Abstract
Mutations in ELA2, encoding the human serine protease neutrophil elastase, cause cyclic and severe congenital neutropenia, and recent evidence indicates that the mutations alter the membrane trafficking of neutrophil elastase. These disorders feature impaired bone marrow production of neutrophils along with excess monocytes—terminally differentiated lineages corresponding to the two alternative fates of myeloid progenitor cells. We utilized a modified yeast two-hybrid system and identified a new, widely expressed gene, N2N, whose product is homologous to Notch2, that interacts with neutrophil elastase. N2N is a 36-kDa protein distributed throughout the cell and secreted. Its amino-terminal sequence consists of several EGF repeats identical to those of the extracellular region of Notch2, and its carboxyl terminus contains a unique 24-residue domain required for interaction with neutrophil elastase. Neutrophil elastase cleaves N2N within EGF repeats in vitro and in living cells, but the C-terminal domain retards proteolysis. In vitro, N2N represses transcriptional activities of Notch proteins. Disease-causing mutations of neutrophil elastase disrupt the interaction with N2N, impair proteolysis of N2N and Notch2, and interfere with Notch2 signaling, suggesting defective proteolysis of an inhibitory form of Notch as an explanation for the alternate switching of cell fates characteristic of hereditary neutropenia.
Neutrophils are phagocytic white blood cells that serve as a first line of defense against bacterial and fungal infection and coordinate inflammatory responses. The surprising finding that mutation of the gene ELA2, encoding neutrophil elastase (NE), is responsible for inherited forms of human neutropenia indicates that this serine protease, primarily found in granules of neutrophils and monocytes, plays an unexpected role in the regulation of hematopoietic stem cell differentiation. The two major forms of human hereditary neutropenia are cyclic neutropenia (CN), characterized by 21-day sinusoidal oscillations in neutrophil production, and severe congenital neutropenia (SCN), comprised of arrested promyelocytic differentiation frequently progressing to myelodysplasia and acute myelogenous leukemia. CN is always (22), and SCN usually (2, 12), the result of autosomal dominant, heterozygous ELA2 mutations.
To date, a total of more than two dozen mutant NE proteins have been reported (21). However, the role of the mutations in the pathophysiology of these disorders and NE's normal function in myeloid differentiation remain unclear. The mutations have variable effects on overall catalytic activity and are not cytotoxic (28). The recent finding that the mutations do affect the subcellular localization of NE (6), and, in particular, their membrane trafficking, suggests the possibility of defective interactions between mutant NE and membrane proteins.
Additionally, two clinical clues suggest how NE might contribute to myelopoiesis. First, it appears that neutropenia results from an aberrant switch in cell fate. A striking feature of CN is that neutrophils and monocytes reciprocally cycle. Thus, when neutrophil production is low, monocyte production is high, and vice versa. And in SCN, neutropenia is accompanied by marked elevation in the number of monocytes. Neutrophils and monocytes represent the two differentiation choices of myeloid progenitor cells. Second, the mutations inhibit granulopoiesis in a cell-intrinsic manner, as demonstrated by a case of germ line mosaicism (3), where the normal father of an SCN patient has bone marrow 50% chimeric for his daughter's ELA2 heterozygous mutation. Although approximately half of his lymphocytes—a lineage unaffected by the disease—contain the mutation, it is nearly absent in his peripheral blood neutrophils, revealing that the mutant NE does not have paracrine effects.
Here we hypothesize that the mutations affect the interaction with key substrates required to execute myeloid differentiation programs. To identify potential substrates, we adopted a recently developed yeast two-hybrid system based on cytoplasmic interactions mediated via the Ras signaling pathway. We screened a human hematopoietic cDNA library and identified a novel member of the cell fate-determining, cell surface receptor Notch family that both specifically interacts with, and serves as a substrate for, NE.
MATERIALS AND METHODS
Cell culture.
KG-1, U937, HL-60, and HeLa cell lines were obtained from American Type Culture Collection. KG-1 and HL-60 cells were maintained and cultured in Iscov's modified Dulbecco's medium containing 20% fetal bovine serum. U937 cells were maintained in RPMI 1640 medium containing 10% fetal calf serum. HeLa cells were maintained in Dulbecco's modified Eagle medium containing 10% fetal calf serum. Cells were grown in a humidified incubator at 37°C with 5% CO2. All media were purchased from Invitrogen and were supplemented with 1% antibiotic-antimycotic solution.
Vectors.
The NE wild-type and various mutant cDNAs were previously cloned into the pCS2+ vector (28). The mRAS bait plasmid (41) was a gift from K. Takemaru (University of Washington). To construct the bait plasmids shown in Fig. 1A, the NE cDNAs were PCR amplified from the pCS2+ plasmids with appropriate primers. All of the constructs were confirmed by DNA sequencing analysis. Myc-NE constructs were generated by amplifying the NE cDNAs from the pCS2+ plasmids with appropriate primers and subsequently cloning into the pCS2+Myc vector. I.M.A.G.E. clone 3623163 was purchased from American Type Culture Collection. Myc-N2N, FLAG-N2N, and glutathione S-transferase (GST)-N2N derivatives were generated by amplifying N2N cDNAs from the I.M.A.G.E. clone with appropriate primers and subsequently subcloning into the pCS2+Myc, pCS2+FLAG, and pGEX4T vectors, respectively. The pMyr vector was from Stratagene. RNA interference (RNAi) constructs were generated based on the pSilencer 1.0-U6 vector (Ambion). Notch1 and Notch2 intracellular domain constructs (39) were provided by D. Small (Maine Medical Center Research Institute). The pGL2-Hes1 luciferase construct (40) was a gift of D. Solecki (Rockefeller University). All constructs were confirmed by DNA sequencing analysis.
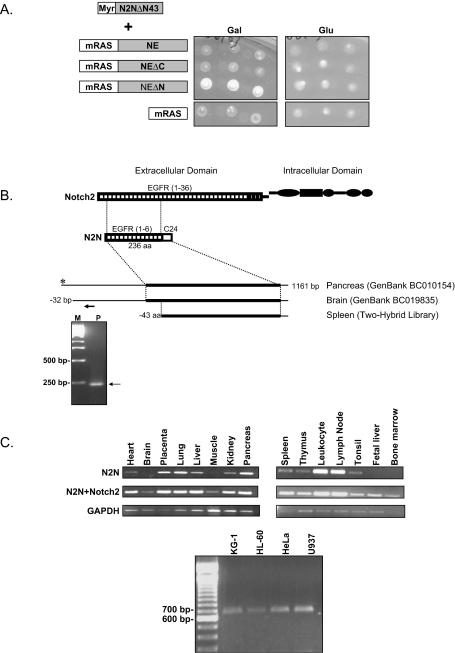
FIG.1.
(A) Specific interaction between N2N and NEΔN in the yeast CytoTrap two-hybrid system. Temperature-sensitive cdc25H yeast cells were cotransformed with the indicated constructs, and the Myr-N2NΔN43 plasmid was isolated in the screening and incubated on minimal galactose or glucose plates at either 25°C (not shown) or 37°C. Three independent clones are shown for each interaction. (B) Cloning of N2N cDNA. Representative results of 5′-RACE (+225 primer) are shown in the lower inset. The alignment of the N2N clone recovered from the screening with the two GenBank clones and the deduced N2N protein sequence and Notch2 are shown in upper inset. The thick lines indicate the open reading frame of N2N. The arrow indicates the position of the primer used in 5′-RACE. (C) Tissue distribution of N2N mRNA revealed by RT-PCR using primers specific for N2N, amplifying both Notch2 and N2N or GAPDH control (upper panels) and myeloid cell line expression with N2N-specific primers (lower panel).
Yeast two-hybrid screening.
Human spleen cDNA library and the CytoTrap yeast two-hybrid system were purchased from Stratagene and used according to manufacturer's instructions. Briefly, cdc25H cells were cotransformed with the pooled bait plasmids and the cDNA library (resulting in ∼104 transformants/plate). The expression of library cDNA was controlled by a galactose-inducible promoter. Transformants were grown on selectable minimal glucose plates for 4 to 6 days at 25°C. Subsequently, colonies were replica plated onto minimal galactose plates and incubated at 37°C for 5 to 7 days. Positive colonies exhibiting efficient growth on galactose plates at 37°C were isolated and tested for galactose-dependent growth at 37°C. Library plasmids were recovered and analyzed by DNA sequencing. The specificity of interaction was tested by retransformation of cdc25H cells.
5′-RACE.
5′ rapid amplification of cDNA ends (5′-RACE) was performed either by using the GeneRacer kit (Invitrogen) with total RNA isolated from HL-60, U937, and KG-1 cells or by using RACE-ready human spleen cDNA (Ambion). The gene-specific primers used in these assays are as follows: N2N(+225), 5′GCCGCCGCCTGGGCAGATCCACATG3′; N2N(+468), 5′GGTTCTTCTCACAGGGGTCTCGAT3′. The RACE products were identified by DNA sequencing analysis.
Western blotting.
Western blots were performed with whole-cell extracts using the ECL Western blot analysis system (Amersham) with antibodies Notch2(25-255) (sc-5545, lot no. C010, 1:500 dilution; Santa Cruz Biotechnology), Myc(9E10) (1:1,000 dilution; Santa Cruz Biotechnology), or FLAG(M2) (1:10,000 dilution; Sigma).
RT-PCR analysis.
Total RNA was prepared using the Absolutely RNA reverse transcription (RT)-PCR miniprep kit (Stratagene). A total of 1 μg of total RNA was used to produce cDNAs with oligo(dT)12 primer by superscript III RNA polymerase (Invitrogen). Human multiple-tissue cDNA panels were purchased from Clontech. Primers were designed based on the cDNA sequences corresponding to each of the genes analyzed by using Primer3 software (Whitehead Institute). The primer sequences are as follows: GAPDH forward, 5′TGAAGGTCGGAGTCAACGGATTTGGT3′; GAPDH reverse, 5′CATGTGGGCCATGAGGTCCACCAC3′; Notch2+N2N forward, 5′CAACATCGAGACCCCTGTGAG3′; Notch2+N2N reverse, 5′GTCCTGGAATGTCACACTCATTG3′; N2N forward, TGTCAACATCGAGACCCCTGTGAG; N2N reverse, 5′CCAGTGTCTAATTCTCATCGTGTT3′. For N2N and Notch2, a total of 30 cycles of PCR were used. For GAPDH, 24 cycles of PCR were used. PCR conditions were determined previously to be in the linear range of amplification. The RT-PCR products were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining.
RNAi assay.
The RNAi plasmids were transfected into HeLa cells using Lipofectamine Plus reagent (Invitrogen) as directed by the manufacturer. The structure of the hairpin RNAs is shown in Fig. 2B. Cells were harvested 65 h posttransfection. Whole-cell lysates and total RNAs were prepared and submitted to Western blot analysis and RT-PCR assays, respectively. Multiple independent experiments were carried out to confirm results.
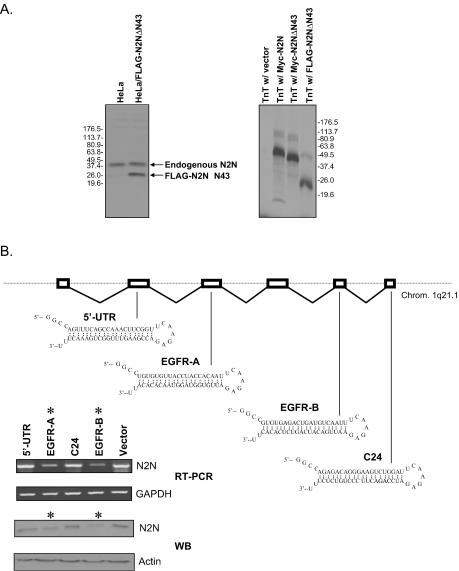
FIG. 2.
Identification of N2N protein product. (A) The Notch2(25-255) antibody recognizes a 36-kDa protein and the shorter, transfected N2N fragment originally recovered in the two-hybrid assay in HeLa cells (left) as well as in vitro-translated (TnT) N2N fragments (right). (B) The expression level of the 36-kDa protein recognized by the Notch2(25-255) antibody is reduced and correlates with the reduction of N2N mRNA in RNAi assays. The chromosomal structure of N2N is indicated. The location and structure of the four hairpins used for RNAi are also shown. The stars indicate the two effective siRNA species.
Immunofluorescent staining and photomicroscopy.
Cells were fixed and stained as described previously (28) using a 1:1,000 dilution of Myc (9E10) antibody (Santa Cruz Biotechnology) and a 1:400 dilution of rhodamine-conjugated anti-mouse serum (Jackson Immunoresearch Laboratories). Nuclei were counterstained for 1 min in 0.5 μg of 4′,6′-diamidino-2-phenylindole/ml. Photomicroscopy utilized a Leica TCS SP/MP confocal microscope with a ×40 planapochromatic objective.
GST pull-down assay.
35S-labeled Myc epitope-tagged NE proteins were in vitro transcribed and translated using the TnT SP6 Quick Coupled transcription/translation system (Promega). GST fusion proteins were expressed in Escherichia coli strain BL21(DE3) under induction by 0.5 mM isopropyl-β-d-thiogalactopyranoside and purified using glutathione-Sepharose 4B beads according to the supplier's instructions (Amersham). About 2 μg of GST or GST-N2N fusion protein was incubated with 1/10 of the in vitro-translated 35S-labeled Myc-tagged NE proteins at 4°C overnight in 1 ml of RIPA buffer (1× phosphate-buffered saline [PBS], 1% Nonidet P40, 0.5% sodium deoxycholate) containing 100 μg of bovine serum albumin. After incubation, 20 μl of glutathione-Sepharose 4B beads was added and incubated for another hour at room temperature. The beads were collected and washed four times with RIPA buffer, and the bound proteins were eluted for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
Coimmunoprecipitation assay.
Forty hours after transient transfection, HeLa cells were harvested and lysed in 1.5 ml of ice-cold RIPA buffer. Cell lysates were cleared by centrifugation at 15,000 × g for 30 min at 4°C. For each assay, 200 μl of the above cell lysates were incubated with 0.4 μg of primary antibody, 100 μg of bovine serum albumin, and 14 μl of protein A or G-Sepharose beads (Jackson Immunoresearch) in 1.4 ml of RIPA buffer at 4°C overnight. Immunoprecipitates were collected and washed four times with 1.5 ml of PBS and then resolved by SDS-PAGE and detected by Western blotting.
NE cleavage assay in vitro.
Proteolysis of GST-N2N fusion proteins was carried out as described previously (36). Briefly, 0.5 to 1 μg of purified GST fusion protein was incubated with purified NE (Calbiochem) at the indicated concentrations in digestion buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 5 mM CaCl2) at 37°C for 30 min. Samples were then resolved by SDS-PAGE and stained with Coomassie blue.
Living cell cleavage by NE.
Living cell treatment by purified NE (18) was performed with modifications. U937, HL-60, or KG-1 cells were washed three times with PBS and resuspended in serum-free RPMI medium (2 × 107 cells/ml). Six hundred microliters of cells were then treated by purified NE at various concentrations with the presence or absence of the specific inhibitor MeOsuc-AAPV-chloromethylketone (Fluka) in 1 ml of digestion buffer at 37°C for 1 h. Cells were then harvested and lysed with 1% Triton X-100 in the presence of protease inhibitor cocktail (Roche). Whole-cell extracts were resolved on SDS-PAGE and analyzed by Western blotting. For radioimmunoprecipitation assays, the cells were first labeled with [35S]methionine for 24 h before being subjected to NE treatment. The whole-cell extracts were immunoprecipitated with the Notch2(25-255) antibody or control rabbit immunoglobulin G. The immunoprecipitates were collected and washed four times with 1.5 ml of PBS and then resolved by SDS-PAGE.
Luciferase reporter assay.
HeLa cells were transiently transfected using Lipofectamine Plus reagent (Invitrogen) as suggested by the manufacturer. Cell lysates were harvested 40 h posttransfection. Luciferase activity assays were performed using the Dual-luciferase reporter assay system (Promega). The results represent the means for multiple independent experiments.
Expression and purification of recombinant NE.
The vector pPIC9WT (13), containing a codon-optimized human NE cDNA fused to a Matα gene leader, capable of directing appropriate prepro-amino-terminal maturation and secretion in Pichia pastoris via Kex 2 cleavage at I16 of NE, was the gift of P. Carter (Genentech). Addition of a His6 affinity tag and deletion of the carboxyl terminus and/or introduction of human neutropenia mutations employed the QuikChange site-directed mutagenesis system (Stratagene). The vectors were introduced into the KM71 strain of P. pastoris as Muts transformants and selected for multiple integrants by growth in high concentrations (3.0 mg/ml) of G418. Culture conditions utilized reagents and methods provided in the Pichia expression kit (Invitrogen). His-tagged proteins were purified from the supernatant by chromatography on Superflow NiNTA resin (Qiagen), followed by concentration with 10,000-kDa cutoff centrifugal filter devices (Amicon), a second His affinity purification with magnetic agarose beads covalently cross-linked to nickel-nitrilotriacetic acid (Qiagen), and repeat centrifugal filtration into storage buffer (50 mM NaAc [pH 5.5], 200 mM NaCl) with storage at −80°C prior to use.
RESULTS
Cloning of N2N cDNA.
To identify proteins that interact with NE, we performed a yeast two-hybrid screen. Since NE is a secreted glycoprotein undergoing extensive subcellular trafficking and compartmentalization that is disrupted by neutropenic mutations (6), we adopted a new yeast two-hybrid technique relying on cytoplasmic protein-protein interaction instead of the conventional assay based on nuclear interactions. (In fact, conventional two-hybrid constructs using intact NE fused to Gal4 activation or DNA binding domains prove toxic to yeast; data not shown.) This system has been productively applied to dissect components of cell signaling pathways (4, 5, 20, 24, 41). The method exploits a temperature-sensitive mutation in the yeast cdc25 gene and the ability of mammalian ras genes to rescue the growth defect. The mutant cells cannot grow at the nonpermissive temperature (37°C) due to the lack of a functional Cdc25 guanyl nucleotide exchange factor. The bait protein is fused with a Ras domain that can only be activated when it is recruited to the plasma membrane. Target library cDNAs are tagged with a myristylation signal sequence. If bait and target interact, then the Ras domain is recruited to the plasma membrane to complement the cdc25 mutation.
NE undergoes posttranslational proteolytic processing of both the amino and carboxyl termini and contains multiple disulfide bonds. Because it is important to reproduce the correctly folded structure of the protein in yeast cytosol, we constructed several different mRAS-NE fusion proteins. We pooled these constructs as bait to screen ∼106 colonies from a library of poly(A)+ human spleen cDNA. Several different clones were found to interact with mRAS-NE fusions, as confirmed by GST pull-down assay and coimmunoprecipitation (see below). However, for only one interacting clone, encoding a carboxyl-terminal fragment of a novel protein resembling Notch2 that we term N2N, did we find that neutropenia-causing mutations of NE disrupted the interaction. We therefore focus on N2N as a good candidate for contributing to the pathogenesis of hereditary neutropenia.
To confirm the interaction, this plasmid was isolated and retransformed into the same yeast strain with either one of the bait constructs or a negative control plasmid. As shown in Fig. 1A, only cdc25H cells coexpressing the N2N fragment and the mRAS-NEΔN fusion protein, in which the amino terminus is processed and the carboxyl terminus remains intact, grow at 37°C, although all of the cells grow at 25°C (data not shown), suggesting that the interaction is specific.
The N2N fragment recovered from the yeast two-hybrid screen matches two GenBank cDNA clones (Fig. 1B). One is from pancreas, and the other is from brain. All three have the same 3′ sequence terminated with poly(A) tails but different 5′ ends. To identify the full-length mRNA of N2N, we performed 5′-RACE and primer extension assays with a variety of cell lines and tissues. A representative 5′-RACE result is shown in Fig. 1B. DNA sequence analysis of the 5′-RACE products indicates that the cDNA from pancreas (BC010154) represents the full-length mRNA of N2N. The deduced open reading frame of N2N encodes a 236-residue protein (I.M.A.G.E. Consortium predicted protein AAH19835) whose first 211 amino acids are identical to part of EGF repeat 1, all of EGF repeats 2 to 5, and a portion of EGF repeat 6 from the extracellular domain of Notch2 (Fig. 1B), a cell surface receptor involved in cell fate specification. The carboxyl terminus of N2N consists of a unique 24-amino-acid sequence (C24) that is not present in Notch2 or elsewhere among other genes. A search of genomic sequence indicates that N2N is a six-exon gene located on chromosome 1q21.1, not overlapping with Notch2. The fragment recovered from the yeast two-hybrid screen lacks the amino-terminal 43 residues.
RT-PCR (Fig. 1C) reveals that N2N is widely expressed in different tissues, with the highest levels in leukocytes and lymph nodes. N2N is also expressed in HeLa cells as well as human hematopoietic cell lines (KG-1, HL-60, and U937), corresponding to myeloid developmental stages during which ELA2 is also expressed.
Identification of the N2N gene product.
N2N transcripts are spliced and polyadenylated; moreover, its exon sequences are nearly completely identical to those of Notch2 while the introns shows greater divergence. Thus, N2N is unlikely to represent a pseudogene. Nevertheless, exclusion of this possibility requires identification of its protein product. In the absence of posttranslational modification, N2N is predicted to encode a 26-kDa protein. EGF repeats characteristic of the extracellular domains of Notch proteins are extensively glycosylated, so the actual molecular weight is probably greater.
Since N2N is identical to Notch2 at its N terminus, we reasoned that polyclonal antibodies specific for the Notch2 N terminus should also recognize N2N. In fact, a commercially available Notch2 polyclonal antibody, Notch2(25-255), does detect a 36-kDa protein in Western blots of lysates from a broad range of cell lines (Fig. 2A, left panel, first lane illustrating HeLa cell lysates.) Given that Notch2 expression is not detectable by Western blotting in HeLa cells, the 36-kDa protein is unlikely to represent a Notch2 degradation product. Furthermore, this antibody identifies both transfected and in vitro-translated N2N fragments in Western blots (Fig. 2A). It is therefore likely that the 36-kDa protein revealed by the Notch2(25-255) antibody is the specific product of N2N. As confirmation, we performed RNAi assays to test whether there is a correlation between levels of the endogenous N2N transcript and the 36-kDa protein. We designed four siRNAs and transfected them into HeLa cells. As shown in Fig. 2B, two of the siRNAs, derived from regions encoding EGF repeats, successfully diminish levels of endogenous N2N mRNA and correspondingly reduce the abundance of the 36-kDa protein recognized by the Notch2(25-255) antibody. Conversely, hsiRNAs derived from unique Notch2 domains that are not present in N2N reduce Notch2 transcript levels but have no effect on the abundance of N2N mRNA or the 36-kDa protein (not shown). These results substantiate this protein as the N2N gene product.
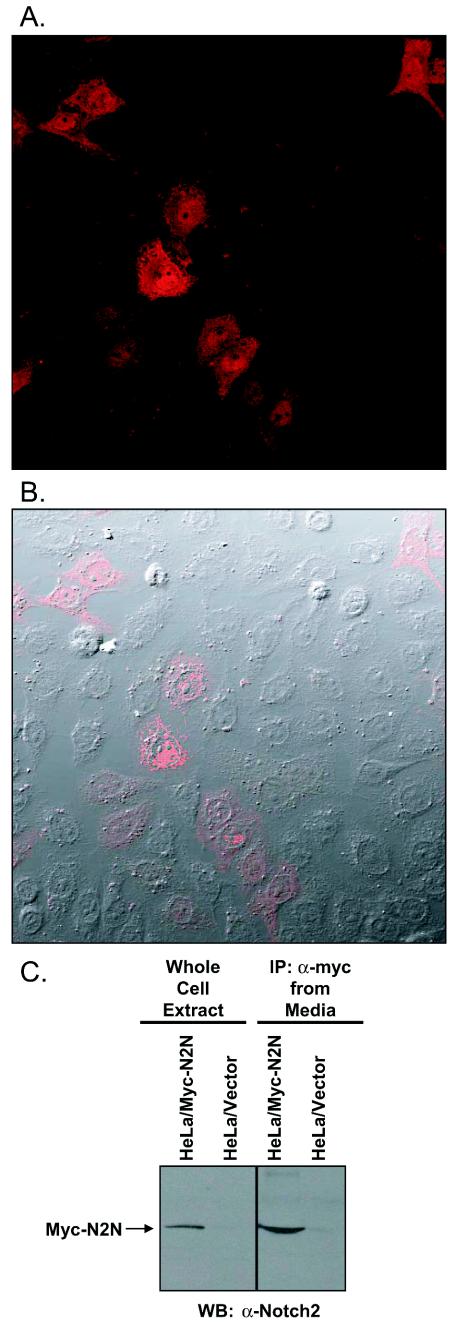
To determine the subcellular localization of N2N, we transiently transfected HeLa cells with N-terminal Myc epitope-tagged constructs. As confocal microscopy of indirect immunofluorescent detection reveals in Fig. 3A and B, N2N is distributed throughout the cell. Controls consisting of detection with nonimmune serum or transfection of the Myc epitope vector by itself produce no signal (not shown). Figure 3C shows that N2N was immunoprecipitated with the Myc antibody from the medium of transfected HeLa cells and detected by Western blotting with the Notch2(25-255) antibody; thus, it is also secreted.
FIG.3.
Subcellular localization of N2N. (A) Indirect immunofluorescence of Myc-tagged N2N transiently transfected in HeLa cells and detected with anti-Myc and rhodamine-conjugated secondary antibody by confocal microscopy. (B) Image superimposed on differential interference contrast transmission photomicrograph. (C) Immunoprecipitation of N2N from culture media of HeLa cells transfected with N2N. Thirteen milliliters of media or whole-cell extracts from 10-cm-diameter plates of HeLa cells transiently transfected 40 h earlier with either Myc-tagged N2N or control expression vectors were immunoprecipitated with anti-Myc and then Western blotted with Notch2(25-255) antibody.
Biochemical interaction between N2N and NE.
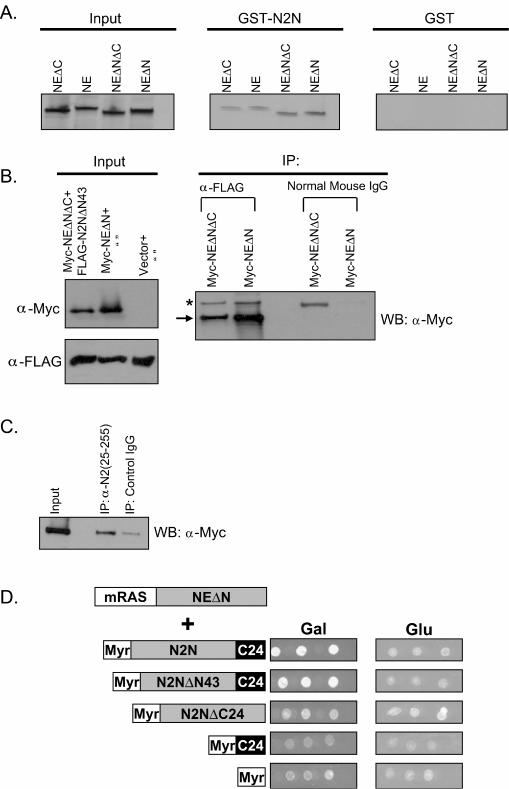
To confirm the in vitro interaction between NE and N2N, we performed GST-pull down assays. Bacterially expressed GST-N2N fusion protein was mixed with 35S-labeled in vitro synthesized NE. The GST fusion protein and interacting proteins were then precipitated with glutathione-conjugated beads, separated by SDS-PAGE, and detected with autoradiography. As shown in Fig. 4A, only GST-fused N2N, but not GST alone, pulls down the radiolabeled in vitro-translated NE, confirming the yeast two-hybrid interaction.
FIG. 4.
Confirmation of the interaction between N2N and NE. (A) N2N directly binds to NE in GST pull-down assays. (B) Cotransfected N2N and NE form an immunoprecipitable complex in HeLa cells. The arrow indicates the NE bands, and the star indicates the immunoglobulin G bands. IP, immunoprecipitation; WB, Western blot; α-Myc, anti-Myc antibody. (C) Endogenous N2N coprecipitates with transfected NE from HeLa cells. (D) The unique C24 domain of N2N is required but not sufficient for interaction with NE in the yeast CytoTrap two-hybrid assay. Three independent clones are shown for each interaction.
NE normally undergoes three proteolytic cleavage events during its maturation (as reviewed in reference 28). First, the 27-residue signal peptide is removed from the preproenzyme. Second, in a step corresponding to zymogen activation, dipeptidyl peptidase I, also known as cathepsin C, cleaves two remaining amino acids from the proenzyme, producing a mature protein containing an N-terminal isoleucine. Third, a 20-residue C-terminal extension is cleaved. Sequential processing of the amino terminus is required for enzymatic activity, but processing of the C terminus has no known influence on catalytic activity and is not required for granule localization, and its function has remained undetermined. In the yeast two-hybrid system, N2N interacts only with the mRAS-NEΔN fusion protein (Fig. 1A), containing a processed amino terminus but an intact carboxyl terminus. Correspondingly, the interaction between N2N and NE in the GST-pull down assay appears stronger with either removal of the amino terminal prepro leader or retention of the C-terminal extension (Fig. 4A), demonstrating the most robust result with NEΔN, which both lacks the prepro leader and retains the C-terminal extension.
To determine if N2N and NE interact in vivo, we performed two different experiments. First, we cotransfected Myc epitope-tagged NE and FLAG epitope-tagged N2N into HeLa cells. Whole-cell extracts were immunoprecipitated with anti-FLAG antibody or control antibody and then subjected to Western blotting with anti-Myc antibody. As shown in Fig. 4B, FLAG-tagged N2N coimmunoprecipitates with Myc-tagged NEΔN and, to a lesser extent, NEΔNΔC, corresponding to processing at both termini. As shown above, HeLa cells express endogenous N2N. Therefore, in the second coimmunoprecipitation experiment, we transfected just Myc-tagged NEΔN into HeLa cells and then performed immunoprecipitation with the Notch2(25-255) antibody. As shown in Fig. 4C, endogenous N2N coimmunoprecipitates with Myc-tagged NEΔN.
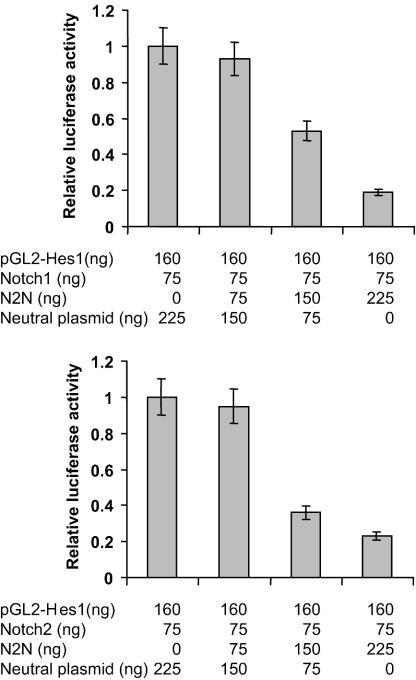
We then employed the yeast two-hybrid assay to map the region of N2N required for interaction with NE. As shown in Fig. 4D, the unique C24 domain of N2N is required, but alone is not sufficient, for interaction with NE. Taken together, these results indicate that the interaction is chiefly mediated by the C24 domain of N2N and the carboxyl terminus of NE.
Cleavage of N2N by NE.
Since NE is a protease, it is possible that N2N is either a substrate for, or an inhibitor of, NE. To discriminate among these possibilities, we first determined if NE cleaves N2N in vitro. NE sensitively cleaves either GST-fused full-length N2N or the shorter fragment recovered in the two-hybrid screen (N2NΔN43), while GST alone is resistant (Fig. 5A). NE therefore cleaves N2N within the EGF repeats. Interestingly, a construct comprised of GST fused with the C24 domain by itself appears resistant to proteolysis, whereas deletion of the C24 domain from N2N renders it more susceptible to cleavage (Fig. 5A). The C24 domain may therefore confer some degree of resistance to proteolysis by NE.
FIG. 5.
Cleavage of N2N by NE. (A) Recombinant GST-N2N fusion proteins cleaved by NE in vitro, Coomassie-stained SDS-polyacrylamide gel. Note that GST itself and the unique C24 domain of N2N are not cleaved by NE. (B) Representative Western blot result of endogenous Notch2 and N2N digested by NE in U937 cells when NE is added to the culture medium. (C) A representative autoradiogram of immunoprecipitation of endogenous Notch2 and N2N digested by NE in 35S-labeled U937 cells when NE is added to the culture medium.
We next tested whether NE can cleave endogenous N2N in living U937, HL-60, and KG-1 myeloid cells. In Fig. 5B, NE was added to the cell culture medium and N2N was quantified by Western blotting with the Notch2(25-255) antibody. In Fig. 5C, cells were first metabolically labeled with [35S]methionine before addition of NE, and N2N was immunoprecipitated with the Notch2(25-255) antibody and then imaged by SDS-PAGE with autoradiography. Both results show that low concentrations of NE cleave endogenous N2N in living cells.
Because NE cleaves N2N within EGF repeats that are identical to those of Notch2, it is possible that NE can also cleave Notch2. Actually, as shown in Fig. 5B and C, endogenous Notch2 appears to be even more vulnerable than N2N to NE-mediated proteolysis, possibly because of Notch2's localization on the cell surface, which conceivably provides easier access for NE added to the cell culture medium.
Relationship between N2N and other Notch family proteins.
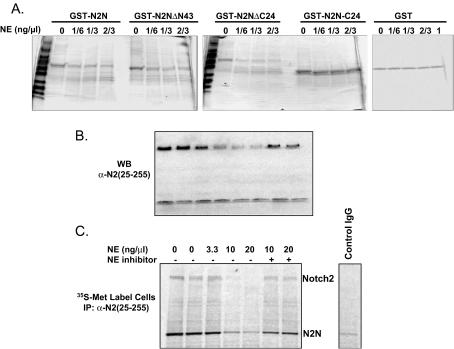
Notch2 is a member of the ligand-activated Notch family of cell surface receptors that function in determination of cell fate. Notch2 has an extracellular domain containing 36 EGF repeats. Upon activation, it undergoes multiple proteolytic cleavage events and releases an intracellular domain, which translocates to the nucleus where it functions as a transcription factor. Since N2N contains sequences identical to EGF repeats 1 to 6 of Notch2, there may be a functional relationship between N2N and other Notch proteins. To determine if expression of N2N influences Notch signaling, we expressed N2N in a transient-transfection assay of HeLa cells employing the promoter of Hes1 as a target of Notch transcriptional activation. Coexpressed N2N represses the transcriptional activities of Notch2 intracellular domain, as well as the Notch1 intracellular domain, in a dose-dependent manner (Fig. 6), indicating that there is a functional relationship between N2N and Notch family proteins with respect to signal transduction pathways.
FIG. 6.
Coexpressed N2N inhibits transactivation activity of Notch2 and Notch1 intracellular domains in a transient reporter system in HeLa cells with a hes1 luciferase reporter. The neutral plasmid, used to keep total plasmid quantity constant, expresses lacZ in the same vector used for Notch constructs.
Disruption of N2N interaction by NE mutations.
Mutation of neutrophil elastase is the primary cause of human hereditary neutropenia. The above-described experiments demonstrate that retention of the C-terminal extension of NE strongly enhances interaction with N2N. Disease-causing mutations are distributed throughout the length of NE, but the C terminus serves as a hot spot for mutation localization. Intriguingly, mutations truncating the protein just prior to the C-terminal extension of NE are the most common cause of SCN. To determine if mutations elsewhere in the molecule also affect its interaction with N2N, we tested several other disease-causing amino acid missense substitutions and deletions (Fig. 7). Immunoprecipitation experiments were performed in which epitope-tagged versions of both proteins were coexpressed in HeLa cells. As shown in Fig. 7, all of the disease-causing NE mutants, except I31T, significantly diminish the interaction with N2N. In contrast, a negative control, S173A, replacing the catalytic serine and abolishing proteolytic activity but not causing disease, has no effect. Similar studies employing the CytoTrap two-hybrid assay yielded equivalent results (not shown).
FIG. 7.
Disease-causing NE mutants disturb the interaction with N2N in HeLa cells in coimmunoprecipitation assays. IP, immunoprecipitation; IgG, immunoglobulin G; α-FLAG, anti-FLAG antibody; α-Myc, anti-Myc antibody.
Proteolysis of N2N and Notch2 by mutant NE.
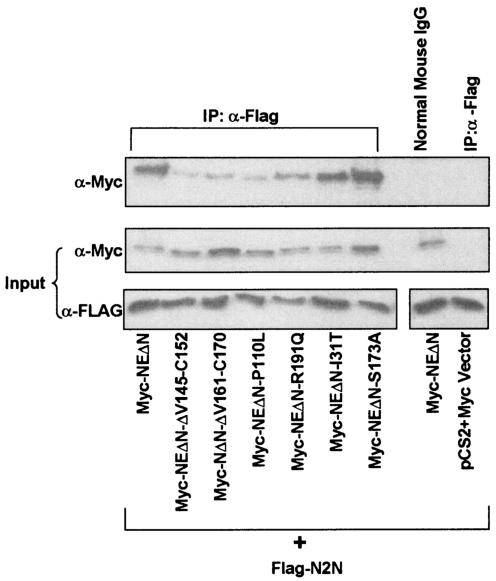
To determine whether neutropenia-causing mutations of NE cleave Notch family members, we purified recombinant mutant enzyme from yeast. We tested two cyclic neutropenia mutations with opposite properties: R191Q, which retains proteolytic activity on model peptide substrates (F.-Q. Li, J. Wechsler, and M. Horwitz, unpublished data) and posttranslationally mislocalizes to membrane compartments (6), and ΔV161-F170, which has minimal residual proteolytic activity on model substrates (28) and traffics excessively to granules (6). Recombinant wild type and the proteolytically active mutant R191Q both digest recombinant N2N in vitro, while ΔV161-F170 does not (Fig. 8A). To test whether the mutants can cleave endogenous Notch2 in living U937 cells, we added these mutant enzymes to the tissue culture media and performed a Western blot of cell lysates (Fig. 8B). Wild-type NE, either commercially obtained or purified from recombinant yeast, digests Notch2 in U937 cells, but the catalytically inactive mutant ΔV161-F170 does not. Surprisingly, the otherwise proteolytically competent mutant R191Q does not digest Notch2 when added to the media. Thus, appropriate binding of Notch2 to NE, probably taking place at the cell surface, appears necessary for its digestion.
FIG. 8.
Cleavage of N2N and Notch2 by mutant NE. (A) Recombinant GST-N2N fusion proteins cleaved by recombinant, wild-type, or mutant NE in vitro, as indicated, Coomassie-stained SDS-PAGE. (B) Western blot result of endogenous Notch2 digested by NE in U937 cells when wild-type or mutant NE is added to the culture medium. p-NE- denotes NE produced recombinantly in P. pastoris; c-NE- denotes commercially obtained NE.
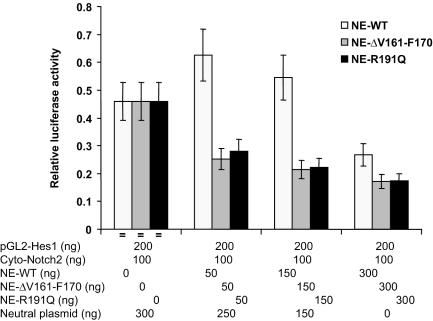
Interference of Notch signaling by mutant NE.
In light of the evidence that the mutations in NE disturb membrane trafficking (6) and the above finding that the NE mutations R191Q and ΔV161-F170 demonstrate defective proteolysis of Notch family members, we determined whether the presence of mutant NE interferes with Notch signaling in cells. We cotransfected a construct expressing the cytoplasmic domain of Notch2 along with either wild-type or mutant NE expression vectors and a Hes1 promoter reporter (Fig. 9). Wild-type NE represses Notch2 transcriptional activation in a dose-sensitive manner; however, both of the mutations, R191Q and ΔV161-F170, more effectively inhibit Notch2 signaling.
FIG. 9.
Effect of wild-type and mutant NE on transcriptional signaling by Notch2 cytoplasmic domain. HeLa cells were cotransfected with the indicated expression vectors along with a hes1 luciferase reporter. The neutral plasmid, used to keep total plasmid quantity constant, expresses lacZ in the same vector used for Notch constructs.
DISCUSSION
The CytoTrap yeast two-hybrid genetic screening method and corroborating biochemical studies indicate that N2N, a novel derivative of Notch2 containing extracellular domain EGF repeats 1 to 6 terminated with a unique 24-residue carboxyl-terminal (C24) domain, interacts with NE. NE cleaves N2N within EGF repeats. The C24 domain of N2N is required, though not sufficient, for interaction with NE and confers relative resistance to proteolysis. N2N appears throughout the cell and is also secreted, and its forced expression interferes with normal Notch1 and Notch2 transcriptional activation. NE requires the processed C-terminal extension for interaction with N2N. Neutropenia-causing NE mutations, including those deleting the C-terminal extension and required for appropriate membrane trafficking, diminish the interaction, implicating N2N and Notch signaling in disease pathogenesis. We observe a specific interaction between NE and N2N, but given N2N′s similarity with Notch2, we cannot exclude the possibility that it is Notch2 that is the actual target of NE. In fact, N2N and Notch2 are both substrates of NE. Mutant forms of NE, including one retaining catalytic activity (R191Q), is defective in proteolysis of Notch2 in living cells. In an in vitro reporter assay, NE, and especially its mutant forms, disrupts transactivation by the Notch2 cytoplasmic domain.
Abnormalities of the Notch signaling pathway offer an appealing explanation for CN and SCN for three reasons. First, a cardinal feature of these human hereditary neutropenia syndromes is monocytosis; neutrophils and monocytes represent the alternate differentiation pathways available to committed myeloid progenitor cells. Second, observations with an individual with germ line mosaicism demonstrating bone marrow chimerism for mutant NE reveals that cells harboring the ELA2 mutation do not develop into neutrophils but also do not inhibit the differentiation of normal cells. Third, the mutations in NE alter its membrane trafficking. It can thus be inferred that hereditary neutropenia is the result of altered membrane proteolysis, thereby disturbing cell-intrinsic mechanisms responsible for binary specification of cell fate—precisely the function of Notch. Supportive evidence for the involvement of Notch pathways in hereditary neutropenia comes from the finding that mutations of WASp are a rare cause of SCN (14), and WASp is required for Notch signaling in Drosophila (7).
The Notch family proteins regulate cell fate decisions for a wide variety of cell types, often inhibiting particular differentiation programs and permitting either self-renewal or differentiation along alternate pathways (reviewed in references 26 and 30). Hematopoietic cells express high levels of Notch receptors and their ligands (32). Notch proteins participate in vitro in myeloid differentiation (9, 27, 33, 42). In particular, activation of Notch1 or Notch2 promotes the entry of myeloid progenitor cells into granulopoiesis (37, 42), but they also appear capable of inhibiting granulocytic differentiation depending upon the cytokine environment (27, 31). In either event, Notch activation diverts myeloid progenitors from the default program of monocyte differentiation (31). Gene targeting studies with mice offer a more circumspect appraisal of the role of Notch family proteins in hematopoiesis. Homozygous disruption of Notch2 is embryonic lethal (19), but a hypomorphic allele of Notch2 achieved by gene targeting did not show hematopoietic defects (29). Mice targeted with a conditional Notch2 disruption revealed a deficiency of B lymphocytes (35). (In both studies, though, it is not clear how extensively the mice were screened for myelopoietic defects.) Homozygous deletion of Notch1, but not Notch2, does cause loss of production of hematopoietic stem cells in the yolk sac and aorta-gonad-mesonephros region of the embryo (35). Finally, there is a question of how well mouse models of hematopoiesis mirror human diseases: for example, gene targeting to “knock-in” human SCN mutations does not reproduce disease in mice (35); mutation of the gene AP3B1, encoding the beta subunit of adaptor protein complex 3, causes static neutropenia in human Hermansky Pudlak syndrome type 2 (23), cyclic pancytopenia in the canine “gray collie syndrome” (6), and only mild static thrombocytopenia without neutropenia in pearl mice (15). Therefore, the contributions of Notch family members to hematopoiesis and, in particular, ongoing myelopoiesis in the postembryonic bone marrow of humans is difficult to assess.
Notch receptors undergo sequential proteolytic processing (16). The first cleavage occurs posttranslationally within the lumen of the trans-Golgi network, where it is broken into separate extracellular and transmembrane/intracellular domains that remain noncovalently associated after trafficking to the plasma membrane. Binding of the Notch ligand, Delta or Jagged, induces cleavage of the extracellular domain by α-secretase. γ-secretase subsequently cleaves the remaining membrane-anchored carboxyl-terminal fragment to liberate an intracellular domain that tracks to the nucleus and functions as a transcriptional activator.
NE has a proclivity for EGF repeats; for example, it cleaves adjacently to two such units in clotting factor IX (36). Here we show that NE cleaves N2N and Notch2 and inhibits Notch2 signaling in a reporter assay. Given the defective proteolysis of Notch family members by mutant forms of NE demonstrated here, it is thus tempting to consider the possibility that proteolysis of Notch receptors by NE regulates hematopoietic differentiation. In particular, NE might function equivalently to α-secretase in the shedding of Notch family extracellular domains. NE would have at least three opportunities to encounter Notch extracellular domains. First, NE undergoes regulated secretion and would be at locally high concentrations in the extracellular environment of the bone marrow. Second, biochemical and immunolocalization studies show that in addition to granules, NE also appears on the plasma membrane and cleaves transmembrane receptors (1, 10, 11, 17, 25, 34). Third, endocytosis of Notch also contributes to Notch regulation (8, 38), and NE is present in endocytically derived granules. We reject the first possibility as being incompatible with the clinically observed absence of paracrine effects in chimeric bone marrow. We favor the second of these possibilities, since our recent results suggest that retention of the C-terminal extension of NE facilitates its localization in the plasma membrane, and we note that the C-terminal extension of NE is required for interaction with N2N (6).
The truncated form of Notch2 represented by N2N could conceivably function as a decoy to compete for Notch ligand and thus inhibit Notch activation. There are at least two other possibilities, however. First, given its interaction with NE and the resistance to proteolysis conferred by the C24 domain, N2N could inhibit cleavage of the extracellular domain of Notch proteins by NE. Second, it is noteworthy that mutant NE, including the nearly catalytically inert mutant ΔV161-F170, inhibited Notch2 signaling more strongly than wild-type NE, raising the possibility that there is an as yet undefined property of NE, not related to proteolysis, that contributes to neutropenia pathogenesis.
Acknowledgments
We thank P. Brunner for assistance with confocal microscopy.
This work was supported by grants (to M.H.) from the NIH (DK55820 and DK58161) and the Burroughs-Wellcome Fund (SATR-1002189).
REFERENCES
- 1.Allen, D. H., and P. B. Tracy. 1995. Human coagulation factor V is activated to the functional cofactor by elastase and cathepsin G expressed at the monocyte surface. J. Biol. Chem. 270:1408-1415. [DOI] [PubMed] [Google Scholar]
- 2.Ancliff, P. J., R. E. Gale, R. Liesner, I. M. Hann, and D. C. Linch. 2001. Mutations in the ELA2 gene encoding neutrophil elastase are present in most patients with sporadic severe congenital neutropenia but only in some patients with the familial form of the disease. Blood 98:2645-2650. [DOI] [PubMed] [Google Scholar]
- 3.Ancliff, P. J., R. E. Gale, M. J. Watts, R. Liesner, I. M. Hann, S. Strobel, and D. C. Linch. 2002. Paternal mosaicism proves the pathogenic nature of mutations in neutrophil elastase in severe congenital neutropenia. Blood 100:707-709. [DOI] [PubMed] [Google Scholar]
- 4.Aronheim, A., Y. C. Broder, A. Cohen, A. Fritsch, B. Belisle, and A. Abo. 1998. Chp, a homologue of the GTPase Cdc42Hs, activates the JNK pathway and is implicated in reorganizing the actin cytoskeleton. Curr. Biol. 8:1125-1128. [DOI] [PubMed] [Google Scholar]
- 5.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, K. F., F. Q. Li, R. E. Person, D. Albani, Z. Duan, J. Wechsler, K. Meade-White, K. Williams, G. M. Acland, G. Niemeyer, C. D. Lothrop, and M. Horwitz. 2003. Mutations associated with neutropenia in dogs and humans disrupt intracellular transport of neutrophil elastase. Nat. Genet. 35:90-96. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Yaacov, S., R. Le Borgne, I. Abramson, F. Schweisguth, and E. D. Schejter. 2001. Wasp, the Drosophila Wiskott-Aldrich syndrome gene homologue, is required for cell fate decisions mediated by Notch signaling. J. Cell Biol. 152:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berdnik, D., T. Torok, M. Gonzalez-Gaitan, and J. A. Knoblich. 2002. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3:221-231. [DOI] [PubMed] [Google Scholar]
- 9.Bigas, A., D. I. Martin, and L. A. Milner. 1998. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol. Cell. Biol. 18:2324-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cepinskas, G., M. Sandig, and P. R. Kvietys. 1999. PAF-induced elastase-dependent neutrophil transendothelial migration is associated with the mobilization of elastase to the neutrophil surface and localization to the migrating front. J. Cell Sci. 112:1937-1945. [DOI] [PubMed] [Google Scholar]
- 11.Clark, J. M., D. W. Vaughan, B. M. Aiken, and H. M. Kagan. 1980. Elastase-like enzymes in human neutrophils localized by ultrastructural cytochemistry. J. Cell Biol. 84:102-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dale, D. C., R. E. Person, A. A. Bolyard, A. G. Aprikyan, C. Bos, M. A. Bonilla, L. A. Boxer, G. Kannourakis, C. Zeidler, K. Welte, K. F. Benson, and M. Horwitz. 2000. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood 96:2317-2322. [PubMed] [Google Scholar]
- 13.Dall'Acqua, W., C. Halin, M. L. Rodrigues, and P. Carter. 1999. Elastase substrate specificity tailored through substrate-assisted catalysis and phage display. Protein Eng. 12:981-987. [DOI] [PubMed] [Google Scholar]
- 14.Devriendt, K., A. S. Kim, G. Mathijs, S. G. Frints, M. Schwartz, J. J. Van Den Oord, G. E. Verhoef, M. A. Boogaerts, J. P. Fryns, D. You, M. K. Rosen, and P. Vandenberghe. 2001. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat. Genet. 27:313-317. [DOI] [PubMed] [Google Scholar]
- 15.Feng, L., A. B. Seymour, S. Jiang, A. To, A. A. Peden, E. K. Novak, L. Zhen, M. E. Rusiniak, E. M. Eicher, M. S. Robinson, M. B. Gorin, and R. T. Swank. 1999. The beta3A subunit gene (Ap3b1) of the AP-3 adaptor complex is altered in the mouse hypopigmentation mutant pearl, a model for Hermansky-Pudlak syndrome and night blindness. Hum. Mol. Genet. 8:323-330. [DOI] [PubMed] [Google Scholar]
- 16.Fortini, M. E. 2002. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat. Rev. Mol. Cell Biol. 3:673-684. [DOI] [PubMed] [Google Scholar]
- 17.Fuks, A., D. Zucker-Franklin, and E. C. Franklin. 1983. Identification of elastases associated with purified plasma membranes isolated from human monocytes and lymphocytes. Biochim. Biophys. Acta 755:195-203. [DOI] [PubMed] [Google Scholar]
- 18.Gardiner, E. E., M. De Luca, T. McNally, A. D. Michelson, R. K. Andrews, and M. C. Berndt. 2001. Regulation of P-selectin binding to the neutrophil P-selectin counter-receptor P-selectin glycoprotein ligand-1 by neutrophil elastase and cathepsin G. Blood 98:1440-1447. [DOI] [PubMed] [Google Scholar]
- 19.Hamada, Y., Y. Kadokawa, M. Okabe, M. Ikawa, J. R. Coleman, and Y. Tsujimoto. 1999. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development 126:3415-3424. [DOI] [PubMed] [Google Scholar]
- 20.Hermand, D., T. Westerling, A. Pihlak, J. Y. Thuret, T. Vallenius, M. Tiainen, J. Vandenhaute, G. Cottarel, C. Mann, and T. P. Makela. 2001. Specificity of Cdk activation in vivo by the two Caks Mcs6 and Csk1 in fission yeast. EMBO J. 20:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horwitz, M., K. F. Benson, Z. Duan, R. E. Person, J. Wechsler, K. Williams, D. Albani, and F. Q. Li. 2003. Role of neutrophil elastase in bone marrow failure syndromes: molecular genetic revival of the chalone hypothesis. Curr. Opin. Hematol. 10:49-54. [DOI] [PubMed] [Google Scholar]
- 22.Horwitz, M., K. F. Benson, R. E. Person, A. G. Aprikyan, and D. C. Dale. 1999. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat. Genet. 23:433-436. [DOI] [PubMed] [Google Scholar]
- 23.Huizing, M., C. D. Scher, E. Strovel, D. L. Fitzpatrick, L. M. Hartnell, Y. Anikster, and W. A. Gahl. 2002. Nonsense mutations in ADTB3A cause complete deficiency of the beta3A subunit of adaptor complex-3 and severe Hermansky-Pudlak syndrome type 2. Pediatr. Res. 51:150-158. [DOI] [PubMed] [Google Scholar]
- 24.Isakoff, S. J., T. Cardozo, J. Andreev, Z. Li, K. M. Ferguson, R. Abagyan, M. A. Lemmon, A. Aronheim, and E. Y. Skolnik. 1998. Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J. 17:5374-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaup, M., K. Dassler, U. Reineke, C. Weise, R. Tauber, and H. Fuchs. 2002. Processing of the human transferrin receptor at distinct positions within the stalk region by neutrophil elastase and cathepsin G. Biol. Chem. 383:1011-1020. [DOI] [PubMed] [Google Scholar]
- 26.Kojika, S., and J. D. Griffin. 2001. Notch receptors and hematopoiesis. Exp. Hematol. 29:1041-1052. [DOI] [PubMed] [Google Scholar]
- 27.Kumano, K., S. Chiba, K. Shimizu, T. Yamagata, N. Hosoya, T. Saito, T. Takahashi, Y. Hamada, and H. Hirai. 2001. Notch1 inhibits differentiation of hematopoietic cells by sustaining GATA-2 expression. Blood 98:3283-3289. [DOI] [PubMed] [Google Scholar]
- 28.Li, F. Q., and M. Horwitz. 2001. Characterization of mutant neutrophil elastase in severe congenital neutropenia. J. Biol. Chem. 276:14230-14241. [DOI] [PubMed] [Google Scholar]
- 29.McCright, B., X. Gao, L. Shen, J. Lozier, Y. Lan, M. Maguire, D. Herzlinger, G. Weinmaster, R. Jiang, and T. Gridley. 2001. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491-502. [DOI] [PubMed] [Google Scholar]
- 30.Milner, L. A., and A. Bigas. 1999. Notch as a mediator of cell fate determination in hematopoiesis: evidence and speculation. Blood 93:2431-2448. [PubMed] [Google Scholar]
- 31.Ohishi, K., B. Varnum-Finney, and I. D. Bernstein. 2002. The notch pathway: modulation of cell fate decisions in hematopoiesis. Int. J. Hematol. 75:449-459. [DOI] [PubMed] [Google Scholar]
- 32.Ohishi, K., B. Varnum-Finney, D. Flowers, C. Anasetti, D. Myerson, and I. D. Bernstein. 2000. Monocytes express high amounts of Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1. Blood 95:2847-2854. [PubMed] [Google Scholar]
- 33.Ohishi, K., B. Varnum-Finney, R. E. Serda, C. Anasetti, and I. D. Bernstein. 2001. The Notch ligand, Delta-1, inhibits the differentiation of monocytes into macrophages but permits their differentiation into dendritic cells. Blood 98:1402-1407. [DOI] [PubMed] [Google Scholar]
- 34.Owen, C. A., M. A. Campbell, P. L. Sannes, S. S. Boukedes, and E. J. Campbell. 1995. Cell surface-bound elastase and cathepsin G on human neutrophils: a novel, non-oxidative mechanism by which neutrophils focus and preserve catalytic activity of serine proteinases. J. Cell Biol. 131:775-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito, T., S. Chiba, M. Ichikawa, A. Kunisato, T. Asai, K. Shimizu, T. Yamaguchi, G. Yamamoto, S. Seo, K. Kumano, E. Nakagami-Yamaguchi, Y. Hamada, S. Aizawa, and H. Hirai. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18:675-685. [DOI] [PubMed] [Google Scholar]
- 36.Samis, J. A., E. Kam, M. E. Nesheim, and A. R. Giles. 1998. Neutrophil elastase cleavage of human factor IX generates an activated factor IX-like product devoid of coagulant function. Blood 92:1287-1296. [PubMed] [Google Scholar]
- 37.Schroeder, T., and U. Just. 2000. Notch signalling via RBP-J promotes myeloid differentiation. EMBO J. 19:2558-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaye, D. D., and I. Greenwald. 2002. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature 420:686-690. [DOI] [PubMed] [Google Scholar]
- 39.Small, D., D. Kovalenko, D. Kacer, L. Liaw, M. Landriscina, C. Di Serio, I. Prudovsky, and T. Maciag. 2001. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J. Biol. Chem. 276:32022-32030. [DOI] [PubMed] [Google Scholar]
- 40.Solecki, D. J., X. L. Liu, T. Tomoda, Y. Fang, and M. E. Hatten. 2001. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron 31:557-568. [DOI] [PubMed] [Google Scholar]
- 41.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149:249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan-Pertel, H. T., L. Walker, D. Browning, A. Miyamoto, G. Weinmaster, and J. C. Gasson. 2000. Notch signaling enhances survival and alters differentiation of 32D myeloblasts. J. Immunol. 165:4428-4436. [DOI] [PubMed] [Google Scholar]