell-free extracts, have found that mitochondrial lysates from apoptotic cells are not able to induce or accelerate apoptosis in S2 extracts [91]. It is important to note that these studies have used different apoptosis inducers, for example expression of exogenous proapoptotic proteins vs.UV, which may have differing effects on mitochondria [51].
Whether Cyt-c is released from mitochondria in Drosophila apoptosis has also been a source of controversy. Earlier studies by Varkey et al. reported exposure of a Cyt-c epitope in apoptotic cells [90]. However, this epitope remains punctate, and the data were interpreted to suggest that Cyt C was still localized to mitochondria. However, newer studies demonstrate that Cyt-c becomes rapidly diffuse in dying cells [51]. These differences may be due to alterations in the antibody staining procedure or differences in antibody epitopes. Subcellular fractionation studies have also provided conflicting evidence for Cyt-c release [37, 51, 90, 92–94]. Again, these apparent contradictions may be due to technical differences. Nevertheless, unlike mammalian cells, studies in Drosophila indicate that Cyt-c release is not the easily detectable indicator of apoptosis initiation in flies.
In mammals, knock-in experiments demonstrate a pro-apoptotic role for Cyt-c in some, but not all, developmental apoptosis [95]. Numerous studies using tissue culture cells did not detect any role for Cyt-c in Drosophila apoptosis [51, 93, 96]. However, genetic data in flies demonstrate a role for Cyt-c in some developmentally important caspase activation. Mutation of one of the two Cyt-c genes, cyt-c-d, and loss of function of the Apaf-1 homolog Dark were found to cause a disruption of caspase-dependent spermatid individualization, suggesting that Dark and cyt-c-d might act together in caspase activation in these cells, similar to the prevailing models for Apaf-1 and Cyt-c in mammals [97–99]. Interommatidial cell death in the developing fly eye is also suppressed by lack of cyt-c-d, as well as by Dark mutation [100]. Moreover, adult flies lacking cyt-c-d also show an extra scutellar bristle, another phenotype that is observed in Dark mutants. Thus, in Drosophila, Cyt-c is apparently important for some apoptosome-dependent caspase activation. Interestingly, similar to mammalian Apaf-1, Drosophila Dark contains the WD40 regulatory region that binds to Cyt-c. However, structural characterization of the Drosophila apoptosome showed that although DarkWD40 region is important for Cyt-c binding, Drosophila apoptosome formation does not appear to require Cyt-c, and forms spontaneously with recombinant proteins in vitro and without additional co-factors [101]. However structural studies may not reflect the active form of the proteins, so this data should be interpreted with some caution.
Studies in other insect models could facilitate understanding of the role of mitochondrial proteins in cell death. A recent study in Lepidopteran sf9 cells identified an important role for cytochrome-c in apoptosis and provided evidence for PTP-independent Cyt-c release in sf9 cells [102]. Caspase activation in sf9 cell extracts was found to be exclusively dependent on Cyt-c release.
The mammalian pro-apoptotic proteins Smac/Diablo, ARTS, and Omi/HTRA2 are released from the mitochondria into the cytoplasm in dying cells, with kinetics similar to that of Cyt-c. These pro-apoptotic proteins can then bind to XIAP, an IAP protein, thereby preventing XIAP from inhibiting caspases [103]. SMAC/Diablo is a putative functional orthologue of the Drosophila RHG proteins. However, activation of SMAC/Diablo activity by release from mitochondria is quite different from the mechanism of activation of the Drosophila IAP inhibitors, which are for the most part transcriptionally upregulated in doomed cells [104].
The mammalian IAP antagonist, Omi/HtrA2, is constitutively expressed and sequestered to the mitochondrial intermembrane space before mitochondrial permeabilization [105, 106]. It contains both IAP binding and serine protease activities and exerts its pro-apoptotic function through binding and cleavage of IAPs upon its release into the cytoplasm following a pro-apoptotic stimulus. In Drosophila, the homolog of this protein, dOmi, was recently described [94, 107]. Upon cellular insults such as UV irradiation, Drosophila dOmi is released from the mitochondria and alleviates DIAP1 inhibition of caspases by proteolytically degrading DIAP1. dOmi induces apoptosis both in cultured cells and in the developing fly eye [94, 108]. Mammalian cell death induced by Omi overexpression can also occur under caspase inhibiting conditions and in Apaf-1 and Caspase-9 null cells [105, 106]. dOmi also has a Caspase-independent pro-apoptotic activity dependent on its serine protease activity [94, 108]. Interestingly, knockdown of dOmi was found to inhibit stress-induced apoptosis in the fly [107], in contrast to knockout mice lacking HtrA2/Omi which shows increased stress-induced apoptosis and neurodegeneration [109].
Apoptosis-inducing factor (AIF) is another mitochondrial protein released during death. AIF can translocate to the nucleus following an apoptotic stimulus and triggers DNA degradation and chromatin condensation in mammals [110]. AIF is one of several related oxido-reductase-like factors encoded from mammals to yeast. Mammalian AIF has dual functions, a pro-apoptotic activity in the nucleus via its DNA binding and an anti-apoptotic activity via the scavenging of free radicals through its oxidoreductase activity [111]. Similarly, knockout of the Drosophila homolog of AIF suggests both pro-survival and proapoptotic functions for this protein [112]. Drosophila AIF knockout causes lethality at early larval stages, with reduced ATP and reduced apoptosis in the developing embryo. Expression of a truncated AIF induced cell death in the eye, however, this phenotype was suppressed by DIAP1 or DIAP2, suggesting that AIF induces cell death through a caspase-dependent mechanism, either directly or indirectly through stress pathways.
Even though apoptosis-related mitochondrial permeabilization is not well understood in C. elegans, several mitochondrial proteins, including WAH-1 and CPS-6, as well as CYC-2.1 and CYC-2.2, the two Cytochromes c of C. elegans, are good candidates for pro-apoptotic factors released from mitochondria during apoptosis. Mutation in the C. elegans cps-6 gene, which encodes the homolog of the mammalian endonuclease G (endoG), causes a delay in apoptosis in C. elegans [113]. Interestingly, CPS-6 has been shown to cooperate with WAH-1, the homolog of the mammalian Apoptosis Inducing Factor (AIF) to degrade nuclear DNA [78]. These two proteins localize to mitochondria. Whereas it has not been determined whether CPS-6 is released from mitochondria during apoptosis, it has been shown that EGL-1 can induce the release of WAH-1 from mitochondria [78]. The mechanism by which WAH-1 is released is currently unknown. However, it is thought to be dependent on the activity of the caspase CED-3, and hence represents a late event that occurs after the cell has committed to die.
How Cytochrome c might function in C. elegans apoptosis remains unclear. Unlike Apaf-1 in mammals and Dark in Drosophila, CED-4 does not contain a WD40 repeat domain, which is responsible for the binding of Cyt-c to Apaf-1. It has therefore always been assumed that Cyt-c does not bind to CED-4 and does not play a role in C. elegans apoptosis. In fact a high resolution structural study of the CED-4 apoptosome was recently reported [29]. The CED-4 apoptosome is a cone-shaped octamer, a fundamentally different structure than the fly or mammalian apoptosome. To our knowledge, it has not been tested whether inactivating the cyc-2.1 and/or cyc-2.2 gene compromises apoptosis, whether the CYC-2.1 and/or CYC-2.2 proteins are released from mitochondria during apoptosis, whether they bind to CED-4 (by a mechanism different from the one discovered in mammals) or whether they can enhance CED-4-induced CED-3 processing. In conclusion, more experiments need to be done to determine whether Cyt-c play a role in C. elegans apoptosis or not.
Whereas two homologs of the Inhibitor of Apoptosis Proteins (IAP) exist in C. elegans (BIR-1 and BIR-2), neither of them play a role in apoptosis [114]. It is therefore not surprising that no homologs of the pro-apoptotic factors Smac/DIABLO and Omi/Htra2 have been identified in C. elegans. However, it remains possible that other mitochondrial proteins play a pro-apoptotic role in C. elegans. These proteins might not have been identified yet if they support an essential mitochondrial function and cause early embryonic lethality when inactivated.
The yeast Saccharomyces cerevisiae encodes homologues of AIF and Htra2/Omi and these factors have been implicated in promoting cell death. Yeast Aif1 (YNR047C), an NADH oxidoreductase, was named after its mammalian orthologue AIF (apoptosis inducing factor). Translocation of yeast Aif1 from mitochondria to the nucleus is suggested to degrade DNA and promote yeast cell death, which is supported by the finding that AIF1 gene deletion leads to increased survival [58]. The Aif1-related yeast protein Ndi1 (internal NADH dehydrogenase), but not the related protein Nde1 (external NADH dehydrogenase), is reported to promote yeast cell death when overexpressed, but deletion of either gene increases chronological life-span [115]. In contrast, expression of yeast Ndi1 in Drosophila neurons was recently reported by two groups to increase Drosophila lifespan [116, 117], and there is interest in exploring the potential application of this single chain NADH-quinone oxidoruductase to the treatment of complex I deficiencies in Parkinson’s disease [118] . While yeast Ndi1 may contribute importantly to yeast cell death, a physiological or evolutionary role in yeast programmed cell death is challenging to prove.
Yeast Nma111 (nuclear mediator of apoptosis) is the yeast orthologue of four mammalian mitochondrial serine proteases including Htra2/Omi. Deletion of the NMA111 gene increases yeast survival following heat shock treatment, while overexpression of Nma111 promotes death in a manner that requires its serine protease activity [119]. Further information about the substrates of the Nma111 protease or how cleavage of these substrates promotes death would help to confirm the importance of Nma111 in programmed yeast death. The yeast mitochondrial nuclease Nuc1, orthologue of the pro-death nuclease EndoG, also may promote death in yeast [120].
V. Alterations in mitochondrial morphology and structure
Mitochondria are highly dynamic organelles that undergo fission and fusion in living cells, with shapes ranging from short isolated organelles to interconnected networks [121]. Morphological changes at the level of single mitochondria, as well as changes in the dynamics of mitochondrial fission and fusion are associated with cell death [122].
In mammals, mitochondrial fission requires several proteins including dynamin-related protein 1 (DRP1/DLP1), Fis1, endophilin B1/Bif-1, MTP18, GDAP1, and DAP3 [123–132]. On the other hand, fusion requires three large GTPases: the mitofusins (Mfns) Mfn1 and Mfn2, and the dynamin-related protein OPA1 [133–136]. Mitochondrial fragmentation has been shown to increase significantly during programmed cell death in mammals, yeast, worms and flies [57, 68, 137–139]. Overexpression of a dominant negative DRP1 or knock down of DRP1 with RNAi or mutation results in decrease mitochondrial fragmentation after apoptosis induction in all four model systems [68, 137–140].
Interestingly, mitochondrial fragmentation in mammalian cells is associated with permeability of the outer mitochondrial membrane (MOMP), a hallmark of apoptosis [141–143]. Mitochondrial swelling is one likely mechanism by which proapoptotic proteins are released in response to the permeabilization of the outer mitochondrial membrane, and can be observed in fly cells as well as in mammalian cells [51, 144]. In mammals, MOMP can occur in a caspase-independent fashion [145], or in a caspase-dependent fashion [146], although there is some controversy in this regard.
Upon apoptotic stimulation, ultrastructural changes, such as swelling, can be detected in both mammalian and Drosophila cells [51, 147]. Interestingly, apoptosis induced by expression of Reaper or Hid causes caspase dependent changes in the mitochondrial ultrastructure [51] . DRP1 inhibition results in mitochondrial fusion and significantly inhibits caspase activation and apoptosis in flies [51], while it delays caspase activation in mammals [148]. One possibility is that the fission machinery may contribute to mitochondrial membrane permeabilization and release of proapoptotic factors [122]. Another explanation is that sites of mitochondrial fission might serve as scaffolding for the localization of other proapoptotic proteins, as seen with Bax in mammalian systems [149]. It is also possible that DRP1 has a role in apoptosis independent of its role in mitochondrial dynamics [139].
In C. elegans, mitochondrial fusion and fission are under the control of these GTPases. FZO-1 and EAT-3, the homologs of mammalian mitofusin and Opa1, are required for the fusion of the outer- and inner-mitochondrial membranes, respectively [150–152]. On the other hand, DRP1, the homolog of mammalian DRP1/DLP1, is required for mitochondrial fission [153]. Jagasia et al. showed that mitochondria undergo DRP1-dependent excessive fission (a process referred to hereafter as ‘mitochondrial fragmentation’) early during the apoptotic process [139]. Interestingly, blocking mitochondrial fragmentation by over-expressing a dominant negative DRP1 protein (DRP1(K40A)) prevents the death of 20% of the cells that are normally programmed to die. Conversely, inducing mitochondrial fragmentation by over-expressing a wild- type DRP1 protein causes the death of cells, which are normally programmed to survive. Consistent with these results, Breckenridge et al. showed that drp-1 loss-of-function mutation (drp-1(tm1108)) enhances the partial block in cell death caused by a weak ced-3 or ced-4 loss-of-function mutation [152] [Y Lu, B. Conradt, unpublished observation]. Together, these studies demonstrate that C. elegans DRP1 has pro-apoptotic activity. However, whether DRP1’s pro-apoptotic activity is related to its ability to cause mitochondrial fission remains unclear. If the mitochondrial fragmentation process per se is required for apoptosis induction, one would expect that blocking mitochondrial fusion causes the same ectopic cell death as observed when inducing mitochondrial fragmentation. In contrast, mutants carrying a loss-of-function mutation in the gene fzo-1 do not show any increase in cell death despite a clear mitochondrial fragmentation phenotype [152]. Similarly, the loss of eat-3 function causes mitochondrial fragmentation but no apparent cell-death phenotype [150]. These observations point toward a role of DRP1 in apoptosis independent of its role in mitochondrial fission. However, the analysis of the latter worm mutations has so far been focused on developmental cell death. In mammals, blocking mitochondrial fusion by inactivating mitofusin does not cause increased apoptosis on its own but sensitizes cells to external apoptotic stimuli [154]. Therefore, to completely rule out a role for mitochondrial fusion proteins in C. elegans apoptosis and consequently rule out a role for the fission activity of DRP1 in apoptosis, it will be necessary to test the effect of external apoptotic stimuli (such as radiation on germ cell death) in mitochondrial fusion defective mutants.
Interestingly, Jagasia et al. showed in C. elegans that the DRP1-dependent mitochondrial fragmentation in apoptotic cells is dependent on CED-9 and EGL-1 [139]. Two apoptotic pathways might therefore be at work in C. elegans (Figure 1B) [30]. On one hand, the binding of EGL-1 to CED-9 causes the release of CED-4, leading to the activation of CED-3 (‘direct’ pathway of apoptosis induction). On the other hand, when bound to EGL-1, CED-9 adopts a pro-apoptotic conformation and stimulates the pro-apoptotic activity of DRP1 through a mechanism that remains to be determined (‘indirect’ pathway of apoptosis induction). Recent work shows that Bcl-2-like proteins can interact with and regulate dynamin-related GTPases [155–159]. Specifically, in C. elegans, CED-9 has been shown to interact with FZO-1 and to promote mitochondrial fusion in healthy cells [157]. Therefore, one possible mechanism is that EGL-1-bound CED-9 directly interacts with and activates DRP1 in dying cells.
Yeast mitochondria, like in other species, undergo dramatic organelle fragmentation during programmed cell death [140, 160, 161]. This fragmentation is mediated in part by the yeast homologue of DRP1, Dnm1, and an additional yeast fission factor for which human/fly/worm orthologues are not known, Mdv1. This WD40-repeat protein is suggested to facilitate the interaction of Dnm1 with the conserved membrane-anchored Fis1 protein [57, 162, 163]. Like other species, Dnm1/Drp1 also promotes yeast cell death because deletion of the DNM1 gene in yeast results in increased survival following a death stimulus, even though this deletion strain grows essentially indistinguishably from wild type under normal conditions [57]. Because Dnm1 deletion strains are resistant to several different death stimuli (including heat, acidic pH, and H2O2), Dnm1 appears to be a common downstream effector of cell death. Overexpression of Dnm1 does not appear to be sufficient to kill yeast, but further addition of a death stimulus, which is apparently required to activate the death function of Dnm1, leads to a striking increase in cell death that can be inhibited by mammalian Bcl-2 family proteins [57]. Also similar to other species, a dominant negative Dnm1 that inhibits yeast mitochondrial fission also protects yeast from cell death.
The yeast cell death mediated by endogenous or overexpressed Dnm1 is partially dependent on the metacaspase Yca1/Mca1, which has distant amino acid sequence similarity to mammalian/worm/fly caspases, but is more closely related to bacterial proteases [164]. Yca1 has been reported to promote yeast cell death after many types of death stimuli, including yeast killer viruses, but neither Yca1 nor yeast Aif1 contribute to cell death that occurs by ammonia-dependent signaling and quorum sensing [40, 47, 165, 166]. Yca1 apparently does not cleave after aspartate residues and therefore does not conform to the definition of a c-asp-ase so strictly adhered to by true caspases, suggesting that alternative non-apoptotic death mechanisms may be involved [167]. The mechanism by which Yca1 mediates yeast cell death is not yet delineated.
Although Fis1 can promote mitochondrial fission and cell death in mammals, the yeast deletion strain is exquisitely sensitive to all cell death stimuli tested thus far. However, this death phenotype is not due to the loss of FIS1 function, at least not directly. Whole genome sequencing identified secondary mutations in three independently derived FIS1 knockout strains, each with a unique inactivating nonsense mutation in WHI2, and this mutation in WHI2 is solely responsible for the cell death phenotype [168]. The hypothesis that deletion of FIS1 is sufficient to drive the selection for WHI2 mutations is supported by the finding that FIS1-only knockouts have a mitochondrial defect resulting in petite formation (presumed loss of respiratory capacity) that is rescued by the WHI2 mutation [168]. However, the cell death inhibitory function of WHI2 is uncharacterized. It is further possible that many other single gene deletions in yeast (and other species) will drive the selection for compensatory mutations that provide small growth and survival advantages and that these have gone unnoticed thus far because programmed cell death is still not a mainstream topic in yeast genetics.
VI. Non-mammalian models of human diseases
Mutations in PINK1, a mitochondrial Ser/Thr kinase, and Parkin, an E3 ubiquitin ligase, were identified as a cause of familial Parkinson’s disease. In Drosophila, Pink1 mutation leads to defects in spermatid individualization, progressive apoptotic death of flight muscles [71, 169, 170] and loss of dopaminergic neurons [71, 170]. Pink1 accumulation on mitochondria was shown to be important for Parkin recruitment to mitochondria [171]. Interestingly, Parkin was found to be involved in targeting of mitochondria to autophagosomes and mitophagy [172]. Tissue-specific phenotypes observed in Drosophila parkin mutants may result from mitochondrial dysfunction [173]. Moreover, Pink1 or Parkin mutants were found to closely interact with the regulators of mitochondrial fission and fusion in Drosophila. For example, expression of DRP1 or knockdown of Drosophila MFN (dMfn or MARF) or Opa1 suppresses Pink1 and Parkin phenotypes. Loss of one copy of DRP1 is lethal in Pink1 and parkin mutant backgrounds [174]. dMfn ubiquitination was suggested to be a mechanism by which terminally damaged mitochondria are labeled and sequestered for degradation by autophagy [175].
Parkinson’s disease has also been associated with point mutations in HtrA2/Omi, and the phosphorylation of HtrA2/Omi was reduced in patients with a PINK1 mutation. In Drosophila, dOmi has been reported to interact with Pink1 or Parkin [176], and loss of dOmi can partially suppress phenotypic changes caused by PINK1 overexpression. In Omi/HtrA2 knockout mouse embryonic fibroblasts, as well as in Omi/HtrA2 silenced human HeLa cells and Drosophila S2R+ cells have elongated mitochondria and abnormal cristae structure [177]. Moreover, PINK1 overexpression can cause a rough eye phenotype that is suppressed by loss of both dOmi and Parkin. These observations contradict a previous report where loss of dOmi did not cause mitochondrial defects and did not suppress Pink1 overexpression phenotypes [176]. Further investigations are required to clarify this contradiction.
Mutation in the leucine-rich repeat kinase 2 (LRRK2) gene, currently recognized as the one of the most common genetic players involved in Parkinsonism, was shown to contribute to specific neurological disorders. Over-expressing various human LRRK2 alleles in flies causes late-onset loss of dopaminergic neurons accompanied by locomotion deficits. Importantly, coexpression of human parkin in LRRK2 G2019S-expressing flies significantly protected against neurodegeneration [178].
The Drosophila gene CG4589 is the ortholog of LETM1, a candidate gene for the haplo-insufficiency seizure phenotype of Wolf-Hirschhorn syndrome (WHS) patients. LETM is required for normal mitochondrial morphology and cell viability [179]. Using RNAi approaches in both Drosophila cultured cells and the adult fly, depletion of DmLETM1 caused roughening of the adult eye, mitochondrial swelling and developmental lethality in third-instar larvae, possibly as a result of deregulated mitophagy [180].
LRK-1 and PINK-1, the C. elegans homologues of the two Parkinson disease-related kinases LRRK2 and PINK1, have been shown to play roles in oxidative and ER stress responses as well as neurite outgrowth [181]. A recent study also showed that over-expression of lrk-1 transgenes carrying Parkinson-disease associated mutations in C. elegans dopaminergic neurons, caused age-dependent neurodegeneration and locomotion defects [182].
Exploitation of yeast as a model system has laid the groundwork for subsequent work on a wide range of human disease genes related to mitochondrial disorders [183, 184], metabolic disorders [185–187], aging [188, 189], neurodegeneration [190, 191], and cancer [192], often leading to major new lines of research. Perhaps the most unexpected application of yeast genetics is in the study of neurodegeneration, where yeast have contributed importantly to our understanding of the mechanisms of protein misfolding and aggregation common to Huntington’s disease (HD), Parkinson’s disease (PD), and Alzheimer’s disease (AD) [193–195]. The analysis of human alpha-synuclein in yeast revealed vesicle trafficking defects and mitochondrial dysfunction as potential mechanisms involved in the neurodegenerative effects of alpha-synuclein in Parkinson’s disease. A small molecule screen using this model system has recently identified a class of small molecules that significantly inhibits the toxic effects of alpha-synuclein on vesicle trafficking and mitochondria in both yeast and neurons [196]. The identification and function of mitochondrial fission and fusion factors in flies and yeast were instrumental to advancements in understanding the mitochondrial defects caused by mutations in their human homologues MFN2 (yeast FZO1) that cause Charcot-Marie-Tooth (CMT) subtype 2A, and OPA1 (yeast MGM1) that causes autosomal dominant optic atrophy (ADOA) [163]. Yeast are being applied to the study of multiple additional human neurological disease genes, such as FRDA (yeast YFH1), the cause of Friedreich ataxia [197], Batten disease caused by mutations in human BAN1 (yeast CLN3) [198, 199], Niemann Pick disease caused by a mutation in NPC1 (yeast NCR1) [200, 201], as well as SOD1 in ALS [202], SPG7 (yeast AFG3 and RCA1) in hereditary spastic paraplegia (HSP) [203], ABCD1 (yeast PXA1/2) in X-linked adrenoleukodystrophy (ALD) [204] and many more. However, few have yet exploited yeast for studying programmed cell death related to cancer or other human disorders.
VII. Conclusions and future prospects
In sum, many conserved mechanisms for mitochondrial involvement in apoptosis have been proposed in non-mammalian eukaryotes. Mitochondrial permeabilization has been suggested to occur in both flies and yeast, however, this finding is still under discussion, and the role of the released Cyt-c in apoptosis in these non-mammalian models remains unclear. Recently, involvement of fusion and fission proteins in the mitochondrial pathway of cell death appears to be a more conserved event between mammals, flies, worms and yeast. Moreover, mitochondrial fragmentation has been documented to be a feature of mammalian and non-mammalian eukaryotic apoptosis. The incomplete state of our knowledge of the mitochondrial death pathways may explain the differences between various species. Thus, more genetic and biochemical analysis might clarify the sophisticated mitochondrial mechanisms regulating the decision whether a eukaryotic cell lives or dies. In addition, the non-mammalian eukaryotes have been shown to be useful models to understand the mechanisms of human disease as they relate to the mitochondrial pathway of cell death. Thus, continued research into the mitochondrial contribution to cell death in model organisms is likely to provide important insight in the future.
Acknowledgements
The authors are supported by NIH grants GM55568 (KW), GM076651 (BC), GM077875 (JMH), ACS grant RSG-06-110-1-CCG (BC), and MGH Fund for Medical Discovery (EA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 2.Miura M, Zhu H, Rotello R, Hartwieg EA, Yuan J. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 3.Hengartner MO, Horvitz HR. C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. 1994;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- 4.Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J. Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 6.Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 7.Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 8.White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 9.Vucic D, Kaiser WJ, Harvey AJ, Miller LK. Inhibition of reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs) Proc. Natl. Acad. Sci. U. S. A. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 11.Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanki T, Klionsky DJ. The molecular mechanism of mitochondria autophagy in yeast. Mol. Microbiol. 2010;75:795–800. doi: 10.1111/j.1365-2958.2009.07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baehrecke EH. Autophagy: dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005;6:505–510. doi: 10.1038/nrm1666. [DOI] [PubMed] [Google Scholar]
- 16.Kornbluth S, White K. Apoptosis in Drosophila: neither fish nor fowl (nor worm) J. Cell. Sci. 2005;118:1779–1787. doi: 10.1242/jcs.02377. [DOI] [PubMed] [Google Scholar]
- 17.Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- 18.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 19.Horvitz HR. Worms, Life and Death (Nobel Lecture) 2003;4:697–711. doi: 10.1002/cbic.200300614. [DOI] [PubMed] [Google Scholar]
- 20.Chinnaiyan AM, O'Rourke K, Lane BR, Dixit VM. Interaction of CED-4 with CED-3 and CED-9: a molecular framework for cell death. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- 21.Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, Horvitz HR. Translocation of C.elegans CED-4 to nuclear membranes during programmed cell death. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- 22.Wu D, Wallen HD, Nunez G. Interaction and regulation of subcellular localization of CED-4 by CED-9. 1997;275:1126–1129. doi: 10.1126/science.275.5303.1126. [DOI] [PubMed] [Google Scholar]
- 23.Spector MS, Desnoyers S, Hoeppner DJ, Hengartner MO. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- 24.Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 25.del Peso L, Gonzalez VM, Nunez G. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- 26.Chinnaiyan AM, Chaudhary D, O'Rourke K, Koonin EV, Dixit VM. Role of CED-4 in the activation of CED-3. 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- 27.Seshagiri S, Miller LK. Caenorhabditis elegans CED-4 stimulates CED-3 processing and CED-3-induced apoptosis. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- 28.Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, Xue D, Shi Y. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- 29.Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, Luo G, Li H, Wang J, Yan N, Shi Y. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Rolland S, Conradt B. The role of mitochondria in apoptosis induction in Caenorhabditis elegans: more than just innocent bystanders? 2006;13:1281–1286. doi: 10.1038/sj.cdd.4401980. [DOI] [PubMed] [Google Scholar]
- 31.Hengartner MO, Horvitz HR. Activation of C. elegans cell death protein CED-9 by an ammo-acid substitution in a domain conserved in Bcl-2. 1994;369:318–320. doi: 10.1038/369318a0. [DOI] [PubMed] [Google Scholar]
- 32.Chen P, Nordstrom W, Gish B, Abrams JM. Abrams, grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 33.Christich A, Kauppila S, Chen P, Sogame N, Ho SI, Abrams JM. The damageresponsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim, and hid. Curr. Biol. 2002;12:137–140. doi: 10.1016/s0960-9822(01)00658-3. [DOI] [PubMed] [Google Scholar]
- 34.Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 35.O'Riordan MX, Bauler LD, Scott FL, Duckett CS. Inhibitor of apoptosis proteins in eukaryotic evolution and development: a model of thematic conservation. Dev. Cell. 2008;15:497–508. doi: 10.1016/j.devcel.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol. Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat. Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 39.Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol. Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- 40.Ivanovska I, Hardwick JM. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 2005;170:391–399. doi: 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breinig F, Sendzik T, Eisfeld K, Schmitt MJ. Dissecting toxin immunity in virus-infected killer yeast uncovers an intrinsic strategy of self-protection. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3810–3815. doi: 10.1073/pnas.0510070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cap M, Vachova L, Palkova Z. Yeast colony survival depends on metabolic adaptation and cell differentiation rather than on stress defense. J. Biol. Chem. 2009;284:32572–32581. doi: 10.1074/jbc.M109.022871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palkova Z, Vachova L. Life within a community: benefit to yeast long-term survival. FEMS Microbiol. Rev. 2006;30:806–824. doi: 10.1111/j.1574-6976.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- 44.Longo VD, Mitteldorf J, Skulachev VP. Programmed and altruistic ageing. Nat. Rev. Genet. 2005;6:866–872. doi: 10.1038/nrg1706. [DOI] [PubMed] [Google Scholar]
- 45.Ameisen JC. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ. 2002;9:367–393. doi: 10.1038/sj.cdd.4400950. [DOI] [PubMed] [Google Scholar]
- 46.Borello ME. The rise, fall and resurrection of group selection. Endvour. 2005;29:43–47. doi: 10.1016/j.endeavour.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Madeo F, Herker E, Maldener C, Wissing S, Lachelt S, Herlan M, Fehr M, Lauber K, Sigrist SJ, Wesselborg S, Frohlich KU. A caspase-related protease regulates apoptosis in yeast. Mol. Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- 48.Sundstrom JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA, Jr, Valineva T, Saarikettu J, Frilander MJ, Suarez MF, Zavialov A, Stahl U, Hussey PJ, Silvennoinen O, Sundberg E, Zhivotovsky B, Bozhkov PV. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 2009;11:1347–1354. doi: 10.1038/ncb1979. [DOI] [PubMed] [Google Scholar]
- 49.Hardwick JM, Youle RJ. SnapShot: BCL-2 proteins. Cell. 2009;138:404. doi: 10.1016/j.cell.2009.07.003. 404.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walter D, Wissing S, Madeo F, Fahrenkrog B. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell. Sci. 2006;119:1843–1851. doi: 10.1242/jcs.02902. [DOI] [PubMed] [Google Scholar]
- 51.Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev. Cell. 2007;12:793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Claveria C, Caminero E, Martinez-A C, Campuzano S, Torres M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J. 2002;21:3327–3336. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olson MR, Holley CL, Gan EC, Colon-Ramos DA, Kaplan B, Kornbluth S. A GH3-like domain in reaper is required for mitochondrial localization and induction of IAP degradation. J. Biol. Chem. 2003;278:44758–44768. doi: 10.1074/jbc.M308055200. [DOI] [PubMed] [Google Scholar]
- 54.Sandu C, Ryoo HD, Steller H. Drosophila IAP antagonists form multimeric complexes to promote cell death. J. Cell Biol. 2010;190:1039–1052. doi: 10.1083/jcb.201004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4936–4941. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freel CD, Richardson DA, Thomenius MJ, Gan EC, Horn SR, Olson MR, Kornbluth S. Mitochondrial localization of Reaper to promote inhibitors of apoptosis protein degradation conferred by GH3 domain-lipid interactions. J. Biol. Chem. 2008;283:367–379. doi: 10.1074/jbc.M708931200. [DOI] [PubMed] [Google Scholar]
- 57.Fannjiang Y, Cheng WC, Lee SJ, Qi B, Pevsner J, McCaffery JM, Hill RB, Basanez G, Hardwick JM. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wissing S, Ludovico P, Herker E, Buttner S, Engelhardt SM, Decker T, Link A, Proksch A, Rodrigues F, Corte-Real M, Frohlich KU, Manns J, Cande C, Sigrist SJ, Kroemer G, Madeo F. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kroemer G, Reed JC. Mitochondrial control of cell death. Nat. Med. 2000;6:513–519. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 60.Tait SW, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Munoz-Pinedo C, Green DR. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev. Cell. 2010;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 62.Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 63.Brachmann CB, Jassim OW, Wachsmuth BD, Cagan RL. The Drosophila bcl-2 family member dBorg-1 functions in the apoptotic response to UV-irradiation. Curr. Biol. 2000;10:547–550. doi: 10.1016/s0960-9822(00)00474-7. [DOI] [PubMed] [Google Scholar]
- 64.Colussi PA, Quinn LM, Huang DC, Coombe M, Read SH, Richardson H, Kumar S. Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J. Cell Biol. 2000;148:703–714. doi: 10.1083/jcb.148.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Igaki T, Kanuka H, Inohara N, Sawamoto K, Nunez G, Okano H, Miura M. Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc. Natl. Acad. Sci. U. S. A. 2000;97:662–667. doi: 10.1073/pnas.97.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 67.Doumanis J, Dorstyn L, Kumar S. Molecular determinants of the subcellular localization of the Drosophila Bcl-2 homologues DEBCL and BUFFY. Cell Death Differ. 2007;14:907–915. doi: 10.1038/sj.cdd.4402082. [DOI] [PubMed] [Google Scholar]
- 68.Galindo KA, Lu WJ, Park JH, Abrams JM. The Bax/Bak ortholog in Drosophila, Debcl, exerts limited control over programmed cell death. Development. 2009;136:275–283. doi: 10.1242/dev.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Huang Q, Ke N, Matsuyama S, Hammock B, Godzik A, Reed JC. Drosophila pro-apoptotic Bcl-2/Bax homologue reveals evolutionary conservation of cell death mechanisms. J. Biol. Chem. 2000;275:27303–27306. doi: 10.1074/jbc.M002846200. [DOI] [PubMed] [Google Scholar]
- 70.Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S, Richardson H. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J. 2003;22:3568–3579. doi: 10.1093/emboj/cdg355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 72.Wu JN, Nguyen N, Aghazarian M, Sevrioukov EA, Monserrate JP, Tang W, Mabuchi M, Tan Y, White K, Brachmann CB. grim promotes programmed cell death of Drosophila microchaete glial cells. Mech. Dev. 2010 doi: 10.1016/j.mod.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Senoo-Matsuda N, Igaki T, Miura M. Bax-like protein Drob-1 protects neurons from expanded polyglutamine-induced toxicity in Drosophila. EMBO J. 2005;24:2700–2713. doi: 10.1038/sj.emboj.7600721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lewis J, Oyler GA, Ueno K, Fannjiang YR, Chau BN, Vornov J, Korsmeyer SJ, Zou S, Hardwick JM. Inhibition of virus-induced neuronal apoptosis by Bax. Nat. Med. 1999;5:832–835. doi: 10.1038/10556. [DOI] [PubMed] [Google Scholar]
- 75.Fannjiang Y, Kim CH, Huganir RL, Zou S, Lindsten T, Thompson CB, Mito T, Traystman RJ, Larsen T, Griffin DE, Mandir AS, Dawson TM, Dike S, Sappington AL, Kerr DA, Jonas EA, Kaczmarek LK, Hardwick JM. BAK alters neuronal excitability and can switch from anti- to pro-death function during postnatal development. Dev. Cell. 2003;4:575–585. doi: 10.1016/s1534-5807(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 76.Sevrioukov EA, Burr J, Huang EW, Assi HH, Monserrate JP, Purves DC, Wu JN, Song EJ, Brachmann CB. Drosophila Bcl-2 proteins participate in stress-induced apoptosis, but are not required for normal development. Genesis. 2007;45:184–193. doi: 10.1002/dvg.20279. [DOI] [PubMed] [Google Scholar]
- 77.Chen P, Ho SI, Shi Z, Abrams JM. Bifunctional killing activity encoded by conserved reaper proteins. Cell Death Differ. 2004;11:704–713. doi: 10.1038/sj.cdd.4401406. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- 79.Shen Q, Qin F, Gao Z, Cui J, Xiao H, Xu Z, Yang C. Adenine nucleotide translocator cooperates with core cell death machinery to promote apoptosis in Caenorhabditis elegans. 2009;29:3881–3893. doi: 10.1128/MCB.01509-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang C, Youle RJ. The role of mitochondria in apoptosis*. Annu. Rev. Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ludovico P, Rodrigues F, Almeida A, Silva MT, Barrientos A, Corte-Real M. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2598–2606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manon S, Chaudhuri B, Guerin M. Release of cytochrome c and decrease of cytochrome c oxidase in Bax-expressing yeast cells, and prevention of these effects by coexpression of Bcl-xL. FEBS Lett. 1997;415:29–32. doi: 10.1016/s0014-5793(97)01087-9. [DOI] [PubMed] [Google Scholar]
- 83.Pereira C, Silva RD, Saraiva L, Johansson B, Sousa MJ, Corte-Real M. Mitochondria-dependent apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1286–1302. doi: 10.1016/j.bbamcr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 84.Pereira C, Chaves S, Alves S, Salin B, Camougrand N, Manon S, Sousa MJ, Corte-Real M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: the role of Pep4 and the ADP/ATP carrier. Mol. Microbiol. 2010;76:1398–1410. doi: 10.1111/j.1365-2958.2010.07122.x. [DOI] [PubMed] [Google Scholar]
- 85.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 86.Heiskanen KM, Bhat MB, Wang HW, Ma J, Nieminen AL. Mitochondrial depolarization accompanies cytochrome c release during apoptosis in PC6 cells. J. Biol. Chem. 1999;274:5654–5658. doi: 10.1074/jbc.274.9.5654. [DOI] [PubMed] [Google Scholar]
- 87.Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10:369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 88.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 89.Korsmeyer SJ, Wei MC, Saito M, Weiler S, Oh KJ, Schlesinger PH. Proapoptotic which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 2000;7:1166–1173. doi: 10.1038/sj.cdd.4400783. [DOI] [PubMed] [Google Scholar]
- 90.Varkey J, Chen P, Jemmerson R, Abrams JM. Altered cytochrome c display precedes apoptotic cell death in Drosophila. J. Cell Biol. 1999;144:701–710. doi: 10.1083/jcb.144.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Means JC, Muro I, Clem RJ. Lack of involvement of mitochondrial factors in caspase activation in a Drosophila cell-free system. Cell Death Differ. 2006;13:1222–1234. doi: 10.1038/sj.cdd.4401821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 2002;156:1089–1098. doi: 10.1083/jcb.200111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zimmermann KC, Ricci JE, Droin NM, Green DR. The role of ARK in stress-induced apoptosis in Drosophila cells. J. Cell Biol. 2002;156:1077–1087. doi: 10.1083/jcb.20112068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Challa M, Malladi S, Pellock BJ, Dresnek D, Varadarajan S, Yin YW, White K, Bratton SB. Drosophila Omi, a mitochondrial-localized IAP antagonist and proapoptotic serine protease. EMBO J. 2007;26:3144–3156. doi: 10.1038/sj.emboj.7601745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hao Z, Duncan GS, Chang CC, Elia A, Fang M, Wakeham A, Okada H, Calzascia T, Jang Y, You-Ten A, Yeh WC, Ohashi P, Wang X, Mak TW. Specific ablation of the apoptotic functions of cytochrome C reveals a differential requirement for cytochrome C and Apaf-1 in apoptosis. Cell. 2005;121:579–591. doi: 10.1016/j.cell.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 96.Dorstyn L, Mills K, Lazebnik Y, Kumar S. The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J. Cell Biol. 2004;167:405–410. doi: 10.1083/jcb.200408054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 98.Huh JR, Vernooy SY, Yu H, Yan N, Shi Y, Guo M, Hay BA. Multiple apoptotic caspase cascades are required in nonapoptotic roles for Drosophila spermatid individualization. PLoS Biol. 2004;2:E15. doi: 10.1371/journal.pbio.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 2006;25:232–243. doi: 10.1038/sj.emboj.7600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A, Steller H, Mollereau B. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep. 2006;7:933–939. doi: 10.1038/sj.embor.7400773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J. Mol. Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 102.Kumarswamy R, Seth RK, Dwarakanath BS, Chandna S. Mitochondrial regulation of insect cell apoptosis: evidence for permeability transition pore-independent cytochrome-c release in the Lepidopteran Sf9 cells. Int. J. Biochem. Cell Biol. 2009;41:1430–1440. doi: 10.1016/j.biocel.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 103.Gottfried Y, Rotem A, Lotan R, Steller H, Larisch S. The mitochondrial ARTS protein promotes apoptosis through targeting XIAP. EMBO J. 2004;23:1627–1635. doi: 10.1038/sj.emboj.7600155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oberst A, Bender C, Green DR. Living with death: the evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ. 2008;15:1139–1146. doi: 10.1038/cdd.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hegde R, Srinivasula SM, Zhang Z, Wassell R, Mukattash R, Cilenti L, DuBois G, Lazebnik Y, Zervos AS, Fernandes-Alnemri T, Alnemri ES. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 2002;277:432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- 106.Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 107.Igaki T, Suzuki Y, Tokushige N, Aonuma H, Takahashi R, Miura M. Evolution of mitochondrial cell death pathway: Proapoptotic role of HtrA2/Omi in Drosophila. Biochem. Biophys. Res. Commun. 2007;356:993–997. doi: 10.1016/j.bbrc.2007.03.079. [DOI] [PubMed] [Google Scholar]
- 108.Khan FS, Fujioka M, Datta P, Fernandes-Alnemri T, Jaynes JB, Alnemri ES. The interaction of DIAP1 with dOmi/HtrA2 regulates cell death in Drosophila. Cell Death Differ. 2008;15:1073–1083. doi: 10.1038/cdd.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alnemri ES. HtrA2 and Parkinson's disease: think PINK? Nat. Cell Biol. 2007;9:1227–1229. doi: 10.1038/ncb1107-1227. [DOI] [PubMed] [Google Scholar]
- 110.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 111.Lipton SA, Bossy-Wetzel E. Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell. 2002;111:147–150. doi: 10.1016/s0092-8674(02)01046-2. [DOI] [PubMed] [Google Scholar]
- 112.Joza N, Galindo K, Pospisilik JA, Benit P, Rangachari M, Kanitz EE, Nakashima Y, Neely GG, Rustin P, Abrams JM, Kroemer G, Penninger JM. The molecular archaeology of a mitochondrial death effector: AIF in Drosophila. Cell Death Differ. 2008;15:1009–1018. doi: 10.1038/cdd.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Mitochondrial endonuclease G is important for apoptosis in C. elegans. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- 114.Fraser AG, James C, Evan GI, Hengartner MO. Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr. Biol. 1999;9:292–301. doi: 10.1016/s0960-9822(99)80137-7. [DOI] [PubMed] [Google Scholar]
- 115.Li W, Sun L, Liang Q, Wang J, Mo W, Zhou B. Yeast AMID homologue Ndi1p displays respiration-restricted apoptotic activity and is involved in chronological aging. Mol. Biol. Cell. 2006;17:1802–1811. doi: 10.1091/mbc.E05-04-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sanz A, Soikkeli M, Portero-Otin M, Wilson A, Kemppainen E, McIlroy G, Ellila S, Kemppainen KK, Tuomela T, Lakanmaa M, Kiviranta E, Stefanatos R, Dufour E, Hutz B, Naudi A, Jove M, Zeb A, Vartiainen S, Matsuno-Yagi A, Yagi T, Rustin P, Pamplona R, Jacobs HT. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9105–9110. doi: 10.1073/pnas.0911539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bahadorani S, Cho J, Lo T, Contreras H, Lawal HO, Krantz DE, Bradley TJ, Walker DW. Neuronal expression of a single-subunit yeast NADH-ubiquinone oxidoreductase (Ndi1) extends Drosophila lifespan. Aging Cell. 2010;9:191–202. doi: 10.1111/j.1474-9726.2010.00546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Marella M, Seo BB, Yagi T, Matsuno-Yagi A. Parkinson's disease and mitochondrial complex I: a perspective on the Ndi1 therapy. J. Bioenerg. Biomembr. 2009;41:493–497. doi: 10.1007/s10863-009-9249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fahrenkrog B, Sauder U, Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell. Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- 120.Buttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, Sigrist S, Madeo F. Endonuclease G regulates budding yeast life and death. Mol. Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 121.Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21:1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martinou JC, Youle RJ. Which came first, the cytochrome c release or the mitochondrial fission? Cell Death Differ. 2006;13:1291–1295. doi: 10.1038/sj.cdd.4401985. [DOI] [PubMed] [Google Scholar]
- 123.Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 126.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell. Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 128.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J. Cell Biol. 2004;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takahashi Y, Karbowski M, Yamaguchi H, Kazi A, Wu J, Sebti SM, Youle RJ, Wang HG. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol. Cell. Biol. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tondera D, Czauderna F, Paulick K, Schwarzer R, Kaufmann J, Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell. Sci. 2005;118:3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- 131.Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J. Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mukamel Z, Kimchi A. Death-associated protein 3 localizes to the mitochondria and is involved in the process of mitochondrial fragmentation during cell death. J. Biol. Chem. 2004;279:36732–36738. doi: 10.1074/jbc.M400041200. [DOI] [PubMed] [Google Scholar]
- 133.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 136.Chan DC. Dissecting mitochondrial fusion. Dev. Cell. 2006;11:592–594. doi: 10.1016/j.devcel.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 137.Goyal G, Fell B, Sarin A, Youle RJ, Sriram V. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev. Cell. 2007;12:807–816. doi: 10.1016/j.devcel.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Frank S, Gaume B, Bergmann-Leitner E, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 139.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 140.Cheng WC, Leach KM, Hardwick JM. Mitochondrial death pathways in yeast and mammalian cells. Biochim. Biophys. Acta. 2008;1783:1272–1279. doi: 10.1016/j.bbamcr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Arnoult D, Grodet A, Lee YJ, Estaquier J, Blackstone C. Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 2005;280:35742–35750. doi: 10.1074/jbc.M505970200. [DOI] [PubMed] [Google Scholar]
- 142.Gao W, Pu Y, Luo KQ, Chang DC. Temporal relationship between cytochrome c release and mitochondrial swelling during UV-induced apoptosis in living HeLa cells. J. Cell. Sci. 2001;114:2855–2862. doi: 10.1242/jcs.114.15.2855. [DOI] [PubMed] [Google Scholar]
- 143.Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- 144.Means JC, Hays R. Mitochondrial membrane depolarization in Drosophila apoptosis. Cell Death Differ. 2007;14:383–385. doi: 10.1038/sj.cdd.4402036. [DOI] [PubMed] [Google Scholar]
- 145.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. 2006;13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 146.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol. Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun MG, Williams J, Munoz-Pinedo C, Perkins GA, Brown JM, Ellisman MH, Green DR, Frey TG. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat. Cell Biol. 2007;9:1057–1065. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- 148.Tanaka A, Youle RJ. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. 2008;29:409–410. doi: 10.1016/j.molcel.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 149.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith E, Buttle KF, McDonald K, Mannella CA, van dB. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. 2008;4 doi: 10.1371/journal.pgen.1000022. e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ichishita R, Tanaka K, Sugiura Y, Sayano T, Mihara K, Oka T. An RNAi screen for mitochondrial proteins required to maintain the morphology of the organelle in Caenorhabditis elegans. 2008;143:449–454. doi: 10.1093/jb/mvm245. [DOI] [PubMed] [Google Scholar]
- 152.Breckenridge DG, Kang BH, Kokel D, Mitani S, Staehelin LA, Xue D. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol. Cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Labrousse AM, Zappaterra MD, Rube DA, van dB. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 154.Sugioka R, Shimizu S, Tsujimoto Y. Fzo1, a protein involved in mitochondrial fusion. inhibits apoptosis. 2004;279:52726–52734. doi: 10.1074/jbc.M408910200. [DOI] [PubMed] [Google Scholar]
- 155.Karbowski M, Norris KL, Cleland MM, Jeong SY, Youle RJ. Role of Bax and Bak in mitochondrial morphogenesis. Nature. 2006;443:658–662. doi: 10.1038/nature05111. [DOI] [PubMed] [Google Scholar]
- 156.Li H, Chen Y, Jones AF, Sanger RH, Collis LP, Flannery R, McNay EC, Yu T, Schwarzenbacher R, Bossy B, Bossy-Wetzel E, Bennett MV, Pypaert M, Hickman JA, Smith PJ, Hardwick JM, Jonas EA. Bcl-xL induces Drp1-dependent synapse formation in cultured hippocampal neurons. 2008;105:2169–2174. doi: 10.1073/pnas.0711647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rolland SG, Lu Y, David CN, Conradt B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. 2009;186:525–540. doi: 10.1083/jcb.200905070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Delivani P, Adrain C, Taylor RC, Duriez PJ, Martin SJ. Role for CED-9 and Egl-1 as regulators of mitochondrial fission and fusion dynamics. 2006;21:761–773. doi: 10.1016/j.molcel.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 159.Tan FJ, Husain M, Manlandro CM, Koppenol M, Fire AZ, Hill RB. CED-9 and mitochondrial homeostasis in C. elegans muscle. 2008;121:3373–3382. doi: 10.1242/jcs.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Carmona-Gutierrez D, Eisenberg T, Buttner S, Meisinger C, Kroemer G, Madeo F. Apoptosis in yeast: triggers, pathways, subroutines. Cell Death Differ. 2010;17:763–773. doi: 10.1038/cdd.2009.219. [DOI] [PubMed] [Google Scholar]
- 161.Hardwick JM, Cheng WC. Mitochondrial programmed cell death pathways in yeast. Dev. Cell. 2004;7:630–632. doi: 10.1016/j.devcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 162.Okamoto K, Shaw JM. Mitochondrial Morphology and Dynamics in Yeast and Multicellular Eukaryotes. 2005 doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 163.Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. 2009;1793:20–26. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 164.Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 165.Reiter J, Herker E, Madeo F, Schmitt MJ. Viral killer toxins induce caspase-mediated apoptosis in yeast. J. Cell Biol. 2005;168:353–358. doi: 10.1083/jcb.200408071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vachova L, Palkova Z. Physiological regulation of yeast cell death in multicellular colonies is triggered by ammonia. J. Cell Biol. 2005;169:711–717. doi: 10.1083/jcb.200410064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Enoksson M, Salvesen GS. Metacaspases are not caspases--always doubt. Cell Death Differ. 2010;17:1221. doi: 10.1038/cdd.2010.45. [DOI] [PubMed] [Google Scholar]
- 168.Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, Hardwick JM. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ. 2008;15:1838–1846. doi: 10.1038/cdd.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 170.Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. U. S. A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 177.Kieper N, Holmstrom KM, Ciceri D, Fiesel FC, Wolburg H, Ziviani E, Whitworth AJ, Martins LM, Kahle PJ, Kruger R. Modulation of mitochondrial function and morphology by interaction of Omi/HtrA2 with the mitochondrial fusion factor OPA1. Exp. Cell Res. 2010;316:1213–1224. doi: 10.1016/j.yexcr.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ng CH, Mok SZ, Koh C, Ouyang X, Fivaz ML, Tan EK, Dawson VL, Dawson TM, Yu F, Lim KL. Parkin protects against LRRK2 G2019S mutant-induced dopaminergic neurodegeneration in Drosophila. J. Neurosci. 2009;29:11257–11262. doi: 10.1523/JNEUROSCI.2375-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dimmer KS, Navoni F, Casarin A, Trevisson E, Endele S, Winterpacht A, Salviati L, Scorrano L. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum. Mol. Genet. 2008;17:201–214. doi: 10.1093/hmg/ddm297. [DOI] [PubMed] [Google Scholar]
- 180.McQuibban AG, Joza N, Megighian A, Scorzeto M, Zanini D, Reipert S, Richter C, Schweyen RJ, Nowikovsky K. A Drosophila mutant of LETM1, a candidate gene for seizures in Wolf-Hirschhorn syndrome. Hum. Mol. Genet. 2010;19:987–1000. doi: 10.1093/hmg/ddp563. [DOI] [PubMed] [Google Scholar]
- 181.Samann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. 2009;284:16482–16491. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yao C, El Khoury R, Wang W, Byrd TA, Pehek EA, Thacker C, Zhu X, Smith MA, Wilson-Delfosse A, Chen SG. LRRK2-mediated neurodegeneration and dysfunction of dopaminergic neurons in a Caenorhabditis elegans model of Parkinson's disease. 2010 doi: 10.1016/j.nbd.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Perocchi F, Mancera E, Steinmetz LM. Systematic screens for human disease genes, from yeast to human and back. Mol. Biosyst. 2008;4:18–29. doi: 10.1039/b709494a. [DOI] [PubMed] [Google Scholar]
- 184.Schwimmer C, Rak M, Lefebvre-Legendre L, Duvezin-Caubet S, Plane G, di Rago JP. Yeast models of human mitochondrial diseases: from molecular mechanisms to drug screening. Biotechnol. J. 2006;1:270–281. doi: 10.1002/biot.200500053. [DOI] [PubMed] [Google Scholar]
- 185.Lai K, Klapa MI. Alternative pathways of galactose assimilation: could inverse metabolic engineering provide an alternative to galactosemic patients? Metab. Eng. 2004;6:239–244. doi: 10.1016/j.ymben.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 186.Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 187.Garbarino J, Sturley SL. Saturated with fat: new perspectives on lipotoxicity. Curr. Opin. Clin. Nutr. Metab. Care. 2009;12:110–116. doi: 10.1097/MCO.0b013e32832182ee. [DOI] [PubMed] [Google Scholar]
- 188.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu. Rev. Cell Dev. Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Roux AE, Chartrand P, Ferbeyre G, Rokeach LA. Fission yeast and other yeasts as emergent models to unravel cellular aging in eukaryotes. J. Gerontol. A Biol. Sci. Med. Sci. 2010;65:1–8. doi: 10.1093/gerona/glp152. [DOI] [PubMed] [Google Scholar]
- 190.Walberg MW. Applicability of yeast genetics to neurologic disease. Arch. Neurol. 2000;57:1129–1134. doi: 10.1001/archneur.57.8.1129. [DOI] [PubMed] [Google Scholar]
- 191.Winderickx J, Delay C, De Vos A, Klinger H, Pellens K, Vanhelmont T, Van Leuven F, Zabrocki P. Protein folding diseases and neurodegeneration: lessons learned from yeast. Biochim. Biophys. Acta. 2008;1783:1381–1395. doi: 10.1016/j.bbamcr.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 192.Nitiss JL, Heitman J SpringerLink (Online service) Yeast as a Tool in Cancer Research. Dordrecht: Springer; 2007. [Google Scholar]
- 193.Lindquist S, Krobitsch S, Li L, Sondheimer N. Investigating protein conformation-based inheritance and disease in yeast. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:169–176. doi: 10.1098/rstb.2000.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tenreiro S, Outeiro TF. Simple is good: yeast models of neurodegeneration. FEMS Yeast Res. 2010 doi: 10.1111/j.1567-1364.2010.00649.x. [DOI] [PubMed] [Google Scholar]
- 195.Auluck PK, Caraveo G, Lindquist S. alpha-Synuclein: Membrane Interactions and Toxicity in Parkinson's Disease. Annu. Rev. Cell Dev. Biol. 2010 doi: 10.1146/annurev.cellbio.042308.113313. [DOI] [PubMed] [Google Scholar]
- 196.Su LJ, Auluck PK, Outeiro TF, Yeger-Lotem E, Kritzer JA, Tardiff DF, Strathearn KE, Liu F, Cao S, Hamamichi S, Hill KJ, Caldwell KA, Bell GW, Fraenkel E, Cooper AA, Caldwell GA, McCaffery JM, Rochet JC, Lindquist S. Compounds from an unbiased chemical screen reverse both ER-to-Golgi trafficking defects and mitochondrial dysfunction in Parkinson's disease models. Dis. Model. Mech. 2010;3:194–208. doi: 10.1242/dmm.004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Knight SA, Kim R, Pain D, Dancis A. The yeast connection to Friedreich ataxia. Am. J. Hum. Genet. 1999;64:365–371. doi: 10.1086/302270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pearce DA, Ferea T, Nosel SA, Das B, Sherman F. Action of BTN1, the yeast orthologue of the gene mutated in Batten disease. Nat. Genet. 1999;22:55–58. doi: 10.1038/8861. [DOI] [PubMed] [Google Scholar]
- 199.Phillips SN, Muzaffar N, Codlin S, Korey CA, Taschner PE, de Voer G, Mole SE, Pearce DA. Characterizing pathogenic processes in Batten disease: use of small eukaryotic model systems. Biochim. Biophys. Acta. 2006;1762:906–919. doi: 10.1016/j.bbadis.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 200.Zhang S, Ren J, Li H, Zhang Q, Armstrong JS, Munn AL, Yang H. Ncr1p, the yeast ortholog of mammalian Niemann Pick C1 protein, is dispensable for endocytic transport. Traffic. 2004;5:1017–1030. doi: 10.1111/j.1600-0854.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 201.Berger AC, Hanson PK, Wylie Nichols J, Corbett AH. A yeast model system for functional analysis of the Niemann-Pick type C protein 1 homolog, Ncr1p. Traffic. 2005;6:907–917. doi: 10.1111/j.1600-0854.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 202.Moore JK, Sept D, Cooper JA. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5147–5152. doi: 10.1073/pnas.0810828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pearce DA. Hereditary spastic paraplegia: mitochondrial metalloproteases of yeast. Hum. Genet. 1999;104:443–448. doi: 10.1007/s004390050985. [DOI] [PubMed] [Google Scholar]
- 204.Shani N, Valle D. A Saccharomyces cerevisiae homolog of the human adrenoleukodystrophy transporter is a heterodimer of two half ATP-binding cassette transporters. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11901–11906. doi: 10.1073/pnas.93.21.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]