Abstract
Increased expression and/or activation of H-Ras are often associated with tumor aggressiveness in breast cancer. Previously, we showed that H-Ras, but not N-Ras, induces MCF10A human breast epithelial cell invasion and migration, whereas both H-Ras and N-Ras induce cell proliferation and phenotypic transformation. In an attempt to determine the sequence requirement directing the divergent phenotype induced by H-Ras and N-Ras with a focus on the induction of human breast cell invasion, we investigated the structural and functional relationships between H-Ras and N-Ras using domain-swap and site-directed mutagenesis approaches. Here, we report that the hypervariable region (HVR), consisting of amino acids 166 to 189 in H-Ras, determines the invasive/migratory signaling program as shown by the exchange of invasive phenotype by swapping HVR sequences between H-Ras and N-Ras. We also demonstrate that the H-Ras-specific additional palmitoylation site at Cys184 is not responsible for the signaling events that distinguish between H-Ras and N-Ras. Importantly, this work identifies the C-terminal HVR, especially the flexible linker domain with two consecutive proline residues Pro173 and Pro174, as a critical domain that contributes to activation of H-Ras and its invasive potential in human breast epithelial cells. The present study sheds light on the structural basis for the Ras isoform-specific invasive program of breast epithelial cells, providing information for the development of agents that specifically target invasion-related H-Ras pathways in human cancer.
Introduction
Ras subfamily proteins, which include H-Ras, N-Ras, and K-Ras, are central signaling molecules that activate downstream signaling networks critical for cellular processes including cell survival, proliferation, motility, and cytoskeletal organization [1]. Thus, general inhibition of Ras activities can be detrimental not only to cancer cells but also to normal cells. A major challenge is to develop drug compounds that target Ras activities that are required for malignant cancer cell responses but are less critical for normal cell functions.
Ras expression has been suggested as a marker for tumor aggressiveness in breast cancer [2,3]. Although mutations are rare, a single H-Ras point mutation that changes Gly to Asp at amino acid codon 12 (G12D) has been found in mammary carcinoma whereas the same mutation in N-Ras is detected in teratocarcinoma and leukemia [4]. To investigate the Ras isoform-specific signaling pathways and the subsequent cellular responses in breast cancer, we previously established the MCF10A human breast epithelial cell system, in which H-Ras or N-Ras is constitutively activated (G12D). We have demonstrated that whereas both H-Ras and N-Ras result in phenotypic transformation of MCF10A cells, only H-Ras induces invasive and migratory phenotypes in these cells [5]. Induction of invasive phenotype by H-Ras, but not N-Ras, was also observed in MDA-MB-453 human breast cancer cell line (unpublished data). In the MCF10A cell system, we showed that H-Ras-induced invasiveness was associated with the activation of p38 mitogen-activated protein kinase (MAPK) and extracellular signal-regulated kinases (ERKs), resulting in induction of matrix metalloproteinases 2 and 9 (MMP-2 and -9). In contrast, N-Ras failed to activate p38 MAPK, and N-Ras-activated ERKs lead to MMP-9 induction with little effect on MMP-2 expression [6–8]. The purpose of the present study was to establish the structural-functional relationships between H-Ras and N-Ras to unveil the sequences of H-Ras that directs the Ras isoform-specific induction of the invasive phenotype in human breast epithelial cells.
Whereas the four closely related Ras isoforms, H-Ras, K(A)-Ras, K(B)-Ras, and N-Ras, share complete sequence identity in the aminoterminal 85 amino acids and the middle 80 amino acids contain 85% homology, the carboxy-terminal hypervariable region (HVR), which consists of residues 166 and 189, is highly divergent as depicted in Figure 1A [1,9–12]. The HVR is composed of a flexible linker domain (residues 167–179) and membrane-targeting or anchor domain-containing residues 180 to 186 [13]. In order for Ras to activate the intracellular signal transduction pathways mediated by growth factors and cytokines, it must associate with the inner surface of the plasma membrane [14]. Two regions in the HVR of Ras were previously suggested to be critical for correct plasma membrane localization [15]. The first region is a C-terminal CAAX box (in which A = aliphatic amino acid) [16,17]. Farnesylation on the cysteine of CAAX is thought to be among the most critical events for Ras activation [14], and the localization of H-Ras and N-Ras is primarily determined by the CAAX motif [18].
Figure 1.
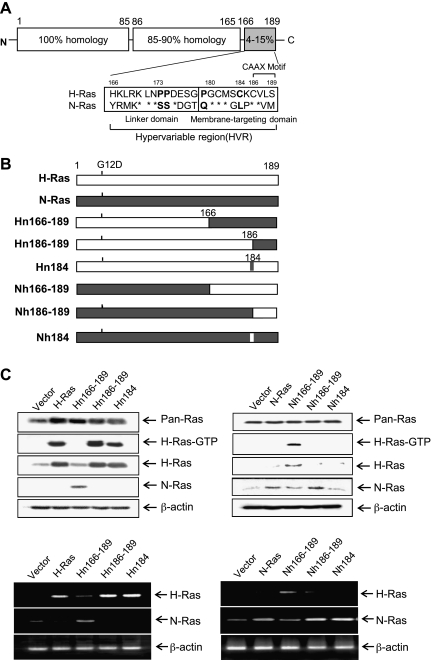
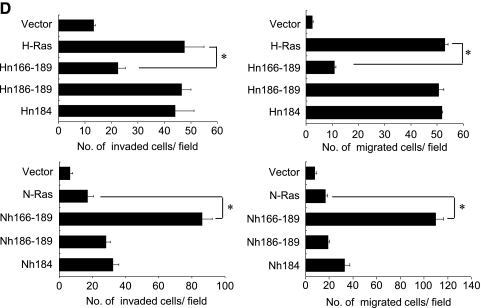
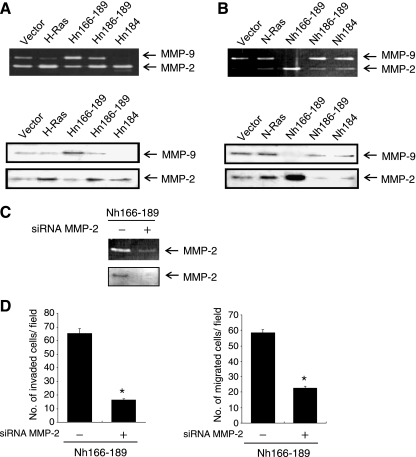
H-Ras HVR determines the invasive/migratory phenotypes of MCF10A cells. (A) Sequence alignment of H-Ras/N-Ras HVR. (B) Schematic outline of chimera. All of the Ras constructs contain an oncogenic mutation at codon 12 (G12→D). (C, upper) Cell lysates were collected, and Ras expression was detected by immunoblot analysis. β-Actin was used as a loading control. H-Ras activity (H-Ras-GTP) was measured as described in Materials and Methods. (C, lower) RT-PCR analysis was performed. (D) Cells were subjected to in vitro invasion assay and Transwell migration assay. The number of invaded or migrated cells per field was counted (x400) in 13 arbitrary visual fields. The results represent means ± SE of triplicates. *Statistically different at P < .01, by 2-tailed Student t test.
The second region is the site for palmitoylation, necessary for proper localization and the oncogenic activity of H-Ras [19–21]. Interestingly, H-Ras is anchored to the plasma membrane by two palmitoylated cysteine residues, Cys181 and Cys184, whereas N-Ras is anchored by a single cysteine, Cys181 [13,22,23].
Despite the vast amount of research on the structural and functional properties of the Ras proteins, little is known regarding the structural basis for H-Ras isoform-specific function in breast epithelial cells. Here, we found that the HVR dictates H-Ras-specific signaling events for breast epithelial cell invasion as demonstrated by swapping the HVR sequences between H-Ras and N-Ras. We also showed that Pro173 and Pro174 within the HVR of H-Ras are critical determinants for the activation of H-Ras and H-Ras-specific invasive program in human breast epithelial cells.
Materials and Methods
Construction of Ras Chimeras
The expression vectors encoding H-Ras (G12D), H-Ras (1–165)/N-Ras (166–189), H-Ras (1–185)/N-Ras (CAAX), H-Ras (G184L), N-Ras (G12D), N-Ras (1–165)/H-Ras (166–189), N-Ras (1–185)/H-Ras (CAAX), and N-Ras (L184G) were constructed and named H, Hn166–189, Hn186–189, Hn184, N, Nh166–189, Nh186–189, and Nh184, respectively. The point mutants were constructed using polymerase chain reaction (PCR) mutagenesis on pcDNA3.1-H-Ras or pcDNA3.1-N-Ras templates. The N-Ras/H-Ras chimeras were constructed using PCR mutagenesis on pcDNA3.1-N-Ras and pcDNA3.1-H-Ras templates. The amplified DNA was ligated into the BamHI and XhoI sites of pcDNA3.1 (Invitrogen, San Diego, CA). All of the Ras chimeric constructs contain an oncogenic mutation at codon 12 (G12→D). The mutations were confirmed by DNA sequence analysis of the plasmids before further study. MCF10A cells (5 x 105) were placed on 6-cm-diameter culture dishes, incubated at 37°C for 24 hours, and then transfected with 4 µg of a mutant complementary DNA (cDNA) or control vector (pcDNA3.1) for 6 hours using Lipofectamine 2000 reagent (Invitrogen) and OPTI-MEMI reduced serum medium (Invitrogen). These cells were selected by G418 (500 µg/ml) for 3 weeks.
Construction of Proline Mutants
Proline mutants were constructed by means of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) using the pcDNA3.1 vector containing H-Ras as a template. Pro179Gln, Pro173Ser, Pro173Ser/Pro174Ser, and Pro173Ser/Pro174Ser/Pro179Gln (also called PPQ, SPP, SSP, and SSQ) were mutated to exchange the proline residues at 173, 174, and 179 to the corresponding amino acids of the N-Ras sequence. The authenticity of all constructs was confirmed by DNA sequencing.
Cell Culture
MCF10A and transfected cells were cultured in Dulbecco modified Eagle medium/F12 supplemented with 5% horse serum, 0.5 µg/ml hydrocortisone, 10 µg/ml insulin, 20 ng/ml epidermal growth factor, 0.1 µg/ml cholera enterotoxin, 100 units/ml penicillin-streptomycin, 2 mM l-glutamine, and 0.5 µg/ml amphotericin. Cells were maintained in a humidified atmosphere with 95% air and 5% CO2 at 37°C. Although it has been shown that the overexpression of oncogenic H-Ras can induce a senescence-like permanent growth arrest in several cell lines [24–26], growth inhibition was not observed in MCF10A cells transfected with oncogenic H-Ras.
Immunoblot Analysis
Anti-N-Ras, anti-H-Ras, anti-MMP-9 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-MMP-2 antibody was from R&D Systems (Minneapolis, MN). β-Actin antibody was from Sigma-Aldrich (St Louis, MO). Anti-p38, anti-phosphorylated p38, anti-phosphorylated MKK3/6, and anti-MKK3 antibodies were from Cell Signaling Technology (Beverly, MA). Anti-Rac1 antibody was from Upstate Biotechnology, Inc (Lake Placid, NY).
Equal amounts of protein extracts in SDS lysis buffer were subjected to 12% SDS-PAGE analysis and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes. An enhanced chemiluminescence (Amersham-Pharmacia, Buckinghamshire, UK) system was used for detection. Relative band intensities were determined by quantitation of each band with an Image Analyzer (Vilber Lourmnat, France).
Reverse Transcription (RT)-PCR
Total RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Incubated RNA/Oligo dT/dNTP mix was reverse-transcribed by RT-Superscript III reverse transcriptase (Invitrogen). Thermocycler conditions were as described previously [27].
Immunofluorescence assay. The cells (1 x 104/well) were plated at Lab-Tek II chamber slide (Nalge Nunc International, Naperville, IL) and then incubated for 1 day at 37°C. Cells were washed three times with PBS and fixed in 4% formaldehyde in PBS for 20 minutes; after washing, cells were permeabilized by treating with 0.5% Triton X-100 for 5 minutes. The fixed cells were washed again with a blocking reagent in PBS. The wells were incubated for 1 hour with an anti-H-Ras or an N-Ras antibody in 1x PBS (1% BSA), washed, and then incubated with anti-IgG-Alexa 488 antibody (Invitrogen, Carlsbad, CA) for 45 minutes. After washing with 1x PBS three times for the second labeling of cells, the cells were incubated with anti-pRac1 antibody and, subsequently, with anti-IgG-Cy5 antibody. Confocal images were taken in an FV-1000 spectral confocal laser scanning microscope (Olympus, Tokyo, Japan). The fluorescence images were acquired with FV10-ASW 1.6 Viewer (Olympus).
Gelatin Zymogram Assay
Cells were cultured in serum-free Dulbecco modified Eagle medium/F-12 for 48 hours. Gelatinolytic activity of the conditioned medium was determined by gelatin zymogram assay as described previously [5]. Areas of gelatinase activity were detected as clear bands against the blue-stained gelatin background.
In Vitro Invasion Assay and Transwell Migration Assay
In vitro invasion assays and Transwell migration assays were performed using a 24-well Transwell unit as described previously [6].
Small Interfering RNA Preparation and Transfection
The small interfering RNA (siRNA) molecules targeting MMP-2, with sequence 5′-AATACCATCGAGACCATGCGG-3′, were purchased from Invitrogen (Carlsbad, CA). Nh166–189 cells were plated in six-well plates at 1.5 x 105 cells/well, grown for 24 hours, then each transfected with 100 pmol of siRNA for 6 hours using Lipofectamine 2000 reagent (Invitrogen) and OPTI-MEMI reduced serum medium (Invitrogen). Control cells were treated with Stealth RNAi Negative Control Duplex (Invitrogen).
Rac1 Activity Assay
The levels of Rac1-GTP were measured by affinity precipitation using PAK-1 p21-binding domain Rac assay reagent (Upstate Biotechnology) following the manufacturer's instructions as previously described [7].
H-Ras Activity Assay
The levels of H-Ras-GTP were measured by affinity precipitation using Raf-1 RBD Ras assay reagent (Upstate Biotechnology) following the manufacturer's instructions. Briefly, cells were lysed in MLB buffer, and Raf-1 RBD-agarose was immediately added to the cell lysate and incubated for 45 minutes at 4°C. The bead pellet was washed three times with MLB buffer. The bead pellet was finally suspended in Laemmli sample buffer. Proteins were separated by 12% SDS-PAGE, transferred to nitrocellulose membranes, and blotted with anti-H-Ras antibody following the manufacturer's instructions.
Results
H-Ras HVR Determines the Invasive and Migratory Phenotypes in MCF10A Cells
To study the molecular mechanism underlying the H-Ras-specific migratory/invasive phenotypes in human breast epithelial cells, we wished to determine the structural-functional properties of H-Ras and N-Ras proteins. Because the amino acid sequences of H-Ras and N-Ras greatly differ in the 166 to 189 HVR (Figure 1A), we tested whether this region is a major contributor to H-Ras-specific signaling for human breast epithelial cell invasion. To this end, we generated chimeric constructs, Hn166–189 and Nh166–189. As depicted in Figure 1 (A and B), Hn166–189 represents a chimera, which is composed of the H-Ras amino acid sequences from 1 to 165 fused to the N-Ras HVR (amino acid residues 166–189). Similarly, Nh166–189 contains residues 1 to 165 of N-Ras fused to the H-Ras HVR (residues 166–189). The stable expression of these chimera in MCF10A cells was confirmed by immunoblot analysis using antibodies against pan-Ras, H-Ras HVR, or N-Ras HVR (Figure 1C, upper) and by RT-PCR analysis (Figure 1C, lower). Importantly, the active form of H-Ras (H-Ras-GTP) was markedly decreased in MCF10A cells transfected with Hn166–189 compared with H-Ras MCF10A cells. Because the anti-H-Ras antibody used in this study recognizes the C-terminal region of H-Ras, the Hn166–189 showed decreased levels of both active H-Ras and total H-Ras compared with the wild-type H-Ras. Conversely, we observed a marked increase in the level of active H-Ras in Nh166–189 cells. These data demonstrate that the HVR is crucial for the activation of H-Ras.
When the effects of chimera protein expression on cell invasion/migration were examined, Hn166–189 drastically reduced the ability of H-Ras to induce invasive and migratory properties in MCF10A cells (Figure 1D and Figure W1). More interestingly, Nh166–189 induced cell invasion and migration as effectively as H-Ras. Similar results were obtained using additional clones expressing chimera constructs (data not shown). These results showed that H-Ras HVR is a critical determinant for the ability of Ras to induce cell invasion and migration in human breast epithelial cells.
Neither the H-Ras-Specific CAAX Motif nor the H-Ras-Specific Palmitoylation Site Alone Plays a Critical Role for H-Ras-Specific Signaling for Cell Invasion/Migration
Mounting evidence has suggested a crucial role for the CAAX motif of Ras proteins in plasma membrane targeting and subsequent activation of intracellular signal transduction pathways [14,28]. Next, we examined whether difference in the CAAX sequences between H-Ras and N-Ras contributes to the differential ability to regulate cell invasion/migration. To this end, we generated two Ras mutants, Hn186–189 and Nh186–189, which represent a H-Ras mutant containing the N-Ras CAAX motif (CVVM at amino acid sequences 186–189) and a N-Ras mutant containing the H-Ras CAAX motif (CVLS), respectively (Figure 1, A and B). Exchange of the CAAX motif between the two Ras isoforms did not alter the level of H-Ras-GTP (Figure 1C) or the invasive/migratory properties of H-Ras and N-Ras (Figure 1D). These data indicate that as long as the farnesylatable site at C186 of CAAX is maintained, the mutation of Ras isoform-specific CAAX motif did not affect the induction of the H-Ras-specific signaling events.
In addition to the CAAX motif, palmitoylation of Ras affects membrane targeting and retention, thereby regulating the intrinsic signaling capacity of Ras proteins [19–21]. Whereas H-Ras is palmitoylated on two cysteines (Cys181 and Cys184), N-Ras has a single palmitoylation site at Cys181. We next questioned whether the additional palmitoylation site at 184 in H-Ras plays a role in H-Ras-specific induction of cell migration or invasion. To address this, we performed site-directed mutagenesis and generated an H-Ras mutant (Hn184) in which Cys184, the second palmitoylation site, was mutated to Leu, resulting the Cys181 monopalmitoylated H-Ras. Similarly, we also generated an N-Ras mutant (Nh184) in which Leu184 was mutated to Cys (Figure 1, A and B). Neither the loss of the second palmitoylation site in H-Ras nor the gain of a potential palmitoylation site in N-Ras affected the level of active H-Ras (H-Ras-GTP; Figure 1C) or the invasive/migratory potentials caused by H-Ras and N-Ras (Figure 1D). These results demonstrate that monopalmitoylation of Cys181 of H-Ras is sufficient for its activation and the induction of invasive/migratory phenotypes, suggesting that the sequence difference between H-Ras and N-Ras in the additional palmitoylation site at Cys184 may not be responsible for their different effects on human breast epithelial cell migration and invasion.
HVR Is Critical for Activation of the Rac-MKK3/6-p38 Pathway and MMP-2 Up-regulation
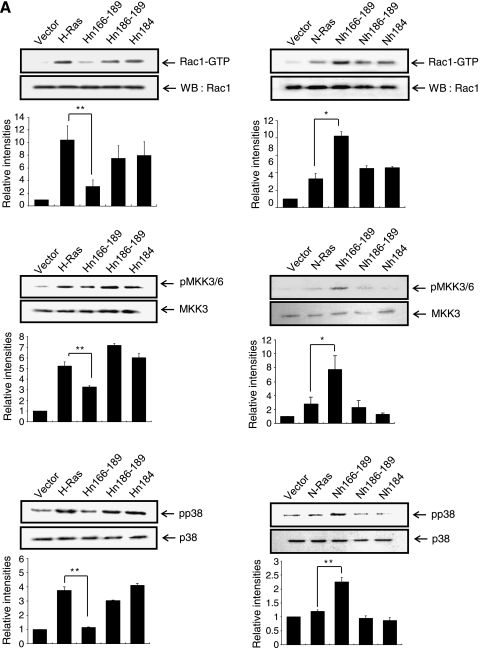
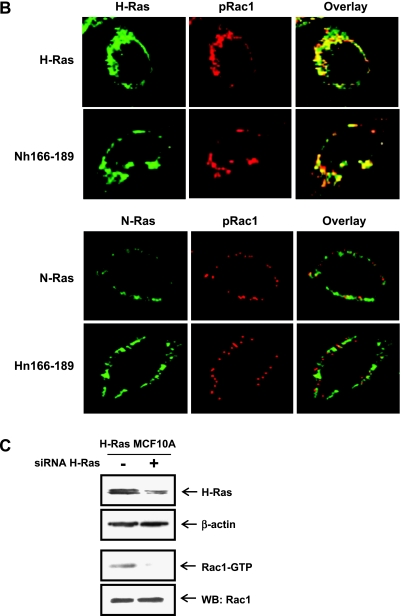
We have previously shown that H-Ras, but not N-Ras, activates the Rac-MKK (MAP kinase kinase) 3/6-p38 pathway, leading to the invasive/migratory phenotype [7]. Here, we further examined how the Ras isoform-specific signaling program is affected by the H-Ras/N-Ras chimeras as well as the H-Ras/N-Ras CAAX and palmitoylation site mutants. As shown in Figure 2A, active Rac1 (Rac1-GTP) and phosphorylated forms of MKK3/6 and p38 were significantly reduced in MCF10A cells transfected with Hn166–189 compared with the H-Ras MCF10A cells. Conversely, we observed a marked increase in the levels of active Rac1 and phosphorylated MKK3/6 and p38 in Nh166–189 cells. Also consistent with the results of H-Ras activation and cell migration/invasion, neither the CAAX mutants Hn186–189 and Nh166–189 nor the palmitoylation site mutants Hn184 and Nn184 showed detectable changes in the activation of Rac1, MKK3/6, or p38. To investigate if localization of H-Ras actually correlates with the activation of Rac1, colocalization of H-Ras, and phospho-Rac1 (pRac1) was examined by confocal microscopy. As shown in Figure 2B (upper), H-Ras colocalized with pRac1 in MCF10A cells transfected with H-Ras and Nh166–189. In contrast, colocalization of N-Ras and pRac1 was barely detected in MCF10A cells transfected with N-Ras and Hn166–189 (Figure 2B, lower). These data indicate that the localization of H-Ras actually correlates with the activation of Rac1 in which H-Ras HVR plays a critical role. To examine if the increase of Rac1 activation in H-Ras MCF10A cells is due to the increase in total H-Ras levels, we knocked down H-Ras expression using an siRNA targeting H-Ras. As shown in Figure 2C, the active form of Rac1 was significantly reduced by siRNA knockdown of H-Ras, indicating that H-Ras is critical for the activation of Rac1 in H-Ras MCF10A cells.
Figure 2.
H-Ras HVRs are critical for the activation of the Rac-MKK3/6-p38 pathway. (A) For Rac1 activity, cell lysates were incubated with GST-PBD fusion protein, and the bound active Rac1-GTPmolecules were analyzed by immunoblot analysis using an anti-Rac1 antibody. The levels of activated MKK3/6, p38 in Ras chimera, and mutant cells were determined by immunoblot analysis of whole cell lysates. Band intensities were quantitated and plotted. *,**Statistically different at P < .05 and P < .01, respectively. (B) Cells were double labeled for H-Ras or N-Ras (green) and pRac1 (red). Immunocytochemical colocalization of H-Ras or N-Ras and flotillin-1 was observed by confocal microscopy. (C) H-Ras MCF10A cells were transfected with control siRNA or siRNA H-Ras. Protein level of H-Ras was detected by immunoblot analysis. β-Actin was used as a loading control. Rac1 activity (Rac1-GTP) was measured as described in Materials and Methods.
Because our previous studies revealed that MMP-2 up-regulation by H-Ras is critical for H-Ras-mediated MCF10A cell invasion and migration [6,8], we next examined the effects of the H-Ras/N-Ras chimeras on MMP-2 expression. As shown in Figure 3A, the ability of H-Ras to induce MMP-2 expression was decreased in Hn166–189, whereas Hn186–189 and Hn184 induced MMP-2 as much or only slightly less than H-Ras. Nh166–189, but not Nh186–189 or Nh184, was observed to induce MMP-2 (Figure 3B). These results demonstrate that the HVR of H-Ras is a major contributor to the regulation of H-Ras-specific signaling that leads to MMP-2 up-regulation in MCF10A cells. These results also indicate that the CAAX motif and the palmitoylation sites of H-Ras and N-Ras are not important for H-Ras-specific MMP induction in these cells.
Figure 3.
H-Ras HVR are necessary for MMP-2 up-regulation. (A, B) Conditioned media were collected and analyzed for expression of MMP-2 and MMP-9 by gelatin zymogram assay (upper) and immunoblot analysis (lower). (C) Knockdown of MMP-2 by siRNA was confirmed by gelatin zymography and immunoblot analysis in Nh166–189 cells. Control cells were treated with Stealth RNAi Negative Control Duplexes. (D) Cells transfected with siRNA targeting MMP-2 was subjected to an in vitro invasion assay (left panel) or a Transwell migration assay (right panel). The number of invaded or migrated cells per field was counted (x400) in 13 arbitrary visual fields. The results represent means ± SE of triplicates. *Statistically different from control at P < .01.
To confirm the functional significance of MMP-2 up-regulation in cell invasion and migration induced by Nh166–189, we knocked down MMP-2 expression using a siRNA targeting MMP-2 mRNA. The expression of MMP-2 was efficiently reduced by this approach, as evidenced by gelatin zymogram assay and immunoblot analysis (Figure 3C). Nh166–189-induced invasion and migration were both significantly inhibited by siRNA knockdown of MMP-2 (Figure 3D), indicating that MMP-2 is a critical mediator of MCF10A cell invasion/migration by H-Ras signaling in its HVR-specific manner.
The Proline-Rich Motif in the Linker Domain of H-Ras Is Critical for H-Ras-Induced Human Breast Epithelial Cell Invasion
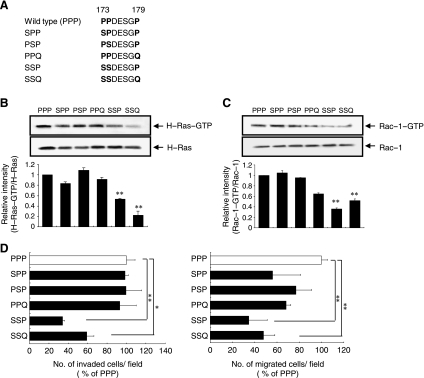
The flexible linker domain of the H-Ras HVR is enriched for proline residues (Pro173, Pro174, and Pro179), whereas the corresponding amino acids in N-Ras are Ser173, Ser174, and Gln179 (Figure 1A). Because the proline-rich motifs have been frequently found in the hinge region and thus have been shown to play a crucial role in protein folding [29–32], we examined the functional significance of these proline residues in H-Ras-specific regulation of breast epithelial cell invasion and migration. To this end, we generated five H-Ras mutants, SPP, PSP, PPQ, SSP, and SSQ, in which each proline residue (P) at positions 173, 174, or 179 of H-Ras was substituted by the corresponding amino acid present in N-Ras, serine (S) or glutamine (Q), as depicted in Figure 4A. The level of active H-Ras (H-Ras-GTP) was measured in MCF10A cells expressing the wild-type (PPP) and the mutant H-Ras proteins. SSP and SSQ, in which the proline residues at 173 and 174 are replaced by serine residues, significantly decreased H-Ras-GTP levels (Figure 4B), demonstrating that Pro173 and Pro174 are required for the activation of H-Ras in breast epithelial cells.
Figure 4.
Pro173 and Pro174 are important for H-Ras-induced invasive/migratory phenotypes. (A) Schematic outline of proline mutants of H-Ras. (B) H-Ras activity (H-Ras-GTP) was measured as described in Materials and Methods. Band intensities were quantitated and plotted. **Statistically different at P < .01. (C) Rac-1 activity (Rac-1-GTP) was measured as described in Materials and Methods. Band intensities were quantitated and plotted. **Statistically different at P < .01. (D) Cells were subjected to the in vitro invasion assay and Transwell migration assay. The number of invaded or migrated cells per field was counted (x400) in 13 arbitrary visual fields. The results represent means ± SE of triplicates. *,**Statistically different from control at P < .05 and P < .01, respectively, by the 2-tailed Student t test.
Active Rac1 (Rac1-GTP), which is activated in a H-Ras-specific manner [7], was also significantly decreased in cells expressing the SSP and SSQ mutants compared with wild-type H-Ras (Figure 4C). Importantly, whereas the SSP and SSQ mutants significantly reduced the invasive and migratory abilities of the cells, the SPP, PSP, and PPQ mutants, in which only one proline residue is mutated, did not significantly inhibit invasion/migration compared with wild-type H-Ras (Figure 4D). Of note, although the SSQ mutant showed a near-complete decrease in H-Ras-GTP, only 40% to 50% inhibition of invasion and migration was observed. Given that invasive/migratory phenotypic changes are complex multistep processes and their genetic and biochemical determinants are still largely unknown, it would be worthwhile to further elucidate the multiple factors other than H-Ras, which are involved in these processes.
We next investigated the invasiveness of two mutant N-Ras constructs, N-Ras(PPQ) and N-Ras(PPP), in which amino acid residues 173, 174, and 179 of N-Ras were substituted to proline. Invasive phenotype was not significantly induced by these mutants (data not shown), indicating that the replacement of amino acid residues 173, 174, and 179 to proline was not sufficient to confer the invasive ability to N-Ras. These data indicate that the consecutive proline residues of H-Ras HVR, Pro173 and Pro174, are essential for H-Ras activity and its induction of invasive/migratory phenotypes in MCF10A cells.
Discussion
Ras is a central signaling molecule that regulates oncogenesis and metastasis of breast cancer. H-Ras and N-Ras differentially regulate the invasive and migratory properties of breast epithelial cells, which led us to investigate which portions of the amino acid sequence are required for H-Ras-specific functions. Here we demonstrate that the HVR, consisting of amino acids 166 to 189 of H-Ras, is essential for H-Ras-specific signaling to induce the invasive phenotype. Consistent with our finding, it has been shown that the HVR of H-Ras is important for its activities [19,33–35]. Similarly to the HVR of H-Ras, the HVR of R-Ras (amino acids 178–218) plays an important role in R-Ras signal transduction and regulation of the morphology and motility of MCF10A cells [36–38]. The present study not only underscores the importance of HVR for H-Ras functions but also reveals a critical role for HVR as a determinant for differences between H-Ras and N-Ras specific signaling events. The present study for the first time shows that noninvasive N-Ras can mediate breast cell invasion if its HVR sequences are replaced with those of H-Ras, whereas H-Ras' ability to induce cell invasion is abolished by the N-Ras HVR sequences. Because the HVR sequences of K-Ras greatly differ from those of H-Ras and N-Ras [39], it would be of interest to further investigate the effect of K-Ras HVR on the breast cell invasion.
The localization of H-Ras and N-Ras is primarily determined by the CAAX motif [18] and the presence and position of the palmitoyl groups [39]. Distinct roles of two palmitoyl residues in H-Ras have been demonstrated; whereas monopalmitoylation of Cys181 is required and sufficient for efficient trafficking of H-Ras from the Golgi apparatus to the plasma membrane, monopalmitoylation of Cys184 supports correct GTP-regulated lateral segregation of H-Ras between microdomains in the membrane [20]. To our surprise, our data show that the loss of second palmitoylation site at amino acid residue 184 did not play a critical role for H-Ras activation and H-Ras-induced specific signaling events, which is inconsistent with the previously suggested role of Cys184 in H-Ras microlocalization and activation [20]. Our findings implicate that monopalmitoylation of Cys181 of H-Ras is sufficient for the H-Ras activation and subsequent cellular responses.
In the HVR of H-Ras, there are three proline residues at positions 173, 174, and 179, whereas N-Ras lacks a proline residue at all of these positions. Pro173 and Pro174 are extensively conserved among species (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/pubmed/), implying that these residues may be important for H-Ras function. Proline-rich sequences, often found in the hinge regions, are known to restrict the conformation of the protein and play an indispensable role in the regulation of various cellular responses. For instance, the proline-rich motifs are important for delivering actin monomers to specific cellular locations where membrane protrusions necessary for cell motility are formed [40]. A recently identified colon cancer-associated gene product, MACC1, induces proliferation and invasion through its proline-rich motif and thereby promotes colon cancer metastasis [41]. Similar to H-Ras, R-Ras contains a polyproline site in the HVR that is required for R-Ras-induced integrin activation [42]. In the present study, we provide evidence suggesting a critical role of the proline-rich motif of the flexible linker domain in the H-Ras HVR for H-Ras activation.
The three-dimensional structure of the soluble part of H-Ras, comprising the amino-terminal 166 residues, has been determined by x-ray crystallography [43]. However, the structure of the membrane-bound HVR of H-Ras is not presently available. Because the three-dimensional structural analysis of the H-Ras HVR has not yet been completed, we can only speculate the potential structural difference between wild-type H-Ras (Pro173/Pro174) and a mutant H-Ras (Ser173/Ser174) proteins and its implication in the regulation of intracellular signal transduction pathways. On the basis of modeling and molecular dynamics simulations of a protein structure for full-length H-Ras in complex with a 1,2-dimyristoylglycero-3-phosphocholine bilayer [12], we propose computational protein structure models of wild-type H-Ras (Pro173/Pro174) and a mutant H-Ras (Ser173/Ser174) as shown in Figure W2. The predicted mutant structure displayed noticeable changes in the HVR, especially in the orientation of the loop containing residues 176 to 182, known to interact with the membrane. This subtle difference may be critical for the activation of H-Ras-specific signal transduction. The structural-functional relationship of H-Ras and the exact molecular mechanisms by which H-Ras induces breast cell invasion awaits the completion of three-dimensional structural analysis of H-Ras that includes the membrane-bound HVR.
This study demonstrated that the HVR (166–189) of H-Ras, especially Pro173 and Pro174, plays a critical role in the activation of H-Ras and H-Ras-specific induction of mammary cell invasion/migration.
The present study sheds light on the structural basis underlying the Ras isoform-specific invasive program of breast epithelial cells. It also provides a rationale for the development of agents that specifically target invasion-related Ras pathways, which may have broad implications in breast cancer.
Supplementary Material
Acknowledgments
The authors thank Yunje Cho at POSTECH, Pohang, South Korea, for helpful discussion.
Footnotes
This work was supported by the Korean Science and Engineering Foundation/National Research Laboratory Program (MEST, no. ROA-2008-000-20070-0) and by the National Research Foundation of Korea (MEST, no. R11-20100001707).
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 2.Clair T, Miller WR, Cho-Chung YS. Prognostic significance of the expression of a ras protein with a molecular weight of 21,000 by human breast cancer. Cancer Res. 1987;47:5290–5293. [PubMed] [Google Scholar]
- 3.Clark GJ, Der CJ. Aberrant function of the Ras signal transduction pathway in human breast cancer. Breast Cancer Res Treat. 1995;35:133–144. doi: 10.1007/BF00694753. [DOI] [PubMed] [Google Scholar]
- 4.Franks LM, Teich NM. Cellular and Molecular Biology of Cancer. New York, NY: Oxford University Press; 1997. [Google Scholar]
- 5.Moon A, Kim MS, Kim TG, Kim SH, Kim HE, Chen YQ, Kim HR. H-ras, but not N-ras, induces an invasive phenotype in human breast epithelial cells: a role for MMP-2 in the H-ras-induced invasive phenotype. Int J Cancer. 2000;85:176–181. doi: 10.1002/(sici)1097-0215(20000115)85:2<176::aid-ijc5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 6.Kim MS, Lee EJ, Kim HR, Moon A. p38 kinase is a key signaling molecule for H-Ras-induced cell motility and invasive phenotype in human breast epithelial cells. Cancer Res. 2003;63:5454–5461. [PubMed] [Google Scholar]
- 7.Shin I, Kim S, Song H, Kim HR, Moon A. H-Ras-specific activation of Rac-MKK3/6-p38 pathway: its critical role in invasion and migration of breast epithelial cells. J Biol Chem. 2005;280:14675–14683. doi: 10.1074/jbc.M411625200. [DOI] [PubMed] [Google Scholar]
- 8.Song H, Ki SH, Kim SG, Moon A. Activating transcription factor 2 mediates matrix metalloproteinase-2 transcriptional activation induced by p38 in breast epithelial cells. Cancer Res. 2006;66:10487–10496. doi: 10.1158/0008-5472.CAN-06-1461. [DOI] [PubMed] [Google Scholar]
- 9.Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 10.McCormick F. Signal transduction. How receptors turn Ras on. Nature. 1993;363:15–16. doi: 10.1038/363015a0. [DOI] [PubMed] [Google Scholar]
- 11.Milburn MV, Tong L, deVos AM, Brünger A, Yamaizumi Z, Nishimura S, Kim SH. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 12.Gorfe AA, Babakhani A, McCammon JA. H-Ras protein in a bilayer: interaction and structure perturbation. J Am Chem Soc. 2007;129:12280–12286. doi: 10.1021/ja073949v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 14.Rowinsky EK, Windle JJ, Von Hoff DD. Ras protein farnesyltransferase: a strategic target for anticancer therapeutic development. J Clin Oncol. 1999;17:3631–3652. doi: 10.1200/JCO.1999.17.11.3631. [DOI] [PubMed] [Google Scholar]
- 15.Willumsen BM, Christensen A, Hubbert NL, Papageorge AG, Lowy D. The p21 ras C-terminus is required for transformation and membrane association. Nature. 1984;310:583–586. doi: 10.1038/310583a0. [DOI] [PubMed] [Google Scholar]
- 16.Hancock JF, Magee AI, Childs JE, Marshall CJ. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 17.Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, Cox AD, Wilson O, Kirschmeier P, Der CJ. Rho family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J Biol Chem. 2008;283:25150–25163. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright LP, Philips MR. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat Cell Biol. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 20.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, Parton RG, Henis YI, Kloog Y, Hancock JF. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Mol Cell Biol. 2005;25:6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draper JM, Smith CD. Palmitoyl acyltransferase assays and inhibitors. Mol Membr Biol. 2009;26:5–13. doi: 10.1080/09687680802683839. [review] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock JF, Peterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 23.Hancock JF, Cadwallader K, Paterson H, Marshall CJ. A CAAX or a CAAX motif and a second signal are sufficient for plasma membrane targeting of ras proteins. EMBO J. 1991;10:4033–4039. doi: 10.1002/j.1460-2075.1991.tb04979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Chen JX, Liao R, Deng Q, Zhou JJ, Huang S, Sun P. Sequential activation of the MEK-extracellular signal-regulated kinase and MKK3/6-p38 mitogen-activated protein kinase pathways mediates oncogenic ras-induced premature senescence. Mol Cell Biol. 2002;22:3389–3403. doi: 10.1128/MCB.22.10.3389-3403.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bihani T, Mason DX, Jackson TJ, Chen SC, Boettner B, Lin AW. Differential oncogenic Ras signaling and senescence in tumor cells. Cell Cycle. 2004;3:1201–1207. [PubMed] [Google Scholar]
- 26.Reddy HK, Graña X, Dhanasekaran DN, Litvin J, Reddy EP. Requirement of Cdk4 for v-H-ras Induced breast tumorigenesis and activation of the v-ras-induced senescence program by the R24C mutation. Genes Cancer. 2010;1:69–80. doi: 10.1177/1947601909358105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon A, Yong HY, Song JI, Cukovic D, Salagrama S, Kaplan D, Putt D, Kim H, Dombkowski A, Kim HR. Global gene expression profiling unveils S100A8/A9 as candidate markers in H-ras-mediated human breast epithelial cell invasion. Mol Cancer Res. 2008;6:1544–1553. doi: 10.1158/1541-7786.MCR-08-0189. [DOI] [PubMed] [Google Scholar]
- 28.Casey PJ, Seabra MC. Protein prenyltransferases. J Biol Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 29.Ishihara N, Yamashina S, Sakaguchi M, Mihara K, Omura T. Malfolded cytochrome P-450(M1) localized in unusual membrane structures of the endoplasmic reticulum in cultured animal cells. J Biochem. 1995;118:397–404. doi: 10.1093/oxfordjournals.jbchem.a124920. [DOI] [PubMed] [Google Scholar]
- 30.Yamazaki S, Sato K, Suhara K, Sakaguchi M, Mihara K, Omura T. Importance of the proline-rich region following signal-anchor sequence in the formation of correct conformation of microsomal cytochrome P-450s. J Biochem. 1993;114:652–657. doi: 10.1093/oxfordjournals.jbchem.a124232. [DOI] [PubMed] [Google Scholar]
- 31.Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, Stout CD. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 Å resolution: evidence for an induced fit model of substrate binding. Biochemistry. 2003;42:9335–9345. doi: 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- 32.Kemper B. Structural basis for the role in protein folding of conserved proline-rich regions in cytochromes P450. Toxicol Appl Pharmacol. 2004;199:305–315. doi: 10.1016/j.taap.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 33.Jaumot M, Yan J, Clyde-Smith J, Sluimer J, Hancock JF. The linker domain of the Ha-Ras hypervariable region regulates interactions with exchange factors, Raf-1 and phosphoinositide 3-kinase. J Biol Chem. 2002;277:272–278. doi: 10.1074/jbc.M108423200. [DOI] [PubMed] [Google Scholar]
- 34.Laude AJ, Prior IA. Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J Cell Sci. 2008;121(pt 4):421–427. doi: 10.1242/jcs.020107. [DOI] [PubMed] [Google Scholar]
- 35.Jaumot M, Hancock JF. Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14-3-3 interactions. Oncogene. 2001;20:3949–3958. doi: 10.1038/sj.onc.1204526. [DOI] [PubMed] [Google Scholar]
- 36.Hansen M, Rusyn EV, Hughes PE, Ginsberg MH, Cox AD, Willumsen BM. R-Ras C-terminal sequences are sufficient to confer R-Ras specificity to H-Ras. Oncogene. 2002;21:4448–4461. doi: 10.1038/sj.onc.1205538. [DOI] [PubMed] [Google Scholar]
- 37.Hansen M, Prior IA, Hughes PE, Oertli B, Chou FL, Willumsen BM, Hancock JF, Ginsberg MH. C-terminal sequences in R-Ras are involved in integrin regulation and in plasma membrane microdomain distribution. Biochem Biophys Res Commun. 2003;311:829–838. doi: 10.1016/j.bbrc.2003.10.074. [DOI] [PubMed] [Google Scholar]
- 38.Jeong HW, Nam JO, Kim IS. The COOH-terminal end of R-Ras alters the motility and morphology of breast epithelial cells through Rho/Rho-kinase. Cancer Res. 2005;65:507–515. [PubMed] [Google Scholar]
- 39.Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signaling. Cell Mol Life Sci. 2007;64:2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holt MR, Koffer A. Cell motility: proline-rich proteins promote protrusions. Trends Cell Biol. 2001;11:38–46. doi: 10.1016/s0962-8924(00)01876-6. [DOI] [PubMed] [Google Scholar]
- 41.Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I, Birchmeier W, Schlag PM. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15:59–67. doi: 10.1038/nm.1889. [DOI] [PubMed] [Google Scholar]
- 42.Wang B, Zou JX, Ek-Rylander B, Ruoslahti E. R-Ras contains a proline-rich site that binds to SH3 domains and is required for integrin activation by R-Ras. J Biol Chem. 2000;275:5222–5227. doi: 10.1074/jbc.275.7.5222. [DOI] [PubMed] [Google Scholar]
- 43.Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.