Abstract
We have shown that steroid hormones coordinately control gene transcriptional activity and splicing decisions in a promoter-dependent manner. Our hypothesis is that a subset of hormonally recruited coregulators involved in regulation of promoter transcriptional activity also directly participate in alternative RNA splicing decisions. To gain insight into the molecular mechanisms by which transcriptional coregulators could control splicing decisions, we focused our attention on a recently identified coactivator, CoAA. This heterogeneous nuclear ribonucleoprotein (hnRNP)-like protein interacts with the transcriptional coregulator TRBP, a protein recruited to target promoters through interactions with activated nuclear receptors. Using transcriptional and splicing reporter genes driven by different promoters, we observed that CoAA mediates transcriptional and splicing effects in a promoter-preferential manner. We compared the activity of CoAA to the activity of other hnRNP-related proteins that, like CoAA, contain two N-terminal RNA recognition motifs (RRMs) followed by a C-terminal auxiliary domain and either have or have not been implicated in transcriptional control. By swapping either CoAA RRMs or the CoAA auxiliary domain with the corresponding domains of the proteins selected, we showed that depending on the promoter, the RRMs and the auxiliary domain of CoAA are differentially engaged in transcription. This contributes to the promoter-preferential effects mediated by CoAA on RNA splicing during the course of steroid hormone action.
Regulation of gene expression is a complex process occurring in several steps, including transcription, splicing, transcript 5′- and 3′-end modification (capping and polyadenylation, respectively), transcript export, and stability. It is now accepted that these different steps are “mechanistically” coupled: transcription is coupled to capping, splicing, and polyadenylation; and splicing is coupled to capping, polyadenylation, and export (9, 30, 32, 40, 44). An emerging view of the coupling among gene expression machines indicates that proteins involved in early steps in the pathway can influence subsequent downstream steps (9, 30, 32). Transcriptional coregulators recruited to target gene promoters by DNA-binding transcriptional factors, including nuclear receptors, could play a major role in this regard (2, 13, 35, 37). Among the more than 100 coregulators identified to date that participate directly in transcriptional regulation, a subset of these proteins are structurally or functionally related to proteins involved in pre-mRNA processing (2, 11, 13, 14, 37).
Functional principles for the “mechanical” coordination between the different steps of the gene expression process are emerging (7, 12, 38). Most human primary transcripts contain several exons separated by introns that are removed during the splicing reaction. Due to the presence of multiple splice sites, the RNA-splicing process can lead to the production of multiple mature transcripts. Alternative splicing is more a rule than an exception since up to 60% of the human gene products are alternatively spliced (7, 12, 15, 38, 48). This mechanism permits diversity of translatable mRNAs, thereby increasing the proteome diversity encoded by a limited number of genes (7, 12, 31, 38). The “mechanical” coupling of transcription to splicing could allow the transcriptional machinery to simultaneously control the amount and the nature (exon content) of the transcript. This hypothesis is supported by reports showing that promoter architecture influences splicing decisions (6, 17, 42). In this context, we also have shown that activated steroid hormone receptors control gene transcription and affect splicing decisions in a promoter-dependent manner (2). Our hypothesis is that hormonally activated nuclear receptors recruit a set of transcriptional coregulators which may participate in the splicing decisions of the neonascent transcripts (2). Supporting this hypothesis, we and others have shown that transcriptional coregulators can indeed affect splicing decisions (2, 11, 14, 37).
To gain insight into the molecular mechanisms by which transcriptional coregulators affect splicing decisions, we focused our attention on a recently identified protein, CoAA (16). CoAA was identified as an interacting protein with the transcriptional coregulator TRBP; TRBP was cloned by several groups as an LXXLL domain-containing protein that interacts with several nuclear receptors (5, 20, 23, 29, 56). CoAA is a heterogeneous nuclear ribonucleoprotein (hnRNP)-like protein similar in structure to a family of proteins involved in pre-mRNA processing (16, 48, 51, 52). CoAA contains two RNA recognition motifs (RRMs) within the N-terminal part of the protein. An auxiliary domain within the C-terminal part contains the TRBP-interacting domain (TRBP-ID [see Fig. 3]) and is rich in glycine and tyrosine residues (16). CoAA enhances the transcriptional activity of several transcription factors, and we have reported that it affects splicing decisions for transcripts from a minigene derived from the human CD44 gene (2, 16).
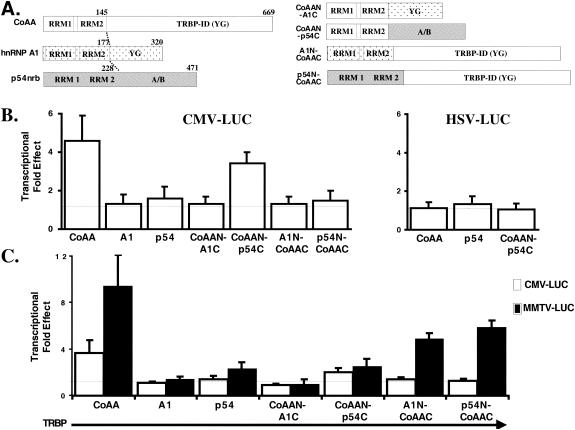
FIG. 3.
The RRMs of CoAA mediate transcriptional effects on CMV-luciferase reporter gene. (A) CoAA, hnRNPA1, and p54nrb contain two N-terminal RNA RRMs and a C-terminal auxiliary domain either rich in glycine (G) and tyrosine (Y) residues (CoAA and hnRNPA1) or rich in acidic and basic residues (A/B, p54nrb). The RRMs and the auxiliary domains of these proteins were exchanged as described in Materials and Methods and as illustrated on the right. (B) CMV- or HSV-luciferase reporter genes were transfected in either the presence or the absence of various protein expression vectors as indicated and under the conditions described for Fig. 2. The histograms represent the effect on luciferase activity of the various proteins as indicated. Means and standard deviations were obtained from at least three separate experiments. (C) CMV-luciferase and MMTV-luciferase were transfected with TRBP expression vector and different protein expression vectors as indicated. The histograms representing the averages of three separate experiments represent the luciferase activity obtained in the presence of the various proteins and TRBP divided by the luciferase activity obtained in the control wells transfected with TRBP alone.
To better understand the function and activity of CoAA, we compared this protein to two other hnRNP-related proteins. We selected hnRNPA1 because it contains two N-terminal RRMs followed by a glycine- and tyrosine-rich auxiliary domain (see Fig. 3) and has been shown to affect CD44 splicing but is not described as being engaged in the transcription process (34, 52). p54nrb is highly similar to the polypyrimidine tract binding protein-associated factor and was selected for its structural and functional similarities to CoAA (see Fig. 3). Like CoAA, p54nrb contains two N-terminal RRMs and is engaged both in transcription and in pre-mRNA processing (3, 28, 33, 43, 45, 55). p54nrb and CoAA have been identified in a nuclear compartment termed paraspeckles, suggesting that these proteins could have similar functions (10).
Transcription factors and transcriptional coregulators contain domains that allow the proteins to be recruited to the promoter through either DNA or protein interactions and an activation domain having either enzymatic activity or allowing the recruitment of other regulatory (or enzymatic) proteins through protein-protein interactions to the promoter (1, 13, 35). Similarly, splicing factors can interact with their RNA substrates via RRM domains and can presumably recruit other proteins through their auxiliary domains (48, 51, 52, 54). Based on these observations, we speculated that the RRMs and the auxiliary domains of these proteins might mediate different activities in terms of both splicing and transcription, allowing for a functional comparison of each of these proteins by exchanging domains between them.
After demonstrating that TRBP is recruited to the progesterone-regulated mouse mammary tumor virus (MMTV) promoter, we demonstrate that both CoAA-mediated transcriptional and splicing effects are enhanced by TRBP, suggesting that CoAA-mediated effects are promoter dependent. Supporting this hypothesis, we observe that CoAA affects both transcription and splicing in a promoter-preferential manner. By swapping the RRMs and the auxiliary domains of CoAA, p54nrb, and hnRNP A1, we demonstrate that depending on the promoter, the RRMs and the auxiliary domain of CoAA are differently engaged in transcription, contributing to the promoter-preferential effects on RNA splicing mediated by CoAA.
MATERIALS AND METHODS
Plasmids.
The cytomegalovirus (CMV) immediate-early enhancer/promoter from pCDNA3.1 (Invitrogen), the herpes simplex virus (HSV) thymidine kinase promoter from pRL-TK (Promega), the MMTV promoter from MMTV-KCR-LUC (Dr. S. Chua, Baylor College of Medicine), the (PRE)2-TATA, and the (ERE)2-TATA promoters were cloned into the pGL3 basic-luciferase reporter (Promega). The above promoters were placed upstream of the previously described CD44 and CT/CGRP test genes (27, 49). The precise cloning steps are available on request. The CoAA, hnRNPA1, and p54nrb cDNAs were cloned into pcDNA3 containing a myc epitope at the 3′ end (Invitrogen). The region corresponding to either the RRMs or the auxiliary domains of the different proteins were amplified by PCR and cloned into pcDNA3-myc. Either at the 3′ end of the RRMs or at the 5′ end of the auxiliary domains, an EcoRV site was inserted, allowing domain switching among the proteins (see Fig. 3A). The precise cloning steps are available on request. All clonings were verified by sequencing and the expression level of the proteins was verified after transfection and Western blot analysis using a myc antibody (data not shown).
Transfection and chromatin immunoprecipitation (ChIP).
The chromatin immunoprecipitation (ChIP) assay was performed with a TRBP antibody using a cell line containing a stably integrated MMTV chloramphenicol acetyltransferase reporter gene as previously described (26). Real-time PCR was performed using CYBR Green PCR master mix from Applied Biosystems. Transfection experiments were done in triplicate using 12-well plates. A transfection master mix was prepared for three wells. Steroid receptors at 5 ng/well were cotransfected with 300 ng of reporter genes per well (except for CMV containing reporter genes, for which 5 ng/well supplemented with 300 ng of pBlueScript vector was used) and 300 ng of different expression vectors per well (except where indicated), using Lipofectamine reagent (Invitrogen) as specified by the manufacturer. After 6 h of incubation, the medium was replaced with medium containing 5% stripped fetal bovine serum and progesterone (Pg; 10−8 M) or estradiol (E2; 10−9 M). After 24 h of incubation at 37°C under 5% CO2, the cells were harvested using either RLT buffer (Promega) for the luciferase assay or 1 ml of TRIzol (Invitrogen) for each set of triplicate wells for RNA isolation as specified by the manufacturer.
DNase treatment and RT-PCR.
A DNase treatment master mix containing AMV/Tfl reaction buffer (Access RT-PCR system; Promega), MgSO4 (2.5 mM final concentration), and RQ1 DNase (1 U; Promega) was prepared and aliquoted to digest plasmid DNA contamination from RNA preparations for 1 h at 37°C, this was followed by 15 min of DNase heat inactivation at 65°C. An aliquot of this reaction mixture was used for reverse transcription-PCR (RT-PCR) using the Access RT-PCR system. RT-PCR master mix containing radiolabeled primers at 1 μM was prepared as specified by the manufacturer. The primers were radiolabeled using [γ-32P]ATP (4,500 Ci/mmol) and T4 kinase (Invitrogen) as specified by the manufacturer. The primers were as follows: sense CD44 primer, AGACACCATGCATGGTGCACC; antisense CD44 primer, CCATAACAGCATCAGGAGTG; sense CT/CGRP primer, CATCGCTGTCTGCGAGGGCC; antisense CT/CGRP (exon 4), GAGTTTAGTTGGCATTCTGG; antisense CT/CGRP (exon5), CTGCTCAGGCTTGAAGGTCC. Radioactive RT-PCR products derived from CT/CGRP minigenes were fractionated on nondenaturing 5% polyacrylamide gels. Radioactive RT-PCR products derived from CD44 minigenes were fractionated on denaturing 5% polyacrylamide gels. Dried gels were exposed to autoradiographic films or placed in PhosphorImager cassettes to allow quantification by the PhosphorImaging system (Molecular Dynamics). DNase heat inactivation at 65°C permits destabilization of RNA secondary structures; for the same reason, the RT step was performed at 48°C. To avoid cross-contamination and variability, we used the Access RT-PCR system (Promega), which allows the RT and PCR steps to be performed in the same tube. Only 20 cycles of PCR (30 s at 94°C, 45 s at 56°C, and 1 min at 68°C) were performed (2).
RESULTS
CoAA-mediated effects on transcription and splicing are enhanced by TRBP.
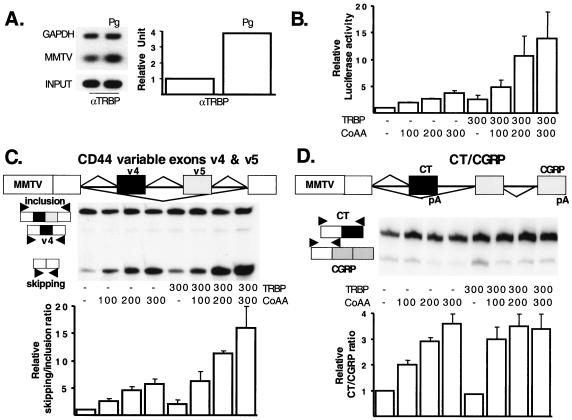
While we had shown earlier that activated steroid hormone receptors simultaneously regulate transcription and splicing decisions in a promoter-dependent manner (2), our hypothesis was that activated nuclear receptors recruit a subset of coregulators participating in both transcription and alternative splicing regulation. To test this hypothesis, we investigated the transcriptional and splicing activity of CoAA, an hnRNP-like protein recently identified as a nuclear receptor coactivator (16). CoAA interacts with the transcriptional coregulator TRBP, which, in turn, interacts with members of the nuclear receptor family, including the progesterone receptor (PR) (5, 16, 20, 23, 29, 56). Using a ChIP assay, we observed that TRBP was recruited to a Pg-activated MMTV promoter (Fig. 1A). Pg treatment enhanced PR recruitment ca. sixfold and TRBP recruitment ca. fourfold to the targeted MMTV promoter (Fig. 1A and data not shown). As a negative control, and as we already have shown, the MMTV promoter was not precipitated in the presence of Pg by an antibody directed against pCAF, a particular coregulator involved in histone acetylation (26).
FIG. 1.
CoAA-mediated effects on transcription and splicing are enhanced by TRBP. (A) Agarose gel-based analysis and quantitative analysis by real-time PCR of Pg-stimulated recruitment of TRBP to a stably integrated MMTV promoter by the ChIP assay (see Materials and Methods). (B) HeLa cells were plated on 12-well plates 24 h before transfection in a 5% stripped serum-based medium. Each condition was used in triplicate wells. Per well, 5 ng of PR and 300 ng of MMTV-luciferase reporter gene were transfected with increasing amounts of pcDNA3-CoAA expression vector in the absence or in the presence of pcDNA3-TRBP expression vector as indicated. The amount of transfected DNA was equilibrated using the pcDNA3-empty expression vector. The serum-free transfection medium was replaced 6 h after transfection by a 5% stripped serum-based medium containing Pg (10−8 M). After 24 h of incubation, transfected cells were harvested for the luciferase assay. The luciferase activities obtained under the different conditions were divided by the luciferase activity obtained in the control wells transfected only with the pcDNA3-empty expression vector (first column). The histogram represents the mean and standard deviation (SD) of three separate experiments. (C) The CD44 minigene gives rise to three spliced variants containing either the two variable exon cassettes, v4 and v5 (inclusion), none of these exons (skipping), or one exon inclusion product (v4). HeLa cells were transfected as described for panel B, except that the reporter gene used was MMTV-CD44. Cells were harvested using 1 ml of TRIzol for each 12-well triplicate before RNA extraction. After DNase treatment, radiolabeled primers were used to amplify CD44 RNA products (see Materials and Methods). Autoradiograms of the radiolabeled-PCR products obtained in a representative experiment are shown. The histogram shows the mean (and SD, n = 3) quantification of the CD44 skipping/inclusion ratio obtained in the presence of different amounts of CoAA and/or TRBP expression vectors, divided by the control skipping/inclusion ratio obtained in the presence of only the empty expression vector (first column). (D) The CT/CGRP minigene contains two polyadenylation sites (pA) in either exon 4, giving rise to the CT product, or exon 6, giving rise to the CGRP product. HeLa cells were transfected as described for panel B, except that the reporter gene used was MMTV-CT/CGRP. Autoradiograms of the radiolabeled PCR products obtained in a representative experiment are shown. The histogram shows the mean (and SD, n = 3) quantification of the CT/CGRP ratio obtained in the presence of different amounts of CoAA and/or TRBP expression vectors, divided by the control CT/CGRP ratio obtained in the presence of only the empty expression vector (first column).
CoAA was shown to enhance the transcriptional activity of several nuclear receptors (16), and, as illustrated in Fig. 1B, the CoAA-mediated transcriptional effect on a PR-activated MMTV-luciferase reporter gene was strongly enhanced by TRBP. Using 300 ng of CoAA or TRBP expression vector, we observed a transcriptional increase of ca. 4- and ca. 2.5-fold respectively, whereas when CoAA and TRBP were expressed together, the MMTV-luciferase activity was increased ca. 15-fold. As reported below, the overexpression of other hnRNP-related proteins did not always enhance the MMTV- and/or the TRBP-activated MMTV-luciferase activity (see Fig. 2A and 3C). Together, these results suggest that the CoAA-mediated transcriptional effects on a PR-regulated MMTV promoter are TRBP dependent. Supporting this hypothesis, a natural spliced variant of CoAA, called CoAM, that lacks the TRBP-interacting domain did not activate transcription and did not enhance TRBP-mediated transcriptional effects (reference 16 and data not shown).
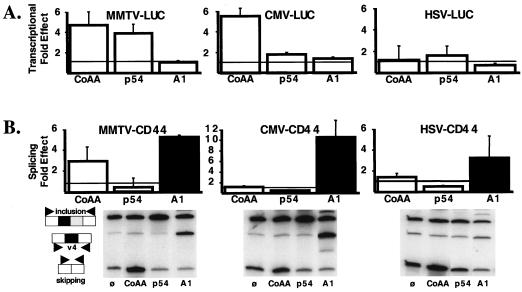
FIG. 2.
CoAA mediates promoter-preferential effects on transcription and alternative splicing. (A) MMTV-, CMV-, or HSV-luciferase reporter genes were transfected in HeLa cells as described in Materials and Methods. The MMTV-luciferase reporter gene, transfected with PR, was activated by Pg. The reporter genes were transfected with 300 ng of pcDNA3-CoAA (CoAA), pcDNA3-p54nrb (p54), pcDNA3-hnRNPA1 (A1), or pcDNA3 empty expression vector. The histograms represent the luciferase activity obtained in presence of either CoAA, p54nrb, or hnRNPA1 divided by the control luciferase activity obtained in presence of the empty expression vector. The means and SD were calculated from three separate experiments. (B) The same conditions as described for panel A were used, except that the reporter genes were MMTV-CD44, CMV-CD44, or HSV-CD44. The histograms represent either the fold effect of CoAA and p54nrb on the skipping/inclusion ratio (open boxes) or the fold effect of hnRNPA1 on the v4/inclusion ratio (solid boxes). The fold effect was obtained by dividing the ratio obtained in the presence of the different protein expression vectors by the ratio obtained in the presence of the empty expression vector (ø). Means and SD were calculated from three separate experiments. Representative autoradiograms of radioactive low-cycle RT-PCR amplification of splicing products are shown at the bottom.
Whereas CoAA can affect splicing decisions, we previously reported that CoAM did not do so (2). Consequently, we asked whether CoAA-mediated effects on splicing were also TRBP dependent. To test this possibility, we used a human CD44-derived minigene (49) driven by the MMTV promoter (Fig. 1C). Normally, the CD44 reporter used produces a mixture of two RNAs that contain either both or neither of the CD44 alternative exons and a minor amount of intermediary RNA containing one of the alternative exons (Fig. 1C). Increasing the concentration of CoAA changed the ratio of the CD44 spliced products to favor the production of RNA lacking both alternative exons. At higher concentrations, CoAA increased the skipping/inclusion ratio ca. fivefold. TRBP increased the skipping/inclusion ratio ca. twofold and enhanced CoAA-mediated splicing effects (Fig. 1C). Transfection of both CoAA and TRBP increased the skipping/inclusion ratio up to ca. 15-fold (Fig. 1C). In the presence of 300 ng of CoAA expression vector or in the presence of both CoAA and TRBP, the overall production of CD44 transcripts was increased compared to the control lane (Fig. 1C, first lane of the autoradiogram corresponding to the transfection of the empty expression vector). Although this observation suggests that the proteins tested have transcriptional effects on the MMTV-CD44 reporter gene, we often observed a less pronounced transcriptional effect of these proteins compared to their transcriptional effect on the MMTV-luciferase reporter gene (Fig. 1B). Since the RT-PCR assay that we used was developed to measure the relative abundance of the CD44 transcripts and not to measure the total amount of the CD44 products synthesized, no firm conclusions can be drawn regarding the transcriptional output from this minigene. It also is possible that the discrepancy observed in the amplitude of the transcriptional effect mediated by the different coregulators on the CD44 and the luciferase reporter gene could be due to different stability of the CD44 and the luciferase transcripts since the luciferase cDNA is designed to produce a very stable mRNA which directs continued synthesis of luciferase products even after 24 h posttransfection. Finally, the overexpression of other hnRNP-related proteins led to no or differential effects on CD44 transcripts splicing (2) (see Fig. 2B), demonstrating the reliability and the specificity of the assay.
We further tested the splicing effects of CoAA by using a second splicing reporter gene derived from the human CT/CGRP gene (27), giving rise to either the CT or to the CGRP RNAs (Fig. 1D). This splicing reporter, like CD44, involves the alternative recognition of a weak internal exon (the black CT exon in Fig. 1D) characterized by a weak 3′-splice site requiring activation by processing enhancer sequences (27, 49). The two systems differ in that the alternative exon in CT/CGRP is a 3′-terminal exon involving differential polyadenylation as well as splicing. In addition, the CD44 alternative splicing decision is regulated by exon-located regulatory sequences whereas the CT/CGRP splicing event has an intron regulator in addition to exon-specific sequences (27, 49). Use of these two systems permits the examination of whether CoAA regulates the splicing of any weak exon to produce more products resulting from exon skipping. As shown in Fig. 1D, increasing the concentration of CoAA changed the ratio of the spliced isoforms. At higher concentrations, CoAA increased the CT/CGRP ratio ca. fourfold (Fig. 1D). Therefore, in contrast to the situation with CD44, CoAA stimulated the inclusion rather than the exclusion of the weak exon, indicating that the CoAA effect is not simply an inhibition of the splicing apparatus to hinder the recognition of weak exons. Interestingly, two- to threefold less CoAA expression vector was required to obtain a maximal effect on splicing in the presence of TRBP (Fig. 1D). The CoAA-mediated effects on splicing of the CT/CGRP products, however, reached a plateau in the presence of TRBP, whereas the transcription response continued in the presence of additional CoAA (Fig. 1B and D). Although we observed a splicing effect on the CD44 transcripts when TRBP was overexpressed alone (Fig. 1C), this coregulator did not affect CT/CGRP splicing in the absence of CoAA (Fig. 1D). This could be because the CoAA/TRBP complex stimulated CT production, which was the major product synthesized in HeLa cells in the absence of any stimulation (Fig. 1D, first lane of the autoradiograph).
Our results demonstrate that both transcriptional and splicing CoAA-mediated effects are enhanced by TRBP (Fig. 1B to D), which interacts with CoAA (16) and which is recruited on the Pg-activated MMTV promoter (Fig. 1A). The data suggest that both transcriptional and splicing CoAA-mediated effects are promoter dependent. To test this possibility further, we asked if the transcriptional and splicing effects mediated by CoAA also would be observed using other promoters including the steroid-independent CMV and HSV promoters.
CoAA mediates promoter-preferential effects on transcription and alternative splicing.
In the luciferase assay, CoAA enhanced the activity of both the PR-activated MMTV promoter and the CMV promoter to a similar extent (Fig. 2A, left and middle graphs). This effect was not a general effect on either transcription or luciferase activity since CoAA had no effect on the luciferase reporter gene activity driven by the HSV promoter (right graph). Using the CD44 reporter gene, we observed that CoAA altered the ratio of CD44 spliced products when CD44 pre-mRNAs were synthesized from the PR-activated MMTV promoter but that it had minimal effects when CD44 pre-mRNAs were synthesized from either the CMV or the HSV promoter (Fig. 2B). In the particular experiments illustrated by the autoradiograms in Fig. 2B, CoAA enhanced the skipping/inclusion ratio by 3.9-, 1.2-, and 1.6-fold when the CD44 pre-mRNAs were synthesized from the MMTV, CMV, or HSV promoters, respectively. As illustrated by the histograms in Fig. 2B, CoAA enhanced the skipping/inclusion ratio by an average of 3 ± 0.9, 1.2 ± 0.2, and 1.4 ± 0.3-fold, respectively, using the MMTV-, CMV-, or HSV-CD44 reporter gene. These results suggest that an RRM-containing protein can affect both transcriptional activity and splicing decisions in a promoter-preferential manner. Importantly, although splicing effects mediated by RNA-binding proteins can show promoter preferences as already reported (6, 42), these promoter-preferential splicing effects are not a direct consequence of transcriptional activation. Indeed, we observed that CoAA enhanced PR-activated MMTV- and CMV-luciferase activity similarly but significantly affected the splicing of the CD44 products only when they were synthesized from the PR-activated MMTV promoter (Fig. 2). The absence of a direct correlation between transcriptional activation and splicing effects is also consistent with our previous finding that different transcriptional coactivators that enhance transcription can alter splicing decisions in an opposite manner and that an ability to alter transcription does not necessarily alter splicing decisions (2).
To test whether the promoter preference observed using CoAA was due to intrinsic properties of the promoter used, we studied the effect of two other hnRNP-related proteins. hnRNP A1, known not to alter transcription output, was chosen as an RNA-splicing factor which has been reported to specifically increase exon v5 skipping on CD44 (34). As expected, hnRNP A1 had no effect on transcription using the luciferase reporter gene driven by either the MMTV, CMV, or HSV promoters (Fig. 2A). In contrast, it enhanced the production of CD44-spliced RNAs containing only exon v4, as a result of exon v5 skipping (Fig. 2B). Using primers localized in either exon v4 or v5, we determined that the levels of the RNA products containing only exon v4 were specifically increased in the presence of hnRNP A1 (data not shown). The effect of hnRNP A1 on the v4/inclusion ratio was ca. 5-, ca. 10-, and ca. 3-fold from the MMTV-, CMV-, and HSV-CD44 minigenes, respectively. Therefore, the inability of CoAA to significantly alter the splicing of transcripts synthesized from the CMV or HSV promoters does not reflect an inherent ability of these promoters to suppress the activity of hnRNP proteins on splicing.
We examined the ability of p54nrb, known as both a transcriptional activator and an RNA-processing factor to produce transcription and splicing effects in our assays. p54nrb activated the transcription of only the MMTV promoter (ca. fourfold [Fig. 2A]) and slightly increased exon inclusion from all three promoters driving the CD44 minigene (Fig. 2B). Consequently, CoAA, hnRNP A1, and p54nrb represent three proteins with similar architecture (Fig. 3A; also see Introduction) that in our assays alter either both transcription and splicing or only splicing. All the proteins or mutants tested in this work (see below) were cloned into the same pcDNA3 expression vector having a myc epitope coding sequence at the 3′ end, allowing Western blot-based analysis of the expression levels of the different proteins (see Materials and Methods). Although CoAA was expressed less than hnRNP A1 or p54nrb, this observation did not alter our conclusions, since we compared the effects only of one protein on different promoters; different proteins were compared based on their respective promoter preferential effects. Since hnRNP A1 had more effect on the CMV-CD44 minigene than on the MMTV-CD44 minigene and CoAA had more effect on the MMTV-CD44 minigene than on the CMV-CD44 minigene (Fig. 2B), we concluded that the splicing effects observed did not reflect an intrinsic ability of the promoters to respond in a particular way to hnRNP-related proteins.
To understand the relationship between the trancriptional coactivation function of CoAA and its splicing activity, we next investigated the structural mechanisms by which CoAA similarly enhanced the transcriptional activity of both CMV and PR-activated MMTV promoters but significantly affected the splicing of CD44 products synthesized only from the PR-activated MMTV promoter.
The RRMs of CoAA mediate its transcriptional effects on a CMV-luciferase reporter gene.
On one hand, CoAA was shown to interact with TRBP through its auxiliary domain (16), suggesting that CoAA could be engaged in transcription through this domain. On the other hand, the RRM motifs of hnRNP-related proteins can bind RNAs as well as single-stranded DNA sequences, potentially allowing the recruitment of RRM-containing proteins to gene promoters (4, 36, 50, 55). In consequence, theoretically, the promoter-preferential transcriptional effects of CoAA could be mediated by either its RRMs or its auxiliary domain. Experiments using deleted CoAAs missing either the RRMs or the auxiliary domain indicated that both domains were necessary for CoAA-mediated transcriptional activity on both the CMV and the MMTV promoters (data not shown). Consequently, we turned to domain swapping between the three utilized hnRNP proteins to determine the role of each domain in mediating promoter-preferential transcriptional effects.
As shown in Fig. 3B (left), the CoAA-mediated transcriptional effect on the CMV promoter was lost when the RRMs from either hnRNPA1 (A1N-CoAAC) or p54nrb (p54N-CoAAC) replaced the RRMs of CoAA, although the three proteins (CoAA, A1N-CoAAC, and p54N-CoAAC) had similar expression levels (data not shown). These results indicated the specific importance of the CoAA RRMs for transcriptional activity on the CMV promoter. Importantly, the CoAA RRMs conferred CMV transcriptional activation when fused to the C terminus of p54nrb (CoAAN-p54C). Because CoAA (but not p54nrb) was active on the CMV promoter, this result again emphasized the importance of CoAA RRMs for transcriptional activation of the CMV promoter. As a control, we observed that CoAAN-p54C was not active on the HSV promoter, which was not activated by either CoAA or p54nrb (Fig. 3B, right). The CoAA RRMs were not sufficient to confer activity to any auxiliary domain since the fusion of these RRMs to the hnRNP A1 auxiliary domain (CoAAN-A1C) produced no transcriptional effect although this mutant had a similar expression level to that of CoAAN-p54C (Fig. 3B, left graph, and data not shown). Consequently, CoAA RRMs require a transcriptionally active auxiliary domain (CoAAC or p54C versus none or A1C) to mediate a transcriptional effect on the CMV promoter. The effects are specific for CoAA RRMs since the RRMs of neither hnRNPA1 nor p54nrb can replace the CoAA RRMs.
Interestingly, it was shown that CoAA interacts with the promoter-recruited TRBP coregulator through its auxiliary domain (16) and that TRBP enhanced CoAA-mediated transcriptional effects on a PR-activated MMTV promoter (Fig. 1B). Together, these observations suggested that CoAA could be engaged through its auxiliary domain interacting with TRBP in PR-activated MMTV transcriptional regulation and could be engaged through its RRMs in CMV transcriptional regulation. To test this hypothesis, we reasoned that CoAA should enhance TRBP-mediated effects on the PR-activated MMTV promoter but not on the CMV promoter. When TRBP was expressed, CoAA enhanced the TRBP-mediated effects on PR-activated MMTV-luciferase by ca. 10-fold but enhanced the TRBP-mediated effects on CMV-luciferase by only 4-fold (Fig. 3C). This contrasted with the equal CoAA-mediated effects previously shown for the two promoters in the absence of TRBP (Fig. 2A). Importantly, we observed that only hybrid proteins containing the auxiliary domain of CoAA (CoAA itself, A1N-CoAAC, or p54N-CoAAC) were able to significantly enhance TRBP-mediated effects on the PR-activated MMTV promoter (Fig. 3C), although the expression levels of these proteins containing the CoAA auxiliary domain were lower than those of the proteins that did not contain the CoAA auxiliary domain (A1, p54, CoAAN-A1C, and CoAAN-p54C [data not shown]). Because the RRMs of either hnRNP A1 or p54nrb can partially replace CoAA RRMs (A1N-CoAAC and p54N-CoAAC, respectively) to mediate the transcriptional effect on a TRBP-activated MMTV promoter, these results emphasized a predominant role of the CoAA auxiliary domain on this promoter. In contrast, and as expected, none of these proteins enhanced the TRBP-mediated effect on the CMV promoter (Fig. 3C).
In summary, our results suggest that CoAA is engaged in regulating steroid-dependent MMTV transcriptional activity through its auxiliary domain and in regulating the steroid-independent CMV transcriptional activity through its RRMs. To compare the transcriptional coactivation activity of CoAA with its splicing activity, the same mutants and hybrid proteins were used next in the splicing assay.
The RRMs of CoAA prevent splicing effects on CMV-CD44 gene products.
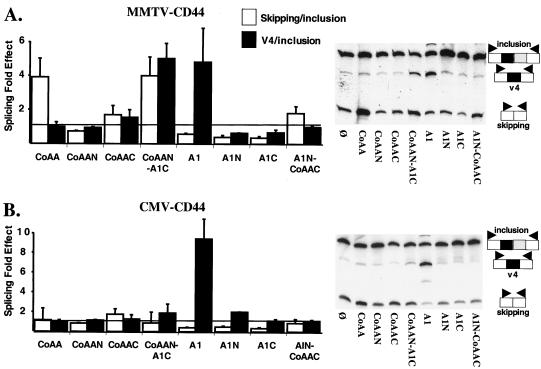
As shown in Fig. 2, CoAA and hnRNP A1 affected CD44 splicing in different ways. CoAA significantly promoted the skipping of both alternative exons, but did so only when transcription occurred from the PR-activated MMTV promoter. In contrast, hnRNP A1 promoted the skipping of only exon v5 of CD44 products synthesized from either the MMTV, CMV, or HSV promoters. Figure 4 compares the ability of natural, deleted, and hybrid proteins to promote the skipping of both exons (skipping/inclusion ratio) and skipping of just exon v5 (v4/inclusion).
FIG. 4.
The RRMs of CoAA prevent splicing effects on CMV-CD44 products. MMTV-CD44 (A) or CMV-CD44 (B) reporter genes were transfected, as described in Materials and Methods, with or without the protein expression vectors as indicated. The histograms represent the fold effect of the different proteins on either the skipping/inclusion ratio or the v4/inclusion ratio. The fold effect was obtained by dividing the ratio obtained in the presence of the various protein expression vectors by the ratio obtained in the presence of the empty expression vector (Ø). Means and standard deviations were calculated from three separate experiments. Representative autoradiograms of radioactive low-cycle RT-PCR amplification of splicing products are shown on the right.
Subdomains of CoAA or hnRNP A1 consisting of either the RRMs or the auxiliary domain were unable to affect splicing decisions (CoAAN, CoAAC, A1N, or A1C [Fig. 4]). Interestingly, addition of the CoAA RRMs to the hnRNP A1 auxiliary domain produced a protein (CoAAN-A1C) able to exert the full activities of both proteins on MMTV directed pre-mRNAs (Fig. 4A). Since both proteins (CoAA and hnRNP A1) can affect the splicing of CD44 products synthesized from the MMTV promoter, no conclusion can be drawn regarding the domain requirement for their splicing activity on this particular promoter.
In contrast, as observed using CoAA, the hybrid protein CoAAN-A1C did not alter splicing decisions on CMV-transcribed CD44 pre-mRNAs even though the expression level of this hybrid protein was higher than that of CoAA (Fig. 4B and data not shown). These results clearly indicate that CoAA RRMs were responsible for the lack of CoAA effects on the splicing of CMV-CD44 gene products (Fig. 2B and 4B).
In conclusion, CoAA RRMs play a major role in mediating a positive transcriptional effect on the CMV promoter (Fig. 3B) and in preventing splicing effects on the CD44 pre-mRNAs transcribed from this promoter (Fig. 4B). These observations raise the hypothesis that CoAA could be engaged in CMV transcriptional regulation through its RRMs, limiting the ability of the protein to be engaged in the splicing of the CD44 pre-mRNAs synthesized from this promoter. In contrast, CoAA could be involved in splicing regulation of CD44 pre-mRNAs synthesized from the PR-activated MMTV promoter because CoAA is engaged in the regulation of this promoter through its auxiliary domains (Fig. 1, 3C, and 4A).
Since CoAA and TRBP have been implicated in steroid receptor-mediated transcription and since CoAA did not behave similarly when a steroid-regulated promoter (MMTV) was compared to a steroid-independent promoter (CMV), we investigated the transcriptional and splicing effects of CoAA by using other steroid-regulated promoters to test whether the ability of CoAA to simultaneously affect transcription and splicing decisions was dependent on an intrinsic ability of steroid-dependent promoters.
CoAA-mediated effects on splicing are inversely correlated with its transcriptional effects.
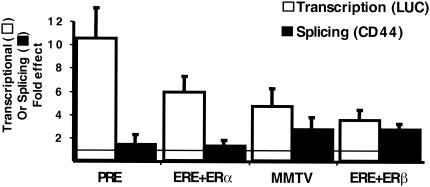
Our results suggested that the role played by CoAA RRMs in transcriptional regulation of the non-steroid-regulated CMV promoter limited the involvement of CoAA in the splicing of the CD44 pre-mRNAs transcribed from this promoter. In contrast, when CoAA was engaged through its auxiliary domain in transcriptional regulation of the steroid-regulated MMTV promoter, the protein could affect splicing. To test whether other steroid-dependent promoters could behave like the MMTV promoter, we tested a PRE-TATA promoter activated by Pg and an ERE-TATA promoter activated by estradiol through the estrogen receptors ERα and ERβ. Interestingly, when CoAA mediated a strong effect in terms of transcription, we observed a modest effect on splicing (Fig. 5). Consequently, the splicing effects mediated by CoAA were inversely correlated with its transcriptional effects on steroid-regulated promoters. These results again demonstrate that the effects on splicing of CoAA are not a direct consequence of its transcriptional effects. Moreover, the results suggest that the promoter-preferential splicing effects mediated by CoAA are not dependent simply on steroid-regulated versus non-steroid-regulated promoters but emphasize again that the precise manner in which CoAA is engaged in transcriptional regulation could have consequences on the subsequent splicing regulatory activity of CoAA.
FIG. 5.
CoAA-mediated splicing effects are inversely correlated with its transcriptional effects on steroid-regulated promoters. PRE-, ERE-, or MMTV-luciferase reporter genes or PRE-, ERE-, or MMTV-CD44 reporter genes were transfected with 5 ng of either PR, ERα, or ERβ per well and with 300 ng of CoAA expression vector or empty expression vector per well. The open boxes represent the fold effect of CoAA on luciferase activity, and the solid boxes represent the fold effect of CoAA on the CD44 skipping/inclusion ratio. Means and SD obtained from at least three separate experiments are shown.
The CMV promoter does not completely abrogate CoAA-mediated splicing effects on CMV-transcribed RNAs.
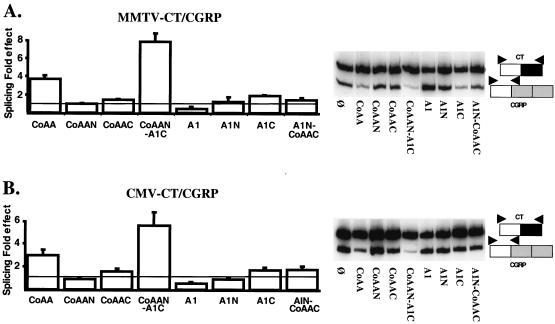
We have shown that the inability of CoAA to alter the splicing of CD44 transcripts synthesized from the CMV promoter does not reflect an inherent ability of this promoter to suppress the splicing activity of hnRNP proteins on CD44 RNAs (Fig. 2B). Moreover, we observed that CoAA can affect the splicing of transcripts synthesized from this promoter. When the PR-activated MMTV promoter was driving the CT/CGRP minigene, CoAA increased the CT/CGRP ratio threefold whereas hnRNPA1 decreased it fourfold (Fig. 6A). Interestingly, the hybrid protein CoAAN-A1C mediated similar effects to CoAA and increased the CT/CGRP ratio ca. sevenfold, suggesting that the CoAA RRMs oriented the splicing decision (Fig. 6A). Importantly, and in contrast to the results obtained using the CD44 minigene (Fig. 4), CoAA and CoAAN-A1C mediated similar but slightly smaller splicing effects on the CT/CGRP products synthesized from the CMV promoter than on products synthesized from the PR-activated MMTV promoter (compare Fig. 6B with 6A). These results suggest that the promoter-preferential effects on splicing mediated by CoAA are gene specific and are also influenced by the transcribed region of the gene. Consequently, the splicing effects mediated by CoAA, an RRM-containing transcriptional coactivator, are simultaneously dependent on both the nature of the promoter and the nature of the RNA product synthesized from this promoter.
FIG. 6.
The CMV promoter does not completely abrogate CoAA-mediated splicing effects on CMV-transcribed RNA. MMTV-CT/CGRP (A) or CMV-CT/CGRP (B) reporter genes were transfected, as described in Materials and Methods, with or without different protein expression vectors as indicated. The histograms represent the fold effect of the various proteins on the CT/CGRP ratio. The fold effect was obtained by dividing the ratio obtained in the presence of the various protein expression vectors by the ratio obtained in the presence of the empty expression vector (Ø). Means and standard deviations were calculated from three separate experiments. Representative autoradiograms of radioactive low-cycle RT-PCR amplification of splicing products are shown on the right.
DISCUSSION
We showed previously that activated steroid receptors can simultaneously affect transcription and splicing decisions in a promoter-dependent manner (2). Our present hypothesis is that activated nuclear receptors recruit a subset of coregulators participating in the regulation of both transcription and alternative splicing (2). Supporting this hypothesis, we showed that CoAA, an hnRNP-like protein identified as a nuclear receptor coactivator, can affect splicing decisions (2, 16). In the present work, we investigated the relationship between CoAA transcriptional coactivation function and its splicing activity. Using reporter genes driven by the steroid receptor-dependent MMTV promoter, we showed that the transcriptional and splicing effects mediated by CoAA were enhanced by TRBP, a protein recruited to the MMTV promoter by interacting with activated nuclear receptors (5, 16, 20, 23, 29, 56) (Fig. 1). These observations suggest that the CoAA-mediated effects on both transcription and splicing are promoter dependent. Supporting this hypothesis, we observed that CoAA demonstrated promoter-preferential effects on both transcription and splicing (Fig. 2). Importantly, CoAA similarly enhanced both PR-activated MMTV and CMV transcriptional activities but significantly affected the splicing of CD44 products synthesized only from the PR-activated MMTV promoter (Fig. 2). These results consequently demonstrate that CoAA-mediated splicing effects clearly are not just a direct consequence of its transcriptional activity. This conclusion was also supported by the inverse correlation observed between transcriptional and splicing effects mediated by CoAA with steroid-regulated promoters (Fig. 5). This absence of a direct correlation between transcriptional and splicing effects is consistent with our previous finding that transcriptional coregulators can affect transcription without affecting splicing and that enhancement of transcription by different transcriptional coactivators can be associated with opposite effects on splicing (2). In conclusion, CoAA-mediated splicing effects are promoter dependent but are not a direct consequence of CoAA effects on the promoter transcriptional activity.
Importantly, the limited splicing effect of CoAA on the CMV-transcribed CD44 RNAs was not due to an intrinsic ability of the CMV promoter to inhibit the splicing activity of hnRNP proteins. Indeed, hnRNP A1 affected the splicing of CMV-transcribed CD44 RNAs (Fig. 2). Moreover, the CMV promoter did not completely abrogate CoAA-mediated splicing effects. Indeed, CoAA affected the splicing of CMV-transcribed CT/CGRP RNAs (Fig. 6). These results are consistent with the recent observation that the splicing activity of SR (serine- and arginine-rich) protein family splicing factors depends on both the promoter and the nature of the splicing reporter used (6, 37, 42).
To explain these promoter-preferential actions of the SR splicing factors, two models have been proposed. PGC-1, structurally related to the SR family of splicing factors, is a transcriptional coregulator recruited to target promoters by the nuclear receptor PPARγ and was shown to affect splicing decisions in a promoter-dependent manner (37). It was proposed that PGC-1, when recruited to a target promoter, moves across the gene due to its interaction with the C-terminal domain of RNA pol II, allowing the interaction of PGC-1 with the neonascent transcript, whose splicing can be affected (37). In this model, the splicing effect of a given regulatory protein would depend on its promoter recruitment. A second model proposed that promoter-dependent effects of SR splicing factors depend on the RNA polymerase II elongation rate. In this model, the nature of a promoter and the nature of the transcriptional regulatory complex buildup on this promoter would control the rate at which the RNA pol II is synthesizing the pre-mRNA. Depending on the elongation rate, SR splicing factors would have more or less time to recognize splicing-regulatory RNA sequences. At a low elongation rate, SR splicing factors would have more time to interact with their RNA substrate than at a high elongation rate, allowing their engagement in the splicing process (17, 39, 42). Consequently, in this model, the action of a given splicing factor would depend on the speed at which the pre-mRNA is synthesized and, very importantly, on the affinity of the splicing factor for its target pre-mRNA (17, 39, 42).
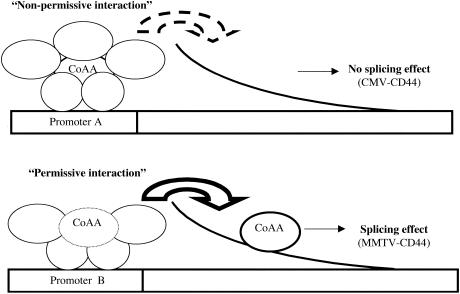
According to these models and based on the results presented here, we propose that promoter-specific splicing effects mediated by a subset of hnRNP-related transcriptional/splicing regulators could depend on the “competitive” recruitment of these proteins by (i) the promoter and (ii) the gene product synthesized from this promoter (Fig. 7). Indeed, we observed that the CoAA RRMs play a major role in mediating a positive effect on CMV transcriptional activity and in preventing the splicing effect on CMV-transcribed CD44 RNAs (Fig. 3B and 4B). Inversely, the CoAA auxiliary domain is important in mediating transcriptional CoAA effects on the PR-activated MMTV promoter that also permits CoAA-mediated splicing effects on the CD44 RNAs (Fig. 3C and 4A). This suggested that the way in which CoAA is engaged in the transcriptional process could affect its involvement in downstream events such as splicing. However, importantly, the engagement of CoAA in the splicing process is not totally dependent on the promoter. Like the CoAA-mediated effect on CD44 RNA splicing, the CoAA-mediated effect on CT/CGRP RNA is enhanced by TRBP when a PR-activated MMTV promoter drives the minigenes (Fig. 1). However, unlike the splicing of CMV-transcribed CD44 RNA, the splicing of CMV-transcribed CT/CGRP RNA is affected by CoAA (Fig. 6). Consequently, the engagement of CoAA in the splicing process could depend on its engagement in transcriptional complexes and probably also on the ability of the transcript to recruit CoAA from these transcriptional complexes.
FIG. 7.
Competitive recruitment of RNA-binding proteins by the promoter and the transcript. A transcriptional/splicing regulator, like CoAA could be recruited to target promoters through either DNA-protein or protein-protein interactions. Depending on the affinity of the regulator for the transcriptional complex and for the transcript synthesized from this promoter, the regulator will or will not be able to interact with the transcript, participating or not participating in the splicing regulation process. For instance, the engagement of CoAA through its RRMs in the CMV promoter regulation (“Non-permissive interaction”) could restrain the ability of CoAA to be engaged in splicing (promoter A), whereas the engagement of CoAA through its auxiliary domain in the PR-activated MMTV promoter regulation (“Permissive interaction”) could allow this engagement (promoter B). In the natural context of a promoter driving the synthesis of an unique transcript, several mechanisms could coexist to modulate this competition either by changing the “strength” of the engagement of a given regulator in the transcriptional complex or by changing the affinity of the regulator for the RNA transcript (see Discussion).
In the natural context of a promoter driving the synthesis of a unique transcript, several mechanisms could coexist to modulate this “competitive” recruitment, either by modulating the “strength” of the engagement of a given splicing regulator in transcriptional complexes or by changing the affinity of the regulator for the RNA transcript. For instance, it is known that transcriptional coregulators are simultaneously engaged in several protein-protein interactions within promoter transcriptional complexes (1, 13, 35). The nature and/or number of interactions, in which a transcriptional/splicing regulator is engaged on a given promoter, could modify the local availability of this regulator for the splicing process. Such interactions might depend either on the nature of the transcriptional stimuli leading to the recruitment of various sets of coregulators or on signaling pathways that are known to affect transcriptional coregulator activities and that could modulate protein-protein interactions (13, 35). Such mechanisms could modify the “strength” at which a regulatory protein is involved in a given transcriptional complex.
Inversely, several mechanisms could also exist to modulate the affinity of these proteins for the RNA transcripts. It is known that transcriptional factors recruit various transcriptional coregulators harboring a variety of enzymatic activities such as acetylation, methylation, ubiquitination, or phosphorylation (13, 35). Interestingly, two RNA-binding proteins were recently identified as substrates for the transcriptional coregulator CARM1, which contains a methyltransferase activity (22, 24). Methylation of RNA-binding proteins is known to affect their activity, notably their interaction with RNAs (18, 19, 46). Several reports also suggest that the binding activity of RNA-binding proteins can be affected by phosphorylation (25, 41). p54nrb can bind either RNA or DNA depending on its phosphorylation status (3, 55). It also was shown that phosphorylation of the IQ domain of the RNA-binding protein EWS, involved in both transcription and splicing regulation, inhibited its interaction with RNA (8). Interestingly, CoAA contains a similar IQ domain (data not shown). Based on these observations, an interesting possibility would be that coregulators harboring enzymatic activities could be recruited to target promoters and could locally affect the fate of RNA-binding proteins, in particular their ability to regulate the RNA-splicing process, by modulating their ability to bind the transcripts.
In conclusion, we propose that activated nuclear receptors recruit a subset of transcriptional coregulators and that their ability to participate in downstream process (including alternative splicing decisions) is influenced by other promoter-recruited coregulators, thereby permitting the promoter to control pre-mRNA-processing events in response to transcriptional stimuli like steroid hormones. Competitive recruitment of RNA-binding proteins by complexes built up on the promoter and complexes built up on the transcripts might have importance beyond splicing regulation. The number of proteins identified harboring RNA-binding domains involved in transcription is increasing rapidly (2, 21, 47, 53). Some of these proteins act as transcription factors and bind to DNA, and others act as transcriptional coregulators and are recruited to target promoters by transcription factors through protein-protein interactions (2, 21, 47, 53). Some of these RNA-binding proteins might act as feedback “sensors” of the amount of gene products synthesized. If coregulator proteins required for the transcriptional activity of a promoter are able also to interact with the gene products synthesized from that promoter, then when more gene products are synthesized, less RNA-interacting protein would be available to coactivate the promoter transcriptional machinery. Such a scenario would lead to a decrease in the promoter activity until the RNA gene products are metabolized, translated, or degraded.
Acknowledgments
This work was supported in part by NIH grant NICHD 08818 NIH-NIDDK Atlas Program (B.W.O.) and the Welch Foundation, GM38526, and GM58019 (S.M.B.).
REFERENCES
- 1.Aranda, A., and A. Pascual. 2001. Nuclear hormone receptors and gene expression. Physiol Rev. 81:1269-1304. [DOI] [PubMed] [Google Scholar]
- 2.Auboeuf, D., A. Honig, S. M. Berget, and B. W. O'Malley. 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298:416-419. [DOI] [PubMed] [Google Scholar]
- 3.Basu, A., B. Dong, A. R. Krainer, and C. C. Howe. 1997. The intracisternal A-particle proximal enhancer-binding protein activates transcription and is identical to the RNA- and DNA-binding protein p54nrb/NonO. Mol. Cell. Biol. 17:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertolotti, A., Y. Lutz, D. J. Heard, P. Chambon, and L. Tora. 1996. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 15:5022-5031. [PMC free article] [PubMed] [Google Scholar]
- 5.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J. A. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308-5317. [DOI] [PubMed] [Google Scholar]
- 6.Cramer, P., J. F. Caceres, D. Cazalla, S. Kadener, A. F. Muro, F. E. Baralle, and A. R. Kornblihtt. 1999. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4:251-258. [DOI] [PubMed] [Google Scholar]
- 7.Cramer, P., A. Srebrow, S. Kadener, S. Werbajh, M. de la Mata, G. Melen, G. Nogues, and A. R. Kornblihtt. 2001. Coordination between transcription and pre-mRNA processing. FEBS Lett. 498:179-182. [DOI] [PubMed] [Google Scholar]
- 8.Deloulme, J. C., L. Prichard, O. Delattre, and D. R. Storm. 1997. The prooncoprotein EWS binds calmodulin and is phosphorylated by protein kinase C through an IQ domain. J. Biol. Chem. 272:27369-27377. [DOI] [PubMed] [Google Scholar]
- 9.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 10.Fox, A. H., Y. W. Lam, A. K. Leung, C. E. Lyon, J. Andersen, M. Mann, and A. I. Lamond. 2002. Paraspeckles: a novel nuclear domain. Curr. Biol. 12:13-25. [DOI] [PubMed] [Google Scholar]
- 11.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 12.Goldstrohm, A. C., A. L. Greenleaf, and M. A. Garcia-Blanco. 2001. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene 277:31-47. [DOI] [PubMed] [Google Scholar]
- 13.Hermanson, O., C. K. Glass, and M. G. Rosenfeld. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 13:55-60. [DOI] [PubMed] [Google Scholar]
- 14.Honig, A., D. Auboeuf, M. M. Parker, B. W. O'Malley, and S. M. Berget. 2002. Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol. Cell. Biol. 22:5698-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 16.Iwasaki, T., W. W. Chin, and L. Ko. 2001. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375-33383. [DOI] [PubMed] [Google Scholar]
- 17.Kadener, S., P. Cramer, G. Nogues, D. Cazalla, M. de La Mata, J. P. Fededa, S. E. Werbajh, A. Srebrow, and A. R. Kornblihtt. 2001. Antagonistic effects of T-Ag and VP16 reveal a role for RNA pol II elongation on alternative splicing. EMBO J. 20:5759-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S., B. M. Merrill, R. Rajpurohit, A. Kumar, K. L. Stone, V. V. Papov, J. M. Schneiders, W. Szer, S. H. Wilson, W. K. Paik, and K. R. Williams. 1997. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry 36:5185-5192. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S., G. H. Park, and W. K. Paik. 1998. Recent advances in protein methylation: enzymatic methylation of nucleic acid binding proteins. Amino Acids 15:291-306. [DOI] [PubMed] [Google Scholar]
- 20.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladomery, M. 1997. Multifunctional proteins suggest connections between transcriptional and post-transcriptional processes. Bioessays 19:903-909. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J., M. T. Bedford, and I. A. Laird-Offringa. 2002. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 3:268-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 24.Li, H., S. Park, B. Kilburn, M. A. Jelinek, A. Henschen-Edman, D. W. Aswad, M. R. Stallcup, and I. A. Laird-Offringa. 2002. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 277:44623-44630. [DOI] [PubMed] [Google Scholar]
- 25.Li, J., T. Kinoshita, S. Pandey, C. K. Ng, S. P. Gygi, K. Shimazaki, and S. M. Assmann. 2002. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature 418:793-797. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2003. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 23:3763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lou, H., D. M. Helfman, R. F. Gagel, and S. M. Berget. 1999. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol. 19:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutz, C. S., C. Cooke, J. P. O'Connor, R. Kobayashi, and J. C. Alwine. 1998. The snRNP-free U1A (SF-A) complex(es): identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor. RNA 4:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis, T., B. Tasic, and K. M. Neugebauer. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 32.Maquat, L. E. 2002. NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes Dev. 16:1743-1753. [DOI] [PubMed] [Google Scholar]
- 33.Mathur, M., P. W. Tucker, and H. H. Samuels. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matter, N., M. Marx, S. Weg-Remers, H. Ponta, P. Herrlich, and H. Konig. 2000. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 275:35353-35360. [DOI] [PubMed] [Google Scholar]
- 35.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 36.Michelotti, G. A., E. F. Michelotti, A. Pullner, R. C. Duncan, D. Eick, and D. Levens. 1996. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol. Cell. Biol. 16:2656-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 38.Neugebauer, K. M. 2002. On the importance of being co-transcriptional. J. Cell Sci. 115:3865-3871. [DOI] [PubMed] [Google Scholar]
- 39.Nogues, G., S. Kadener, P. Cramer, D. Bentley, and A. R. Kornblihtt. 2002. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 277:43110-43114. [DOI] [PubMed] [Google Scholar]
- 40.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 41.Ostrowski, J., Y. Kawata, D. S. Schullery, O. N. Denisenko, Y. Higaki, C. K. Abrass, and K. Bomsztyk. 2001. Insulin alters heterogeneous nuclear ribonucleoprotein K protein binding to DNA and RNA. Proc. Natl. Acad. Sci. USA 98:9044-9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pagani, F., C. Stuani, E. Zuccato, A. R. Kornblihtt, and F. E. Baralle. 2003. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 278:1511-1517. [DOI] [PubMed] [Google Scholar]
- 43.Peng, R., B. T. Dye, I. Perez, D. C. Barnard, A. B. Thompson, and J. G. Patton. 2002. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8:1334-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 45.Shav-Tal, Y., D. Zipori, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2002. PSF and p54(nrb)/NonO--multi-functional nuclear proteins. FEBS Lett. 531:109-114. [DOI] [PubMed] [Google Scholar]
- 46.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siomi, H., G. Dreyfuss, and D. R. Storm. 1997. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 7:345-353. [DOI] [PubMed] [Google Scholar]
- 48.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25:381-388. [DOI] [PubMed] [Google Scholar]
- 49.Stickeler, E., S. D. Fraser, A. Honig, A. L. Chen, S. M. Berget, and T. A. Cooper. 2001. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 20:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tolnay, M., L. A. Vereshchagina, and G. C. Tsokos. 1999. Heterogeneous nuclear ribonucleoprotein D0B is a sequence-specific DNA-binding protein. Biochem. J. 338:417-425. [PMC free article] [PubMed] [Google Scholar]
- 51.Varani, G. 2001. Delivering messages from the 3′ end. Proc. Natl. Acad. Sci. USA 98:4288-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varani, G. 1998. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 27:407-445. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson, M. F., and A. B. Shyu. 2001. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23:775-787. [DOI] [PubMed] [Google Scholar]
- 54.Will, C. L., R. Luhrmann, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol. 9:320-328. [DOI] [PubMed] [Google Scholar]
- 55.Yang, Y. S., J. H. Hanke, L. Carayannopoulos, C. M. Craft, J. D. Capra, and P. W. Tucker. 1993. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 13:5593-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, Y., L. Kan, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510-13516. [DOI] [PubMed] [Google Scholar]