Abstract
Copper is essential to mammalian physiology and its homeostasis is tightly regulated. In humans, genetic defects in copper excretion result in copper overload and Wilson's disease (WD). Previous studies in the mouse model for WD (Atp7b-/-) revealed copper accumulation in hepatic nuclei and specific changes in the mRNA profile prior to pathology onset. To find a molecular link between nuclear copper elevation and changes in hepatic transcriptome we utilized quantitative ionomic and proteomic approaches. X-ray fluorescence and ICP-MS analysis indicate that copper in Atp7b-/- nucleus, while highly elevated, does not markedly alter nuclear ion content. Widespread protein oxidation is also not observed, although glutathione reductase SelH is upregulated, likely to maintain redox balance. We further demonstrate that accumulating copper affects abundance and/or modification of a distinct subset of nuclear proteins. These proteins populate pathways most significantly associated with RNA processing. An alteration in the splicing pattern was observed for hnRNP A2/B1, itself the RNA shuttling factor and spliceosome component. Analysis of hnRNP A2/B1 mRNA and protein revealed an increased retention of exon 2 and a selective 2-fold upregulation of a corresponding protein spliced variant. Mass-spectrometry measurements suggest that the nucleo-cytoplasmic distribution of RNA binding proteins, including A2/B1, is altered in the Atp7b-/- liver. We conclude that remodeling of RNA processing machinery is an important component in cells’ response to elevated copper that may guide pathology development in early stages of WD.
Keywords: ATP7B, copper, Wilson's disease, hnRNP A2/B1, RNA, nucleus, liver
Introduction
Wilson disease (WD) is a severe disorder of copper metabolism. WD is caused by mutations in ATP7B, a gene encoding a copper transporting ATPase. The loss of ATP7B expression or function disrupts copper homeostasis, particularly in the liver, by greatly diminishing hepatocytes’ ability to export excess copper into the bile. Copper accumulates to high levels in the liver causing severe morphological and functional changes, including cirrhosis, hepatitis, and liver failure. Although phenotypic manifestations of WD have been well described, detailed mechanistic understanding of copper-induced pathology in WD is still lacking. To obtain such mechanistic insight we have utilized Atp7b-/- mice, an animal model for hepatic course of WD 1, 2. Similarly to WD patients, the Atp7b-/- mice accumulate copper in the liver, fail to incorporate copper into the ferroxidase ceruloplasmin, and develop marked liver pathology2.
In WD, accumulating copper was proposed to mediate its toxic effects through the generation of reactive oxygen species and a widespread damage to proteins, DNA, and lipids 3, 4. These reactions take place at the height of liver disease 5-8 and whether they represent the cause of the disease or secondary pathology manifestations is not clear. The initial stages of pathology development in WD are characterized by specific and limited changes in the mRNA profile that are observed prior to the development of noticeable morphological changes in the liver 9. The observed changes in the liver transcriptome involve distinct pathways, predominantly cell cycle (up-regulated) and lipid metabolism (down-regulated), and are accompanied by corresponding metabolic changes 9. The mechanism through which copper induces these early effects is unknown.
We have previously shown that in Atp7b-/- hepatocytes accumulating copper distributes non-uniformly between cellular compartments 2. Early in the disease, copper levels increase predominantly in the nuclei and cytosol 2, and only subsequently copper migrates to other cell compartments. In the cytosol, accumulating copper binds to metallothioneins (MTs), well known metal scavengers. MT1 and MT2 are highly elevated in Atp7b-/- animals 9; by binding copper tightly, MTs most likely decrease copper availability and reactivity in the cytosol.
Essentially no information is available on the physical form and the activity of elevated copper in the WD nuclei. The nuclei are responsible for all initial mRNA processing (transcription, splicing, polyadenylation) that determines mRNA abundance and stability. Since selective changes in the liver mRNA profile are detected in the Atp7b-/- mice 9, it seems likely that these changes are triggered by elevated copper entering the Atp7b-/- nuclei. To test this hypothesis and understand the mechanism behind copper activity in WD nuclei, we asked the following questions: (i) does copper entry into the nucleus alter nuclear metal ion content and/or protein machinery? (ii) are changes broad or limited/selective? (iii) can we identify nuclear functions most significantly affected by copper overload? (iv) is there a link between changes in the affected nuclear processes and the observed attenuation of liver transcriptome?
To answer these questions, it was important to evaluate many potential targets of elevated copper in an unbiased manner. Consequently, we utilized the ionomics (metal ion analysis) and proteomics approaches and compared the control and Atp7b-/- nuclei. We demonstrate that copper entering hepatic nuclei does not cause significant changes in the elemental content of the nuclei, nor does it induce a widespread cysteine oxidation or tyrosine nitration. Instead, copper selectively alters protein machinery involved in RNA biogenesis. In particular, copper elevation is associated with a distinct modification of a splicing pattern (due to selective exon 2 retention) for hnRNP A2/B1, an important nuclear protein involved in mRNA trafficking and stability. This new and unanticipated role of copper in RNA biogenesis facilitates understanding of WD pathology, as well as regulatory effects of copper in normal tissues.
Results
Elevated copper does not significantly alter nuclear metallome of Atp7b-/- liver
In Atp7b-/- mice hepatic copper accumulation reaches its highest level approximately 6 weeks after animal birth 2. At this time, liver pathology (ballooning hepatocytes, inflammation, necrosis) is not yet evident 2. Therefore, we reasoned that if molecular changes are observed in the 6 week old Atp7b-/- nuclei, they are likely be directly induced by copper, as opposed to being secondary manifestations of the disease. Since neither the scope of the effects caused by copper nor specific molecular targets in the nucleus are known, we considered several possibilities and initially evaluated whether nuclear copper overload has widespread or more limited consequences.
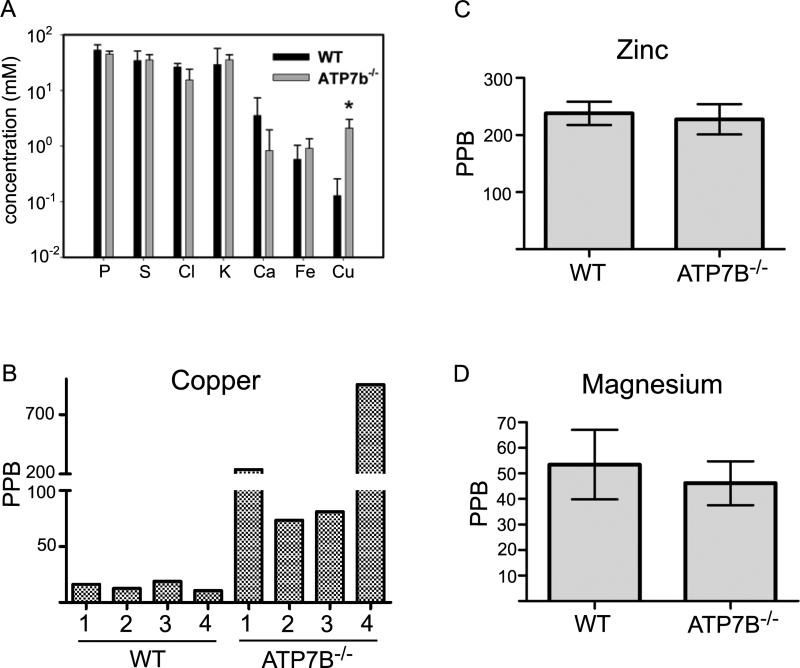
Cu(I), a likely form of copper in cells, and Cu(II) both bind to proteins with high affinity 10, 11 and compete can with other metals 12. Thus, it is possible that excess copper in nuclei may displace other metals from their binding sites. To test whether copper elevation in the nucleus is associated with the corresponding decrease of other metals we first carried out analysis of elemental concentration using synchrotron radiation x-ray fluorescence (SXRF). This methodology allows quantitative measurements of elemental content in nuclei and cytosol directly in tissue, which alleviates concerns about potential losses of elements during fractionation. Figure 1A illustrates that copper was the only element significantly elevated in the Atp7b-/- nuclei. A trend towards the decrease was observed for calcium (although not statistically significant), while other elements were unchanged.
1. Ion composition of Atp7b-/- nuclei is not significantly affected by elevated copper.
(A) Elemental concentration in the wild type and Atp7b-/- nuclei in situ was determined by SXRF following rapid freezing of liver tissue and sectioning. Statistically significant change is indicated by a star (Two independent biological replicates were used for each control and Atp7b-/- samples, with 4 and 3 scans per sample, respectively. The SD values were determined using the sample mean average for all scans). (B) Copper concentration in purified wild type and Atp7b-/- hepatic cell nuclei, as measured by ICP-MS; values for individual animals are shown. (C,D) The ICP-MS results for zinc and magnesium in purified wild type and Atp7b-/- cell nuclei. Two independent experiments (with the same results) were done using 4 animals of each genotype (4 controls and 4 Atp7b-/-) per experiment.
These results were confirmed using inductively coupled plasma mass-spectrometry (ICP-MS) of purified nuclei, which allowed us to analyze a larger spectrum of elements (Table 1). The nuclear pores have a resting diameter of 10 nm and can further dilate to become permeable for RNA and proteins 13. Therefore, it was unexpected and interesting that relative concentrations of elements in purified nuclei were similar to those in situ despite the anticipated loss of ions from nuclei during purification (see more in discussion). Copper was highly elevated in all Atp7b-/- samples of purified nuclei (Figure 1B) also illustrating the lack of easy loss. Similarly to the SXRF data, ICP-MS detected the decrease in mean Ca2+ levels, however variation between samples was high and the difference was not statistically significant (Table 1).
Table 1. Elemental analysis of isolated wild type and Atp7b-/- liver nuclei.
Normalized means in measured parts per billion and standard deviations are shown for indicated elements.
| Wild type | Atp7b-/- | |||
|---|---|---|---|---|
| Element | mean | st. dev. | mean | st. dev. |
| P31 | 56,576 | 5,628 | 51,476 | 4,816 |
| K39 | 28,477 | 2,182 | 32,317 | 2,579 |
| S34 | 14,374 | 506 | 14,443 | 638 |
| Ca44 | 317 | 194 | 164 | 42 |
| Zn66 | 266 | 15 | 232 | 44 |
| Fe57 | 133.8 | 68.3 | 154.8 | 40 |
| Mg25 | 58.2 | 22 | 47 | 15.2 |
| Cu63 | 14.6 | 3.7 | 336.6 | 41.8 |
| Cu65 | 14.2 | 3.6 | 336.2 | 42 |
| Ni60 | 4.5 | 1.6 | 5.2 | 5.1 |
| Rb85 | 1.6 | 0.1 | 1.8 | 0.1 |
| Mn55 | 1.4 | 1.1 | 0.7 | 0.1 |
| Mo98 | 0.4 | 0.1 | 0.6 | 0.3 |
Cell nuclei are rich in proteins that bind zinc via the Cys-based “zinc fingers”. Cys residues are excellent ligands for copper, which can compete with zinc for its binding sites 14, 15. DNA-associated magnesium is another important metal in cell nuclei. Although copper concentration was well above mean magnesium concentration, and close to zinc concentration, there was no marked decrease of either metal in Atp7b-/- nuclei (Figure 1C, D). Since accumulating copper does not significantly modify the elemental content of the nuclei, it seems likely that copper mediates its effects through mechanisms other than a large-scale metal displacement. However, presently we cannot exclude that the metal buffering capacity of nuclei may be sufficient to absorb metals displaced by copper
Accumulation of copper in the nucleus is not associated with global protein oxidation
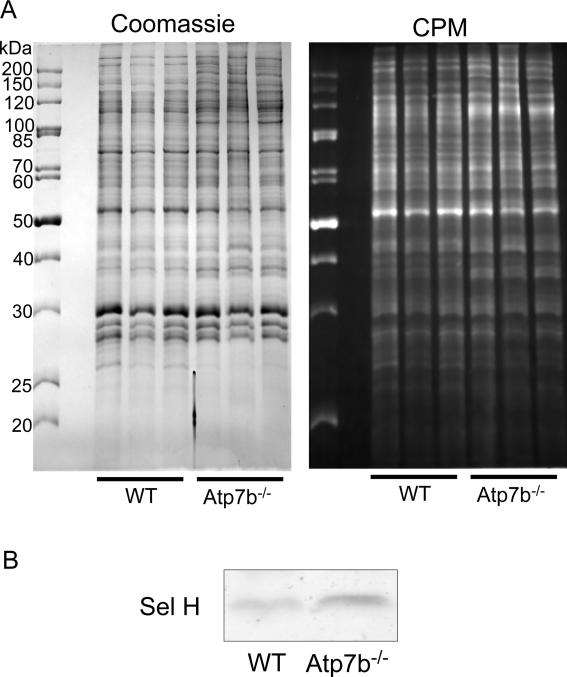
Copper is a redox active metal capable of inducing formation of reactive oxygen species, which in turn catalyzes nitration of tyrosine residues 16. To determine whether these processes occur to a significant degree in the Atp7b-/- nuclei, we compared protein nitration in control and Atp7b-/- nuclei using anti-nitrotyrosine antibody. After separating nuclear extracts by SDS PAGE and western blotting, no detectable difference was found (Supplementary Figure S1, panel A). To determine whether accumulating copper causes significant cysteine oxidation we labeled nuclear proteins with the thiol-reactive fluorescent reagent CPM and analyzed them by SDS page (Figure 2A). Although some differences in CPM labeling were detected, subsequent staining of the same gel with Coomassie revealed that changes in fluorescence correlated with the changes in protein levels, i.e. they were likely due to changes in protein expression. Proteins showing changes in CPM/protein levels were in-gel-digested with trypsin and identified by mass-spectrometry as containing RNA binding proteins hnRNP A2/B1, U2 snRNP, and hnRNP A3.
2. Copper does not induce a widespread oxidation of cysteines in Atp7b-/- nuclei.
Nuclear protein extracts were labeled with the Cys directed fluorescein coumarin maleimide CPM, separated by standard Laemmli gel and then stained by Coomassie. (A) Left: Comparison of protein patterns in control and Atp7b-/- nuclei by Coomassie staining, Right: CPM fluorescent image of the same gel. (B) Immunodetection of Sel H in purified nuclei from the wild type and Atp7b-/- livers. Three of each control and Atp7b-/- livers were used for the experiment.
The above results did not exclude possibility of oxidation for low abundance proteins or partial (10-20%) oxidation. Nevertheless it was apparent that copper did not induce a widespread oxidative modification. This conclusion was further confirmed by the lack of significant changes in protein glutathionylation (Supplementary Figure S1, panel B).
Since oxidation is a commonly assumed consequence of copper overload, this apparent lack of oxidation-induced protein modifications was unexpected and suggested involvement of compensatory mechanisms. We have measured glutathione levels in the Atp7b-/- nuclei (Supplementary Figure S2) and found them consistently elevated, although more samples are needed for the difference to be statistically significant; lengthy isolation procedure precluded reliable estimates of glutathione oxidation. To identify candidate protein(s) with a potential compensatory redox function, we analyzed our previously generated datasets of the mRNA levels in Atp7b-/- liver 9. In our earlier studies we characterized the most populated and significantly changed pathways 9; in the current work we searched for the upregulated mRNAs encoding proteins with known redox activity. We found that the Atp7b-/- livers had a significant increase in the mRNA levels for the glutathione reductase SelH 9. This change was interesting because the recombinant SelH was shown to be targeted to the nucleus and protect cells against oxidative stress 17. The localization of the endogenous SelH in tissues is not known. Consequently, to test whether the upregulation of SelH mRNA levels results in the increased abundance of SelH protein in the nucleus, we measured SelH in purified nuclear fractions. A significant increase of SelH protein in Atp7b-/- nuclei was observed (Figure 2B).
Copper accumulation in Atp7b-/- nuclei is associated with changes in the abundance and post-translational modification of nuclear proteins
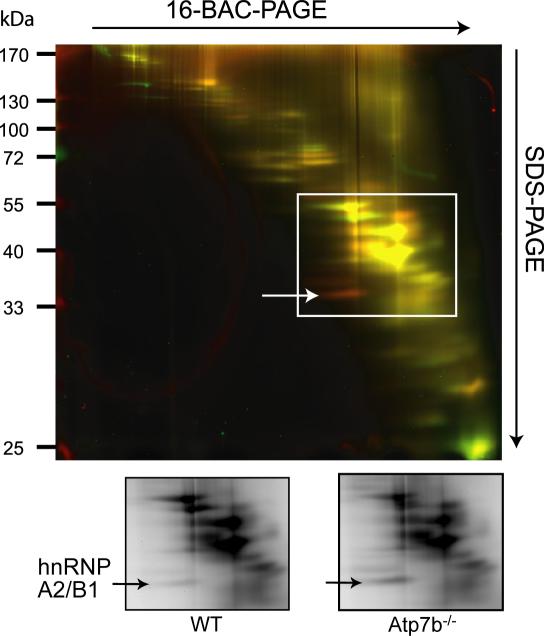
Up to this point, our data have made it clear that copper in the nucleus is likely to have limited number of targets, and that methods with higher resolution are needed to identify them. Consequently, we utilized 2-dimensional differential gel electrophoresis (2D-DIGE) as a quantitative unbiased approach for analysis of complex protein mixtures. Initial optimization experiments were performed on a small scale (see supplement for details). These experiments determined that the two-dye analysis combined with dye-reversal allowed for highest confidence of data. These experiments also identified several proteins with a significantly changed abundance in Atp7b-/- nuclei (these proteins include RNA/DNA binding proteins TDP-43, HNRNP U, Mad1-like 1, a component of the NuA4 histone acetyltransferase complex RUVB1, and KHSRP). To separate proteins with high PI, we also applied the low pH 16-BAC/SDS-PAGE version of 2D gels; these experiments identified the RNA binding protein hnRNPA2/B1 as significantly elevated in Atp7b-/- nuclei (Figure 3). The initial experiments also revealed that a larger scale separation is needed to improve protein identification.
3. The RNA binding protein hnRNP A2/B1 is significantly changed in Atp7b-/- nuclei.
Top: 16-BAC/SDS-PAGE 2D gel separation of proteins from Cy3-labeled extract of wild type (green) and Cy5-labeled Atp7b -/- (red) nuclei. (B) Grayscale image of the gel with differentially abundant spot indicated. The protein in the spot was extracted, digested and identified by mass-spectrometry as hnRNP A2/B1. Two independent experiments were performed with the same result, the results of one of them is shown.
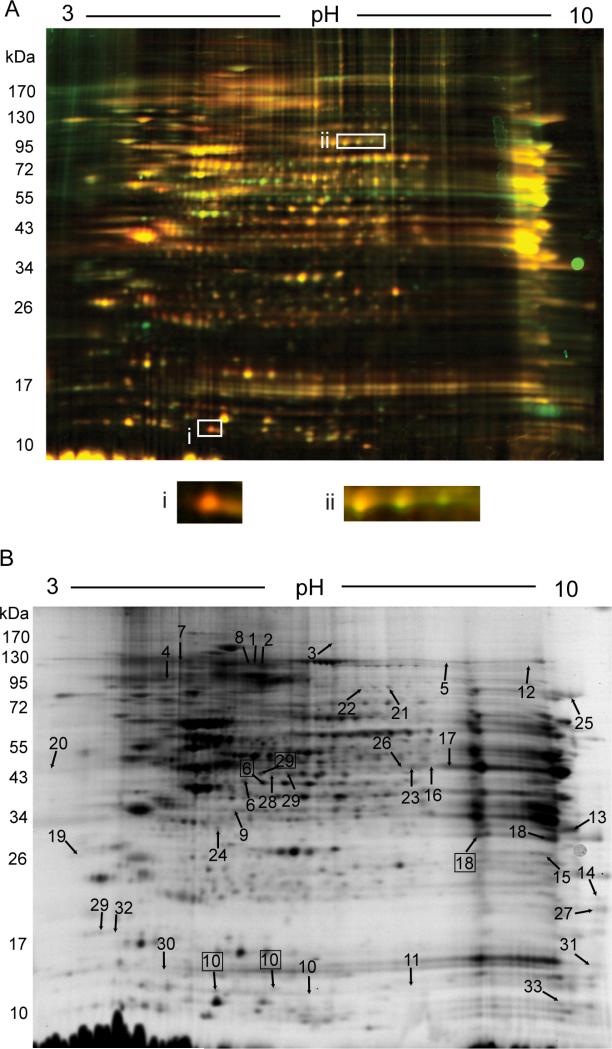
The Ettan-DALT (24 cm × 20 cm) gel format greatly expanded the resolving capacity and reproducibility of 2D gels. Over 1000 spots were resolved on each gel (Figure 4A). Most of the spots showed similar intensity (Comparison of average normalized density of Atp7b-/- and control spots is shown in Supplementary Figure S3). However, 33 spots reproducibly showed more than 1.5-fold difference in fluorescence intensity between the control and Atp7b-/- samples (p-value <0.05). Mass-spectrometry of trypsin digests from these 33 spots identified 25 unique proteins (Table 2). Thus, less than 5% of detectable nuclear proteins are affected by copper accumulation pointing to a selective effect of copper on nuclear proteome.
4. Remodeling of nuclear proteome in response to copper elevation.
(A) Representative DIGE gel comparing the wild type (Cy3, green) and Atp7b -/- (Cy5, red) samples: box (i) illustrates apparent change in protein abundance, box (ii) - change in modification. (B) Grayscale image of the representative DIGE gel with differentially changed spots indicated by arrows. Numbers identify spots changed in abundance. Boxed numbers indicate the same protein as the one indicated by number but without change in abundance. The presence of the same protein in two spots suggests changes in posttranslational modification. (6 independent biological replicates of each genotype/per gel was used in analytical experiments; 4 independent biological replicates of each genotype per gel was used for the large-scale experiment. Fold-difference and t-test were employed to identify all changed spots)
Table 2.
Proteins with altered abundance in 2D DIGE analysis
| Label | Accession | Name |
|---|---|---|
| 1 | AAH39939 | Ccar1: cell cycle and apopotosis regulatory protein 1 |
| 2 | Q99P88 | nuclear pore complex NU155 |
| 3 | AAH57983 | Pzp |
| 4 | AAH18353 | HnrpU |
| 5 | - | Not identified |
| 6 | AK083312 | TAR DNA binding protein isoform 1 (TDP-43) |
| 7 | Q04857 | Collagen alpha 1(VI) chain precursor |
| 8 | - | Not identified |
| 9 | P62137 | Serine/threonine-protein phosphatase PP1-alpha catalytic subunit (PP-1A) |
| 10 | P62806 | Histone H4 |
| 11 | P62806 | Histone H4 |
| 12 | AAB01163 | Transcription factor C1 (HCF-1) |
| 13 | P49312 | HnRNP A1 |
| 14 | XP 484160 | OSCP conserved domain |
| 15 | P57784 | U2 small nuclear ribonucleoprotein A' |
| 16 | P10126 | EF1A1 |
| 17 | P10126 | EF1A1 |
| 18 | O88569 | HnRNPA2/B1 |
| 19 | P57784 | U2 small nuclear ribonucleoprotein A' |
| 20 | Q8CCS6 | Polyadenylate-binding protein 2 (PABII) |
| 21 | Q922D8 | C-1-tetrahydrofolate synthase, cytoplasmic |
| 22 | Q922D8 | C-1-tetrahydrofolate synthase, cytoplasmic |
| 23 | Q9R0H0 | Acyl-coenzyme A oxidase 1, peroxisomal |
| 24 | Q9QZH3 | PPIase E |
| 25 | AAH89305 | Splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| 26 | P10126 | EF1A1 |
| 27 | P35979 | 60S ribosomal protein L12 |
| 28 | AAH04649 | Interferon inducible GTPase 1 |
| 29 | Q8BG32 | 26S proteasome non-ATPase regulatory subunit 11 |
| 30 | Q9ERR7 | 15 kDa selenoprotein precursor SEP15 |
| 31 | Q9QXA5 | U6 snRNA-associated Sm-like protein LSm4 |
| 32 | P60605 | Ubiquitin conjugating enzyme UB2G2 |
| 33 | AAH11436 | Mitochondrial import inner membrane translocase subunit TIM13 A |
The changes appear to involve both changes in protein abundance as well as posttranslational modification (examples are shown in Figure 4A, small panels). This latter conclusion is supported by the identification of histone H4 as well as U2 snRNP in several “changed” spots, whereby these proteins have the same apparent mass but different pI values. To determine if other proteins with altered abundance can also be post-translationally modified we analyzed, by mass-spectrometry, spots that (i) had a pattern suggesting a change in pI due to modification of one or more charged residues and (ii) were located in proximity to differentially changed spots (Figure 4B). These experiments revealed additional modifications of histone H4 (spot 10) and identified potential modifications of TDP-43 (spot 6) and hnRNP A2/B1 (spot 18).
RNA processing is the most significantly changed processes in Atp7b-/- nuclei
To determine whether the identified proteins belong to distinct pathways, we analyzed data using DAVID gene ontology and clustering algorithm with high stringency. This analysis revealed that the largest fraction of changed proteins involves proteins that play an important role in pre-mRNA processing, particularly RNA splicing (The cluster with the highest enrichment score is shown in Table 3). Other “significant” clusters (P<0.05) have more general gene ontology descriptors, i.e. ”cellular metabolic process”; “intracellular membrane-bound organelle” and “nucleic acid transport/RNA transport”.
Table 3.
The most populated DAVID ontology cluster.
| Ontology Term | Proteins | Enrichment Score |
|---|---|---|
| • Polyadenylate-binding protein 2 (PABII) | ||
| • mRNA processing | • SFPQ Splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) | |
| • mRNA metabolic process | ||
| • hnRNP A1 | ||
| • mRNA splicing | • U2 small nuclear ribonucleoprotein A' | 4.95 |
| • RNA processing | • PPIase E | |
| • Spliceosome | • TAR DNA binding protein isoform 1 (TDP-43) | |
| • RNA metabolic process | • U6, snRNA-associated Sm-like protein LSm4 hnRNP A2/B1 |
The role of copper in regulating RNA processing machinery was completely novel; therefore, we verified this observation independently using quantitative mass-spectrometry. Dried purified control and Atp7b-/- nuclei were digested with trypsin and analyzed by LC MS/MS with an LTQ quadrupole linear ion trap; protein abundance was compared by spectral counting. Altogether, 6729 peptides passed thresholds and 456 proteins were identified. These experiments confirmed the lack of significant changes in many nuclear proteins (both abundant and less abundant) such as heterochromatin binding proteins, DNA binding proteins, DNA lyase, Histone 1.1, Histone 2.1, nucleophosmin, small nuclear ribonuclear proteins, DEAD box proteins and others, (Supplementary Table S1). By contrast, notable differences were detected for several hnRNPs, KH splicing regulatory protein, RNA helicases, RNA binding protein EWS, nucleolar protein NOP57, nucleolin, Arg/Ser-rich splicing factor (Supplementary Table S1). Many of these proteins are involved in RNA binding and processing. Importantly, such proteins as TDP-43, Arg/Ser rich splicing factor, protein phosphatase 2, polypyrimidine tract binding protein 1, HCF1, and U2 snRNP were found as changed in both the 2D DIGE and mass-spectrometry experiments, validating our 2D DIGE data.
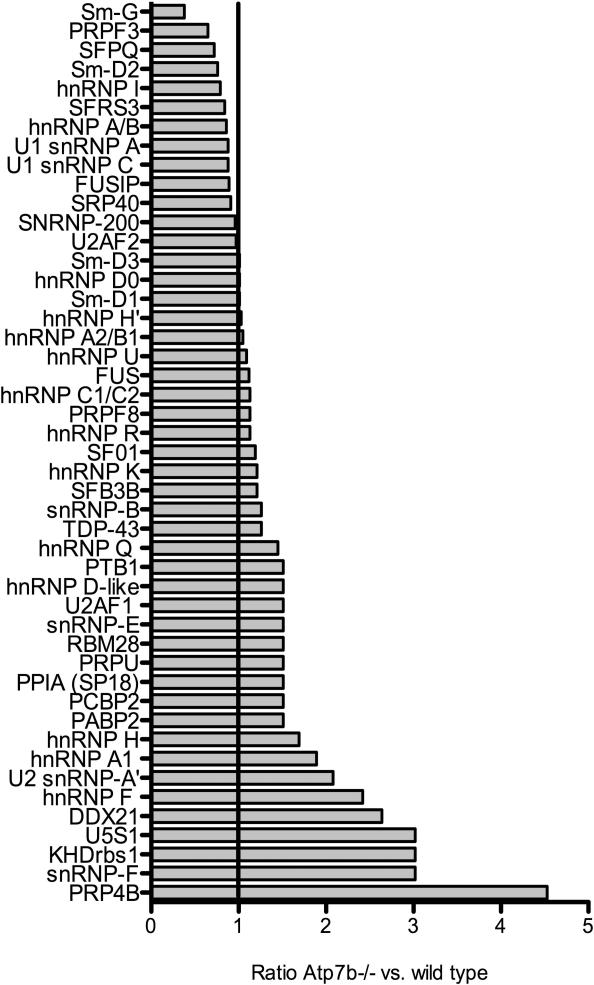
The changes in the RNA processing machinery are not uniform, i.e. we have observed substantial increases in some factors, decreases in others, as well no changes (Figure 5). Thus, copper does not simply inhibit RNA biogenesis, rather it alters ratio of the components of RNA-processing machinery, adjusting it to changed cellular environment. Further analysis revealed that copper modifies the RNA processing machinery through at least two mechanisms.
5. Relative abundance of RNA binding proteins in the Atp7b-/- nuclei compared to wild type.
Fold change of the RNA binding proteins in Atp7b-/- sample compared to wild type is indicated. Abundance was determined by spectral counting.
Copper modulates nucleo-cytoplasmic trafficking of RNA-binding proteins
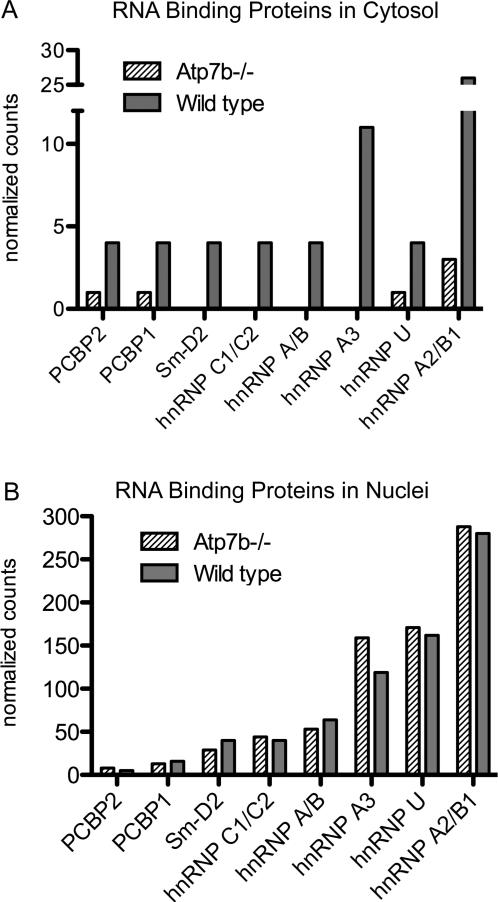
A number of RNA binding proteins have both nuclear and cytosolic roles in RNA processing and can traffic between nucleus and cytosol. To determine whether copper altered the nuclear RNA machinery by inducing nuclear-cytoplasmic trafficking of protein components, we analyzed by mass-spectroscopy cytosolic fractions from the same liver samples for which nuclear proteins had been characterized (Supplementary Table S1). As expected, in the cytosol we observed a significant increase in metallothionein 1 (MT-1). This result is consistent with the previous transcript and protein analysis data for MT 2, 9 and thus validates our mass-spectrometry approach. Eight RNA binding proteins were differentially changed in the Atp7b-/- cytosol compared to control (Figure 6A), all were down-regulated. For several proteins, the decrease in the abundance in the cytosol was accompanied by a reciprocal increase in the nuclear fraction (Figure 6B), suggesting preferential retention in the nuclei. Specifically, hnRNP A2/B1 showed a decrease in the cytosolic fraction and a corresponding increase in the nuclear pool. The nuclear pools of hnRNP U and hnRNP A3 were also increased, while their cytosolic amounts were decreased. The latter result was particularly interesting because these three proteins (hnRNP A2/B1, and A3 and hnRNP U) are known to form a complex 18. We also found an increase in a number of glutathione S-transferase isoforms, which is again consistent with the earlier mRNA profiling data 9 and implies additional buffering capacity for oxidation in Atp7b-/- hepatocytes.
6. Nucleo-cytoplasmic distribution of RNA binding proteins in Atp7b-/- and wild type samples.
Normalized MS/MS spectral counts illustrate (A) the decreased amount of several RNA binding proteins in Atp7b-/- cytosol and (B) reciprocal increase of hnRNP A3, hnRNPU, and hnRNP A2/B1 in Atp7b-/- nuclei
The increase in the nuclear abundance of hnRNPA2/B1 in the Atp7b-/- nuclei is due to selective up-regulation of the B0b splice variant
In the DIGE experiments, as well as in experiments separating nuclear proteins using 16-BAC/SDS-PAGE 2D gels, the hnRNP A2/B1 protein was among the most significantly changed (Figure 3). Its nucleo-cytoplasmic distribution was also affected. To identify potential reasons for this behavior we characterized hnRNPA2/B1 in more detail. We noticed that in mass-spectrometry experiments, the difference between the control and Atp7b-/- nuclei for hnRNP A2/B1 was small (Figure 5), compared to a marked change seen in gels. We hypothesized that this could be due to the fact that mass-spectrometry identifies the most abundant peptides from the entire protein population (i.e. from the mixture of all variants of the same protein generated by alternative splicing), while on gels we separate and detect individual protein variants. Since the RNA splicing machinery is altered in Atp7b-/- nuclei and hnRNPA2/B1 mRNA is known to be alternatively spliced, we compared the patterns of the hnRNPA2/B1 in control and Atp7b-/- nuclei.
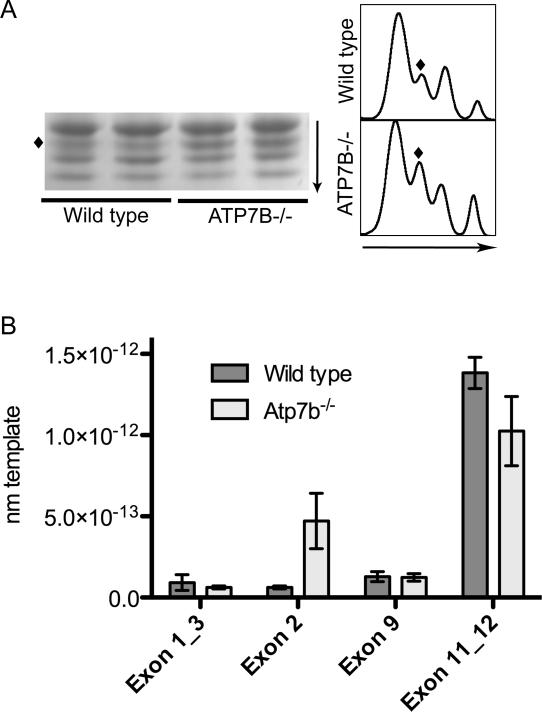
The hnRNP A2/B1 has 4 major forms (B1, A2, B0b, and B0a) that are produced through alternative splicing of exon 2 and/or exon 9 of the hnRNP A2/B1 pre-mRNA19. Intron 11 may also be retained; however, it is unclear if this transcript is translated 19. To determine whether all or some of these splice variants changed in Atp7b-/- nuclei, we first analyzed the hnRNP A2/B1 composition in the nuclei using an antibody directed against the portion of the molecule common for all isoforms. Figure 7A illustrates that the intensity of three hnRNP A2/B1-related bands are similar, while one band is increased at least 2 fold in the Atp7b-/- sample. Based on the electrophoretic mobility, the changed band could be an A2 isoform (second largest among the 4 variants). However, previous studies indicated that the largest B1 protein has low abundance and is difficult to detect, while A2 is the most abundant variant followed by B0b and B0a19. Thus, if the top most abundant band is A2, the changed isoform could be B0b (retaining exon 2 and lacking exon 9). To clarify this issue, we isolated mRNA fraction from control and Atp7b-/- livers and compared the molar abundance of different exons (the exon1-3 splice junction, exon 2, exon 9 and the exon 11-12 splice junction) in the hnRNP A2/B1 transcript by quantitative PCR (Figure 7B). These experiments revealed an increased retention of exon 2 in the hnRNP A2/B1 transcript from Atp7b-/- samples compared to the wild type with little change in other exons. These results are consistent with a specific upregulation of the B0b isoform of hnRNP A2/B1.
7. Increased retention of exon 2 is associated with the up-regulation of a single splice variant of hnRNPA2/B1.
(A) Left: SDS-PAGE and immunodetection of hnRNP A2/B1 variants (one of 6 independent experiments is shown). Right: Signal intensity as measured by densitometry. Arrows indicate direction of migration in electrophoresis. The diamond symbol (◆) indicates the upregulated variant. (B) The hnRNP exon abundance was determined by quantitative PCR and normalized to GAPDH. Mean values (generated using 3 of each independent control and Atp7b-/- samples) and standard deviation are shown.
Discussion
In this study, we utilized the Atp7b-/- mice to identify molecular events behind initial stages of pathology development in WD. Our results demonstrate that copper entry into hepatic nuclei induces specific changes in the mRNA processing machinery, while nuclear structural components and transcriptional apparatus are less affected. A significant number of altered proteins are directly involved in regulating mRNA trafficking and stability. Therefore, the changes in mRNA profile observed at the initial stages of WD are likely to be mediated, at least in part, through changes in RNA biogenesis. Our data do not exclude oxidation of selected targets, however marked oxidative modification of nuclear proteins is not observed, and the nuclear content for such metals as zinc, iron, or magnesium is not greatly affected by accumulating copper.
The apparent lack of marked protein oxidation (despite significant increase in copper content) is likely due to cells’ capacity to combat oxidative stress by upregulating nuclear factors such as glutathione reductase SelH, which we observed as increased in Atp7b-/- nuclei. Our current results do not exclude oxidative modification of selected susceptible targets. In fact, the SelH upregulation and slight increase in total glutathione suggests that copper may alter redox balance (perhaps by changing the GSH:GSSG ratio). Further analysis of the redox state for nuclear proteins may identify such susceptible targets; one candidate is a transcription factor MTF1, which regulates expression of MTs and SelH 20 and is known to be sensitive to oxidation 21.
How excess copper enters the Atp7b-/- nuclei is unclear. Neither Atox1 nor metallothioneins are elevated in Atp7b-/- nuclei and preliminary protein fractionation shows more than one copper-containing peak (our data, not shown). It is also intriguing that copper is retained in nuclei during purification. It would be tempting to conclude that copper is present in a non-diffusible form, however our results with other nuclear metal ions caution against such a seemingly logical conclusion. The ICP-MS data on elemental composition of nuclei are consistent with the results of SXRF imaging performed in situ, providing evidence towards separate metal ion pools and an apparent lack of a simple diffusion-mediated metal ion equilibrium between the nucleus and cytosol. Thus, a yet to be identified mechanism might be involved in maintaining the precise elemental content of mammalian nuclei.
Our data indicate that changes in the RNA processing machinery are not uniform, confirming specific effects of copper in Atp7b-/- nuclei. The mechanisms through which this machinery is altered involve posttranslational modification, alteration in the pattern of alternatively spliced variants, as well as the redistribution of RNA binding proteins between the cytosol and the nucleus. These events are likely to be coupled. For example, we observed that the component of the nuclear protein export machinery eF1A22 was changed in 2D analysis apparently by posttranslational modification. Significantly, one of the targets of for eF1A-mediated export is the polyadenylate binding protein (PABP2, PABN1), which we also found as significantly altered in our datasets.
The most interesting and highly reproducible response to elevated copper was observed for hnRNP A2/B1 This RNA binding protein is a component of the spliceosome C complex. It also functions in nuclear-cytoplasmic mRNA shuttling through interactions with A2RE (hnRNP A2 response elements)23. The hnRNP A2/B1 protein is present as 4 variants (A2/B1/B0a/B0b) generated through the alternative splicing of corresponding mRNA. Our data indicate that elevated copper induces a distinct modification in the pattern of these variants. Higher abundance of exon 2 and the lack of changes in other examined exons suggest that the hnRNP B0b variant is selectively upregulated. Interestingly, hnRNP A2/B1 and hnRNP A1 (also significantly changed in our 2D DIGE analysis) are associated with exon exclusion during alternative splicing 24-26, thus hinting to possible self-regulation of RNA splicing machinery and/or copper-dependent feed-back loops.
Specific mRNA targets of the B0a variant of hnRNPA2/B1 are not known. In our previous studies, the most significant group of upregulated transcripts in Atp7b-/- mice was associated with cell cycle progression. Considering that numerous reports show upregulation of hnRNP A2/B1 in proliferating cells 27-33, it is tempting to speculate that the upregulation and change in the nucleo-cytoplasmic distribution of hnRNPA2/B1 observed in our study may represent the mechanism through which copper influences cell cycle and proliferation in the liver.
In conclusion, we have uncovered a novel and important connection between copper homeostasis and RNA biosynthetic machinery. This finding contributes to better understanding of RNA changes observed at the early stages of WD and offers potential early markers of disease development. Furthermore, it is highly significant that such affected proteins as TDP-43 and hnRNP A2/B1 are also known to play an important role in neuronal degeneration/myelination. So far, there has been no common molecular link between hepatic and neurological manifestations of WD. The effect of copper on RNA biogenesis could be the first common feature of these two phenotypic manifestations of WD.
Materials and Methods
Animal handling
The generation of Atp7b-/- mice has been described previously 1. The Atp7b-/- and wild type mice were housed at the Oregon Health & Science University Department of Comparative Medicine according to the National Institutes of Health guidelines on the use of laboratory and experimental animals. Mice were maintained on strain C57BL × 129S6/SvEv, in shoebox cages on rodent chow (Lab Diet 5001) and water ad libitum, with a 12-hour light/dark cycle. Mice of both sexes were used for metal measurements and proteomic studies. Animals were euthanized at six weeks of age by CO2 inhalation followed by cervical dislocation. Animals were immediately perfused with 0.9% saline and livers placed on ice before isolation of hepatic nuclei.
Isolation of hepatic nuclei from mouse liver
All procedures were performed in a cold room at 4°C. Excised liver tissue was minced with a razor blade and homogenized in 3.0 mL HB (10 mM HEPES pH 7.6, 25 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1.0 mM EDTA, 2.0 M sucrose, 10% glycerol, 1.0 mM DTT and Complete Protease Inhibitor Cocktail (Roche, Nutley, New Jersey)) per gram of tissue by six passes with a motor-driven Potter-Elvejem homogenizer at approximately 200 rpm. The homogenate was layered over a 1.0 mL HB cushion, balanced with HB and centrifuged for 45 minutes at 30,000 RPM in a Beckman SW50.1 rotor. The nuclear pellet was resuspended in 1 ml of RB buffer (10 mM HEPES pH 7.6, 25 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, 1 mM EGTA, 20% glycerol, 1 mM DTT, and protease inhibitors). The supernatant was separated further on the sucrose density gradient by diluting to 0.25 M sucrose in the RB buffer and centrifuging for 1 hour at 100,000g. The resulting supernatant was considered the cytosolic fraction. For enrichment and detection of hnRNP A2/B1, nuclear extracts were prepared as previously described 2. For pilot experiments using small quantities of tissue, nonionic detergent was used to purify nuclei as previously described 34.
Synchrotron radiation X-ray fluorescence (SXRF)
SXRF microscopy was performed on beamlines 2-IDE and 2-ID-D of the Advanced Photon Source at the Argonne National Laboratory (APS, Argonne, IL). The APS storage ring operates at 7 GeV with an injection current of 100 mA. An incident X-ray energy of 10 keV is produced by an undulator source. X-rays are monochromatized by a double-crystal Si(111) monochromator (Kohzu) and a Fresnel zone plate focuses the X-ray beam on the sample to a spot size of ≤0.5 (h) × 0.5 (v) μm2 35. Samples were raster-scanned and the resulting fluorescence of each element was measured simultaneously at each pixel (dwell time: 1sec per pixel) using an energy-dispersive silicon drift detector (SII NanoTechnology). Samples were mounted onto a kinematic specimen holder and the scan regions of interest selected on the sample using a Leica light microscope with an attached precision x/y-stage (Ludl Bioprecision). The specimen were then transferred to the x-ray microprobe where the same regions are located using the x/y-coordinates derived from the light microscope.
For quantification, full x-ray fluorescence spectra were collected at every position during the scan. The spectra were individually fitted using modified Gaussians for fluorescence peak descriptions and an adapted version of the SNIP algorithm for the determination of background 36. Calibration and conversion of fluorescence counts to area concentrations of elements (μg/cm2) were done using a calibration curve based on the measured X-ray fluorescence of the thin film NBS standards 1832 (Cu) and 1833 (Zn) (National Institute of Standards and Technology (NIST), Gaithersburg, MD). For the analysis of the elemental content for specific regions of interest (ROIs), spectra for the respective ROIs are extracted from original dataset, added up, and fitted and quantified as described above.
Copper concentration in the nuclei by SXRF
Healthy hepatic nuclei are smaller than 10 μm (tissue thickness) in diameter, and therefore comprise only a fraction of the total amount of copper in the corresponding “nuclear” ROI. Consequently, we calculated the volume of the nucleus (approximately 10% of the total hepatocellular volume) and subtracted it from the total cylindrical volume of the selected ROI. We then determined the amount of copper in the remaining (cytoplasmic) volume using the cytoplasmic Cu concentration, subtracted this contribution from the total amount of copper in the cylinder to yield the amount and concentration of copper in the nuclei.
Elemental analysis by ICP-MS
Elemental analysis was performed by the Center for Environmental Stress Physiology at Purdue University (West Lafayette, IN). Each sample was digested with 0.12 ml of c. HNO3 (Mallinckrodt, AR Select grade) at 110°C for 3 h, in a 96-well Teflon plate. Each sample was diluted to 1.2 ml with 18 MΩ water and analyzed by Inductively-Coupled Plasma Mass Spectrometry (ICP-MS) on a PerkinElmer Elan DRCe ICP-MS instrument. Indium (EM Science, Gibbstown, NJ) was used as an internal standard. National Institute of Standards and Technology traceable calibration standards (ULTRA Scientific, N. Kingstown, RI) were used for the calibration. Elements measured included Na, Mg, P, S, K, Ca, Mn, Fe, Co, Ni, Cu, Zn, and Se. Elements were normalized to MS signal for P, S, K, Zn, Rb, Mo using the INFACT algorithm developed by the Center for Environmental Stress Physiology. All of these elements had relative standard deviations of less than 11% of mean value.
CPM labeling
Soluble nuclear extracts were prepared as described 2. 10 μg of each sample was labeled for 20 min with 18 μM of the thiol-reactive fluorescent dye CPM (7-diethylamino-3-(4'-maleimidylphenyl)-4-methylcoumarin) (Invitrogen, Carlsbad, CA) under non-reducing conditions. The labeling reaction was quenched by adding DTT to 100 mM. Samples were loaded onto a 12.5% Laemmli SDS-PAGE gel. The fluorescent image was captured with a camera-equipped UV transilluminator. The same gel was then stained with Coomassie G-250 for imaging of total protein.
Immunodetection of glutathionylated and nitrosylated proteins
30 μg of protein for each sample was loaded on a 12.5% polyacrylamide gel in 62.5 mM Tris-HCl pH 6.8, 3.5 M urea 5% SDS, 10 mM DTT and trace bromophenol blue. Proteins were transferred to PVDF in 10 mM CAPS pH 11.0 and 10% methanol at 180 mA for 2 hours. The blot was blocked with 5% nonfat dry milk in PBS. Immunodetection of glutathionlylated proteins was performed with mouse anti-GSH (Virogen, Waterton, MA) at 1:5000 dilution. HRP-conjugated goat anti-mouse (eBioscience, San Diego, CA) secondary antibody was used at a dilution of 1:10,000. Anti-nitrotyrosine (Millipore, Billerica, MA) was used at 1:1000 dilution, HRP-conjugated goat anti-mouse (eBioscience) was used at 1:10,000 dilution in PBS-T. Visualization was with Western Lightning ECL substrate (Perkin Elmer, Waltham, MA).
Immunodetection of SelH
. 15 μg of soluble nuclear proteins were separated on a 15% Laemmli gel. Proteins were transferred to nitrocellulose in 10% CAPS pH 11.0 and 10% methanol at 180 mA for 2 hours. The blot was blocked with 5% nonfat dry milk in PBS. Rabbit anti-SelH (a kind gift from Dr. Vadim Gladyshev) was used at a dilution of 1:1000, HRP-conjugated goat anti-rabbit (eBioscience) was used at 1:10,000 dilution. Detection and visualization was the same as above.
2D-DIGE
Purified nuclei were solublized in 7 M urea, 2 M thiourea, 30 mM Tris-HCl pH 8.3 and 2% w/v dodecylmaltoside. After 1 hour of solublization on ice, samples were centrifuged at 20,000g, 4°C for 30 min. Supernatants were used for DIGE CyDye labeling. Protein concentration was determined using Bradford assay 37. Initially, small-scale experiments were performed to optimize conditions. The details of these pilot experiments and their results are described in Supplementary materials. For the large-format 2D-gels, 50 μg of wt and Atp7b-/- sample (four wt and four Atp7b-/-) in 20 μL of buffer was labeled on ice for 45 min with 400 pmol of Cy3 or Cy5, respectively, as suggested by the manufacturer (GE Healthcare, Piscataway, NJ). Reactions were quenched by addition of 1 μL of 10 mM lysine. 150 μg of corresponding unlabeled protein mixture was then added to each labeled sample, the wt and Atp7b-/- samples were mixed (total protein per gel 400 μg), and loaded on 24 cm, linear pH 3-10 IPG strips (GE Healthcare). To avoid spectral artifacts, half of the samples were labeled with dyes reversed.
Isoelectric focusing was performed in the above solublization buffer; 12 μL Destreak™ Reagent (GE Healthcare) and 0.5% pH 3-10 carrier ampholytes were added to a final volume of 450 μL. The focusing procedure was at 30 mA/strip in the following steps: 30 V for 6 hours, 60 V for six hours, 200 V for 1 hour, 500 V for 1 hour, 1000 V for 1 hour, linear gradient from 1000 V to 8000 V over 30 minutes then 8000 V until 60,000 V-hours is reached. Strips were stored at -80°C after focusing.
For second dimension (SDS-PAGE), IEF strips were treated 15 minutes in equilibration buffer (50 mM Tris-HCl pH 8.8, 6 M urea, 30% (w/v) glycerol, 2% (w/v) SDS and 0.002% bromophenol blue) with 10 mg/mL DTT, followed by 15 minutes in equilibration buffer with 25 mg/mL iodoacetamide. Each strip was placed on top of a 10-20% linear gradient gel in low fluorescence glass plates (Jule, Inc., Milford, CT). Strips were sealed in place with 0.5% agarose and gels were run using the ETTAN DALT 12 system. Cathode buffer was 50 mM Tris, 384 mM glycine, 0.2% SDS. Anode buffer was 25 mM Tris, 192 mM glycine, 0.1% SDS. Gels were run at 16°C as follows: 20 W for 30 minutes then 160 W until the bromophenol blue dye ran off the gel (about 4 hours).
Cy3 and Cy5 images were acquired by scanning with a Fuji FLA-5000 flatbed scanner. Images were analyzed with Delta2D 5.0 (generously provided by Decodon, Greifswald, Germany). Spots were delineated on each gel image and matched across all gel images. Normalization was to the total spot volume in a given image using spots common to all gels; the intensity of each spot (spot volume) was expressed as a percentage of total spot volume. Spots were considered changed if they met the following criteria: identified in all samples, showed greater than 1.5-fold change in mean intensity when comparing Atp7b-/- and wild type, and a p-value of <0.05 by Student's T-test. Gels were then stained with Coomassie G-250 and imaged with an Odyssey Scanner (Licor, Lincoln, NE). Coomassie images were matched to CyDye images and spots of interest were cut from stained gels.
16-BAC /SDS-PAGE DIGE
To separate proteins that are not well resolved in the pH-3-10 range, we utilized an 8.5% acidic denaturing gel in the first dimension in the presence of the detergent 16-benzyldimethyl-n-hexadecyl ammonium chloride (16-BAC) and 10% Laemmli SDS-PAGE in the second dimension 38. For each gel, 50 μg of wild type and Atp7b-/- nuclei in RB (described above) were treated with 2 units of DNase 1 for 2 hours, labeled with Cy3 or Cy5 as described above, and solublized in the described acidic sample loading buffer 38. Cy3 and Cy5 images were compared to identify proteins with substantial changes in abundance. This analysis was done twice with two wt and to Atp7b-/- samples for each experiment. The gels were silver stained and bands cut for identification by mass spectrometry. Silver-stained gel slices were destained with 15 mM potassium ferricyanide and 50 mM sodium thiosulfate for 15 minutes before proceeding with the in-gel digestion procedure.
In-gel protein digestion
For in-gel digests a previously published procedure 9 was followed. Briefly, the spots of interest are excised, washed with doubly deionized water followed by an acetonitrile/NH4CO3 (50 mM) mixture. After brief incubation with neat acetonitrile the liquid is removed and the gel pieces are air dried until white in appearance. The pieces were rehydrated in 20 μl digestion buffer (50 mM NH4CO3 and 50 mM CaCl2) with trypsin (10% w/v (Princeton)) and incubated at 37ºC overnight after adding 60 μl of digestion buffer. The digestion was halted by adding 98% formic acid to a final concentration of 5% to the digest and frozen at -80 °C until further use.
Protein identification by mass spectrometry
45 μl aliquots were analyzed by liquid chromatography electrospray ionization tandem mass spectroscopy (LC-MS) using an ion trap mass spectrometer (LCQ Deca XP Plus, or LTQ, Thermo Electron Corporation, Waltham, MA). The analysis by LC-MS was performed using 180 μm reversed phase capillary columns packed in house with stable-bond packing material (C18 reversed phase material, 5-μm particle size, 100 Å pore size, ZORBAX, Agilent Technologies, Santa Clara, CA). The samples were applied to the column through a trap column (180 μm × 2.5 cm, packed in house with the same packing material) and the peptides separated using a linear gradient changing the solvent composition from 2% to 30% acetonitrile over a 30 min period with a constant flow rate of 1.8 μl/min (supplied by an Agilent Cap Pump 1100, Agilent Technologies). MS data were acquired in data dependent mode in which a single survey scan (MS) is followed by up to 4 sequential data-dependent MS/MS scans for the 4 most intense peptide ions. A dynamic exclusion feature was used to extend the analysis to less abundant ions. The peptides were identified using SEQUEST® 39, from within the Trans Proteomic Pipeline (TPP, V2.9.5 Gale) 40. Scaffold (Proteome Software Inc.) was used to validate protein identifications derived from MS/MS sequencing results using ProteinProphet 41 and PeptideProphet 42. Relative quantification of proteins was performed by the method of spectral counting 43.
Quantitative mass spectrometry of cytosolic and nuclei fractions
Nuclear and cytosolic fractions from one control and one Atp7b-/- animals were digested with trypsin and analyzed by LC MS/MS with an LTQ quadrupole linear ion trap. The combined dataset was 73,791 MS/MS spectra created from BioWorks v3.2 (Thermo) using the ZSA charge state determination algorithm. A MOUSE species subset of the current Sprot (v57.2) protein database (16,123 proteins) was prepared with concatenated REVERSED entries (and common contaminants) and searched with SEQUEST®. A parent ion mass tolerance of 2.5 Da was used with average parent ion mass and monoisotopic fragment ion masses. Cys had a static modification mass of +57 Da and Met had a variable modification mass of +16 Da. An in-house suite of programs was used to provide a Peptide Prophet-like discriminant function scoring to identify “correct” peptides and discard “incorrect” peptides using sequence-reversed matches to estimate false discovery rates. Protein identification lists were prepared using DTASelect v1.9 with post processing of results to improve spectral count accuracy and allow more strict protein identification criteria. Proteins were considered present in a given sample if they had two or more peptides with distinct sequences, having a unique count greater than or equal to one in the respective sample. For protein identification, peptides had to be consistent with fully tryptic cleavage and modified peptides were excluded.
In spectral counting studies, modified and non-fully-tryptic peptides were allowed to contribute to spectral count quantities, and samples were normalized to an identical number of “correct” matches after removing any spectra matched to contaminants. For proteins with a large numbers of shared peptides such as actin, spectral counts of peptides were assigned to the individual proteins on the basis of relative unique peptide evidence to have a more accurate total spectral count.
Scaffold Version 2.01.01 (Proteome Software) was used for protein identifications. Proteins were considered as “identified” when probability scores were ≥0.999 (protein) and ≥0.90 (peptide) with a minimum of two peptides identified. Protein probabilities were assigned by the Protein Prophet algorithm 44. Similar proteins (such as splice variants) that could not be differentiated based on MS/MS analysis alone were grouped.
Ontology analysis and pathway analysis of quantitative proteomics by 2D-DIGE
DAVID (Database for Annotation, Visualization and Integrated Discovery 45, 46) ontology and clustering analysis was used to evaluate changed proteins and identify enriched biological terms. Gene identifiers for each protein were clustered according to ontology. An enrichment score was generated for each cluster by comparing identified genes (proteins) against the mouse genome 45, 47. The enrichment score is the geometric mean of all P-values generated by EASE score: EASE score is the enrichment with a modified Fisher's Exact T-test. This compares the percent of input genes in a cluster to the percentage of those genes in the genome. For clustering analysis in the DAVID application, stringency was set to “high” and all other parameters were set at default values.
Detection of hnRNP A2/B1 splicing variants
For detection of hnRNP A2/B1 proteins by western blot and immunodetection, 5 μg of extracted nuclear protein for each sample was loaded on a 12.5% polyacrylamide gel in 62.5 mM Tris-HCl pH 6.8, 3.5 M urea 5% SDS, 10 mM DTT and trace bromophenol blue. Proteins were transferred to PVDF in 10 mM CAPS pH 11.0 and 10% methanol at 180 mA for 2 hours. The blot was blocked with 5% nonfat dry milk in PBS. Goat anti-hnRNP A2/B1 (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:5000 dilution in PBS-T and anti-goat IgG-HRP conjugate was used at 1:10,000. Detection was by chemiluminescence with Western Lightning substrate (PerkinElmer, Waltham, MA).
The splicing regime for hnRNP A2/B1 is described in 19. To detect changes in hnRNP A2/B1 mRNA splicing, RNA was isolated from small pieces of liver tissue from three of each control and Atp7b-/- 6-week-old animals using the TRIzol method followed by the RNeasy RNA isolation kit (Qiagen, Hilden, Germany). Poly(A)+ RNA was reverse transcribed using with M-MuLV reverse transcriptase and a 21-mer oligo(dT). Each qPCR reaction utilized first-strand cDNA from 0.16 μg of total RNA. Glyceraldehyde 3-phosphate dehydrogenase and the Exon 1-3 splice junction were detected by dye-quencher labeled (Taqman-style) probes. Probe-based reactions used the Perfecta Universal FastMix (Quanta Biosciences, Gaithersburg, Maryland), with 5 minutes of denaturation at 95°C, and 45 cycles of 95°C for 10 seconds and 60°C for 30 seconds. Exon 2, exon 9, and Exon 11-12 junctions were detected by SYBR green intercalating dye. SYBR green reactions utilized the SYBR Advantage SYBR green master mix (Clontech, Mountain View, California), with 5 minutes at 95°C denaturation, followed by 45 cycles of 95°C for 15 seconds, and 57°C for 60 seconds. Primers and probes used in the reactions are shown in Supplementary Table S2. Transcript abundance was determined by real-time PCR. Standard curve templates were generated by 35 cycles of PCR using a mix of wild type and Atp7b-/- cDNA as a template. PCR products were separated on a 1.5% agarose gel, extracted from the agarose gel, and quantified using the fluorescent Hoechst 33258 dye and a double-stranded DNA standard. Serial dilutions over at least three orders of magnitude were analyzed by real-time PCR to generate the standard curve. The standard curve was analyzed by linear regression to generate a standard curve equation. Molar amounts of each amplified template as calculated from the standard curve were determined. Each target was normalized to the GAPDH control amplified from the same sample.
Supplementary Material
Acknowledgements
This work was supported by the National Institute of Health grant R21 DK075659 to SL. MR and LD were supported through the Metal Ion Core funded by the National Institute of Health grant P01 GM 067166 to SL. We thank Lawrence Gray and Tony Capps for their help with collecting and analyzing liver samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buiakova OI, Xu J, Lutsenko S, Zeitlin S, Das K, Das S, Ross BM, Mekios C, Scheinberg IH, Gilliam TC. Null mutation of the murine ATP7B (Wilson disease) gene results in intracellular copper accumulation and late-onset hepatic nodular transformation. Hum. Mol. Genet. 1999;8:1665–1671. doi: 10.1093/hmg/8.9.1665. [DOI] [PubMed] [Google Scholar]
- 2.Huster D, Finegold MJ, Morgan CT, Burkhead JL, Nixon R, Vanderwerf SM, Gilliam CT, Lutsenko S. Consequences of copper accumulation in the livers of the Atp7b-/- (Wilson disease gene) knockout mice. Am. J. Pathol. 2006;168:423–434. doi: 10.2353/ajpath.2006.050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris ZL, Gitlin JD. Genetic and molecular basis for copper toxicity. Am J Clin Nutr. 1996;63:836S–841S. doi: 10.1093/ajcn/63.5.836. [DOI] [PubMed] [Google Scholar]
- 4.Waggoner DJ, Bartnikas TB, Gitlin JD. The role of copper in neurodegenerative disease. Neurobiol Dis. 1999;6:221–230. doi: 10.1006/nbdi.1999.0250. [DOI] [PubMed] [Google Scholar]
- 5.Carmichael PL, Hewer A, Osborne MR, Strain AJ, Phillips DH. Detection of bulky DNA lesions in the liver of patients with Wilson's disease and primary haemochromatosis. Mutat Res. 1995;326:235–243. doi: 10.1016/0027-5107(94)00177-7. [DOI] [PubMed] [Google Scholar]
- 6.Gu M, Cooper JM, Butler P, Walker AP, Mistry PK, Dooley JS, Schapira AH. Oxidative-phosphorylation defects in liver of patients with Wilson's disease. Lancet. 2000;356:469–474. doi: 10.1016/s0140-6736(00)02556-3. [DOI] [PubMed] [Google Scholar]
- 7.Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C, Hohler T, Galle PR, Phillips DH, Markin R, Marrogi AJ, Harris CC. Increased p53 mutation load in nontumorous human liver of wilson disease and hemochromatosis: oxyradical overload diseases. Proc. Natl. Acad. Sci. U S A. 2000;97:12770–12775. doi: 10.1073/pnas.220416097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagasaka H, Inoue I, Inui A, Komatsu H, Sogo T, Murayama K, Murakami T, Yorifuji T, Asayama K, Katayama S, Uemoto S, Kobayashi K, Takayanagi M, Fujisawa T, Tsukahara H. Relationship between oxidative stress and antioxidant systems in the liver of patients with Wilson disease: hepatic manifestation in Wilson disease as a consequence of augmented oxidative stress. Pediatr Res. 2006;60:472–477. doi: 10.1203/01.pdr.0000238341.12229.d3. [DOI] [PubMed] [Google Scholar]
- 9.Huster D, Purnat TD, Burkhead JL, Ralle M, Fiehn O, Stuckert F, Olson NE, Teupser D, Lutsenko S. High copper selectively alters lipid metabolism and cell cycle machinery in the mouse model of Wilson disease. J. Biol. Chem. 2007;282:8343–8355. doi: 10.1074/jbc.M607496200. [DOI] [PubMed] [Google Scholar]
- 10.Harrison MD, Jones CE, Dameron CT. Copper chaperones: function, structure and copper-binding properties. J Biol Inorg Chem. 1999;4:145–153. doi: 10.1007/s007750050297. [DOI] [PubMed] [Google Scholar]
- 11.Linder MC, Hazegh-Azam M. Copper biochemistry and molecular biology. Am J Clin Nutr. 1996;63:797S–811S. doi: 10.1093/ajcn/63.5.797. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Otvos JD. HPLC characterization of Ag+ and Cu+ metal exchange reactions with Zn- and Cd-metallothioneins. Biochemistry. 1996;35:13937–13945. doi: 10.1021/bi961402f. [DOI] [PubMed] [Google Scholar]
- 13.Kramer A, Ludwig Y, Shahin V, Oberleithner H. A pathway separate from the central channel through the nuclear pore complex for inorganic ions and small macromolecules. J. Biol. Chem. 2007;282:31437–31443. doi: 10.1074/jbc.M703720200. [DOI] [PubMed] [Google Scholar]
- 14.Hutchens TW, Allen MH, Li CM, Yip TT. Occupancy of a C2-C2 type ‘zinc-finger’ protein domain by copper. Direct observation by electrospray ionization mass spectrometry. FEBS Lett. 1992;309:170–174. doi: 10.1016/0014-5793(92)81088-4. [DOI] [PubMed] [Google Scholar]
- 15.Predki PF, Sarkar B. Metal replacement in “zinc finger” and its effect on DNA binding. Environ Health Perspect 102 Suppl. 1994;3:195–198. doi: 10.1289/ehp.94102s3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubbo H, Radi R. Protein and lipid nitration: role in redox signaling and injury. Biochim. Biophys. Acta. 2008;1780:1318–1324. doi: 10.1016/j.bbagen.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Novoselov SV, Kryukov GV, Xu XM, Carlson BA, Hatfield DL, Gladyshev VN. Selenoprotein H is a nucleolar thioredoxin-like protein with a unique expression pattern. J. Biol. Chem. 2007;282:11960–11968. doi: 10.1074/jbc.M701605200. [DOI] [PubMed] [Google Scholar]
- 18.Raju CS, Goritz C, Nord Y, Hermanson O, Lopez-Iglesias C, Visa N, Castelo-Branco G, Percipalle P. In cultured oligodendrocytes the A/B-type hnRNP CBF-A accompanies MBP mRNA bound to mRNA trafficking sequences. Mol. Biol. Cell. 2008;19:3008–3019. doi: 10.1091/mbc.E07-10-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatfield JT, Rothnagel JA, Smith R. Characterization of the mouse hnRNP A2/B1/B0 gene and identification of processed pseudogenes. Gene. 2002;295:33–42. doi: 10.1016/s0378-1119(02)00800-4. [DOI] [PubMed] [Google Scholar]
- 20.Stoytcheva ZR, Vladimirov V, Douet V, Stoychev I, Berry MJ. Metal transcription factor-1 regulation via MREs in the transcribed regions of selenoprotein H and other metal-responsive genes. Biochim. Biophys. Acta. 2010;1800:416–424. doi: 10.1016/j.bbagen.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X, Ma Q. Induction of metallothionein I by arsenic via metal-activated transcription factor 1: critical role of C-terminal cysteine residues in arsenic sensing. J. Biol. Chem. 2009;284:12609–12621. doi: 10.1074/jbc.M901204200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khacho M, Mekhail K, Pilon-Larose K, Pause A, Cote J, Lee S. eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell. 2008;19:5296–5308. doi: 10.1091/mbc.E08-06-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munro TP, Magee RJ, Kidd GJ, Carson JH, Barbarese E, Smith LM, Smith R. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 1999;274:34389–34395. doi: 10.1074/jbc.274.48.34389. [DOI] [PubMed] [Google Scholar]
- 24.Matter N, Marx M, Weg-Remers S, Ponta H, Herrlich P, Konig H. Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J. Biol. Chem. 2000;275:35353–35360. doi: 10.1074/jbc.M004692200. [DOI] [PubMed] [Google Scholar]
- 25.Rooke N, Markovtsov V, Cagavi E, Black DL. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol. Cell. Biol. 2003;23:1874–1884. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu D, Xu G, Ghandhi S, Hubbard K. Modulation of the expression of p16INK4a and p14ARF by hnRNP A1 and A2 RNA binding proteins: implications for cellular senescence. J. Cell. Physiol. 2002;193:19–25. doi: 10.1002/jcp.10147. [DOI] [PubMed] [Google Scholar]
- 27.Hiraki A, Murakami T, Aoe K, Sueoka E, Sueoka N, Taguchi K, Kamei T, Sugi K, Ueoka H, Kishimoto T. Heterogeneous nuclear ribonucleoprotein B1 expression in malignant mesothelioma. Cancer Sci. 2006;97:1175–1181. doi: 10.1111/j.1349-7006.2006.00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsimpoula S, Patrinou-Georgoula M, Makrilia N, Dimakou K, Guialis A, Orfanidou D, Syrigos KN. Overexpression of hnRNPA2/B1 in bronchoscopic specimens: a potential early detection marker in lung cancer. Anticancer Res. 2009;29:1373–1382. [PubMed] [Google Scholar]
- 29.Peebles KA, Dwyer-Nield LD, Malkinson AM. Altered expression of splicing factor, heterogeneous nuclear ribonucleoprotein A2/B1, in mouse lung neoplasia. Mol Carcinog. 2007;46:887–900. doi: 10.1002/mc.20321. [DOI] [PubMed] [Google Scholar]
- 30.Satoh H, Kamma H, Ishikawa H, Horiguchi H, Fujiwara M, Yamashita YT, Ohtsuka M, Sekizawa K. Expression of hnRNP A2/B1 proteins in human cancer cell lines. Int J Oncol. 2000;16:763–767. doi: 10.3892/ijo.16.4.763. [DOI] [PubMed] [Google Scholar]
- 31.Tauler J, Mulshine JL. Lung cancer and inflammation: interaction of chemokines and hnRNPs. Curr Opin Pharmacol. 2009;9:384–388. doi: 10.1016/j.coph.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Wu S, Sato M, Endo C, Sakurada A, Dong B, Aikawa H, Chen Y, Okada Y, Matsumura Y, Sueoka E, Kondo T. hnRNP B1 protein may be a possible prognostic factor in squamous cell carcinoma of the lung. Lung Cancer. 2003;41:179–186. doi: 10.1016/s0169-5002(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 33.Yan-Sanders Y, Hammons GJ, Lyn-Cook BD. Increased expression of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP) in pancreatic tissue from smokers and pancreatic tumor cells. Cancer Lett. 2002;183:215–220. doi: 10.1016/s0304-3835(02)00168-4. [DOI] [PubMed] [Google Scholar]
- 34.Guillemin I, Becker M, Ociepka K, Friauf E, Nothwang HG. A subcellular prefractionation protocol for minute amounts of mammalian cell cultures and tissue. Proteomics. 2005;5:35–45. doi: 10.1002/pmic.200400892. [DOI] [PubMed] [Google Scholar]
- 35.Yun W, Lai B, Cai Z, Maser J, Legnini D, Gluskin E, Chen Z, Krasnoperova AA, Vladimirsky Y, Cerrina F. Nanometer focusing of hard x rays by phase zone plates. Review of Scientific Instruments. 1999;70:2238. [Google Scholar]
- 36.Van Espen P. Van Grieken R, Markowicz A, editors. Spectrum Evaluation. Handbook of X-ray Spectrometry: Revised and Expanded. 2002.
- 37.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 38.Hartinger J, Stenius K, Hogemann D, Jahn R. 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal. Biochem. 1996;240:126–133. doi: 10.1006/abio.1996.0339. [DOI] [PubMed] [Google Scholar]
- 39.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 40.Keller A, Eng J, Zhang N, Li X, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Molecular systems biology. 2005;1 doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. ANALYTICAL CHEMISTRY-WASHINGTON DC- 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 42.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 43.Carvalho PC, Hewel J, Barbosa VC, Yates JR., III Identifying differences in protein expression levels by spectral counting and feature selection. Genetics and molecular research: GMR. 2008;7:342. doi: 10.4238/vol7-2gmr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 45.Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 46.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Dennis GJ, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.