Abstract
Members of the Argonaute protein family have been linked through a combination of genetic and biochemical studies to RNA interference (RNAi) and related phenomena. Here, we describe the characterization of the first Argonaute protein (AGO1) in Trypanosoma brucei, the earliest divergent eukaryote where RNAi has been described so far. AGO1 is predominantly cytoplasmic and is found in a ribonucleoprotein particle with small interfering RNAs (siRNAs), and this particle is present in a soluble form, as well as associated with polyribosomes. A genetic knockout of AGO1 leads to a loss of RNAi, and concomitantly, endogenous retroposon-derived siRNAs as well as siRNAs derived from transgenic double-stranded RNA are reduced to almost undetectable levels. Furthermore, AGO1 deficiency leads to an increase in retroposon transcript abundance via mechanisms operating at the transcriptional level and at the RNA stability level. Our results suggest that AGO1 function is required for production and/or stabilization of siRNAs and provide the first evidence for an Argonaute protein being involved in the regulation of retroposon transcript levels.
In a variety of eukaryotic organisms the introduction of synthetic double-stranded RNA (dsRNA), or the production of dsRNA from transgenes, transposons, viruses, or transgenic gene arrays, induces silencing of cognate genes (8, 19). Originally, the term RNA interference (RNAi) was introduced to indicate the experimental application of synthetic or transgene-derived dsRNA to down regulate a gene of interest (11). However, it has become evident that proteins involved in RNAi also function in other dsRNA-dependent gene silencing pathways operating at both the transcriptional and the posttranscriptional levels. The hallmarks of RNAi and related phenomena are small RNAs of approximately 21 to 26 nucleotides (nt), named small interfering RNAs (siRNAs), which, in association with protein factors, function as guides to trigger degradation of target transcripts (19). The search for proteins involved in RNAi through a combination of genetic and biochemical analyses has uncovered two gene families that appear to be ancient and evolutionarily conserved, namely, Dicer and Argonaute (4, 8). Whereas Dicer, an RNase III-related enzyme, is required for processing of the dsRNA to siRNAs, members of the Argonaute family have yet to be assigned a biochemical function(s).
The Argonaute gene family was first defined in Arabidopsis thaliana through a genetic screen (3), and proteins belonging to this family are characterized by the presence of two motifs: a PAZ domain found near the middle of the protein and a Piwi domain located close to the C terminus of the protein (6). Whereas the PAZ domain is poorly conserved, the 300-amino-acid Piwi domain of unknown function displays a high degree of similarity among Argonaute family members (4). The genomes of most multicellular organisms code for several Argonaute proteins. For instance, in Drosophila melanogaster there are four characterized Argonaute genes, namely, AGO1, AGO2, Piwi, and Aubergine, all of which have been implicated in RNAi and related silencing phenomena, and a fifth Argonaute member of unknown function was predicted from genomic DNA (4). In Arabidopsis, the Argonaute family consists of 10 members, but only three have been studied in detail, and Caenorhabditis elegans has more than 20 predicted Argonaute-like genes. In contrast to the complexity of AGO gene families in multicellular organisms, the genomes of single-cell eukaryotes analyzed so far encode one (Schizosaccharomyces pombe and Tetrahymena thermophila) or two (Neurospora crassa) Argonaute genes.
Despite the lack of a biochemical function(s) for Argonaute proteins, phenotypic analysis has implicated members of this family in a wide variety of biological processes, including silencing phenomena. For instance, RDE-1 is required for RNAi in C. elegans (13), QDE-2 is essential for quelling (posttranscriptional gene silencing) in N. crassa (5), and AGO1 and AGO2 are essential for RNAi in Drosophila (18, 35). At present the precise function of these proteins in the RNAi pathway remains to be elucidated, but it is known that mutations in RDE-1 or QDE-2 do not affect the accumulation of siRNAs (5, 28), and Drosophila AGO1 is not required for the generation of siRNAs in vitro (35). Silencing phenomena linked to Argonaute proteins include transcriptional and posttranscriptional gene silencing in Drosophila (27) and programmed gene rearrangements in Tetrahymena (24). Furthermore, RNAi genes in fission yeast, including the single member of the Argonaute family, have been shown to be required for epigenetic gene silencing at centromeres (33), the initiation of heterochromatin formation at the silent mating type region (15), and chromosome dynamics during mitosis and meiosis (14). Despite the emerging view that members of the Argonaute gene family play an essential role in a variety of silencing phenomena, there is at present no evidence that the control of mobile genetic elements, proposed to be one of the biological functions of RNAi (20, 31, 37), is linked to this protein family.
Trypanosoma brucei represents the earliest divergent eukaryote for which RNAi-mediated degradation of mRNA has been demonstrated (26). Previously, we reported that retroposon-derived siRNAs are constitutively expressed in trypanosomes, hinting that the RNAi mechanism is involved in maintaining low levels of retroposon transcripts (10). More recently, we have provided evidence that 10 to 20% of siRNAs are found associated with translating polyribosomes in the form of a ribonucleoprotein complex with an apparent molecular mass of 70 kDa (9). Here, we show that this RNP contains a member of the Argonaute family (AGO1) which is essential for RNAi. Cells deficient in AGO1 not only failed to accumulate siRNAs but also revealed an increased steady-state level of retroposon transcripts that was brought about by a combination of transcriptional activation of retroelements and increased stability of retroposon transcripts.
MATERIALS AND METHODS
Trypanosome cell lines.
Procyclic T. brucei was transfected as previously described (26). Cell lines constitutively expressing green fluorescent protein (GFP) dsRNA as a hairpin RNA were constructed by inserting a pXS-based construct in the tubulin locus of wild-type and AGO1−/− cells. The AGOT7 cell line was generated by inserting nt 8 to 469 of the AGO1 translated region between two opposing tetracycline (TET)-inducible T7 RNA polymerase promoters of pZJM (34). The construct was linearized with NotI for integration at the ribosomal DNA nontranscribed spacer region of strain 29.13.6, expressing the TET repressor and T7 RNA polymerase (36). In the AGO1-knockout cell line, we deleted both alleles by homologous recombination with PCR-generated cassettes, encoding either the blasticidin (BSR) or the hygromycin resistance marker gene (30), and the cell line was maintained in the presence of 10 and 50 μg of BSR and hygromycin/ml, respectively.
Antibodies used and polyclonal antibody production.
The anti-protein A antibody (Sigma) was used at a dilution of 1:75,000. The BB2 epitope tag corresponds to 10 amino acids from the immunologically well-characterized major structural protein of the Saccharomyces cerevisiae Ty1 virus-like particle, and mouse monoclonal antibodies to this epitope were generated in Keith Gull's laboratory (2). To produce anti-AGO1 antibodies, amino acids 569 to 860 of the T. brucei AGO1 protein, corresponding to the region between the PAZ and Piwi domains, were expressed as a His-tagged protein in Escherichia coli BL21 by using the vector pET-28a (Novagen), and polyclonal rabbit antibodies were raised against the purified recombinant protein.
Immunoprecipitations and Western blot analysis.
Cell extracts were prepared as described previously (9). Anti-BB2 antibodies bound to protein G-Sepharose beads in NET-2 buffer (150 mM NaCl, 50 mM Tris [pH 7.5], 0.05% NP-40) were mixed with cell extracts and incubated for 4 h at 4°C. The beads were then washed seven times with NET-2, and equivalent amounts of supernatants and beads were processed for Western blot analysis.
Proteins were transferred from polyacrylamide gels to nitrocellulose membranes by electrophoretic transfer, and membranes were blocked in saline solution (0.9% NaCl, 0.05% MgCl2, 3% bovine serum albumin, 10 mM Tris [pH 8.0], and 10% calf serum). Primary and secondary antibody incubations were done in the same solution. Membranes were washed (10 min each) two times in phosphate-buffered saline (PBS), two times in PBS containing 0.3% Tween 20, and two times in PBS after each antibody incubation. The horseradish peroxidase-conjugated secondary antibody (Roche) was used at a 1:6,000 dilution. Blots were then developed using ECL reagents (Amersham Biosciences) and exposed to Kodak RX film.
Other procedures.
Cells expressing tandem affinity purification (TAP)-tagged AGO1 were processed for double-label indirect immunofluorescence as described previously (26). RNA extraction, dsRNA transfection, and Northern blot analysis were performed as described previously (10). Cell fractionation, sucrose density gradient analysis of cytoplasmic extracts, and gel filtration on a Superdex 200 column (Amersham Biosciences) were carried out as described previously (9).
RESULTS
T. brucei AGO1 is a member of the Argonaute family.
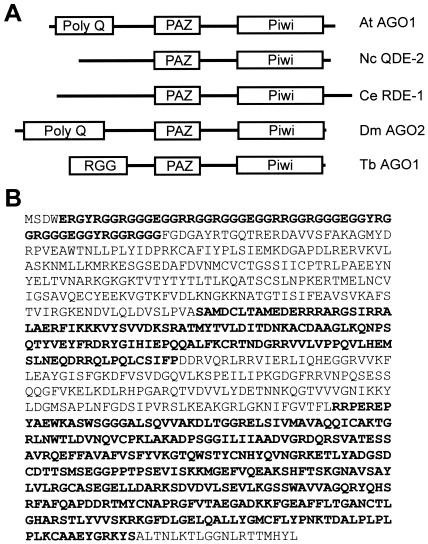
Through database mining we have identified a single-copy T. brucei gene with the signature motifs of the Argonaute family of proteins, namely, the PAZ and Piwi domains. We have named this gene AGO1. Automatic annotation by the Sanger Centre predicts a protein of 94.4 kDa (Sanger Centre number TRYP10.0.000587_99). However, BLAST searches indicated that the homology to other Argonaute proteins extended beyond the predicted termination codon. Indeed, cloning and sequencing of the AGO1 gene and mRNA revealed one discrepancy with the predicted gene, thus increasing the open reading frame to encode a protein with a predicted size of 98 kDa and a predicted pI of 9.2 (Fig. 1). So far, only one member of this family is present in the current T. brucei database. Similar to other Argonaute proteins, in AGO1 the well-conserved Piwi domain of approximately 330 amino acids is located close to the C terminus and the less well conserved PAZ domain of about 120 amino acids is found near the middle of the protein. The very 5′ end of the gene consists of five 33-nt direct repeats, and translation of these direct repeats results in a region rich in arginine and glycine residues with several arginine-glycine-glycine (RGG) repeats (Fig. 1).
FIG. 1.
Argonaute family members. (A) Domain structure of selected members of the Argonaute family. Abbreviations: At AGO1, A. thaliana Argonaute 1 (accession number U91995); Nc QDE-2, Neurospora crassa QDE-2 (accession number AF217760); Ce RDE-1, C. elegans RDE-1 (accession number AF180730); Dm AGO2, D. melanogaster Argonaute 2 (accession number AE003530); Tb AGO1, T. brucei Argonaute 1. The drawing is not to scale. (B) Protein sequence of T. brucei Argonaute 1. The domains schematically indicated in panel A are highlighted.
AGO1 is required for RNAi.
To investigate whether the AGO1 gene product functions in the RNAi pathway, we inserted a portion of the AGO1 coding region in the vector pZJM between opposing and TET-inducible T7 RNA polymerase promoters (34) and generated the stable cell line AGOT7. Upon induction with TET, the AGO1 mRNA declined in abundance, without any noticeable effects on cell viability (data not shown). We then tested for RNAi by assaying the ability of dsRNA corresponding to the α-tubulin gene to produce the FAT phenotype (cells with multiple nuclei, flagella, and mitochondrial genomes [27]) following electroporation into uninduced and TET-induced cells. By the use of nonsaturating amounts of α-tubulin dsRNA, approximately 39 to 56% of the cells became FAT in the uninduced culture (Table 1). In contrast, the percentage of FAT cells was significantly decreased in induced AGOT7 cells, with only 10 and 2% of the cells exhibiting the FAT phenotype following 1 and 4 days of induction, respectively. From this we concluded that the T. brucei AGO1 gene was likely to be required for the RNAi response.
TABLE 1.
Down regulation of AGO1 leads to a decrease in RNAi
| AGOT7 cell line | % FAT cells at day of growth:
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| −TET | 56 | 40 | 39 | 40 |
| +TET | 10 | 4 | 5 | 2 |
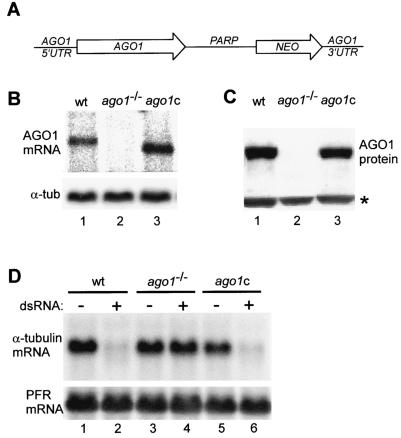
To begin to examine the role that AGO1 plays in RNAi in T. brucei, we next generated a cell line deficient in the AGO1 gene. We deleted both alleles by homologous recombination with PCR-generated cassettes, encoding either the BSR or the hygromycin resistance marker gene (30). As expected, we were not able to detect either the AGO1 mRNA (Fig. 2B, lane 2) or the protein in ago1−/− cells (Fig. 2C, lane 2). Confirming our results above (Table 1), ago1−/− cells did not respond to challenge with α-tubulin dsRNA, i.e., the mRNA remained intact (Fig. 2D, lane 4), and no FAT phenotype was apparent following electroporation of different amounts of α-tubulin dsRNA (data not shown). These results were consistent with an essential role of the AGO1 protein in the RNAi pathway.
FIG. 2.
The AGO1 gene is essential for RNAi. (A) Schematic representation of the complementation cassette, which contains in a 5′-to-3′ direction 330 nt of AGO1 5′-flanking sequences including processing signals for 5′-end formation of the AGO1 mRNA (AGO1 5′UTR), the AGO1 open reading frame of 2,712 bp (AGO1), the procyclic acidic repetetive protein intergenic region of 990 bp (PARP), the neomycin resistance gene (NEO), and finally 780 bp of AGO 3′-flanking sequences (AGO1 3′UTR). (B) Northern blot analysis. RNA isolated from wild-type cells (wt; lane 1), AGO1-knockout cells (ago1−/−; lane 2), and ago1−/− cells complemented with the AGO1 gene (ago1c; lane 3) was probed for AGO1 mRNA (upper panel). α-Tubulin mRNA served as a control for RNA recovery and loading (lower panel). (C) Western blot. Total cell extracts from wild-type cells (wt; lane 1), AGO1-knockout cells (ago1−/−; lane 2), and ago1−/− cells complemented with the AGO1 gene (ago1c; lane 3) were analyzed by Western blotting for the level of AGO1 protein by using anti-AGO1 polyclonal antibodies. An immunologically cross-reacting protein was used as a loading control (indicated by an asterisk). (D) Assay for RNAi. Wild-type cells (wt; lanes 1 and 2), AGO1-knockout cells (ago1−/−; lanes 3 and 4), and ago1−/− cells complemented with the AGO1 gene (ago1c; lanes 5 and 6) were challenged with poly(dI-dC) (lanes 1, 3, and 5) or α-tubulin dsRNA (lanes 2, 4, and 6), and the level of α-tubulin mRNA was monitored by Northern blotting (upper panel). Paraflagellar rod (PFR) mRNA served as a control for RNA recovery and loading (lower panel).
To validate the notion that the loss of RNAi was a specific effect of AGO1 elimination, we reintroduced a copy of the AGO1 gene into ago1−/− cells. By a combination of PCR and restriction fragment assembly we generated a complementation cassette designed to integrate by homologous recombination at one of the two ago1 knockout alleles (Fig. 2A). Thus, a restriction fragment harboring the cassette was electroporated into ago1−/− cells and Neor cells were selected. The resulting cells were shown to express AGO1 mRNA, which is slightly smaller than that in wild-type cells due to a different and shorter 3′ untranslated region (UTR) (Fig. 2B, lane 3), as well as AGO1 protein (Fig. 2C, lane 3). Importantly, the levels of both AGO1 mRNA and protein were comparable in the complemented and wild-type cells. When the complemented cells were challenged with α-tubulin dsRNA, the RNAi response was restored, since the α-tubulin mRNA was degraded (Fig. 2D, lane 6), and the FAT phenotype was manifested in the majority of the cells (data not shown). Taken together, these results unequivocally established an essential role for AGO1 in the T. brucei RNAi pathway.
AGO1 is cytoplasmic and is found both soluble and cosedimenting with polyribosomes.
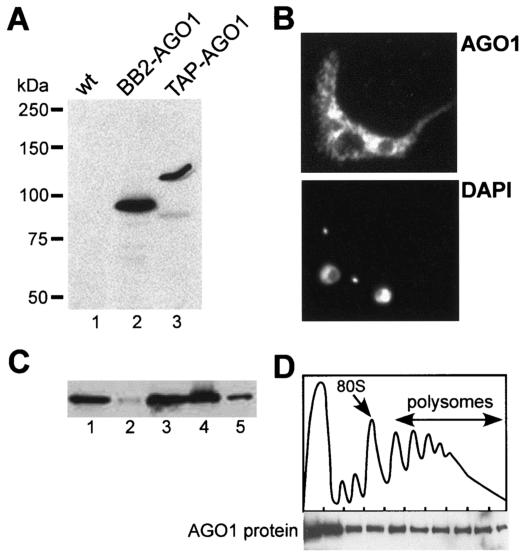
To analyze the cellular distribution of AGO1, we epitope tagged the AGO1 protein by inserting in the N-terminal RGG domain of the complementation cassette either a BB2 tag (BB2-AGO1) or a triple tag (TAP-AGO1), namely, the BB2 tag followed by the TAP tag, consisting of two protein A domains and a calmodulin binding domain (29). After transfection of the corresponding constructs into ago1−/− cells, the expression of BB2- or TAP-AGO1 was confirmed by Western blot analysis with an anti-BB2 antibody (Fig. 3A). Moreover, both tagged AGO1 proteins were functional in RNAi, since challenging the cells with α-tubulin dsRNA resulted in the degradation of α-tubulin mRNA (data not shown).
FIG. 3.
Cellular localization of AGO1. (A) Western blot with anti-BB2 antibodies of extracts from wild-type (lane 1), BB2-tagged AGO1 (lane 2), and TAP-tagged AGO1 (lane 3) cells. (B) Cells expressing TAP-tagged AGO1 were processed for indirect immunofluorescence as described previously (26) by staining with a rabbit anti-protein A antibody (subpanel AGO1). DNA was stained with 4′,6′-diamidino-2-phenylindole (subpanel DAPI). The small dots represent the kinetoplast DNA. (C) Western blot of TAP-tagged AGO1 with a rabbit anti-protein A polyclonal antibody of equivalent amounts of total extract (lane 1), postnuclear pellet (lane 2), supernatant (lane 3), S100 fraction (lane 4), and ribosomal pellet (lane 5). (D) Sucrose density gradient analysis of a cytoplasmic extract from T. brucei cells expressing TAP-tagged AGO1 (9). The upper panel shows the absorbance profile at 254 nm, and the positions of the 80S monosome and polyribosomes are indicated. The lower panel shows a Western blot analysis of the sucrose density gradient fractions using a rabbit anti-protein A polyclonal antibody.
Trypanosomes expressing TAP-AGO1 were then used for immunofluorescence analysis (Fig. 3B) with a rabbit anti-protein A antibody. AGO1 was predominantly cytoplasmic but was not homogeneously distributed in the cytoplasm. In particular, the fluorescence signal appeared more intense in the perinuclear region and there were “clusters” of fluorescence superimposed on a diffuse cytoplasmic signal.
Next, we analyzed the distribution of TAP-AGO1 in cell extracts. Figure 3C shows the results of a fractionation by differential centrifugation of a whole-cell extract prepared by detergent lysis. By this analysis TAP-AGO1 was mostly found in the postnuclear supernatant (lane 3) and in the S100 fraction (lane 4). However, there was a proportion of AGO1 that sedimented at 100,000 × g as large complexes (lane 5). To further evaluate the sedimentation characteristics of these large AGO1 complexes, a cytoplasmic extract was centrifuged through a 15 to 50% sucrose gradient and individual fractions were analyzed by Western blotting with the anti-protein A antibody (Fig. 3D). AGO1 was found near the top of the gradient, where soluble material and small ribonucleoprotein particles sediment, as well as heterodispersed throughout the gradient fractions, where polyribosomes sediment. A similar distribution of the AGO1 protein was detected in wild-type cells by using a polyclonal anti-AGO1 antibody (data not shown).
AGO1 is associated with siRNAs.
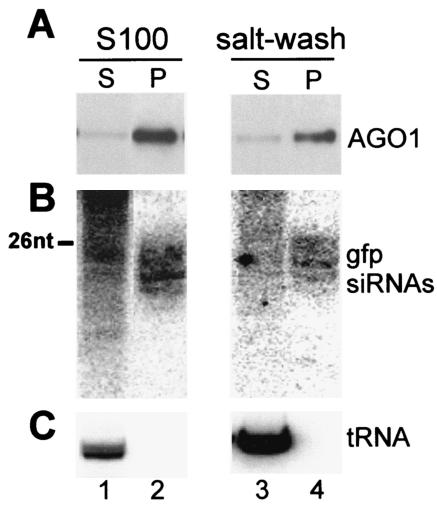
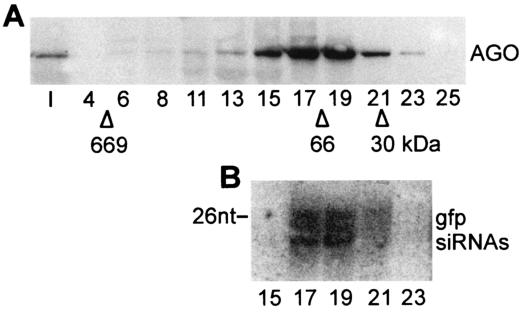
We have recently shown that 10 to 20% of siRNAs are associated with translating polyribosomes and that the polysome-associated siRNAs can be released by salt extraction as a ribonucleoprotein particle with an apparent molecular mass of approximately 70 kDa (9). The sedimentation profile of AGO1 in the sucrose density gradient of Fig. 3D paralleled the distribution of siRNAs in trypanosome extracts, and in preliminary experiments we found that, similarly to the siRNAs, AGO1 present in a polysomal pellet was rendered soluble upon salt extraction (data not shown). To test whether siRNAs are in a complex with the AGO1 protein, we prepared an S100 and a corresponding ribosome salt-wash fraction from a cell line expressing BB2-AGO1, as well as GFP dsRNA. Next, we immunoprecipitated BB2-AGO1 from these two fractions by using the monoclonal BB2 antibody coupled to protein A beads. Western blot analysis revealed that most of the AGO1 protein was immunoprecipitated from the S100 and the ribosome salt-wash fractions (Fig. 4A). Since Northern blot analysis showed that a significant proportion of GFP siRNAs were enriched in the respective immunoprecipitates (Fig. 4B), we concluded that AGO1 is part of a complex containing siRNAs. Further fractionation of the ribosome salt-washed material on a Superdex 200 gel filtration column revealed that the majority of BB2-AGO1 cofractionated with GFP siRNAs (Fig. 5) with an apparent molecular mass of 70 kDa, as previously described for siRNAs (9). Similar results were obtained with the S100 fraction (data not shown).
FIG. 4.
AGO1 is associated with siRNAs. S100 and ribosome salt-washed fractions from cells expressing BB2-tagged AGO1 and GFP dsRNA were subjected to immunoprecipitations with anti-BB2 antibodies, and supernatant (S) and pellet fractions (P) were processed for Western blot analysis for AGO1 (A) and for Northern blot analysis for GFP siRNAs (B), as well as for initiator methionyl tRNA to control for immunoprecipitation specificity (C). A DNA size marker of 26 nt is indicated in panel B.
FIG. 5.
AGO1 cofractionates with GFP siRNAs. The salt-extracted material from the ribosome pellet of cells expressing BB2-tagged AGO1 and GFP dsRNA was loaded onto a Superdex 200 column, and selected fractions were subjected to Western blot analysis with anti-BB2 antibodies (A) and Northern blotted for the presence of GFP siRNAs (B). The elution positions of carbonic anhydrase (30 kDa), bovine serum albumin (66 kDa), and thyroglobulin (669 kDa) are indicated. A DNA size marker of 26 nt is indicated in panel B.
AGO1 plays a role in the accumulation of siRNAs.
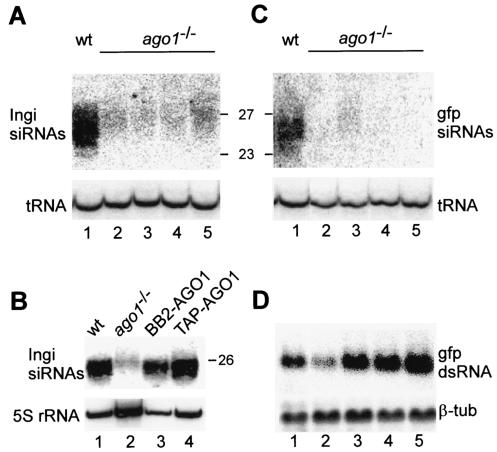
To begin to examine the function of AGO1 in the RNAi mechanism, we analyzed the steady-state levels of siRNAs in ago1−/− cells. First, we monitored endogenous siRNAs homologous to the Ingi retroposon transcripts by Northern blot hybridization with RNA derived from wild-type cells and four independently cloned ago1−/− cell lines (Fig. 6A). This experiment showed that the accumulation of Ingi siRNAs was severely reduced in ago1−/− cells compared to wild-type cells (compare lane 1 to lanes 2 to 5). Interestingly, the residual small RNAs present in ago1−/− cells were a few nucleotides longer than the average siRNAs present in wild-type cells. Complementation of the RNAi deficiency with the BB2- or TAP-AGO1 cassette restored both the abundance and the size of Ingi siRNAs (Fig. 6B, lanes 3 and 4).
FIG. 6.
siRNA accumulation is dependent on AGO1. (A) The level of Ingi siRNAs was probed by Northern blotting in wild-type cells (wt; lane 1) and four independently cloned cell lines deficient in the AGO1 protein (ago1−/−; lanes 2 to 5). RNA size markers in nucleotides are indicated on the right. The hybridization to initiator methionyl tRNA (tRNA) served as a loading control. (B) Abundance of Ingi siRNAs in wild-type (lane 1), AGO1-deficient (lane 2), BB2-AGO1-complemented (lane 3), and TAP-AGO1-complemented (lane 4) cells. The level of 5S rRNA was used as a loading control. (C) Northern blot analysis for GFP siRNAs of RNA isolated from wild-type cells expressing GFP dsRNA (lane 1) and four independent clonal ago1−/− cell lines expressing GFP dsRNA (lanes 2 to 5). RNA size markers in nucleotides are indicated on the left. The hybridization to initiator methionyl tRNA (tRNA) served as a loading control. (D) Detection of GFP dsRNA in the cell lines described in panel C, with β-tubulin mRNA (β-tub) serving as a loading control.
Second, we examined whether ago1−/− cells were impaired in the accumulation of siRNAs derived from a transgene. To this end, we generated ago1−/− cell lines constitutively expressing GFP dsRNA as a hairpin RNA. All four cell lines tested expressed transcripts diagnostic of GFP dsRNA, albeit to different levels (Fig. 6D). This variable level of expression most likely reflects integration of the GFP hairpin transgene at different and perhaps multiple sites in the tubulin gene array. In contrast, GFP siRNAs were almost undetectable in cells expressing GFP dsRNA (Fig. 6C; compare lane 1 to lanes 2 to 5). Similarly to what we observed for the retroposon-derived siRNAs, at least in one cell line the residual GFP siRNAs were of the “long” kind (Fig. 6C, lane 3). In conclusion, ago1−/− cells are deficient in the accumulation of siRNAs derived from endogenous retroposon transcripts, as well as from a transgene, and the residual amounts of small RNAs still detected in ago1−/− cells are of a distinct size class.
AGO1 affects the accumulation of retroposon transcripts.
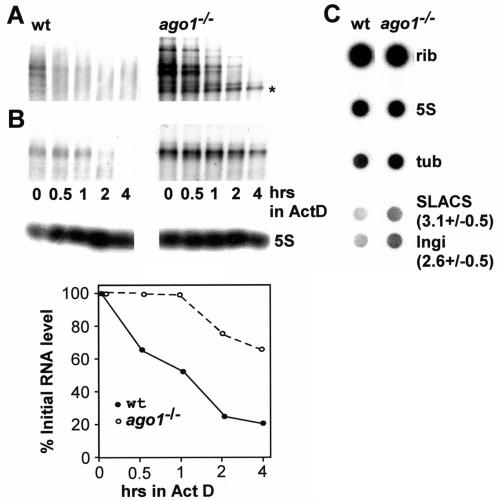
Our previous sequence analysis of siRNAs in T. brucei revealed a large proportion of siRNAs derived from Ingi and spliced leader-associated conserved sequence (SLACS) retroposon elements and suggested that RNAi down regulates the steady-state levels of retroposon transcripts (10). Consequently, we would predict that elimination of the RNAi response would result in an increased steady-state level of retroposon transcripts. To test this, total RNA from wild-type and ago1−/− trypanosomes was analyzed by Northern blot hybridization (Fig. 7A and B). Indeed, we found that Ingi and SLACS transcripts were approximately three- and fivefold more abundant, respectively, in ago1−/− cells than in wild-type cells. To address the origin of this difference, we first evaluated the decay rate of Ingi (Fig. 7A) and SLACS (Fig. 7B) transcripts in wild-type and ago1−/− cells over a period of 4 h after blocking of transcription with actinomycin D. The pattern of Ingi hybridization was quite complex with several discrete bands superimposed on a smear (Fig. 7A). This is consistent with previous reports by others (25) and reflects the highly dispersed genomic organization of the Ingi retroposon family (22). We observed that the decay rate of the majority of the discrete transcripts visible by Northern blotting was similar in ago1−/− and wild-type cells, with the exception of one transcript, indicated by an asterisk in Fig. 7A, which appeared to be longer lived in ago1−/− cells than in wild-type cells. In contrast, the SLACS element, which is a site-specific retroposon inserted in the spliced leader RNA gene cluster (1), generated a major transcript whose decay was at least four times slower in ago1−/− cells than in wild-type cells (Fig. 7B). Next, we asked whether AGO1 deficiency had any effect on the transcription of Ingi or SLACS elements. To this end we synthesized radiolabeled RNA in permeabilized trypanosome cells and analyzed the abundance of newly synthesized Ingi or SLACS transcripts by dot blot hybridization (Fig. 7C). Using the large rRNA and 5S RNA hybridization values as standards for normalizing the dot blots, we determined that transcription of Ingi and SLACS elements was increased approximately 2.6 (±0.5)- and 3.1 (±0.5)-fold, respectively, in ago1−/− cells.
FIG. 7.
AGO1 affects the accumulation of Ingi and SLACS transcripts. (A) Northern blot of Ingi transcripts in wild-type (wt) and ago1−/− cells over a period of 4 h following the addition of actinomycin D. The asterisk indicates a transcript which appeared to be more long lived in ago1−/− cells than in wild-type cells. (B) Northern blot of SLACS transcripts in wild-type and ago1−/− cells over a period of 4 h following the addition of actinomycin D (upper panel). Hybridization to 5S RNA served as a loading control (middle panel). SLACS transcripts were quantitated relative to 5S RNA (lower panel). (C) Newly synthesized RNA from wild-type and ago1−/− cells was hybridized to the following DNAs spotted onto a nitrocellulose filter: large rRNAs (rib), 5S rRNA (5S), α-tubulin (tub), SLACS, and Ingi. The numbers next to the SLACS and Ingi hybridizations indicate the fold increase in ago1−/− cells relative to wild-type cells. The experiment was repeated three times, and the standard deviation is indicated.
DISCUSSION
In this paper we describe the characterization of the first component of the RNAi machinery in T. brucei, namely, a member of the Argonaute gene family. Like other members of this family, the T. brucei AGO1 protein is a highly basic protein and contains the signature domains PAZ and Piwi (6), which at present do not have a defined function. One characteristic feature of AGO1 is the presence of an N-terminal domain rich in arginine and glycine residues with a high abundance of RGG repeats, resembling the RGG RNA binding motif. Similar repeats, although not as prevalent, are found in two members of the Arabidopsis Argonaute family (accession no. NP_565662 and NP_175274). Arginine residues present within RGG domains are often asymmetrically dimethylated, and it is intriguing that the N terminus of AGO1 has several repeats that are preferred for asymmetric arginine dimethylation, namely, G/FGGRGGG/F, making it a likely candidate for this posttranslational modification (21).
Our genetic analysis demonstrated that AGO1 is required for RNAi in T. brucei. Cells lacking both copies of the AGO1 gene are deficient in sequence-specific mRNA degradation when challenged with dsRNA. These cells did not show a substantial growth defect, suggesting that the RNAi mechanism is dispensable for cell growth in the laboratory-adapted insect-form trypanosomes. However, since RNAi deficiency in T. brucei leads to upregulation of retroposon transcripts, it is possible that mobilization of retroelements is occurring in ago1−/− cells, and thus, we cannot exclude the possibility that the loss of RNAi might have detrimental effects on the genome integrity in the long term.
By immunofluorescence and cell fractionation experiments we found that AGO1 is mainly localized in the cytoplasm. However, the immunofluorescence pattern is not typical of a soluble protein. Rather, a proportion of AGO1 appears to be compartmentalized in some subcellular compartment, whose identity is at present uncertain. It is possible that a proportion of AGO1 is associated with endoplasmic reticulum-bound polyribosomes translating mRNAs coding for membrane or secretory proteins. Although we do not have biochemical evidence that AGO1 is directly associated with membranes, it is worth mentioning that EIF2C1/hAGO1, also known as GERp95, is a peripheral membrane protein that localizes primarily to the Golgi complex or to the endoplasmic reticulum, depending on the cell type (7).
Previous studies in Drosophila revealed that the RNA-induced silencing complex, containing AGO2 and most siRNAs, can be salt extracted from a 200,000 × g pellet fraction from S2 cell extracts (17). Moreover, affinity purification of QDE-2, a Neurospora AGO family member, uncovered a complex containing siRNAs (5), and affinity purification of human RNA-induced silencing complex showed that EIF2C1 and EIF2C2 are part of a complex containing siRNAs (23). Here, we have provided evidence that in trypanosomes siRNAs are also in a complex containing AGO1. This is probably not surprising, considering the AGO2-QDE-2-EIF2C results. What is quite unique to trypanosomes is the small size of about 70 kDa of the ribonucleoprotein particle, where siRNAs and AGO1 reside. We now know that this measurement, which was obtained on a gel filtration column, is an underestimate, since the particle contains at least one AGO1 molecule of about 98 kDa plus siRNAs, which contribute an additional 8 to 16 kDa depending on whether one or both strands of siRNAs are present. Notwithstanding this uncertainty about the mass of the AGO1 ribonucleoprotein particle, it is tempting to speculate, as suggested previously (18), that one of the functions of AGO1 is to bind siRNAs. Similar to what we described for siRNAs (9), we found that a proportion of the 70-kDa AGO1 RNP cosediments with polyribosomes and can be released from polyribosomes by salt extraction. It could be argued that the highly basic AGO1 protein binds to ribosomes nonspecifically. However, we think that this interpretation is unlikely, because we recently determined that a mutant protein lacking the RGG domain does not bind to polyribosomes and its calculated pI is only slightly lower than that of wild-type AGO1 (H. Shi, A. Djikeng, C. Tschudi, and E. Ullu, unpublished data).
At the RNA level ago1−/− cells accumulate significantly reduced amounts of endogenous siRNAs and almost undetectable levels of transgene-derived GFP siRNAs. These observations are consistent with the possibility that AGO1 is required to produce and/or stabilize siRNAs. In this respect the siRNA phenotype of ago1−/− trypanosomes resembles that of Piwi mutant flies (27), as well as that of the alg-1/alg-2 mutant worms (12), but is different from that of rde-1 and qde-2 mutations in C. elegans (28) and Neurospora (5), respectively. Thus, our results support the notion that there are functional differences between AGO family members in terms of their role in production and/or stabilization of siRNAs.
The residual small RNAs homologous to the Ingi retroposon that we detected in ago1−/− cells had a size distribution a few nucleotides longer than that of wild-type Ingi siRNAs. These “longer” small RNAs might result from a defect in processing of dsRNA or might be produced by a different “Dicer” activity and might have a different function than siRNAs. The presence of different size classes of small RNAs has been reported previously in plants, where short (21- to 23-nt) siRNAs are required for mRNA degradation and long (25-nt) siRNAs correlate with systemic silencing (16), and their identity as bona fide siRNAs has been confirmed (32). More recently, inactivation of the Arabidopsis AGO4 gene has been shown to control accumulation of 25-nt-long siRNAs corresponding to the retroposon AtSN1 (38).
The second RNA phenotype that we uncovered in ago1−/− cells concerns the increased accumulation of Ingi and SLACS retroposon transcripts in steady-state RNA. Our results further suggest that retroposon transcript abundance is regulated at both the transcriptional (Ingi and SLACS) and posttranscriptional (SLACS) levels. This is to our knowledge the first report that links a member of the Argonaute gene family to the expression of retroposon transcripts. This observation ties in well with our previous results demonstrating the presence of constitutively expressed retroposon-derived siRNAs (10) and provides further evidence that RNAi is involved in regulating the abundance of retroposon transcripts in trypanosomes. It will be interesting to determine whether retroposition is activated in AGO1-deficient cells and in what cellular compartment RNAi-mediated degradation of retroposon transcripts takes place.
Acknowledgments
This work was supported by NIH grant AI28798 to E.U. A.D. was supported in part by a James Hudson Brown-Alexander Brown Coxe Fellowship, and C.T. is the recipient of a Burroughs Wellcome Fund New Investigator Award in Molecular Parasitology.
We are grateful to Paul Englund and Keith Gull for their generous supply of reagents. We thank Susan Baserga and Sandy Wolin for critical comments on the manuscript.
REFERENCES
- 1.Aksoy, S., T. M. Lalor, J. Martin, L. H. Van der Ploeg, and F. F. Richards. 1987. Multiple copies of a retroposon interrupt spliced leader RNA genes in the African trypanosome, Trypanosoma gambiense. EMBO J. 6:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastin, P., Z. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235-239. [DOI] [PubMed] [Google Scholar]
- 3.Bohmert, K., I. Camus, C. Bellini, D. Bouchez, M. Caboche, and C. Benning. 1998. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17:170-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmell, M. A., Z. Xuan, M. Q. Zhang, and G. J. Hannon. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16:2733-2742. [DOI] [PubMed] [Google Scholar]
- 5.Catalanotto, C., G. Azzalin, G. Macino, and C. Cogoni. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerutti, L., N. Mian, and A. Bateman. 2000. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25:481-482. [DOI] [PubMed] [Google Scholar]
- 7.Cikaluk, D. E., N. Tahbaz, L. C. Hendricks, G. E. DiMattia, D. Hansen, D. Pilgrim, and T. C. Hobman. 1999. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell 10:3357-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denli, A. M., and G. J. Hannon. 2003. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 28:196-201. [DOI] [PubMed] [Google Scholar]
- 9.Djikeng, A., H. Shi, C. Tschudi, S. Shen, and E. Ullu. 2003. An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA 9:802-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djikeng, A., H. Shi, C. Tschudi, and E. Ullu. 2001. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA 7:1522-1530. [PMC free article] [PubMed] [Google Scholar]
- 11.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 12.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 13.Grishok, A., H. Tabara, and C. C. Mello. 2000. Genetic requirements for inheritance of RNAi in C. elegans. Science 287:2494-2497. [DOI] [PubMed] [Google Scholar]
- 14.Hall, I. M., K. Noma, and S. I. Grewal. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 16.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 18.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 19.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 20.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk. 1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S., B. M. Merrill, R. Rajpurohit, A. Kumar, K. L. Stone, V. V. Papov, J. M. Schneiders, W. Szer, S. H. Wilson, W. K. Paik, and K. R. Williams. 1997. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry 36:5185-5192. [DOI] [PubMed] [Google Scholar]
- 22.Kimmel, B. E., O. K. ole-MoiYoi, and J. R. Young. 1987. Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology with mammalian LINEs. Mol. Cell. Biol. 7:1465-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez, J., A. Patkaniowska, H. Urlaub, R. Luhrmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 24.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a Piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 25.Murphy, N. B., A. Pays, P. Tebabi, H. Coquelet, M. Guyaux, M. Steinert, and E. Pays. 1987. Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J. Mol. Biol. 195:855-871. [DOI] [PubMed] [Google Scholar]
- 26.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pal-Bhadra, M., U. Bhadra, and J. A. Birchler. 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell 9:315-327. [DOI] [PubMed] [Google Scholar]
- 28.Parrish, S., and A. Fire. 2001. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7:1397-1402. [PMC free article] [PubMed] [Google Scholar]
- 29.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 30.Shen, S., G. K. Arhin, E. Ullu, and C. Tschudi. 2001. In vivo epitope tagging of Trypanosoma brucei genes using a one step PCR-based strategy. Mol. Biochem. Parasitol. 113:171-173. [DOI] [PubMed] [Google Scholar]
- 31.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello. 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 32.Tang, G., B. J. Reinhart, D. P. Bartel, and P. D. Zamore. 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17:49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 35.Williams, R. W., and G. M. Rubin. 2002. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc. Natl. Acad. Sci. USA 99:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 37.Wu-Scharf, D., B. Jeong, C. Zhang, and H. Cerutti. 2000. Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290:1159-1162. [DOI] [PubMed] [Google Scholar]
- 38.Zilberman, D., X. Cao, and S. E. Jacobsen. 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716-719. [DOI] [PubMed] [Google Scholar]