Abstract
Glyconanomaterials, nanomaterials carrying multiple carbohydrate ligands, provide an excellent platform for sensitive protein recognition. Using nanomaterials as the scaffold, multivalent interactions between glycan ligands and proteins have been demonstrated. However, the quantitative analysis of the binding affinity of these glyconanomaterials has been lacking. In this article, we report a new method to measure the binding affinity of glyconanoparticle (GNP)-protein interactions based on a fluorescent competition binding assay, which yielded the apparent dissociation constant (Kd) of GNPs with the interacting protein. Au nanoparticles conjugated with underivatized mono-, oligo-, poly-saccharides were synthesized using our recently developed photocoupling chemistry. The affinities of these GNPs with lectins were measured and were several orders of magnitude higher than the corresponding free ligands with lectins. The effect of ligand display on the binding affinity of GNPs was furthermore studied where GNPs of varying linker type, spacer length, ligand density, and nanoparticle size were prepared and Kd values determined. The long spacer linker containing hydrocarbon and ethylene oxide units gave the highest binding affinity as well as assay sensitivity. The binding affinity increased with ligand density in general, showing a drastic increase in affinity at low ligand density. In addition, the affinity enhancement was more pronounced on smaller NPs than the larger ones. These results not only demonstrate that the binding affinity of GNPs is highly influenced by how the ligands are presented on the nanoparticles, but also pave the way for tailor-made glyconanomaterials with tunable affinity by way of ligand display.
Introduction
Biofunctionalized nanomaterials constitute an emerging class of entities with unique applications in a variety of areas, such as sensing, imaging, targeting, delivery, diagnostics and therapy.1–6 As a subset of this class, carbohydrate-conjugated nanomaterials – glyconanomaterials – are witnessing increasing interest as a new platform for probing and controlling biological functions.7 Carbohydrates mediate numerous recognition events through their interactions with proteins and other biological entities, and complex carbohydrate structures are involved in, for example, cell communication and trafficking, tumor genesis and progression, immune responses, fertilization, apoptosis, and infection.8–14 A general feature of carbohydrate-protein interactions relates to their inherent complexity, and multivalent ligand expression is frequently used to achieve sufficiently high affinities required for sensitive recognition.15–17 In analogy to ligand clustering at cell surfaces, nanomaterials can function as efficient nano-sized scaffolds for multiple carbohydrate presentation. The functionality and biocompatibility of such nanomaterials open up a wide range of biological applications, where the unique physical and chemical properties of nanomaterials can be interfaced with biological systems.
Strong binding enhancements resulting from ligand presentation at the surfaces of nanomaterials have been demonstrated in numerous studies.18–20 However, the current investigations are generally short of in-depth characterization of the structures, compositions and densities of the surface ligands, as well as the biological activities of the constructs, since conventional surface analysis methods have been less optimized for nanomaterials. Owing to the increasing interest in nanomaterials research, recent development in spectroscopic and microscopic techniques has afforded detailed structural and compositional information for nanomaterials.21–23 Nevertheless, quantitative analysis of multivalent biological affinity at nanoparticle surfaces still represents a considerable challenge. Furthermore, very limited methods were available to measure the binding affinity of glyconanoparticles with proteins.24–26
In this article, we present detailed quantitative analyses on nanoparticle-based multivalent carbohydrate-protein interactions. The effects resulting from the nanoparticle size was investigated, and the ligand presentation with respect to ligand density, surface environment, and linker spacer was studied. The well-established interaction system involving the lectin Con A, together with a panel of carbohydrate species, was chosen as the target. This lectin is present as a tetramer at pH > 7, and possesses specific affinities to α-D-mannopyranoside, α-D-glucopyranoside and their derivatives.27,28 The system constitutes an excellent model for investigating the multivalency effect,29 and studies on mannose–Con A interactions have established that multivalent effects are highly sensitive to a range of factors, including the number of binding sites, the ligand density, the structure of the ligand linkers, and the coupling chemistry of the ligand attachments.30–33 In the present study, a recently developed photochemical carbohydrate immobilization technique was adopted,34,35 and an array of GNPs functionalized with different mono-, oligo-, and poly-saccharides were prepared and ligand densities determined. A fluorescence-based ligand competition assay was employed for the quantitative analysis of the binding affinities of the GNPs where the apparent Kd values of the resulting GNPs were determined. The binding affinity of GNPs with respect to the ligand density, spacer length, linker structure, as well as the nanoparticle size was furthermore investigated.
Experimental Section
Materials
Hydrogen tetrachloroaurate (III) hydrate (HAuCl4·XH2O, 99.9%-Au) was purchased from Strem Chemicals (Newburyport, MA). Sodium citrate was obtained from Mallinckrodt. 1-Ethyl-3-(3- dimethylaminopropyl)carbodiimide (EDAC) hydrochloride, D-(+)-mannose (Man), methyl α-D-mannopyranoside (MeMan), D-(+)-glucose (Glc), D-(+)-galactose (Gal), maltopentaose (Glc5), dextran 40 (Dex, MW ca. 40,000), 1-hexanethiol (>96%), Tween 20 were obtained from TCI America. (1-Mercaptoundec-11-yl)tetra(ethylene glycol), 4-dimethyl aminopyridine (DMAP), 6-mercapto-1-hexanol (97%), 11-mercapto-1-undecanol (97%), zinc powder, anthrone (97%), FITC conjugated Con A (lectin from Canavalia ensiformis (Jack bean), Type IV) (Con A-FITC), bovine serum albumin (BSA) were purchased from Sigma-Aldrich. 2-O-α-D-Mannopyranosyl-D-mannopyranose (Man2) and 3,6-di-O-(αD-mannopyranosyl)-D-mannopyranose (Man3) were obtained from V-Labs Inc (Covington, Louisiana). All chemicals were used as received without purification. Water used was from a Milli-Q ultrapure water purification system. Dialysis tubes (G-Biosciences Tube-O-dialyzer, 15K, medium) were purchased from VWR International. The phosphate buffer solution in this study was prepared by dissolving a phosphate buffered saline tablet (Sigma) in 200 mL Milli-Q water to yield pH 7.4 phosphate buffer (0.01 M) containing KCl (0.0027 M) and NaCl (0.137 M).
1H and 13C NMR spectra were collected on a Bruker 400 MHz NMR spectrometer. Infrared spectra were measured on a Perkin Elmer 2000 Fourier transform spectrometer. UV-vis spectra were recorded on a Perkin Elmer Lambda 45 UV-vis spectrophotometer. Fluorescence measurements were conducted on a PTI spectrofluorometer (Photon Technology International). TEM images were obtained on a JEOL 100CX transmission electron microscope operating at an accelerating bias voltage of 100 kV. The specimens were prepared by dropping nanoparticles suspension (10 μL) onto a 200 mesh copper grid (coated with carbon supporting film, Electron Microscopy Sciences). Dynamic light scattering (DLS) experiments were carried out on Horiba LB-550 Dynamic Light Scattering Nano-Analyzer.
Preparation of carbohydrate-conjugated gold nanoparticles
Citrate-protected Au NPs, ~22 nm in diameter, were prepared by adding sodium citrate (1 wt%, 1.8 mL) to a boiling solution of HAuCl4 (0.25 mM, 100 mL) under vigorous stirring, and the boiling was continued for an additional 5 min when the solution became purple and finally light red.36 PFPA-thiol 3a–3d were synthesized following the procedures in Scheme 1S (see Supporting Information for detailed synthesis and characterization). In a 250-mL flask, the Au NP solution prepared above (50 mL) was mixed with a solution of PFPA-thiol in ethanol (2.5 mM, 50 mL), and the resulting solution was stirred for 3 h. The solution was centrifuged at 10,000 rpm for 15 min, and the solid obtained was re-dispersed in ethanol (10 mL). The centrifugation and re-dispersion process was repeated for 3 times to remove excess PFPA-thiol, and the resulting PFPA-functionalized Au NPs was finally dispersed in acetone (10 mL). The concentration of the Au NP solution, about 10 nM, was determined by drying the sample under reduced pressure for 3 h and weighing. The carbohydrate coupling was carried out following the procedure reported previously.34 A solution of PFPA-functionalized Au NPs in acetone (10 mL) was placed in a flat-bottomed dish, and an aqueous solution of carbohydrate (1 mM, 0.5 mL) was added. The mixture was covered with a 280-nm long-path optical filter (WG-280, Schott Glass) and was irradiated with a 450-W medium pressure Hg lamp (Hanovia) for 10 min under vigorous stirring. Centrifugation of the solution at 12,000 rpm for 15 min separated the carbohydrate-attached Au NPs as precipitates. Excess carbohydrate was removed by membrane dialysis in water for 24 hours. Before binding experiments, the nanoparticles were incubated in the pH 7.4 PBS buffer solution containing 0.01% Tween 20 and 3% BSA for 30 min, centrifuged, and incubated in a pH 7.4 PBS solution without BSA for further use.37
Determination of carbohydrate ligand density on GNPs
A previously-developed colorimetry method was used to determine the ligand density on Au NPs.34 Calibration curves were obtained for each carbohydrate where carbohydrate solutions of various concentrations were incubated with anthrone/sulfuric acid and the absorbances at 620 nm were measured (See Figure 1S, Supporting Information for calibration curves for all carbohydrates used in the study). Carbohydrates coupled on nanoparticles were subjected to the same assay where solutions of the GNPs in Milli-Q water (30–50 μg/0.5 mL) were treated with anthrone/H2SO4. Background absorption due to Au NPs themselves was accounted for by treating citrate-protected Au NPs solution of the same concentration with anthrone/H2SO4, and the absorbance at 620 nm was subtracted from that of the GNPs. The amount of surface-bound carbohydrate was then computed from the corresponding calibration curve.
Preparation of GNPs of varied carbohydrate density
Mixed thiol solutions were prepared from PFPA-thiol and 1-hexanethiol in ethanol, with the mole percentage of PFPA-thiol varying from 10% to 98%. The functionalization of Au NPs with mixed thiols followed the general functionalization procedure described above, except that the pure thiol solution was replaced by the mixed thiol solution when treating citrate-protected Au NPs. The subsequent carbohydrate coupling was carried out following the same procedure described in detail above.
Fluorescence competition binding assay
AuNP-a-Man solutions of various concentrations (1 × 10−8 –5 nM) were prepared from the stock solution (10 nM). The Con A-FITC solution (190 nM) was prepared in pH 7.4 PBS buffer containing MnCl2 (1 mM) and CaCl2 (1 mM). To the AuNP-a-Man solution (1 mL) in a 1.5-mL microcentrifuge tube, D-mannose (1.44 mM, 0.1 mL) and Con A-FITC (0.1 mL) were added. The total volume of the final solution was 1.20 mL, where the concentration of D-mannose and Con A-FITC was 120 μM and 16 nM, respectively. The solutions were shaken for 1 h, which was sufficient for reaching equilibrium as shown in a time-based study, and centrifuged at 12,000 rpm for 30 min where nanoparticles precipitated to the bottom of the tube. The supernatants were transferred to a quartz cuvette for fluorescence measurements at 480 nm excitation, and the emissions at 517 nm were recorded using Felix32 software. The incubation time was determined from a time study where GNPs were incubated with Con A-FITC for varying amount of time. The fluorescence of the supernatant was monitored, and after 1-hour incubation, the intensity no longer changed indicating that the reaction had reached equilibrium. Measurement at each concentration was repeated 5 times, and the mean value of the emission intensities was used for the analysis. For all other GNPs, the same procedure was followed except that the concentrations of the GNPs were varied.
Results and Discussions
Synthesis of GNPs and ligand density determination
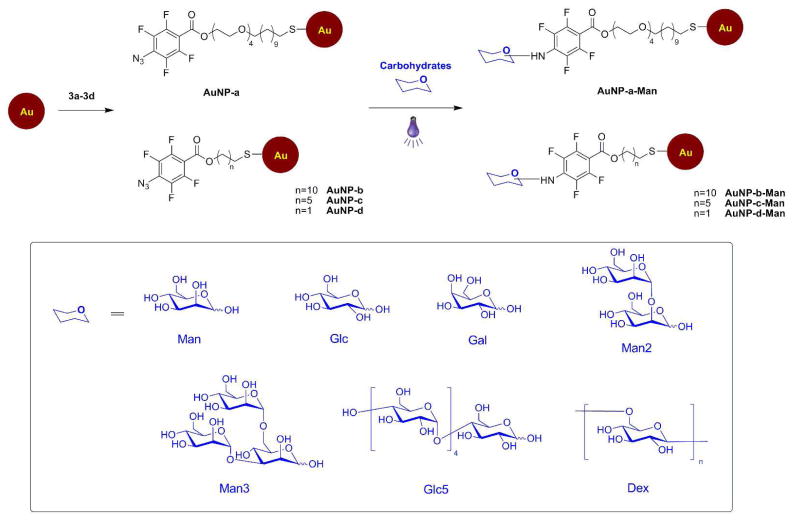
GNPs were synthesized by coupling carbohydrate ligands to PFPA-functionalized Au NPs. PFPA-thiol compounds of varying spacer lengths (a–d) (Scheme 1) were synthesized following the procedure shown in Scheme 1S (Supporting Information). PFPA-thiols b–d contain varying lengths of methylene spacer linkage, whereas PFPA-thiol a has four ethylene oxide (EO) in addition to the 11 methylene units. These PFPA-thiols were used to investigate the impact of spacer linkage on the ligand density and binding affinity of the corresponding GNPs.
Scheme 1.
Functionalization of Au NPs with PFPA-thiol and subsequent coupling of carbohydrates.
Preparation of GNPs followed the photocoupling method developed previously (Scheme 1).34 Citrate- protected Au NPs were prepared from HAuCl4 and sodium citrate. The size and uniformity were examined by DLS and TEM, which showed the spherically-shaped Au NPs of 22 ± 2.6 nm in diameter (see Figures 7S&8S, Supporting Information, for DLS data and TEM images). PFPA-thiols were then introduced to the NP surface via a ligand-exchange reaction, and the presence of PFPA on Au NPs was confirmed by 1H NMR and FTIR. Subsequent carbohydrate immobilization was carried out by a photocoupling reaction, and the unattached carbohydrates were removed by membrane dialysis. The resulting GNPs exhibited excellent solubility in water. Furthermore, the centrifuged nanoparticle pellet was easily dissolved in water, and the solution showed no change in the optical property. The GNPs dispersed well in the PBS buffer and the solutions were stable for weeks at 4 °C.
In order to quantitatively analyze GNPs–protein interactions, it is essential to determine the carbohydrate ligand density. The anthrone-H2SO4 assay, a colorimetric method that was widely used to measure carbohydrate concentrations in solution38 and on solid surfaces39 was employed to measure the carbohydrate ligand density on the GNPs.34 Calibration curves were obtained by treating various concentrations of each carbohydrate with anthrone/sulfuric acid, and the absorption at 620 nm was plotted against the carbohydrate concentration (Figure 1S, Supporting Information). Au NPs with immobilized carbohydrate ligands were then subjected to the same assay, and the amount of ligand on the nanoparticles was subsequently derived by comparing with the calibration curve. The coupling yield was estimated from the theoretically calculated maximal amount of each ligand that can occupy the 22-nm Au NP assuming a close-packed arrangement of the ligand on the NP (see Table 1S, Supporting Information, for detailed calculation). Table 1 summarizes the coupling yields of mono-, oligo- and poly-saccharides on Au NPs functionalized with PFPA-thiol a. The results show that the coupling yield increased with the size of the carbohydrate, which was anticipated since the probability of attaching the ligand via CH insertion reactions increases with the number of available CH bonds on the ligand.
Table 1.
Coupling yield of mono-, oligo-, and poly-saccharides on Au NPs.
| Carbohydrates | Coupling yield (%)a |
|---|---|
| Man | 63 ± 4.2 |
| Glc | 57 ± 5.8 |
| Man2 | 67 ± 6.1 |
| Man3 | 73 ± 5.4 |
| Glc5 | 79 ± 7.2 |
| Dex | 77 ± 16 |
Each data was the average of 5 samples.
Detection limit
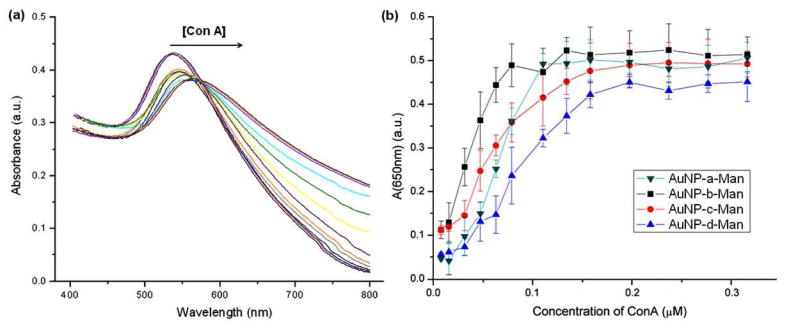
The unique optical property of metal nanoparticles, plasmon resonance absorption, offers a simple and attractive means to study molecular interactions with high sensitivity.40,41 The binding events occurring at the surface of the Au NPs result in a red shift in the plasmon resonance band, which can be conveniently monitored by UV-vis spectroscopy.42 To determine the sensitivity of the GNPs in detecting lectins, D-mannose-coupled Au NPs were titrated with Con A, and the UV-vis spectra recorded (Figure 1a). The absorbance at 650 nm vs. Con A concentration was then plotted (Figure 1b), and the dynamic linear range of each curve was used to determine the limit of detection (LOD), which was calculated to be 6.2 nM, 7.4 nM, 10 nM, and 22 nM for AuNP-a-Man, AuNP-b-Man, AuNP-c-Man and AuNP-d-Man, respectively (see Supporting Information for detailed experimental procedures and calculations). The results showed that the sensitivity of the GNPs increased with the spacer length, and the lowest LOD was obtained for GNPs prepared from PFPA-thiol a that contains the long and flexible spacer linkage.
Figure 1.
(a) UV-vis spectra of AuNP-a-Man upon addition of increasing concentration of Con A. (b) Absorbance of GNPs at 650 nm vs. Con A concentration.
Determination of binding affinity by fluorescence competition assay
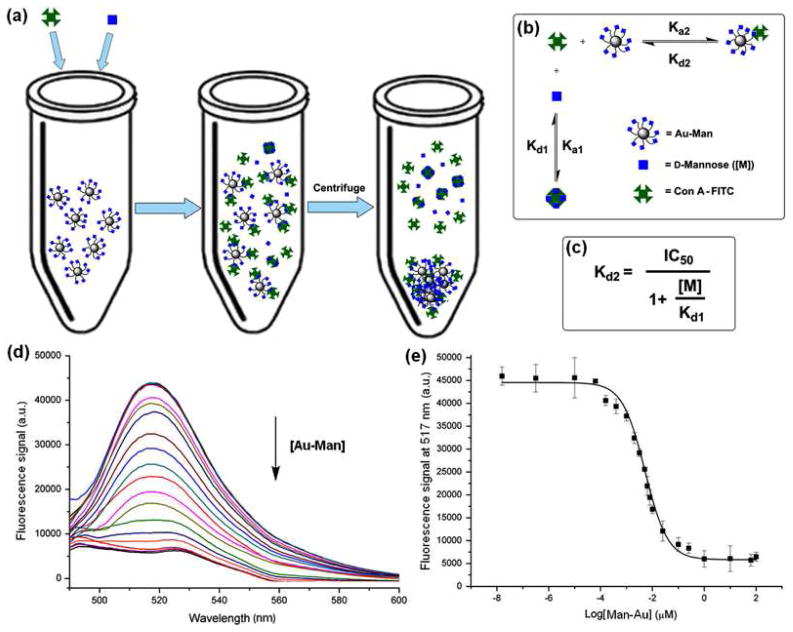
We developed a fluorescence competition assay to determine the binding affinity of GNPs with lectins using a fluorescently-labeled lectin and a free competing ligand. In a typical assay, GNPs of varying concentrations and a fixed concentration of a free competing ligand were incubated with Con A-FITC (Figure 2a). Two equilibria, Con A with GNPs and Con A with the free ligand, were established in the system (Figure 2b). Because a relatively low concentration of Con A was used, no agglomeration was observed in the assay. After the solution was incubated for 1 hour, it was centrifuged, bringing down GNPs including those bound to Con A-FITC. The unbound Con A-FITC and free ligand-Con A conjugate remained in the supernatant, corresponding to the amount of Con A-FITC that did not bind GNPs. The fluorescence of the supernatant was measured (Figure 2d), and the intensity at 517 nm was plotted as a function of the ligand density on GNPs (Figure 2e). From this concentration response curve, the IC50 value was determined and the apparent dissociation constant Kd was computed according to the Cheng-Prusoff equation43 (Figure 2c, Kd = Kd2).
Figure 2.
(a) The fluorescence-based competition binding assay; (b) two equilibriums established in the system; (c) Cheng-Prusoff equation, where IC50 = concentration of ligand displaying 50% of specific binding; [M] = concentration of free ligand, Kd1 = dissociation constant of the free ligand with Con A; and Kd2 = dissociation constant of GNPs with Con A; (d) fluorescence spectra of the supernatant as a function of increasing concentration of AuNP-b-Man; (e) concentration response curve, where the IC50 value was obtained.
This method was furthermore validated by several control experiments. First, different concentrations of Con A-FITC, 18 nM, 40 nM and 80 nM, were used in the assay and the binding affinity determined. The results obtained were similar for all three concentrations with less than 5% variation among the three Kd values (Table 4S, Supporting Information). Secondly, different competing ligands, e.g., MeMan, Man2, and Man3, were used in addition to Man. Although the binding affinities of these ligands with Con A were significantly different from that of Man, when they were used as the competing ligand to determine the Kd value of the same GNPs, the results were consistent and did not show significant variations (Table 5S, Supporting Information). Thirdly, AuNP-a-Man was incubated with Con A-FITC in the absence of the competing ligand. In this case the fluorescence intensity decreased with the concentration of Con A-FITC, and the typical IC50 concentration response curve was not observed (Figure 3S, Supporting Information). Lastly, Au NPs coupled with Gal, a carbohydrate that does not bind Con A, was tested to ensure that the binding was due to the specific interactions of the surface-bound ligands with the lectin. No significant changes in the fluorescence intensity were observed after varying concentrations of GNPs were incubated with Con A-FITC (Figure 4S, Supporting Information), demonstrating that the strong affinity of AuNP-a-Man with Con A was indeed due to the specific binding of surface-bound Man with Con A. This result furthermore revealed that the non-specific adsorption of lectin to non-lectin binding carbohydrates was minimal. With the methodologies developed, we next tested the impact of nanoparticles as the scaffold on the affinity ranking of various carbohydrate ligands with lectins. We chose Man, Man2, and Man3, which have at least several-fold differences in binding affinity between each ligand with Con A (Table 2). These ligands were attached to Au NPs using PFPA-thiol 3a, and the apparent Kd values of the resulting GNPs with Con A were determined by the fluorescence competition assay. An increase in the binding affinity of 4.5-fold and 33-fold was observed for AuNP-a-Man2 and AuNP-a-Man3 in comparison with AuNP-a-Man, respectively. Since the number of ligands on each GNPs was different, when taking into consideration the ligand density, the affinity increase was 12-fold and 141-fold, respectively. This compares well with the affinity ranking of the corresponding free ligands, which was 20 and 158 times higher affinity of Man2 and Man3 than Man with Con A in solution.44,45
Table 2.
Binding affinity of different GNPs with Con A
| GNPs | Number of ligands/NP | Kd (nM) | KD (μM)a |
|---|---|---|---|
| AuNP-a-Man | 3,991 | 0.43 ± 0.044 | 47048 |
| AuNP-a-Man2 | 1,450 | 0.095 ± 0.008 | 24.045 |
| AuNP-a-Man3 | 937 | 0.013 ±0.002 | 2.9745 |
| AuNP-a-Glc | 3,641 | 12.7 ± 2.5 | 1,78648 |
KD: dissociation constant of the corresponding free ligand with Con A.
In addition, the binding affinity of AuNP-a-Glc was determined, and the result showed over 4 orders of magnitude higher affinity than that of the free ligand with Con A (Table 2). Compared with the affinity enhancement in the case of Man, however, the binding of AuNP-a-Man with Con A was over 30 times higher than that of AuNP-a-Glc. Considering that the affinity of free Man with Con A was only 4 times higher than Glc, the affinity enhancement when the ligands were attached to NPs was significantly higher. This observation, i.e., the amplification of binding affinity difference due to multivalent interactions, has also been observed for carbohydrate ligands on neoglycopolymers46 and dendrimers.47
Binding affinity with respect to ligand presentation
Unlike the free ligand that has the translational and rotational freedom in solution, the surface-bound ligand is no longer an un-restricted entity. Each ligand becomes a member of the nanomaterial carrier and can act cooperatively when interacting with their binding partners. The binding affinity is sensitive to a number of factors including the coupling chemistry, the size of the nanomaterial scaffold, the type and length of the spacer linkage connecting the ligand and the nanomaterial, the flexibility/rigidity of the spacer, the density of ligands and the distance between them. In the present study, the ligand density, linker length, and nanoparticle size were varied, and their impacts on the binding affinities of the resulting GNPs were investigated.
Ligand density
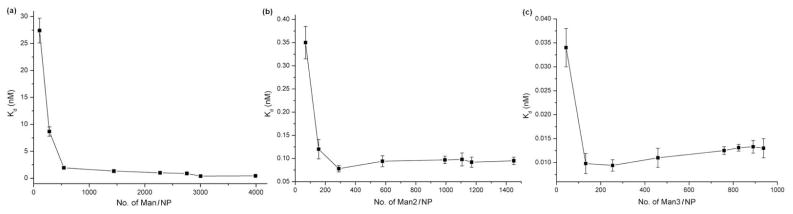
To control the surface ligand density, we employed the mixed SAM approach where a non-photoactive thiol together with PFPA-thiol was used to functionalize Au NPs. Solutions containing PFPA-thiol a or b and 1-hexanethiol at varying mole ratios were used to treat Au NPs. Man was subsequently coupled and the density of attached ligand measured (Table 3). The apparent Kd value of the resulting GNPs was then determined using the fluorescence competition assay described above. Results showed that the binding affinity generally increased with the ligand density. For AuNP-a-Man, however, there seems to be a maximal affinity for Au NPs treated with 98% PFPA-thiol a at the ligand density of 3,004 Man/NP. This maximal affinity was not observed in the case of AuNP-b-Man, likely due to the lower ligand density even at 100% PFPA-thiol treatment (2,824 Man/NP). To investigate the generality of this observation, Man2 and Man3 were coupled on Au NPs treated with mixed SAM of 1- hexanethiol and PFPA-thiol a, and the Kd values were measured with respect to the ligand density. The highest binding affinity of the resulting GNPs occurred at 50% and 30% of PFPA-thiol a in the mixed SAM, corresponding to the ligand density of 289 Man2/NP and 132 Man3/NP, respectively (Figure 3).
Table 3.
Binding affinity as a function of ligand density.
| PFPA-thiol in mixed SAM (mole%) | AuNP-a-Man |
AuNP-b-Man |
||
|---|---|---|---|---|
| Number of Man/NP | Kd (nM)a | Number of Man/NP | Kd (nM)a | |
| 10% | 107 | 27.4 ± 2.3 | 99 | 92 ± 12 |
| 30% | 283 | 8.67 ± 0.87 | 260 | 16.1 ± 5.5 |
| 50% | 544 | 1.93 ± 0.25 | 549 | 12.3 ± 4.1 |
| 70% | 1,444 | 1.33 ± 0.31 | 1,196 | 10.8 ± 1.8 |
| 90% | 2,275 | 1.01 ± 0.22 | 1,726 | 9.6 ± 3.0 |
| 95% | 2,756 | 0.88 ± 0.14 | 2,090 | 7.1 ± 1.7 |
| 98% | 3,004 | 0.39 ± 0.09 | 2,474 | 5.0 ± 1.0 |
| 100% | 3,991 | 0.43 ± 0.044 | 2,824 | 4.0 ± 0.7 |
Each data was the average of 3 measurements each on 5 samples.
Figure 3.
Binding affinity vs. ligand density for Man (a), Man2 (b), and Man3 (c) with Con A. Each data point was the average of 3 measurements each on 5 samples.
A few general observations can be drawn from the ligand density studies. For all three ligands, there was a sudden increase in binding affinity, and after which, the Kd values remained more or less constant. The ligand density at which the drastic change in binding affinity occurred was estimated from the intercept of the two relatively linear curves in each graph, which was 370 Man/NP, 166 Man2/NP and 138 Man3/NP, respectively. Assuming that the ligands were evenly distributed on the GNPs, the footprint occupied by each ligand on the nanoparticle surface was calculated to be 4.1 nm2, 9.2 nm2 and 11.0 nm2, and the distance between neighboring ligands were approximately 2.0 nm, 3.0 nm and 3.3 nm for Man, Man2 and Man3, respectively (see Supporting Information for calculations). Note that the distance between each binding site on Con A is 6.5 nm as determined by X-ray structural analysis.49 This distance is larger than the ligand spacing on NPs indicating that the binding is not monovalent and the higher ligand density is necessary for the enhanced affinity. The internal diffusion mechanism, which states that a lectin molecule “binds and jumps” from ligand to ligand along the scaffold,17 can be applied to explain the results. According to the mechanism, the more ligands there are on the scaffold, the longer the dwelling time of the lectin on the ligands, and the slower the lectin would dissociate. This affinity increase was observed for all three ligands where the Kd values decreased drastically at lower ligand density (Figure 3). The affinity started to decrease after the maximal affinity was reached, and this decrease in affinity was more pronounced as the size of the ligand increased. This was likely due to the steric effect where the larger ligand hinders the lectin binding.50
Spacer
PFPA-thiols a, b, c and d were used as the coupling agent to evaluate the effect of spacer linker on the binding affinity of the resulting GNPs. PFPA-thiols c, d contain a shorter spacer of six and two methylene units, and the Kd values of the corresponding GNPs were 15 nM and 19 nM, respectively (Table 4). When Au NPs were functionalized with PFPA-thiol b having 11 methylene units, Kd decreased to 4.0 nM, which represents 4–5 times increase in binding affinity as compared to the shorter spacer c and d. When PFPA-thiol a was used, the binding affinity increased an additional order of magnitude in comparison to PFPA-thiol b. Because the ligand density increases with the spacer length, it could also contribute to the observed enhancement in the binding affinity. However, the density increase alone could not account for the magnitude of the affinity enhancement. At the similar ligand density, for example, 544 and 549 Man/NP for AuNP-a-Man and AuNP-b-Man, respectively, the binding affinity was 6 times higher for AuNP-a-Man than AuNP-b-Man (Table 4). This affinity increase was therefore solely caused by the difference in the spacer linkage between the two GNPs. In order to quantitatively assess the binding affinity enhancement of different GNPs, an affinity enhancement factor (EF) was used, as defined in Eq. 1,
Table 4.
Binding affinity of GNPs with Con A.
| GNPs | Number of Man/NP | Kd (nM)a | EF |
|---|---|---|---|
| AuNP-a-Man | 3,991 | 0.43 ± 0.044 | 274 |
| AuNP-b-Man | 2,824 | 4.0 ± 0.72 | 42 |
| AuNP-c-Man | 1,959 | 15 ± 2.0 | 16 |
| AuNP-d-Man | 1,590 | 19 ± 2.2 | 16 |
Each data was the average of 5 samples, 3 measurements each.
| (1) |
where KD is the dissociation constant of free ligand with Con A, which is 470 μM for Man,48 Kd is the apparent dissociation constant of GNPs with Con A, and N is the number of ligands on each GNP. Here, the ligand density is taken into consideration, and the EF value can therefore be used to rank binding affinity with respect to the difference in the spacer linkage. Results in Table 4 show that the EF values for AuNP-d-Man and AuNP-c-Man were similar, ~16, whereas that of AuNP-b-Man was 2.6 times higher at 42, and that of AuNP-a-Man was in addition 17 times higher at 274. These results clearly demonstrate that longer spacers led to enhanced binding affinity of GNPs with lectins. The spacer elevates the ligand further from the solid substrates. On curved surfaces such as nanoparticles, a longer spacer would also result in a larger distance between the ligands at the same ligand density. Both events would reduce the steric hindrance when the lectin approaches the ligands, making the ligands more accessible for interacting with the lectin.
Nanoparticle size
It is well-established that the physical and chemical absorption, and the catalytic property of gold nanoparticles are highly size-dependent.51–53 In this study, the effect of nanoparticle size on binding affinity was investigated by varying the diameter of Au NPs. In addition to the 22-nm nanoparticles, Au NPs of 7 nm, 14 nm, and 30 nm in diameter were synthesized using the same protocol (see procedure in Supporting Information, Figures 7S&8S for DLS and TEM characterization), and Man was subsequently conjugated using PFPA-thiol b. The ligand densities of the resulting GNPs and their binding affinities were measured (Table 5). As expected, the number of ligands attached on Au NPs increased with the size of the nanoparticles. The binding affinity of GNPs with particle sizes of 7 nm, 14 nm, and 22 nm were similar, whereas a decrease of about 6-fold in binding affinity was observed for the 30-nm GNPs. The EF values calculated for each GNPs showed a considerable size-dependent effect, with the EF value increasing with decreasing particle size. Smaller nanoparticles yielded the highest affinity enhancement, likely due to their large surface-to-volume ratio and higher mobility in solution.
Table 5.
Binding affinity of GNPs of varying sizes
| Au NP Diameter (nm) | Number of Man/NP | Kd (nM) | EF |
|---|---|---|---|
| 7.2 ± 1.8 | 297 | 3.38 ± 0. 67 | 468 |
| 14 ± 2.6 | 1,127 | 3.14 ± 0.49 | 132 |
| 22 ± 3.3 | 2,824 | 3.99 ± 0. 81 | 42 |
| 30 ± 4.0 | 4,486 | 24.8 ± 3.1 | 4.2 |
Conclusions
In conclusion, we developed a fluorescence-based competition assay to determine the apparent dissociation constants of GNPs with lectin. The assay was successfully used to determine the Kd values and to evaluate the binding affinity of GNPs. When carbohydrate ligands were attached on the Au NP scaffold, their interactions with lectins were drastically enhanced, and several orders of magnitude increases in binding affinity were observed between GNPs and lectin. Systematic studies were conducted to investigate the impacts of nanoparticle size, spacer length, ligand size and density on the binding affinity of GNPs. Results show that the lectin binding to ligands on GNPs is profoundly affected by how the ligands are displayed on the NP surface. Findings from this study are important that GNPs with controlled binding affinity can be readily synthesized by varying ligand density, spacer linker, and scaffold configuration. This will open up immense opportunities for tailor-made glyconanomaterials where the ligand display can be exploited to tune their bioaffinities.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of General Medical Science (NIGMS) under NIH Award Numbers R01GM080295 and 2R15GM066279, and in part by the European Commission (MRTN-CT-19561).
Footnotes
Supporting Information Available: Synthesis of PFPA compounds, calibration curves for ligand density measurements, detection limit calculations, IC50 validation studies and fitting curves, ligand distance calculations, and nanoparticle synthesis with characterizations. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Nat Biotechnol. 2003;21:47. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- 2.Rojo J, Díaz V, de la Fuente JM, Segura I, Barrientos AG, Riese HH, Bernade A, Penadés S. ChemBioChem. 2004;5:291. doi: 10.1002/cbic.200300726. [DOI] [PubMed] [Google Scholar]
- 3.Cipolla L, Peri F, Airoldi C. Anti-Cancer Agents Med Chem. 2008;8:92. doi: 10.2174/187152008783330815. [DOI] [PubMed] [Google Scholar]
- 4.De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225. [Google Scholar]
- 5.Sperling R, Rivera GP, Zhang F, Zanella M, Parak W. Chem Soc Rev. 2008;37:1896. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- 6.Giljohann DA, Mirkin CA. Nature. 2009;462:461. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Liu LH, Ramstrom O, Yan M. Exp Biol Med. 2009;234:1128. doi: 10.3181/0904-MR-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dube DH, Bertozzi CR. Nat Rev Drug Discov. 2005;4:477. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 9.Fuster MM, Esko JD. Nat Rev Cancer. 2005;5:526. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 10.Liu FT, Rabinovich GA. Nat Rev Cancer. 2005;5:29. doi: 10.1038/nrc1527. [DOI] [PubMed] [Google Scholar]
- 11.Szymanski CM, Wren BW. Nat Rev Microbiol. 2005;3:225. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsubo K, Marth JD. Cell. 2006;126:855. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Sharon N. Biochim Biophys Acta, Gen Subj. 2006;1760:527. doi: 10.1016/j.bbagen.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Crocker PR, Paulson JC, Varki A. Nat Rev Immunol. 2007;7:255. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 15.Lee YC, Lee RT. Acc Chem Res. 1995;28:321. [Google Scholar]
- 16.Mammen M, Choi SK, Whitesides GM. Angew Chem Int Ed. 1998;37:2754. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Dam TK, Brewer CF. Glycobiology. 2010;20:270. doi: 10.1093/glycob/cwp186. [DOI] [PubMed] [Google Scholar]
- 18.Drechsler U, Erdogan B, Rotello VM. Chem Eur J. 2004;10:5570. doi: 10.1002/chem.200306076. [DOI] [PubMed] [Google Scholar]
- 19.Huskens J. Curr Opin Chem Biol. 2006;10:537. doi: 10.1016/j.cbpa.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Jayaraman N. Chem Soc Rev. 2009;38:3463. doi: 10.1039/b815961k. [DOI] [PubMed] [Google Scholar]
- 21.Roy D, Fendler J. Adv Mater. 2004;16:479. [Google Scholar]
- 22.Grainger DW, Castner DG. Adv Mater. 2008;20:867. [Google Scholar]
- 23.Rao CNR, Biswas K. Annu Rev Anal Chem. 2009;2:435. doi: 10.1146/annurev-anchem-060908-155236. [DOI] [PubMed] [Google Scholar]
- 24.Lin CC, Yeh YC, Yang CY, Chen GF, Chen YC, Wu YC, Chen CC. Chem Commun. 2003;23:2920. doi: 10.1039/b308995a. [DOI] [PubMed] [Google Scholar]
- 25.Lyu YK, Lim KR, Lee BY, Kim KS, Lee WY. Chem Commun. 2008:4771. doi: 10.1039/b807438k. [DOI] [PubMed] [Google Scholar]
- 26.Tsai CS, Yu TB, Chen CT. Chem Commun. 2005:4273. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie GH, Sawyer WH, Nichol LW. Biochim Biophys Acta. 1972;263:283. doi: 10.1016/0005-2795(72)90081-5. [DOI] [PubMed] [Google Scholar]
- 28.Huet M. Eur J Biochem. 1975;59:627. doi: 10.1111/j.1432-1033.1975.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 29.Lis H, Sharon N. Chem Rev. 1998;98:637. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 30.Kanai M, Mortell KH, Kiessling LL. J Am Chem Soc. 1997;119:9931. [Google Scholar]
- 31.Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. J Am Chem Soc. 2002;124:1615. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]
- 32.Jason EG, Laura ES, Laura LK. Angew Chem Int Ed. 2000;39:4567. [Google Scholar]
- 33.Barrientos AG, Fuente JMdl, Jiménez M, Solís D, Cañada FJ, Martín-Lomas M, Penadés S. Carbohydrate Res. 2009;344:1474. doi: 10.1016/j.carres.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Ramstrom O, Yan M. J Mater Chem. 2009;19:8944. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Ramström O, Yan M. Adv Mater. 2010;22:1946. doi: 10.1002/adma.200903908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turkevich J, Stevenson PC, Hollier J. Discuss Faraday Soc. 1951;11:55. [Google Scholar]
- 37.Pei ZC, Yu H, Theurer M, Walden A, Nilsson P, Yan M, Ramström O. ChemBioChem. 2007;8:166. doi: 10.1002/cbic.200600447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koehler LH. Anal Chem. 1952;24:1576. [Google Scholar]
- 39.Chien Y, Jan M, Adak A, Tzeng H, Lin Y, Chen Y, Wang K, Chen C, Chen C, Lin C. ChemBioChem. 2008;9:1100. doi: 10.1002/cbic.200700590. [DOI] [PubMed] [Google Scholar]
- 40.El-Kouedi M, Keating CD. In: Nanobiotechnology. Niemeyer CM, Mirkin CA, editors. Wiley-VCH; Weinheim: 2004. p. 429. [Google Scholar]
- 41.Homola J. In: Springer Series on Chemical Sensors and Biosensors. Urban G, editor. Vol. 4. Springer-Verlag GmbH; Heidelberg: 2006. p. 232. [Google Scholar]
- 42.Hone DC, Haines AH, Russell DA. Langmuir. 2003;19:7141. [Google Scholar]
- 43.Cheng YC, Prusoff WH. Biochem Pharmacol. 1973;22:3099. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 44.Sanders JN, Chenoweth SA, Schwarz FP. J Inorg Biochem. 1998;70:71. doi: 10.1016/s0162-0134(98)00016-6. [DOI] [PubMed] [Google Scholar]
- 45.Mandal DK, Kishore N, Brewer CF. Biochemistry. 2002;33:1149. doi: 10.1021/bi00171a014. [DOI] [PubMed] [Google Scholar]
- 46.Mortell KH, Weatherman RV, Kiessling LL. J Am Chem Soc. 1996;118:2297. [Google Scholar]
- 47.Wolfenden ML, Cloninger MJ. J Am Chem Soc. 2005;127:12168. doi: 10.1021/ja053008n. [DOI] [PubMed] [Google Scholar]
- 48.Schwarz FP, Puri KD, Bhat RG, Surolia A. J Biol Chem. 1993;268:7668. [PubMed] [Google Scholar]
- 49.Bittiger H, Schnebli HP. Concanavalin A as a Tool. John Wiley and Sons; London: 1976. [Google Scholar]
- 50.Dhayal M, Ratner DM. Langmuir. 2009;25:2181. doi: 10.1021/la8031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell CT, Parker SC, Starr DE. Science. 2002;298:811. doi: 10.1126/science.1075094. [DOI] [PubMed] [Google Scholar]
- 52.Hill HD, Millstone JE, Banholzer MJ, Mirkin CA. ACS Nano. 2009;3:418. doi: 10.1021/nn800726e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De M, Miranda OR, Rana S, Rotello VM. Chem Commun. 2009:2157. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.