Abstract
Non-long terminal repeat (non-LTR) retrotransposons, most of which carry two open reading frames (ORFs), are abundant mobile elements that are distributed widely among eukaryotes. ORF2 encodes enzymatic domains, such as reverse transcriptase, that are conserved in all retroelements, but the functional roles of ORF1 in vivo are little understood. We show with green fluorescent protein-ORF1 fusion proteins that the ORF1 proteins of SART1, a telomeric repeat-specific non-LTR retrotransposon in Bombyx mori, are transported into the nucleus to produce a dotted localization pattern. Nuclear localization signals N1 (RRKR) and N2 (PSKRGRG) at the N terminus and a highly basic region in the center of SART1 ORF1 are involved in nuclear import and the dotted localization pattern in the nucleus, respectively. An in vivo retrotransposition assay clarified that at least three ORF1 domains, N1/N2, the central basic domain, and CCHC zinc fingers are required for SART1 retrotransposition. The nuclear import activity of SART1 ORF1 makes it clear that the ORF1 proteins of non-LTR retrotransposons work mainly in the nucleus, in contrast to the cytoplasmic action of Gag proteins of LTR elements. The functional domains found here in SART1 ORF1 will be useful for developing a more efficient and target-specific LINE-based gene delivery vector.
Retrotransposable elements can be classified into two subclasses, long terminal repeat (LTR) and non-LTR retrotransposons, according to whether they retain LTRs at their ends. LTR retrotransposons usually resemble retroviruses in both their structure and their integration processes in cells. In contrast, the retrotransposition mechanisms of non-LTR retrotransposons, which are also called long interspersed nuclear elements (LINEs) in mammals, are poorly understood at present, even though they are major components of the widespread eukaryotic genomes, from protozoa to vertebrates. Recent reports showed that one LINE, L1, occupies approximately 16% of the human genome (20), and a few copies of L1 are still active (17). Most non-LTR retrotransposons insert into random genomic locations, sometimes causing de novo disease by the destruction of gene function (16) and sometimes generating genome rearrangement (12, 35).
Considerable numbers of non-LTR retrotransposons, including human L1, have two putative open reading frames (ORFs), whereas some elements have only one ORF (22). In the former non-LTR retrotransposon group, the second ORF (ORF2) encodes endonuclease (EN) and reverse transcriptase (RT) domains. In ORF2, EN and RT are the only characterized domains and are essential for retrotransposition of non-LTR retrotransposons with two ORFs. However, there is very little functional information available on the first ORF (ORF1), because of low levels of sequence similarity among non-LTR retrotransposons. In ORF1, CCHC zinc finger motifs are considered essential (37), although a leucine zipper motif may substitute for the zinc fingers in ORF1 of the human L1 element (15). Moran et al. showed that three regions conserved in the ORF1 sequences of mouse, rat, and rabbit L1s are essential for in vivo retrotransposition, but their functional roles were not yet clarified (26). Moreover, previous studies indicated that ORF1 of non-LTR retrotransposons binds to nucleic acids in vitro (7, 14, 18, 19), multimerizes to form ribonucleoprotein particles (RNPs) (13, 28), acts as a chaperone to nucleic acids to facilitate stable reverse transcription (7, 25), and transports RNPs into the nucleus (31, 32). There are a few results to show which structure in ORF1 is responsible for specific functions (13, 24, 26). At present, however, these roles of ORF1 protein have been demonstrated mostly by in vitro methods and are insufficiently understood.
The ORF1 and ORF2 proteins of non-LTR retrotransposons are translated from their mRNAs, although it is not yet known how the translation of two independent units of overlapping ORFs is regulated. Two putative ORF proteins and the mRNA are assembled into RNPs, and then the RNPs are transported into the nucleus, make a nick in the target DNA by using the EN domain in ORF2, and start the reverse transcription of RNA by using the nick as primer. These processes in the nucleus that are specific to non-LTR retrotransposons are called target-primed reverse transcription (TPRT) (21). There are two hypotheses explaining reverse transcription in the nucleus. One states that the integration event occurs during mitosis, when nuclear transport itself is unnecessary. The other states that active transport of RNPs is controlled by unidentified functional domains in the ORFs. The nuclear transport mechanism of RNPs of non-LTR elements, however, is largely unknown.
To clarify the functional roles of ORF1 in the retrotransposition of non-LTR retrotransposons, we used telomere-specific SART1 of the silkworm Bombyx mori. Recently, we established the retrotransposition system of SART1 in vivo, using baculovirus-mediated expression (37), and therefore it is possible to identify every domain essential for SART1 retrotransposition with mutation-based analyses. SART1 is a sequence-specific non-LTR retrotransposon that integrates into insect telomeric repeats (TTAGG)n (36). Because telomerase activity cannot be detected in the silkworm B. mori (33), SART1 may contribute to telomere maintenance or back up the undetectable levels of telomerase activity in this insect (11).
In this study, by combining an in vivo retrotransposition assay and green fluorescent protein (GFP) fusion analyses, we demonstrate that three domains of ORF1 are required for the retrotransposition of SART1. Each of these domains is essential for either active transport into the nucleus, control of the dotted localization pattern in the nucleus, or putative RNP formation. Furthermore, we also show that baculovirus-expressed ORF1 protein rescues the ORF1-deficient mutants of SART1 in trans in cells, suggesting that the ORF1 protein works as an independent functional unit in multiple forms. This report demonstrates directly that nuclear import signals in ORF1 of SART1 are required for retrotransposition and that several functional domains involved in retrotransposition are clearly localized to ORF1 of non-LTR retrotransposons.
MATERIALS AND METHODS
Plasmid construction.
SART1 DNAs for plasmid construction were amplified by PCR from the genomic library clone BS103 (36) or from SART1WT-pAcGHLTB (37) with the Pfu Turbo DNA polymerase (Stratagene).
(i) Mutation introduction.
The deletion and substitution mutants were constructed by inverse PCR and self-ligation with 5′-phosphorylated primers (Table 1). After the inverse PCR, the products with the phosphorylated 5′ ends were self-ligated with the TaKaRa DNA ligation kit (version 2) for 16 h. To introduce various mutations, one primer with a 3- or 12-base substitution at its 5′ end was used in inverse PCR. Double-mutated constructs were generated from a single mutated construct as template DNA by inverse PCR and self-ligation as described above. Phosphorylated primers were made with T4 polynucleotide kinase (TaKaRa) and ATP (Roche). Deletion mutants for in vivo retrotransposition assays were subcloned between NcoI and BglII sites of the pAcGHLT-B plasmid (PharMingen). SART1ORF1pWT was amplified by PCR with SART1S880 and SART1A3029NotI and subcloned between NcoI and NotI sites of pAcGHLT-B.
TABLE 1.
Primers
| Function and primer | Sequence (5′ to 3′)a | Construct(s) produced or use |
|---|---|---|
| Subcloning to pAcGHLTB | ||
| S1S1306NcoI | AAAAAACCATGGTTGGCCTCGCCGC | SART1 Δ1-142 |
| S1S1036NcoI | AAAAAACCATGGTGTCCGCGAAGGACGGAGAA | SART1 Δ1-52 |
| S1S1195NcoI | AAAAAACCATGGAGTGCTCAGAGAGCAGCTCC | SART1 Δ1-105 |
| S1S880NcoI | AAAAAACCATGGGCAGTTATAAAGAAGAATTACCCCAG | SART1 ORF1pWT |
| S1A6704BgIII | AAAGAAGATCTTTTTTTTTTTTTTTTTTTTGGTATCGATGGGG | SART1 Δ1-52, Δ1-105, Δ1-142 |
| Subcloning to pHEGFPB | ||
| S1S1306BgIII | GAAGATCTGTTGGCCTCGCCGCC | SART1 143-284, 143-447 |
| S1S880BgIIINcol | AAAAAAAGATCTACCATGGGCAGTTATAAAGAAGAATTACCCC | SART1 1-712, 1-567, 1-447, 1-353, 1-284, 1-142, 1-93 |
| S1S1724BgIII | GAAGATCTGTCCCCGCCCCCGTAC | SART1 285-447, 285-712 |
| S1S1156BgIII | AAAAAAAGATCTTTGACCGGAAGCCGAAG | SART1 92-142 |
| S1S2617BgIII | AAAAAAGATCTCAGGTCTTGGTCAGATGCC | SART1 580-712 |
| S1S2221BgIII | AAAAAAGATCTTCGCAGGCAGTTATTGTCAAG | SART1 448-712, 448-579 |
| S1A1731NotI | AAAAAAAAAAGCGGCCGCTTAGGCTGCCGGCGATCCC | SART1 1-284, 143-284 |
| S1A2220NotI | AAAAAAAAAAGCGGCCGCTTAGCGGGGTGGCCGGAG | SART1 1-447, 143-447, 285-447 |
| S1A1305NotI | AAAAAAAAAAGCGGCCGCTTAGTACTGCCCGGTTGTGGG | SART1 1-142, 92-142 |
| S1A2616NotI | AAAAAAAAAAGCGGCCGCTTAGCCAGTGCCTCCGGG | SART1 448-579 |
| S1A1248NotI | AAAAAAAAAAGCGGCCGCTTAGGCTCTTCGGGCTGAGC | SART1 1-123 |
| S1A1158NotI | AAAAAAAAAAGCGGCCGCTTAGGTCAACGACGAACGGC | SART1 1-93 |
| S1A2505NotI | GATTCGTCGCGGCCGCTCAGTCTGACAGC | SART1 1-541 |
| S1A1935NotI | AAAAAAAAAAGCGGCCGCTTACGGCGTCTTTTCTTGACC | SART1 1-353 |
| Subcloning to pAcGHLTB and pHEGFPB S1A3029NotI | TTTTTGCGGCCGCGCTGCTGGTCATTATTCGTCGTCCATTGGTGT | SART1 1-712, 580-712, 285-712 (pHGFPB), SART1 and ORF1pWT (pAcGHLTB) |
| Introduction of mutation | ||
| S1S1165N1>A | GCAGCAGCTGCGCTGAAGGGCTCCAGCACG | SART1 N1>A |
| S1A1164 | GCTTCCGGTCAACGACGAAC | SART1 N1>A |
| S1S1267N2>A | GCAGCAGCTGCAGGAAGACCGCCCACAACCG | SART1 N2>A |
| S1A1266 | TGACGGGGTCTTGGAGGCG | SART1 N2>A |
| S1S2221 | TCGCAGGCAGTTATTGTCAAGCTGC | SART1 Δ440-447, Δ354-447 |
| S1A2196 | CTTTTCCTTCAGGGGCGGTG | SART1 Δ440-447, K440A |
| S1A1935 | CGGCGTCTTTTCTTGACC | SART1 Δ354-447 and to detect virus genotype |
| S1A2025 | TTTCTGCGGGCCACCACC | SART1 K383A |
| S1S2199K>A | GCGAAGAGACTCCGGCCACCCC | SART1 K440A |
| S1S2028K>A | GCGAAGGCGCCGAAAAGCAGG | SART1 K383A |
| Generation of pHEGFPB vector | ||
| EGFP A813 | CTTGTACAGCTCGTCCATGC | pHEGFPB vector |
| EGFP S814BgIII | AGATCTAAAAAAGCGGCCGCGACTCTAGATC | pHEGFPB vector |
| Other | ||
| S1S6311 | TGCCTACCTCACGAAGAAGTTGCGGTCA | To detect retrotransposition |
| CCTAA + T | CCTAACCTAACCTAACCTAACCTAACCTTTTTT | To detect retrotransposition |
| pS2941 | AGCCCAGGACTCGATGGCATATATG | To detect virus genotype |
| S1S1153 | TGACCGGAAGCCGAAGAA | To detect virus genotype |
| S1A3235 | TCGAATTCGGGCGGAGC | To detect virus genotype |
Underlined nucleotides indicate restriction sites. Sequences in boldface are mutagenized.
(ii) GFP fusion plasmids.
Plasmids for the GFP expression experiment were constructed as follows. The Drosophila HSP promoter was subcloned into the HindIII site of pEGFP-1 (Clontech). The BglII site of the plasmid was removed by BglII digestion and T4 DNA polymerase (TaKaRa) treatment in the presence of deoxynucleoside triphosphates (TaKaRa). Next, to make a vector adapted for GFP fusion at the C terminus, we introduced a BglII site instead of the stop codon (TAA) of the GFP-coding region in the above plasmid. By using EGFPS814BglII and A813 primers, inverse PCR and self-ligation were performed, and the resulting plasmid was named pHEGFPB. By using the BglII and NotI sites of pHEGFPB, various regions of SART1 ORF1 were subcloned and expressed as GFP fusion protein at the C terminus in frame. Each 3′ primer (S1AXXXXNotI) (Table 1) used for inserting a SART1 ORF1 region was designed to include the stop codon (TAA) just before the NotI site. The inserted DNA and junction regions in all plasmids were sequenced with the Big Dye terminator cycle sequencing kit (Applied Biosystems) on an automatic DNA sequencer (ABI 310 Genetic Analyzer). Plasmid DNAs were purified with the Qiagen plasmid minikit. Junction sequences of all constructs are given at http://www.biol.s.u-tokyo.ac.jp/users/animal/mansumoto/supplement.html. All primers used in this study are listed in Table 1 and at http://www.biol.s.u-tokyo.ac.jp/users/animal/matsumoto/supplement.html.
Generation of recombinant Autographa californica nuclear polyhedrosis virus (AcNPV).
Sf9 cells were cultured at 27°C in TC-100 medium containing 10% fetal bovine serum (Nihon-Nosankougyou Co., Yokohama, Japan) and penicillin-streptomycin (Gibco). The recombinant baculovirus was generated by cotransfection of the wild-type (WT) and mutant SART1-pAcGHLT-B plasmids together with BaculoGold DNA (PharMingen) into the Sf9 cells by using the Tfx-20 lipofection reagent (Promega). Four days later, the recombinant baculovirus was plaque purified and amplified from a single plaque, according to the manufacturers' instructions.
In vivo retrotransposition assay.
The in vivo retrotransposition assay of the SART1 element was performed essentially as described by Takahashi and Fujiwara (37). Sf9 cells (106) were infected with a SART1 ORF-containing AcNPV at a multiplicity of infection of 10 PFU. In the trans-complementation experiments, Sf9 cells were infected with two AcNPVs at a multiplicity of infection of 5 PFU. At 24, 36, and 48 h after infection, cells were collected, and the total genomic DNAs were purified with the Puregene DNA isolation kit (Gentra Systems). PCR amplification was carried out with LA-Taq DNA polymerase (TaKaRa) and the primer set SART1S6311-CCTAA+T (Table 1) in the presence of anti-Taq antibody (Clontech) and 10 ng of Sf9 DNA. The reaction mixture was denatured at 96°C for 2 min, followed by 35 cycles of 98°C for 20 s, 62°C for 30 s, and 72°C for 30 s. The PCR products were electrophoresed on 2% agarose gels in Tris-borate-EDTA buffer and visualized by ethidium bromide staining. PCR products were cloned into the pGemT-Easy vector (Promega) and sequenced with the Big Dye terminator cycle sequencing kit (Applied Biosystems) on an ABI 310 genetic analyzer.
GFP microscopy.
Sf9 cells were transfected with GFP-expressing plasmids by using the Tfx-20 lipofection reagent. After 72 h, transfected cells were gathered, adhered to coverslips, and then fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). The slips were washed with PBS and mounted by using Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) (Vector). The cellular localization of GFP-fused proteins was analyzed and photographed with UV illumination on a Zeiss Axioscop instrument, and GFP images were imported into Adobe Photoshop Elements.
Immunoblot detection of GST-His6 fusion protein.
Glutathione S-transferase (GST)-His6-fusion proteins were detected by immunoblot analysis. Total proteins were extracted from Sf9 cells at various times after the AcNPV infection and subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis. Electrophoresed proteins were transferred to a polyvinylidene difluoride membrane (Nippon Genetics). The membrane was immersed in 4% skim milk in PBS-T (PBS plus 0.1% Tween 20), washed several times with PBS-T, and incubated with anti-His antibody (Amersham) in PBS-T. Band detection was carried out with the ECL Plus Western blotting system (Amersham).
RESULTS
SART1 ORF1 protein has two basic domains containing putative NLS-like motifs.
SART1 is a typical non-LTR retrotransposon with two ORFs (Fig. 1A). We first studied the structural features of ORF1 of SART1. Within the 712-amino-acid sequence of ORF1 (Fig. 1B), we found several clusters of basic amino acids near the N-terminal region and the central region. Three CCHC zinc finger motifs have been identified near the C-terminal region (Fig. 1B) (36). For the sake of simplicity, we divided the whole ORF1 region into five subregions (Fig. 1C). The first and third regions include strongly basic amino acid clusters, and the last region includes zinc finger motifs. When each isoelectric point (pI) was estimated for these three regions (Fig. 1C), all were shown to be highly basic. We further screened for nuclear localization signals (NLSs) by computer-based analysis with the PSORT program (GenomeNet, Tokyo, Japan). As shown in Fig. 1B, we found four putative NLS-like motifs: RRKR (amino acids 96 to 99, N1), PSKRGRG (amino acids 128 to 134, N2), PQKKKAP (amino acids 379 to 385, N3), and KKKR (amino acids 439 to 442, N4). N1 and N2 occur in the first region, and N3 and N4 occur in the third region.
FIG. 1.
Structural features of ORF1 of SART1. (A) Schematic structure and sequence specificity of SART1. SART1 inserts between T and A of the telomeric repeats (TTAGG)n. The 5′ and 3′ untranslated regions (UTR) are shown in gray, and the ORFs are shown as open boxes. APE/RT, endonuclease/RT domain. Vertical lines in ORF1 and ORF2 represent the zinc finger motifs. ORF1 terminates at codon 713. (B) Amino acid sequence of the putative ORF1 protein of SART1 (accession number D85594.1). Asterisks indicate the NLS-like motifs predicted by the PSORT program. Three CCHC-type zinc finger motifs at the C-terminal end are underlined. (C) Electrical properties of the SART1 ORF1 protein. ORF1 is subdivided into five regions. The regional isoelectric points were calculated with the sequence analysis software Vector NTI and are indicated in the open boxes. The amino acid numbers are shown below the diagram. Arrowheads represent the positions of putative NLSs. There are four putative NLSs, N1, N2, N3, and N4.
Residues 354 to 447 of SART1 ORF1 are responsible for nuclear localization with a dotted pattern.
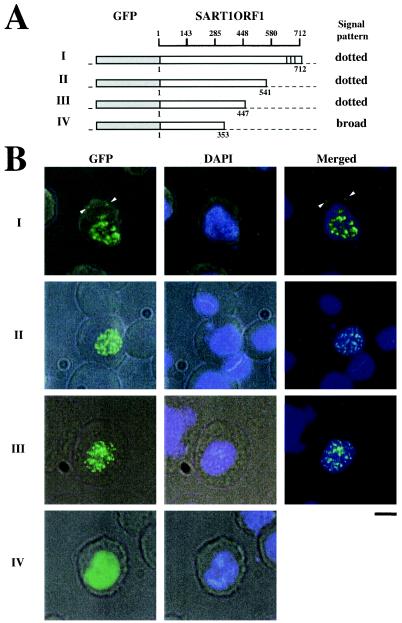
We analyzed the cellular localization of SART1 ORF1 by monitoring the expression of GFP fusion proteins under the control of the Drosophila heat shock protein 70 (hsp70) promoter. The GFP was stably expressed in cells of Spodoptera frugiperda origin (Sf9 cells) (see Fig. 3) or of B. mori origin (BmN cells) at 24 h after transfection. The localization of GFP signals was identical in Sf9 and BmN cells in all experiments (data not shown; see http://www.biol.s.u-tokyo.ac.jp/users/animal/matsumoto/supplement.html). The in vivo retrotransposition assay was performed with heterologous Sf9 cells because it is difficult to discriminate the newly retrotransposed SART1 from the endogenous copies in BmN cells. To compare the results of cellular localization and retrotransposition assay directly, we show the data for Sf9 cells only in this paper. Cells transfected with the GFP-reporter region alone showed GFP signals distributed in both the nucleus and the cytoplasm (see Fig. 3). However, the full-length SART1 ORF1 fused to the GFP reporter was localized only in the nucleus with a dotted pattern, although a few signals were detected in the cytoplasm (Fig. 2B, panels I). These dotted signals in the cytoplasm, which were named cytoplasmic clusters previously (30, 32), were abundantly observed in full-length constructs containing the C-terminal Zn finger motifs (Fig. 2B, panels I; Table 2 [see Fig. 4B]). When the C-terminal region of ORF1 including residues 448 to 712 was deleted from the GFP fusion vector, a similar dotted pattern of nuclear localization was observed (Fig. 2B, panels II and III). Deletion of residues 354 to 712, however, produced a homogeneous and broad distribution of signals in the nucleus (Fig. 2, panels IV). Table 2 summarizes the results of the GFP localization experiments described above. Seventy-five to 85% of cells transfected with the WT, 1-541, and 1-447 proteins showed a nuclear dotted pattern, but 100% of cells with the 1-353 protein showed only a nuclear broad pattern (Table 2). These results suggest that amino acid residues 1 to 353 in SART1 ORF1 are responsible for nuclear localization and that residues 354 to 447, which include the N3 and N4 motifs (Fig. 1B), are responsible for the dotted localization pattern.
FIG. 3.
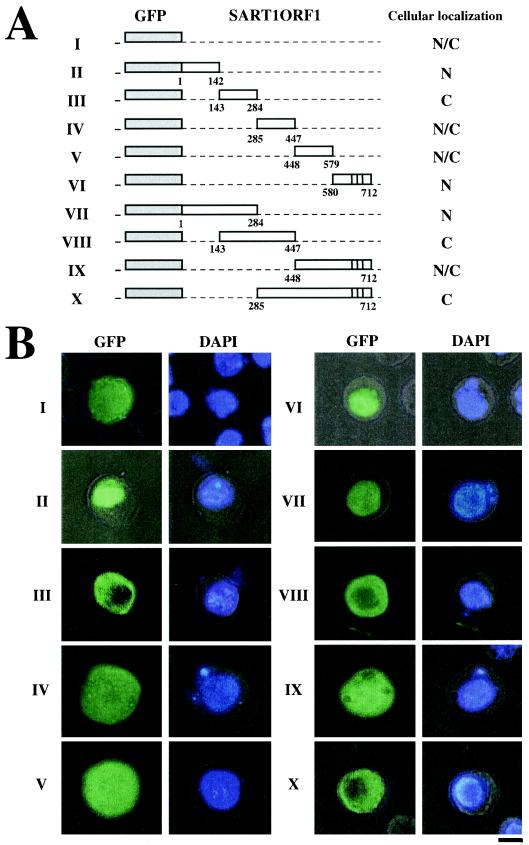
The N terminus of SART1 ORF1 protein has a karyophilic domain. (A) Expression constructs in which GFP was fused with each region of SART1 ORF1. The nuclear localization ability of each region in SART1 ORF1 was examined with GFP fusion proteins. Open boxes indicateeach region of SART1 ORF1, and gray boxes indicate the GFP region. The vertical lines in constructs VI, IX, and X represent the zinc finger motifs. Amino acid numbers for the ORF1 region used for each construct are shown below the open boxes. N, nucleus; C, cytoplasm; N/C, both nucleus and cytoplasm. I, GFP without SART1 ORF1; II to VI, the five ORF1 regions (see Fig. 1) fused to GFP; VII to X, large ORF1 regions fused to GFP. (B) Cellular localization of the GFP fused with each region of SART1 ORF1 in Sf9 cells. The left and right pictures show cellular localizations of GFP fusion protein and DNA stained with DAPI, respectively. Bar, 10 μm.
FIG. 2.
SART1 ORF1 proteins localize in the nucleus in a dotted pattern. (A) Constructs for analysis of the localization of GFP-SART1 ORF1 fusion protein. Open boxes represent the SART1 ORF1 protein and gray boxes represent the GFP region. Amino acid numbers for the ORF1 deletions are shown below constructs I to IV. The results from panel B are summarized on the right. Vertical lines represent zinc finger motifs. (B) Cellular localization of the GFP fused with each region of SART1 ORF1 in Sf9 cells. Intact SART1 ORF1 protein is localized in the nucleus in a dotted pattern. The left-hand pictures show the localization of GFP fusion protein analyzed by fluorescence microscopy. The center pictures show DNA localization by staining with DAPI. Merged pictures of GFP-fused protein and DNA (for panels I to III) are shown on the right. In panels I, arrowheads indicate GFP signals in the cytoplasm (cytoplasmic cluster). Bar, 10 μm.
TABLE 2.
Localization pattern of GFP-fused SART1 ORF1
| Construct | na | No. with the following pattern:
|
% Nuclear dotted | Cytoplasmic cluster
|
|||
|---|---|---|---|---|---|---|---|
| Nuclear dotted | Nuclear broad | Cytoplasmic broad | No. | % | |||
| WT | 48 | 36 | 7 | 5 | 75.0 | 15 | 31.3 |
| 1-541 | 53 | 45 | 8 | 0 | 84.9 | 5 | 9.4 |
| 1-447 | 57 | 45 | 12 | 0 | 78.9 | 3 | 5.3 |
| 1-353 | 39 | 0 | 39 | 0 | 0 | 0 | 0 |
| NLS-less | 31 | 0 | 0 | 31 | 0 | 15 | 48.4 |
Number of independent transfections observed.
FIG.4.
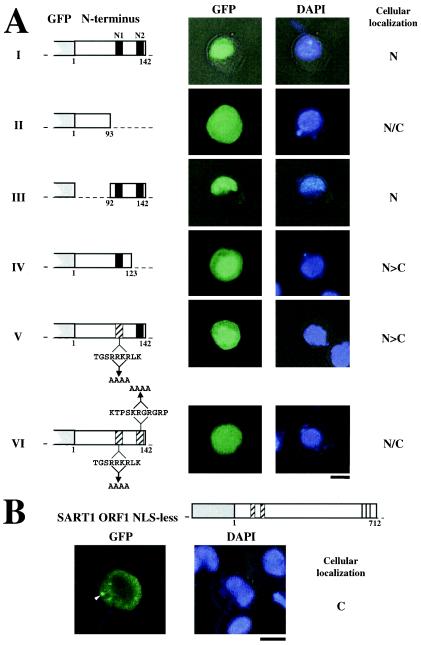
Mutational analysis of NLS activity in the N-terminal domain of SART1 ORF1. (A) NLS activity was examined by mutational analysis of SART1 ORF1. Expression constructs in which GFP was fused with each region of SART1 ORF1 are shown on the left. The left and right pictures show the localizations of GFP fusion protein and DNA stained with DAPI, respectively. N, nucleus; N/C, both nucleus and cytoplasm; N>C, many signals in the nucleus. The two putative NLSs (N1 and N2) identified by a computer-based search are represented by closed boxes. In panels V and VI, hatched boxes indicate the alanine substitution (N1, RRKR to AAAA; N2, KRGR to AAAA) within the putative NLS sequences. Bars, 10 μm. (B) Cellular localization of the NLS-less and full-length SART1 ORF1 proteins fused with GFP. The expression construct is shown at the top. Open boxes represent the SART1 ORF1 protein, and gray boxes represent the GFP region. Amino acid numbers for the ORF1 region are shown below the open box. Hatched boxes indicate the alanine substitution (N1, RRKR to AAAA; N2, KRGR to AAAA) within the NLS sequences, exactly as in panels VI in panel A. Vertical lines show zinc finger motifs. The left and right pictures show the localization of GFP fusion protein and DNA stained with DAPI, respectively. The arrowhead indicates the cytoplasmic cluster. Bar, 10 μm.
The N terminus of SART1 ORF1 includes authentic signals for transporting large proteins into the nucleus.
To obtain more information on the control of nuclear localization by ORF1, we examined GFP expression in detail (Fig. 3). In addition to mock experiments without any contribution from ORF1 (Fig. 3, panels I), GFP fused to residues 285 to 447 (IV) or to 448 to 579 (V) of SART1 ORF1 were localized to both the nucleus and cytoplasm. GFP fused to residues 285 to 447 were distributed with broad localization pattern, while this region includes residues 354 to 447 which are required for the dotted pattern. This result suggests that the N-terminal 1-284 region is also necessary for nuclear dotted pattern. The signals of GFP fusion proteins containing residues 143 to 284 were found in the cytoplasm (Fig. 3, panels III). In contrast, GFP fused to residues 1 to 142 (Fig. 3, panels II) and residues 580 to 712 (panels VI) of ORF1 were localized only in the nucleus. These results suggest that two regions, the first (residues 1 to 142) and fifth (residues 580 to 712) regions of the subdivided ORF1, have putative nuclear localization activity. In general, the nuclear pore complex, which traverses the nuclear envelope, serves as the gate through which small proteins of less than 40 kDa pass independently, whereas molecules larger than 40 kDa cannot pass through the nuclear pore complex without an NLS (27). Therefore, to ascertain whether the two small GFP molecules described above, fused to the first and fifth regions of ORF1 (the GFP fusion proteins used for panels II to VI are about 45 kDa, on average), include actual NLS activity, we tested the localization of larger fusion proteins.
The GFP fusion protein containing residues 1 to 284 of ORF1 was localized only in the nucleus (Fig. 3, panels VII). However, other larger fused proteins, containing residues 143 to 447 (Fig. 3, panels VIII), 448 to 712 (panels IX), or 285 to 712 (panels X), were found only in the cytoplasm (panels VIII and X) or in the whole cellular region (panels IX). This observation indicates that the first region of SART1 ORF1, which includes residues 1 to 284, possesses an authentic NLS. The small GFP molecule fused with residues 580 to 712 of ORF1 entered the nucleus by passive diffusion and was localized in the nucleus. Since a longer GFP molecule fused with residues 285 to 712 distinctly localized in the cytoplasm (Fig. 3, panels X), residues 580 to 712 do not have obvious activity to import the ORF1 protein into the nucleus. It is surprising that there was no NLS activity in the regions 285-447 (Fig. 3, panels IV), 143-447 (panels VIII), and 285-712 (panels X), all of which include the putative NLSs, N3 and N4 (see above). This indicates that N3 and N4, which appear to be typical NLSs with strongly basic domains, do not act as signals for nuclear translocation in SART1.
Localization in the nucleus requires both N1 and N2 signals near the N terminus of SART1 ORF1.
Two putative NLSs, N1 and N2, were identified in the N-terminal region by a computer-based search (Fig. 1B). Therefore, we used mutational analysis to study these putative NLSs, which occur in residues 96 to 134 (Fig. 4). When this region was completely deleted from the WT construct (Fig. 4A, panels I), nuclear localization was blocked and GFP signals were observed in both the nucleus and the cytoplasm (Fig. 4A, panels II). In contrast, a minimal region including N1 and N2 (92 to 142) was sufficient to localize GFP signals in the nucleus (Fig. 4A, panels III). These results strongly suggest that the nuclear import activity is encoded within the region from residue 92 to 142. To clarify whether N1 and N2 are essential for nuclear localization, we deleted the N2 region (residues 124 to 142) (Fig. 4A, panels IV) or introduced severe substitution mutations into the basic region of N1 (changing RRKR to AAAA) (Fig. 4A, panels V). In both cases, the GFP signals were observed predominantly in the nucleus, with few in the cytoplasm. When double mutations were introduced into both N1 and N2 (RRKR to AAAA in N1 and KRGR to AAAA in N2), the nuclear localization of GFP was abolished (Fig. 4A, panels VI). These results demonstrate that N1 and N2 work independently as weak NLSs but that both signals are required for nuclear transport activity. It seems that localization of some of the GFP signals shown in Fig. 4A is affected by passive diffusion because the GFP-ORF1 proteins are relatively small.
Furthermore, we analyzed localization of a full-length ORF1 construct without an NLS (Fig. 4B). This construct includes the alanine substitution in both N1 and N2, which is exactly the same as the substitution construct in Fig. 4A, panel VI. GFP signals fused to the full-length ORF1 lacking the NLS were observed only in the cytoplasm and not in the nuclear portion. When 31 examples of this construct were tested (Table 2), all GFP signals were localized only in the cytoplasm, and half of them showed dotted signals in the cytoplasm (Fig. 4B; Table 2). Improper cytoplasmic accumulation of GFP-fused protein is possibly due to blocking of localization into the nucleus. These data demonstrate that N1 and N2 control the nuclear localization of SART1 ORF1 protein.
An N-terminal domain containing NLSs is essential for in vivo retrotransposition of SART1.
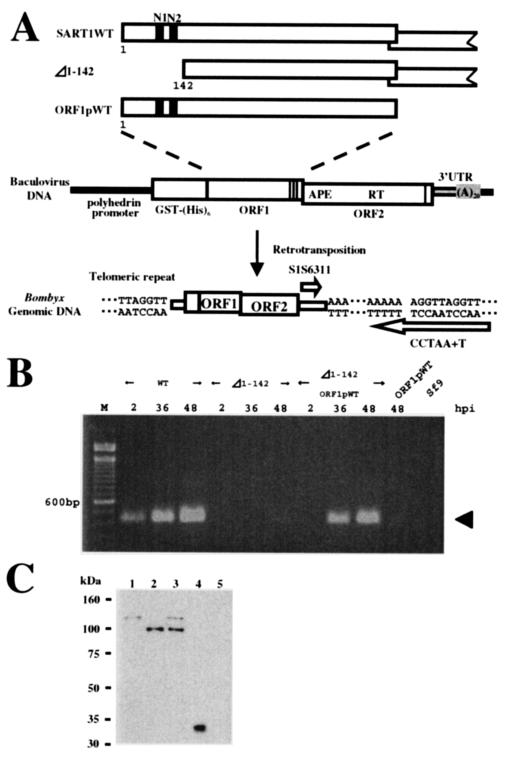
Next, we examined the requirement for the NLSs at the N terminus of ORF1 in the retrotransposition of SART1, using a recently developed in vivo retrotransposition assay (37). Highly expressed SART1 under control of the polyhedrin promoter of AcNPV in Sf9 cells can transpose into specific sites at telomeric repeats (TTAGG)n. This event is detectable by PCR with a specific primer set (Fig. 5A). Since the genome of Sf9 cells does not have the Bombyx SART1, the retrotransposed SART1 copy expressed by AcNPV infection could be detected by this analysis. First, we constructed an NLS-deficient mutant of SART1 (Δ1-142 construct) in the AcNPV vector and infected Sf9 cells with it. At 24 to 48 h after infection, we extracted the genomic DNA and used PCR to amplify the boundaries between the retrotransposed SART1 3′ ends and the telomeric repeats. Although retrotransposition of wild-type SART1 occurred actively and increased from 24 to 48 h postinfection (hpi), Δ1-142 did not show a significant PCR band even at 48 hpi (Fig. 5B). The above result, therefore, shows that the N-terminal region of SART1 ORF1 is essential for the efficient retrotransposition of the SART1 element.
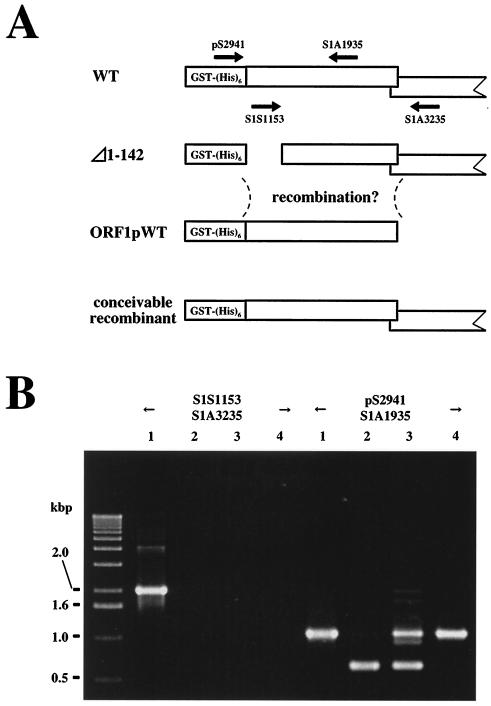
FIG. 5.
In vivo retrotransposition assay of WT SART1 and Δ1-142 SART1. (A) Schematic overview of constructs and primers used for the in vivo retrotransposition assay of SART1. The SART1 ORF1, ORF2, and 3′ untranslated region (UTR) were expressed downstream from the GST-His6 tag under control of the polyhedrin promoter of AcNPV (37). PCR amplification was performed with two primers (open arrows) (see Table 1) between the 3′ UTR of retrotransposed SART1 and the telomeric repeats in the Sf9 host genome. In Δ1-142 SART1, the ORF1 N-terminal residues 1 to 142, which include both N1 and N2, were deleted from WT SART1 AcNPV. Therefore, in Δ1-142, all regions other than residues 1 to 142 of ORF1 are the same as those of the WT construct. ORF1pWT AcNPV includes only the ORF1 region of SART1 and not ORF2 and the 3′ UTR. (B) Results of the in vivo retrotransposition assay. Lane M, size marker with 100-base ladder; Sf9, intact Sf9 genome with no recombinant AcNPV infection. The PCR band, which represents the exact retrotransposition, is 423 bp in length (arrow). (C) Immunoblot detection of ORF1 proteins N-terminally conjugated with the GST-His6 tag in the in vivo retrotransposition assay (see panel A). Total proteins from Sf9 cells infected with each recombinant AcNPV were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblot assay with anti-His6 antiserum to detect His-tagged proteins. Lanes: 1, WT SART1; 2, Δ1-142; 3, coinfection of Δ1-142 and ORF1pWT; 4, mock infection; and 5, noninfected Sf9 cells. The mock infection should express only GST-His6.
We also investigated whether the retrotranspositional activity of the Δ1-142 mutant could be rescued by normal ORF1 protein by trans-complementation. When the AcNPV virus that expresses only the ORF1 protein of SART1 (Fig. 5A, ORF1pWT) was coinfected with Δ1-142 mutant virus, a strong PCR band representing highly active retrotransposition was observed 36 to 48 h after coinfection, which seemed slightly delayed compared with WT infection (Fig. 5B). This time lag for retrotransposition between infection with WT SART1 only and ORF1pWT/Δ1-142 coinfection may indicate that multiple ORF protein complexes are formed less stably among WT ORF1 and N-terminally deficient Δ1-142 ORF1 proteins or that cis preference is more efficient than trans-complementation in the SART1 element (39). The result clearly shows that the loss of the N-terminal region of Δ1-142 can be compensated for by WT ORF1 protein.
ORF1 protein of SART1 acts as an independent functional unit.
The above result of the trans-complementation experiment also suggests another important concept described below. Until now, it has not been demonstrated that ORF1 protein in non-LTR retrotransposon acts as an independent functional unit. We usually envisage the ORF1 and ORF2 units based only on sequence information, because we do not know the exact translation mechanisms for producing the two overlapping ORFs in non-LTR retrotransposons. In this study, therefore, we expressed the longest ORF1, which starts at ATG+1 (+880 from the SART1 5′ end) and ends at putative stop codon TAA+713 (+3021), conjugated with the GST-His6 tag in the baculovirus expression system (Fig. 5A). This putative ORF1 protein from ORF1pWT complemented the NLS-deficient element Δ1-142 in trans (Fig. 5B).
To confirm this hypothesis, we further studied this trans-complementation event at the protein level by immunoblotting analyses (Fig. 5C). The total size of the GST-His6 tag fused to ORF1 1-712 of SART1 expressed in the AcNPV system is estimated to be 106 kDa. When the proteins expressed from the WT SART1-AcNPV hybrid element were analyzed by Western blotting with anti-His antibody, we detected a band of about 115 kDa, with no other bands (Fig. 5C, lane 1). Although some readthrough products caused by frameshift translation are usually observed in retroviruses and LTR-type retrotransposons, such a frameshift event did not occur, at least in the non-LTR element SART1. We observed about a 9-kDa increase in size in the actual ORF1 protein, which is probably caused by some modification. This proposition is supported by the observation that the ORF1 protein of Δ1-142 (calculated to be 91 kDa) was detected as a protein of about 100 kDa (lane 2). Because similar amounts of ORF1 proteins were expressed in both the WT and the Δ1-142 mutant (Fig. 5C, lanes 1 and 2), a loss of ORF1 function but not a decrease in ORF1 quantity was responsible for the block of retrotransposition in Δ1-142 (Fig. 5B). When the Δ1-142 mutant was rescued with ORF1pWT by trans-complementation, two bands of 100 and 115 kDa were expressed (Fig. 5C, lane 3). The band in lane 1 (WT SART1 origin) and the uppermost band (ORF1pWT origin) in lane 3 seem to be almost the same in molecular mass (115 kDa). This indicates that WT SART1 ORF1 (lane 1) is actually translated from the presumed ORF to 712Glu (Fig. 1B).
In this type of trans-complementation experiment, however, it is possible that a WT retrotransposon unit may be produced by recombination (Fig. 6A). To understand what occurs in the trans-complementation of Δ1-142 and ORF1pWT, we studied the viral genotypes in Sf9 cells by PCR analysis (Fig. 6B). After 72 h of infection with WT (Fig. 6B, lane 1), Δ1-142 (lane 2), Δ1-142 plus ORF1pWT (lane 3), or ORF1pWT (lane 4), genomic DNAs were extracted and PCR was performed with two different primer sets. With the primer set pS2941 and S1A1935, the PCR bands were expected to appear in all cases, and in fact we found an approximately 1.0-kb band for the WT ORF1 genome (lanes 1, 3, and 4) and a band of about 0.6 kb for Δ1-142 genome (lanes 2 and 3). PCR analysis with another primer set, consisting of S1S1153 in the N-terminal region of ORF1 and S1A3235 in the ORF2 region, produced a band only in the WT (lane 1) and not in Δ1-142 (lane 2), Δ1-142 plus ORF1pWT (lane 3), or ORF1pWT (lane 4). The result in lane 3 showed that there was no recombination event producing the WT SART1 element when Δ1-142 and the ORF1pWT virus were coinfected. These results indicate that the translation of SART1 ORF1 ends at a stop codon of TAA713 and that its product has the functional roles in retrotransposition of SART1.
FIG. 6.
Identification of the viral genotype in coinfection. (A) Schematic model for the possible recombinant that could appear in the trans-complementation experiment with Δ1-142 plus ORF1pWT. Arrows indicate the primers used to analyze the viral genotype that appears during the coinfection (see panel B). The PCR products for the WT genotype should be 2,082 bp for the primer set S1S1153-S1A3235 and 1,006 bp for pS2941-S1A1935. The PCR product for the Δ1-142 genotype with the pS2941-S1A1935 primer set should be 580 bp. (B) Identification of the viral genotype by PCR. Lanes: 1, WT SART1; 2, Δ1-142; 3, coinfection of Δ1-142 and ORF1pWT; and 4, ORF1pWT. Size markers are shown on the left.
NLSs N1 and N2 of ORF1 in SART1 control retrotransposition activity.
We analyzed the retrotransposition activities of various mutants in which mutations were introduced into the putative NLSs, N1 and N2 (Fig. 7). When the extreme N-terminal region of SART1 ORF1, which is upstream from N1 and N2, was deleted (Δ1-52) (Fig. 7A), in vivo retrotransposition occurred from 24 hpi and increased gradually (Fig. 7B). The similar retrotransposition activities of WT and the Δ1-52 mutant indicate that about 50 amino acids at the N-terminal end of SART1 ORF1 are not essential under the conditions used in the assay. However, an N1 deletion mutant, Δ1-105, showed a dramatic decrease in retrotransposition activity, similar to that of Δ1-142, in which both N1 and N2 are deleted. In these deletion mutants, there is a possibility that incorrect folding of ORF1 may cause the block of their retrotransposition. We then tried to test the substitution mutants. An N1 mutant, N1>A, in which RRKR in the N1 sequence was replaced by AAAA, also exhibited a dramatic decrease in the retrotransposition activity. As described above, the same mutation (with RRKR in the N1 sequence replaced by AAAA) was studied in a GFP fusion experiment and resulted in a weakened nuclear localization signal (Fig. 4, panels V). Therefore, degeneration of the nuclear localization signal in SART1 ORF1 strongly affects the retrotransposition activity of the SART1 element. Similarly, an N2 mutant, N2>A (KRGR to AAAA), and an N1/N2 double mutant (RRKR to AAAA in N1 and KRGR to AAAA in N2) also lacked the retrotransposition activity. In order to confirm that the loss of retrotransposition activity in these N-terminal mutants is not due to destabilization of the ORF1 proteins, we analyzed the proteins expressed in the mutants by the Western blotting assay. In all N-terminal mutants, we identified ORF1 proteins the expected sizes (Fig. 7C, lanes 2 to 7). Moreover, the retrotransposition activity of the N-terminal mutants (lanes 2 to 7) was rescued by coinfection of ORF1pWT expressing only WT SART1 ORF1 (Fig. 7D). This demonstrates that all mutations and deletions at the N terminus of ORF1 did not affect the mRNA stability, ORF2 functions, and RNP formation. These data indicate that the functional defect but not the instability of ORF1 proteins causes the block of retrotransposition for these mutants. In conclusion, N1 and N2 act as NLSs to control the retrotransposition activity of SART1.
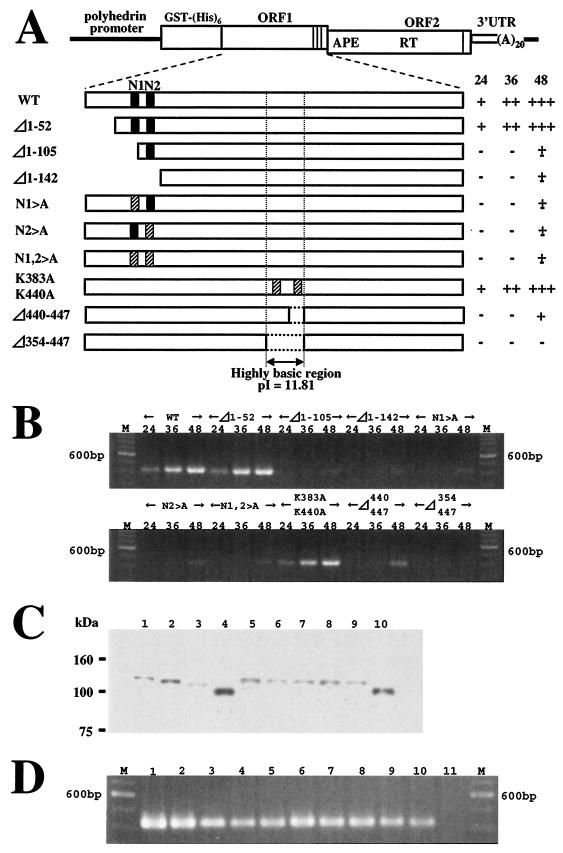
FIG. 7.
Retrotransposition activity of SART1 with mutations in the N-terminal and central regions of ORF1. (A) Constructs of various SART1 mutants. WT and mutated SART1 ORF1, ORF2, and 3′ untranslated region (UTR) were expressed downstream from the GST-His6 tag by AcNPV-recombinant virus infection of Sf9 cells. Closed boxes indicate putative NLSs. Hatched boxes indicate the substituted regions where several amino acids were replaced by alanine. In constructs N1>A, N2>A, and N1,2>A, RRKR in N1 and KRGR in N2 were replaced with AAAA. The K residues at 383 (K383A) and 440 (K440A) were replaced with A. The results of the in vivo retrotransposition assay (see panel B) are summarized on the right. The numbers at the top represent hpi. +++, very strong signal; ++, strong signal; +, weak signal; ±, very weak signal; −, no signal. (B) PCR amplification of the 3′ junctions of the retrotransposed WT SART1 or various mutants. The PCR band, which represents the SART1 retrotransposition, is 423 bp in length. (C) Western blots of mutant proteins. Protein production in mutants was analyzed by immunoblot assay with anti-His6 antiserum to detect His-tagged proteins. Lanes: 1, WT SART1; 2, Δ1-52; 3, Δ1-105; 4, Δ1-142; 5, N1>A; 6, N2>A; 7, N1,2>A; 8, K383,440A; 9, Δ440-447; 10, Δ353-447. (D) Rescue of mutant constructs by trans-complementation. At 36 h after coinfection of AcNPV viruses expressing various mutants with a construct producing only SART1 ORF1 (ORF1pWT), Sf9 genomic DNA was extracted and assayed by PCR detection (see Fig. 5B). Lanes: M, size marker (100-base ladder); 1, WT SART1; 2, Δ1-52 plus ORF1pWT; 3, Δ1-105 plus ORF1pWT; 4, Δ1-142 plus ORF1pWT; 5, N1>A plus ORF1pWT; 6, N2>A plus ORF1pWT; 7, N1,2>A plus ORF1pWT; 8, K383,440A plus ORF1pWT; 9, Δ440-447 plus ORF1pWT; 10, Δ353-447 plus ORF1pWT; 11, ORF1pWT.
A highly basic region in the center of ORF1 is essential for SART1 retrotransposition.
There is a highly basic region (residues 285 to 447; pI, 11.55) in the center of SART1 ORF1 (Fig. 1C). In particular, the pI of amino acid residues 354 to 447 is estimated to be 11.81, and in this region we found two putative NLSs, N3 and N4. Based on the results of the GFP fusion experiment, however, this highly basic region does not to act in nuclear localization (Fig. 3, panels IV, VIII, and X). Rather, amino acid residues 354 to 447 seem to be responsible for the dotted localization pattern of SART1 ORF1 (Fig. 2; see “Residues 354 to 447 of SART1 ORF1 are responsible for nuclear localization with a dotted pattern” above). Therefore, to ascertain whether this area is essential for SART1 retrotransposition, we used in vivo retrotransposition analysis of several mutants to block its function (Fig. 7). When two lysine (K) residues at 383 and 440 were replaced by A (Fig. 7A), retrotransposition occurred as in WT infection (Fig. 7B). Deletion of residues 440 to 447(N4 deletion mutant) produced a dramatic decrease in retrotransposition activity. Furthermore, a mutant with a deletion of residues 354 to 447, in which both basic amino acid clusters, N3 and N4, were deleted, completely lacked retrotransposition activity (Fig. 7B). It is of interest that the K383A-K440A substitution mutant retrotransposed more efficiently than deletion mutants (Δ354-447). This implies that the deletion may cause incorrect folding of proteins and that a conformation including the highly basic region is important for SART1 retrotransposition. These constructs normally expressed mutated SART1 ORF1 proteins of the expected sizes (Fig. 7C, lanes 8 to 10) and also could retrotranspose to the telomeric repeats by trans-complementation with ORF1pWT (Fig. 7D, lanes 8 to 10). These results show that a highly basic region in the center of ORF1, residues 354 to 447, is essential for SART1 retrotransposition.
DISCUSSION
Functional map of SART1 ORF1.
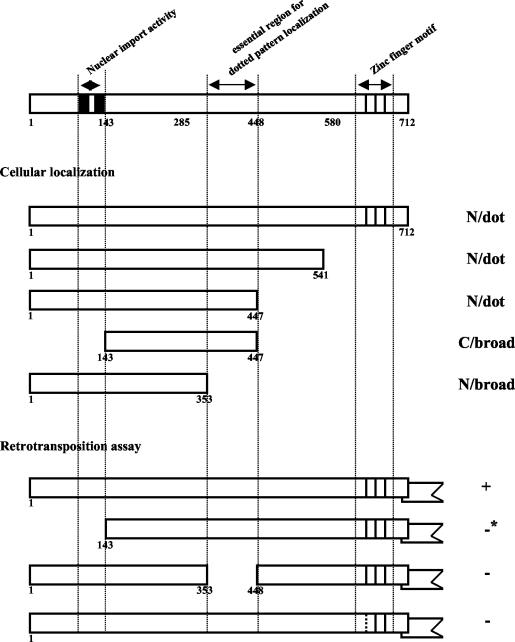
In this study, we show that N1 and N2 near the N terminus of SART1 ORF1 are involved in nuclear import and that the highly basic region consisting of residues 354 to 447 (which includes N3 and N4) is involved in localizing the ORF1 protein as a dotted pattern in the nucleus (Fig. 8). The loss of these nuclear localization-controlling signals resulted in a deficiency in SART1 retrotransposition (Fig. 8). We previously showed that the replacement of His (LGH626VS) in the CCHC zinc finger motif with Pro caused a loss of the retrotransposition activity of SART1 (37), suggesting that the zinc finger motif in ORF1 is also involved in the SART1 retrotransposition event. Therefore, there are at least three functional domains in ORF1 that are necessary for SART1 retrotransposition.
FIG. 8.
Three essential domains in ORF1 required for the retrotransposition activity of SART1. This figure summarizes the results in this paper, indicating the essential ORF1 regions involved in cellular localization and retrotransposition of SART1. N/dot, nuclear and dotted signal distribution; C/broad, cytoplasmic and broad signal distribution; N/broad, nuclear and broad signal distribution. +, active retrotransposition; −, no retrotransposition; −*, very weak retrotransposition (see Fig. 5). Closed boxes and vertical lines indicate N1 and N2 NLSs and the three CCHC-type zinc finger motifs, respectively. The broken line in the first zinc finger motif in the bottom construct shows the substitution mutation site (1H626P, LGHVS to LGPVS), which results in complete loss of retrotransposition activity (37). Deletion of residues 354 to 447 inhibited both the dotted nuclear localization and retrotransposition of SART1.
(i) N1 and N2 regions as NLSs.
The NLSs so far characterized in various proteins are categorized into two classes: the single type and the bipartite type, which includes two clusters composed of about 10 basic amino acids (27). In SART1 ORF1, the length of the interval between the N1 and N2 clusters is 28 amino acids, which is longer than that in the standard bipartite type. The respective N1 and N2 clusters are categorized as single type by computer-based estimations (Fig. 1B). However, the present results show that mutations in even one of them caused the loss of the nuclear import activity (Fig. 4) and a deficit in SART1 retrotransposition (Fig. 7). This indicates that N1 and N2 function cooperatively and are similar to bipartite-type NLSs; they are classified as NLSs of a novel type.
(ii) Central highly basic regions for dotted pattern localization.
As shown in Fig. 2, residues 354 to 447 are involved in the formation of the dotted pattern. This localization activity in the nucleus seemed to be dependent on the NLS signals of N1 and N2, because the region from residue 143 to 447, which contains residues 354 to 447 but not N1 and N2, showed only broad cytoplasmic localization for the SART1 ORF1 protein (Fig. 3, panels VIII). A recent report demonstrated that the ORF1 protein of HeT-A and TART, the telomere-specific non-LTR retrotransposons of Drosophila melanogaster, are localized in a similar dotted pattern in the nucleus, like those in this study, whereas the ORF1s of the other three elements (jockey, Doc, and I) in Drosophila showed cytoplasmic localization (30). Colocalization of the ORF1s of HeT-A and the telomere-binding protein HOAP suggests that the ORF1s of these elements play a role in localization to the telomere, although there is no information on the responsible structure in the ORF1s (31). Combining the observations on telomere-specific retrotoransposons in Drosophila, the dotted localization pattern of ORF1 of the telomeric repeat-specific retrotransposon SART1 may also reflect telomere or telomeric repeat binding. Our preliminary results from Southwestern hybridization analysis and GST pull-down assays indicated that full-length ORF1 protein binds specifically to (TTAGG)n repeats (data not shown), although it is unclear at present whether the 354-447 region is involved in the (TTAGG)n binding.
In several site-specific non-LTR retrotransposons, it has been demonstrated that EN domains in ORF2 specifically nick the target DNA sequence (4, 8, 9). We reported that EN of TRAS1, another type of telomeric repeat-specific non-LTR retrotransposon, has a specific nicking activity for (TTAGG/CCTAA)n (1). However, we found that EN of SART1 has a relatively lower specificity for the telomeric repeats (T. Anzai and H. Fujiwara, unpublished data). Basically, a specific enzymatic activity of EN plays a central role in TRAS1 integration into the telomeric repeat, while the SART1 retrotransposition into telomeres seems to be dependent both on the SART1 EN activity in ORF2 and on the telomere-targeting ability in ORF1.
(iii) Zinc finger motif.
Most non-LTR retrotransposons, except mammalian LINEs, retain one or more zinc finger motifs in ORF1 (7). At present, however, there is little functional implication of the zinc finger motifs in non-LTR retrotransposons. The zinc finger motifs are also found among Gag proteins in other retroelements, i.e., the LTR-type retrotransposons and retroviruses. These zinc finger motifs (CXXCXXXXHXXXXC, where X is a space), which are an unusual type not seen among many other zinc finger proteins, are specifically conserved among retroelements (7). In human immunodeficiency virus type 1 (HIV-1), these zinc finger motifs in the nucleocapsid are involved in the recognition and packaging of the viral RNA genome (29). In retroviruses and LTR retrotransposons such as Ty1, it is believed that nucleic acid chaperone and matchmaker properties are derived from CCHC zinc fingers or basic amino acid clusters (5, 10). In this study, we found that the SART1 ORF1 protein was localized in the cytoplasm as cytoplasmic clusters, which were observed abundantly in the WT and in the full-length construct lacking an NLS but rarely in the zinc finger-deficient constructs (Fig. 2B [panels I] and 4B and Table 2). Recently, Rashkova et al. have reported that zinc finger-containing regions in ORF1 of the Drosophila telomeric non-LTR retrotransposons were localized with a body or cluster pattern, indicating protein-protein interactions (32). These findings imply that the domain containing zinc fingers in ORF1 of non-LTR retrotransposon has the functional roles in RNP formation, although further studies are necessary to confirm the hypothesis.
ORF1 protein in the process of retrotransposition.
Most non-LTR retrotransposons encode two putative ORFs, but there is no report that the ORFs work as substantial and independent units. One reason for this is that we do not yet understand the exact translation mechanisms that produce ORF proteins from the bicistronic non-LTR retrotransposon mRNA. In the SART1 element, ORF2 overlaps ORF1 by 54 nucleotides in the −1 reading frame (36). In retroviruses and LTR retrotransposons, −1 (or rarely +1) translational frameshifting usually produces Gag-Pol fusion proteins from the mRNA. In non-LTR retrotransposons, however, it is unclear whether the two ORFs are translated into one fusion protein by −1 frameshifting or into two independent proteins by specific mechanisms.
Selem et al. (34) detected the putative ORF1 protein of a Drosophila non-LTR element, I factor, by Western blotting and indirect immunofluorescence labeling. Here, we have demonstrated not only that ORF1s of the expected sizes expressed from their mRNAs by both the WT and deletion mutant Δ1-142 (Fig. 5C) but also that ORF1 protein itself can rescue the NLS-less Δ1-142 function by trans-complementation (Fig. 5B). This is the direct evidence showing that ORF1 of SART1 is an independent functional unit and is translated by using the putative stop codon (+713; TAA) and not by translational frameshifting. SART1 ORF1 protein expressed with baculovirus in Sf9 was observed to have a 9-kDa increase in size, and we speculate that this is caused by some modification such as glycosylation.
The human L1 element shows cis preference for the ORF proteins produced from its own mRNA (39). SART1 showed trans-complementation, but its lower retrotranspositional efficiency also suggests a cis preference in the SART1 element. These observations indicate that ORF1 and ORF2 of non-LTR retrotransposons are assembled with their mRNAs immediately after translation in the cytoplasm. By analogy with other retroelements, multimerized ORF1 may act as the Gag protein does, to surround and enclose ORF1 and mRNA, but its RNP structure remains unclear. Although the ORF1 proteins of SART1 (this study) and Drosophila TART and HeT-A (30) are actually transported into the nucleus, there is no report of nuclear localization of ORF1 in some other elements, such as mouse LINE-1 (23, 38) and Drosophila I factor (30, 34). Therefore, it is not currently clear whether RNP transport into the nucleus is generally conserved among non-LTR retrotransposons. Compared with that in general non-LTR retrotransposons, the capacity for nuclear transport may be elevated in telomere-specific non-LTR elements, such as SART, HeT-A, and TART, resulting in nuclear localization during the interphase of cell division when large proteins without NLS cannot pass the nuclear membrane.
Comparison of nuclear transport mechanisms in various retroelements.
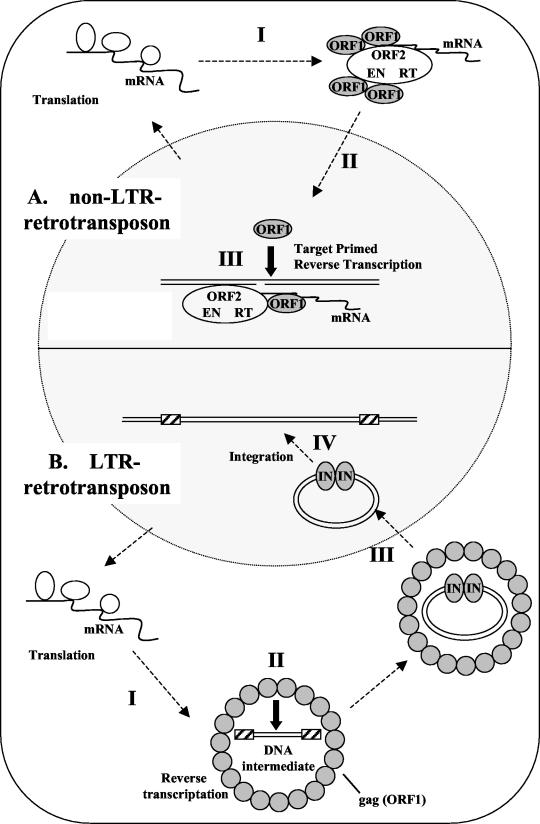
There is a critical difference between non-LTR retrotransposons and other retroelements, such as LTR retrotransposons and retroviruses, in the process of retrotransposition (Fig. 9). The latter elements are reverse transcribed and formed into double-stranded cDNA in the cytoplasm, whereas the non-LTR elements are reverse-transcribed in the nucleus, in a process called TPRT (21). During the TPRT event in the nucleus, the non-LTR retrotransposon machinery includes mRNA, the ORF2 unit (EN and RT), and ORF1 protein, which is required for nuclear localization (Fig. 9A, step III), as shown in this study. In addition to the NLS function, ORF1 may be generally required in the retrotransposition machinery during TPRT. Martin and Bushman (25) reported that the ORF1 protein from mouse LINE-1 promotes the annealing of the cDNA strand and mediates nucleic acid transfer steps during TPRT. Several other studies also reported that the ORF1 proteins from non-LTR retrotransposons have roles in nucleic acid binding activity (7, 14, 18, 19). These data indicate that the ORF1 protein has nucleic acid chaperone activity and brings mRNA and ORF2 together into the RNP in the non-LTR retrotransposition machinery. Three essential domains, i.e., the NLS, telomere-binding, and zinc finger domains, in SART1 ORF1 support this hypothesis that ORF1 protein is necessary in the nucleus. The Gag protein of LTR retroelements and retroviruses also has nucleic acid chaperone activity during reverse transcription in the cytoplasm (5, 10).
FIG. 9.
Comparison of the life cycles of non-LTR retrotransposons and LTR retrotransposons. (A). Non-LTR retrotransposons. I, translated ORF proteins and their own mRNAs assemble to form RNPs. II, RNP moves to the nucleus. SART1 has a remarkable NLS in the ORF1 protein, which results in efficient import of the element into the nucleus, leading to its efficient retrotransposition. III, RNP retrotransposes into the target site of the host genome by TPRT. In TPRT, the ORF1 protein may be required for nucleic acid chaperone function to promote the formation of stable double-stranded DNA, as suggested by Martin and Bushman (25). (B). LTR retrotransposons. I, translated ORF proteins and their own mRNAs form virus-like particles. II, reverse transcription occurs in the cytoplasm to generate a DNA intermediate in the virus-like particle. In this process, Gag protein is thought to exhibit the nucleic acid chaperone activity. III, the DNA intermediate and integrase (IN) from a complex and are imported into the nucleus by the NLS of IN. VI, the IN-DNA complex moves to an insertion site in the host genome, and the insertion event is catalyzed by IN. Closed arrows represent the role of ORF1 (or Gag) as the nucleic acid chaperone in both non-LTR and LTR elements.
However, in non-LTR elements, LTR elements, and retroviruses, the insertional event occurs in the host genome, and the protein-nucleic acid complex required for integration must move to the target DNA. The preintegration complexes (PICs) of many retroviruses, such as murine leukemia virus, cannot enter the nucleus but instead wait for the breakdown of the nuclear envelope during mitosis (6). Although murine leukemia virus is unable to replicate in nondividing cells, the HIV-1 PIC can enter the intact nucleus and productively replicate in nondividing cells (3). Active nuclear import of the PIC is essential for HIV-1 infection of dividing and nondividing cells (2). Like HIV-1, the non-LTR element SART1 is actively transported into the nucleus, which allows its efficient retrotransposition into the telomeric repeat. The functional domains in ORF1 of SART1 found here will be used for making a more efficient gene delivery vector, which will enable delivery of genes into specific genomic locations.
Acknowledgments
This work was supported by grants from the Ministry of Education, Science and Culture of Japan (MESCJ); by a Grant-in-Aid from the Research for the Future Program of the Japan Society for the Promotion Science (JSPS); and by research fellowships of the Japan Society for Promotion of Science for Young Scientists (to H.T.).
REFERENCES
- 1.Anzai, T., H. Takahashi, and H. Fujiwara. 2001. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)n by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell. Biol. 21:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, S., G. Pont-Kingdon, and D. Carroll. 2000. Target specificity of the endonuclease from the Xenopus laevis non-long terminal repeat retrotransposon, Tx1L. Mol. Cell. Biol. 20:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cristofari, G., D. Ficheux, and J. L. Darlix. 2000. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 275:19210-19217. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, B. R. 2001. Journey to the center of the cell. Cell 105:697-700. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, A., E. Hartswood, T. Paterson, and D. J. Finnegan. 1997. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 16:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, Q., J. V. Moran, H. H. Kazazian, Jr., and J. D. Boeke. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905-916. [DOI] [PubMed] [Google Scholar]
- 9.Feng, Q., G. Schumann, and J. D. Boeke. 1998. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. USA 95:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng, Y. X., S. Campbell, D. Harvin, B. Ehresmann, C. Ehresmann, and A. Rein. 1999. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 73:4251-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara, H., Y. Nakazato, S. Okazaki, and O. Ninaki. 2000. Stability and telomere structure of chromosomal fragments in two different mosaic strains of the silkworm, Bombyx mori. Zool. Sci. 17:743-750. [Google Scholar]
- 12.Gilbert, N., S. Lutz-Prigge, and J. V. Moran. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315-325. [DOI] [PubMed] [Google Scholar]
- 13.Hohjoh, H., and M. F. Singer. 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15:630-639. [PMC free article] [PubMed] [Google Scholar]
- 14.Hohjoh, H., and M. F. Singer. 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16:6034-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, S. E., M. F. Singer, and G. D. Swergold. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267:19765-19768. [PubMed] [Google Scholar]
- 16.Kazazian, H. H., Jr., C. Wong, H. Youssoufian, A. F. Scott, D. G. Phillips, and S. E. Antonarakis. 1988. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 322:164-166. [DOI] [PubMed] [Google Scholar]
- 17.Kazazian, H. H., Jr., and J. L. Goodier. 2002. LINE drive: retrotransposition and genome instability. Cell 110:277-280. [DOI] [PubMed] [Google Scholar]
- 18.Kolosha, V. O., and S. L. Martin. 1997. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl. Acad. Sci. USA 94:10155-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolosha, V. O., and S. L. Martin. 2003. High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1). J. Biol. Chem. 278:8112-8117. [DOI] [PubMed] [Google Scholar]
- 20.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 21.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 22.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 23.Martin, S. L., and D. Branciforte. 1993. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cell. Biol. 13:5383-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, S. L., J. Li, J. A. Weisz. 2000. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 304:11-20. [DOI] [PubMed] [Google Scholar]
- 25.Martin, S. L., and F. D. Bushman. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 27.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 28.Pont-Kingdon, G., E. Chi, S. Christensen, and D. Carroll. 1997. Ribonucleoprotein formation by the ORF1 protein of the non-LTR retrotransposon Tx1L in Xenopus oocytes. Nucleic Acids Res. 25:3088-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon, D. T., G. Li, and A. Aldovini. 1998. Nucleocapsid and matrix protein contributions to selective human immunodeficiency virus type 1 genomic RNA packaging. J. Virol. 72:1983-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashkova, S., S. E. Karam, and M. L. Pardue. 2002. Element-specific localization of Drosophila retrotransposon Gag proteins occurs in both nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA 99:3621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashkova, S., S. E. Karam, and M. L. Pardue. 2002. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J. Cell Biol. 159:397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashkova, S., A. Athanasiadis, and M. L. Pardue. 2003. Intracellular targeting of Gag proteins of the Drosophila telomeric retrotransposons. J. Virol. 77:6376-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki, T., and H. Fujiwara. 2000. Detection and distribution patterns of telomerase activity in insects. Eur. J. Biochem. 267:3025-3031. [DOI] [PubMed] [Google Scholar]
- 34.Selem, M., I. Busseau, S. Malinsky, A. Bucheton, and D. Teninges. 1999. High-frequency retrotransposition of a marked I factor in Drosophila melanogaster correlates with a dynamic expression pattern of the ORF1 protein in the cytoplasm of oocytes. Genetics 151:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Symer, D. E., C. Connelly, S. T. Szak, E. M. Caputo, G. J. Cost, G. Parmigiani, and J. D. Boeke. 2002. Human L1 retrotransposition is associated with genomic instability in vivo. Cell 110:327-338. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi, H., S. Okazaki, and H. Fujiwara. 1997. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res. 25:1578-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, H., and H. Fujiwara. 2002. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 21:408-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trelogan, S. A., and S. L. Martin. 1995. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl. Acad. Sci. USA 92:1520-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, W., N. Gilbert, S. L. Ooi, J. F. Lawler, E. M. Ostertag, H. H. Kazazian, Jr., J. D. Boeke, and J. V. Moran. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]