Preface
Regulatory machinery is focally organized within the interphase nucleus. Information contained in these nuclear microenvironments must be inherited during cell division to sustain physiologically responsive gene expression in progeny cells. Recent results suggest that focal mitotic retention of phenotypic transcription factors at promoter elements, together with histone modifications and DNA methylation, a mechanism collectively known as gene bookmarking, is a novel parameter of inherited epigenetic control that sustains cellular identity after mitosis. The epigenetic signatures imposed by bookmarking poise genes for activation or suppression following mitosis. We discuss the implications of phenotypic transcription factor retention on mitotic chromosomes in biological control and disease.
Keywords: nuclear organization, cancer, mitosis, Runx, epigenetic mechanism, histone modifications, DNA methylation
Introduction
Understanding the relationship of cellular phenotype with mechanisms that are DNA encoded (genetic) and those that are conveyed to progeny cells through non-genetic means (epigenetic) is an important and rapidly evolving biological question. Each cell lineage exhibits distinct epigenetic signatures, which can be reversibly modified to regulate gene expression. A subset of epigenetic modifications is inheritable through successive cycles of cell division, which permits the maintenance of cellular identity 1, 2. The post-translational modification of nucleosomal histone proteins and the methylation of gene promoters are two extensively characterized epigenetic mechanisms that regulate gene expression, influence cellular phenotypes without altering the genotype, and are inheritable through successive cell divisions 3–18 (BOX I). In addition to a prominent role in regulating gene expression during development and under normal physiological conditions, epigenetic control is important in cancer 12–14.
Box I. Established mechanisms of epigenetic regulation.
The information dictated by epigenetic signatures is equally important as the sequence information encoded by nucleic acids for regulating gene expression 1. Here we outline these established parameters of epigenetic control. For more extensive insights, we refer to in-depth reviews discussing multiple dimensions to epigenetic control 3–14.
DNA methylation
DNA methylation is a covalent modification that occurs at CpG dinucleotides and can be catalyzed by three different DNA methyl transferases (DNMT): DNMT1, DMNT3a, and DNMT3b 12. DNA methylation plays a crucial role in long-term silencing of transcription and in heterochromatin formation, either by directly interfering with the binding of transcription factors to their target sites or by altering chromatin structure through affecting histone modifications and nucleosome occupancy at gene promoter regions 13. During the development of cancer, many CpG islands undergo hypermethylation, which leads to changes in chromatin structure and causes silencing of tumor suppressor genes and genome instability; examples include the genes encoding p16 and Runx3 13, 14. CpG methylation is also important for X-chromosome inactivation and for establishing and maintaining the expression of ‘imprinted’ genes that are thought to be related to the onset and progression of tumor phenotype, such as Oct4 14.
Histone Modifications
More than 60 different residues in nucleosomal histones that undergo post-translational modifications have been identified 2–9. These modifications can either disrupt contacts between nucleosomes to increase chromatin accessibility and the recruitment of non-histone proteins or increase nucleosome-DNA interactions to render a closed chromatin conformation 2–9. For example, methylation of Lysine residue at position 9 (K9) of Histone 3 (H3) protein (i.e., H3K9 methylation) plays an important role in the transmission of heterochromatin to the progeny cells. This modification, which in turn recruits Heterochromatin Protein 1 (HP1), leads to additional H3K9-methylating activity that modifies nucleosomes, thus ensuring the transmission of the H3K9 methylation mark to the progeny cells 11.
Non-coding RNA molecules
Non-coding RNAs, including micro RNAs, and small nucleolar RNAs, also regulate gene expression at the epigenetic level 67. Examples include dosage compensation mechanisms mediated by rox RNA in Drosophila and by XIST RNA in mammals, and the silencing of genes and repetitive DNA sequences by RNA interference-related (such as microRNA and siRNA) pathways in most eukaryotes 68. These non-coding RNAs often act in concert with components of chromatin and the DNA methylation machinery to establish and/or sustain silencing of gene expression. MicroRNA- and siRNA-mediated gene silencing is often short-term and may not be inherited 69. However, XIST RNA in mammals, and small RNAs in Schizosaccharomyces pombe induce long-term inheritable gene silencing 70. Epigenetic mechanisms mediated by non-coding RNAs may also be important in development, transformation and tumor progression.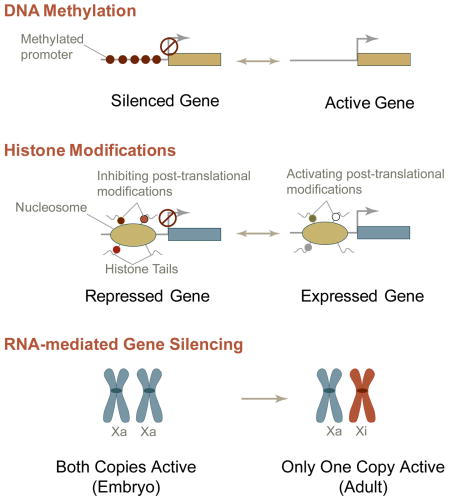
Gene bookmarking is emerging as a novel epigenetic mechanism that controls cell fate and lineage commitment 19. Although the term bookmarking was initially used to describe nuclease hypersensitivity of gene promoters in mitotic cells 19–21, it is now used to describe the retention of phenotype-specific transcription factors at target gene loci on mitotic chromosomes, allowing the necessary information to be conveyed to progeny cells. Several advances in understanding the composition and organization of regulatory complexes at chromosomal gene loci that include high-resolution microscopy, in vivo genomic occupancy assays, as well as genome-wide expression analyses, together with inheritable histone marks and DNA methylation, have added a novel epigenetic dimension to bookmarking. We use the term ‘architectural epigenetics’ to describe a combination of mechanisms, such as histone modifications and the binding of transcription factors and co-regulatory factors that mark the structural and functional state of a gene for inheritance following mitosis.
In this Opinion article, we evaluate the evidence for gene bookmarking, with an emphasis on mitotic retention of transcriptional regulatory machinery. Several lines of evidence suggest that a variety of regulatory and co-regulatory proteins associate with mitotic chromatin 15–18. We will focus on retention of transcription factors, with an emphasis on phenotypic master regulators, with target genes during mitosis. We first discuss the role of bookmarking during development, including aspects such as control of cell growth, maintenance of lineage commitment, cell survival and asymmetric cell division. We then discuss the potential clinical implications of deregulated gene bookmarking, highlighting recent evidence that this mechanism might be important in tumorigenesis. Finally, we put forward a model drawing together the various elements involved in gene bookmarking and suggest that this mechanism plays a crucial role in ensuring that cell retain memory of phenotype-specific gene expression after cell division.
Interphase Nuclear Microenvironments
To understand how mitotic gene bookmarking controls gene expression, it is important to appreciate how regulatory proteins are organized in the interphase nucleus prior to and following mitosis. Temporal and spatial assembly of the transcriptional regulatory machinery is a pre-requisite for physiologically responsive gene expression. The compartmentalization of the multifunctional protein complexes that support transcription, replication and repair is illustrated by focal organization of both regulatory proteins and nucleic acids in nuclear microenvironments These include RNA polymerase I and II machinery, Runx transcription factors, SC35 splicing speckles, steroid hormone receptors, and chromatin remodeling machinery 22–27. This focal organization is quantifiable by bioinformatics approaches that include intranuclear informatics, which examines the organization of subnuclear domains.
Nuclear microenvironments must be dynamic to accommodate protein-protein and protein-DNA interactions in response to physiological cues. Subnuclear domains are assembled by scaffolding proteins (e.g., runt-related transcription factor (Runx) proteins) at the site of their target-gene promoters, where they recruit co-regulatory proteins. There are defined mechanisms that regulate the targeting of scaffolding proteins to nuclear microenvironments; for example, in osteoblasts, myeloid cells and lymphoid cells, Runx transcription factors are directed to transcriptionally active subnuclear domains by a nuclear matrix targeting signal (NMTS;22, a functionally autonomous motif present in the carboxyl-terminus of all Runx proteins. Furthermore, genetic knock-in studies with mice expressing either Runx1 or Runx2 proteins that have impaired subnuclear targeting functions show similar phenotypes to Runx-null mice, indicating that the assembly and organization of nuclear microenvironments by Runx proteins is required for their biological activity 28, 29.
The in vivo requirement for proper subnuclear targeting of regulatory proteins for physiological functions emphasizes the importance of preserving subnuclear organization following mitosis when the entire nucleus is remodeled. There must be mechanisms to restore organization of the regulatory machinery following mitosis to sustain fidelity of gene regulation in progeny cells.
Mitotic Gene Bookmarking
Global remodeling of cellular and nuclear architecture as well as a general repression of transcription during mitosis 30 leads to dynamic re-localization of many transcription factors (such as Ets1, Oct2, B-Myb, Sp1) and their displacement from mitotic chromosomes in mammalian cells. In addition, several key regulatory proteins, that include cyclins, are degraded at the onset of mitosis and re-synthesized as soon as cells exit mitosis 20, 31, 32(Figure 1A). Transcription factors that are displaced from mitotic chromosomes or are degraded at the onset of mitosis resume target gene occupancy or are re-synthesized at a varied time-scale following mitosis that depends upon their requirements for cellular functions. However, it is also necessary to sustain the structural and functional integrity of the core regulatory machinery following a mitotic event to accommodate requirements for gene expression in progeny cells to maintain cell growth potential and lineage identity; gene bookmarking during mitosis may be an important mechanism that enables this to occur.
Figure 1. Distinct mechanisms control protein content of the cell during mitosis.
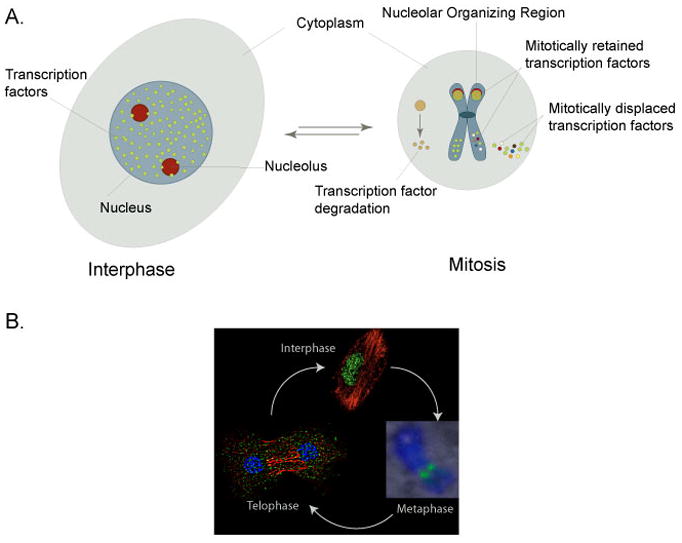
(A) Examples of three distinct mechanisms that regulate mitotic presence of proteins are depicted. The cytoplasm is illustrated by a light blue oval, the nucleus is a blue circle, and nucleoli are red circles. Three types of nuclear proteins are shown: 1) proteins that are destined to be degraded during mitosis, 2) proteins that are displaced from mitotic chromosomes, and 3) transcription factors that are retained on target gene loci through sequence-specific protein-DNA interactions during mitosis. As cells enter mitosis, many nuclear proteins are either displaced from mitotic chromosomes to be re localized in G1 nuclei or are degraded to be re synthesized in progeny cells. Several phenotypic transcription factors are retained on mitotic chromosomes and localize to either RNA Pol I transcribed nucleolar organizing regions or RNA Pol II regulated genes involved in cell proliferation and differentiation. (B) Mitosis of a mammalian cell is depicted. Chromosomes are schematically represented in light and dark blue colors. Microtubules and spindle are drawn as green lines, while the centromeres are shown as red color circles (Metaphase).
Gene bookmarking may include several mechanisms for retention of structural information (e.g., specific histone modifications or non-genetic DNA alterations) and functional state (e.g., repression or activation marked by transcription factors and co-regulatory proteins) of a gene 15–18. These parameters of bookmarking a gene are not mutually exclusive. One gene may be marked by all, some or only one of these mechanisms. Experimentally, it is possible to identify genes that are bookmarked by any of these mechanisms through techniques that include immunofluorescence microscopy and chromatin immunoprecipitation during mitosis using specific antibodies against proteins of interest. Finally, interference with gene bookmarking should lead to perturbed gene expression in progeny cells. Recent studies have shown that mitotic gene bookmarking is indeed a general mechanism to sustain cellular identity following mitosis. Here, we provide examples of specific biological functions in which phenotypic transcription factors as well as components of the general transcriptional and chromatin remodeling machineries remain associated with target genes during cell division.
Bookmarking functions during development
Control of cell growth
Studies using biochemical techniques and in situ immunofluorescence microscopy suggest that selected regulatory proteins involved in critical cellular functions (including proliferation, growth and differentiation) remain associated with target gene loci on mitotic chromosomes 33–38. These gene-protein associations are DNA sequence specific and mark target genes for transcription following mitosis. An example of gene bookmarking is the Myc gene promoter, which is rapidly re-activated as cells transition to the G1 phase. Levens and colleagues have shown that that this gene is permanganate sensitive, an indication of accessibility to single-strand nucleases in the promoter regions only during mitosis 39, 40. It is known that many DNA-binding proteins (including the sequence-specific single-stranded DNA-binding proteins hnRNP K and FBP that occupy the gene promoter region during mitosis) can modulate transcription from the Myc promoter 41, 42. However, it remains unclear how these factors interact preferentially with single-stranded DNA. The mitotic sensitivity of the promoter to permanganate suggests that the “bookmark” – that is, the mitotically bound transcription factors hnRNP K and FBP – modifies its interaction with promoters following mitosis, and can re-associate prior to the next round of cell division.
Another example is provided by Runx proteins, master regulators of osteogenesis, hematopoeisis, neurogenesis and gastrointestinal development 43–47. As noted above, the localization of Runx proteins to subnuclear domains is required for their biological activity 28, 29 as disruption of this nuclear localization causes cell-specific effects; a transformed phenotype is observed in myeloid progenitors, whereas there is an inhibition of osteolytic disease caused by breast cancer cells that metastasize to bone 48, 49. During mitosis, all Runx proteins (Runx1, 2 and 3) associate with RNA Pol I-transcribed ribosomal RNA genes and RNA Pol II-transcribed phenotype-specific genes that are involved in control of the cell cycle and differentiation 33, 34, 50, 51. Runx transcription factors are equally partitioned in progeny cells at the completion of cell division 52(Figure 1B). The association of Runx factors with ribosomal and cell-cycle regulatory genes (such as the cell-cycle inhibitor p21) during mitosis ‘marks’ these genes for repression during the early G1 phase of the cell cycle 33, 34. In addition, the occupancy of differentiation-related genes (such as Mad-related proteins (Smads), effectors of the transforming growth factor beta/bone morphogenetic protein signaling pathway) by Runx proteins during mitosis provides a mechanistic basis for lineage-restricted transcriptional memory in progeny cells. The occupancy and regulation of RNA Pol I- and RNA Pol II-transcribed genes by Runx proteins during interphase and mitosis enables them to coordinate cell proliferation, growth and differentiation by acting at both the genetic and epigenetic levels.
Maintenance of lineage commitment
The developmentally regulated and globin gene loci contain several functionally related genes arranged in tandem 53. In hematopoietic progenitor cells, erythroid specific DNase I hypersensitive sites (HSs) have been detected in the distal regulatory regions of the mouse globin gene cluster 54. Progeny cells can inherit these hypersensitive sites over at least 20 generations 21. Recent studies have provided mechanistic insight into inheritance of globin gene transcription status through mitoses 55. The lineage-restricted expression of globin genes is controlled mainly by the transcription factors NF-E2 and GATA 56, 57. NF-E2 remains bound to mitotic chromosomes, while GATA1 is dissociated from the condensed chromatin during mitosis 55. This observation suggests that NF-E2 is an epigenetic marker that maintains the locally hypersensitive state of the globin gene clusters. NF-E2 can also recruit the coactivators TAFII130 and CBP to the globin gene locus, further supporting its role in rapid re activation of the gene post mitotically. In addition, the distal regulatory regions of transcriptionally competent globin gene loci are marked during mitosis by active histone modifications such as H3 acetylation, H3 K4 dimethylation and K79 dimethylation 55.
The basic helix-loop-helix myogenic regulatory factors (including MyoD, Myf5, and MRF4) bind to sequences called E-boxes in target gene promoters, and play crucial roles in skeletal-muscle development (reviewed in 35, 58, 59). Together with Mef2 proteins and E-box factors, these transcription factors are responsible for coordinating muscle-specific gene expression by negatively regulating proliferation and promoting differentiation 60, 61. During interphase, muscle-regulatory proteins are organized in punctate nuclear microenvironments, where control of muscle gene expression takes place. During the proliferative stage of uncommitted mesenchymal cells, MyoD is localized to mitotic chromosomes and associates with ribosomal RNA genes and nucleolar-organizing regions (NORs). The association of MyoD with the interphase nucleolus in early stages of myogenesis, and its replacement by myogenin in later stages, results in the repression of ribosomal RNA genes, concomitant with the initiation of the skeletal-muscle differentiation program 36. These observations are consistent with a gene bookmarking mechanism.
CCAAT/enhancer-binding proteins α and δ (C/EBPα and δ) are early markers in the adipocyte differentiation program and provide another example of gene bookmarking 62. As adipocyte differentiation proceeds, both C/EBPα and δ become capable of interacting with the C/EBP regulatory element in the C/EBPβ gene promoter. The expression of C/EBPβ, in turn, is upregulated and this protein then acts as a transcriptional activator of late-adipocyte genes. During mitotic clonal expansion, pre-adipocytes enter S phase synchronously, concomitant with the localization of C/EBPα and δ to centromeres through C/EBP consensus-binding sites in centromeric satellite DNA. As mitotic expansion of pre-adipocytes ceases and the differentiation program ensues, C/EBPβ, which has anti-proliferative activity, becomes centromere-associated 37. Recent studies from our lab show that, in addition to occupying centromeric repeats, C/EBP transcription factors also occupy ribosomal RNA genes during mitosis. As pre-adipocytes complete cell division, C/EBP proteins repress ribosomal RNA genes, consistent with their role in initiating adipocyte differentiation when ribosomal gene expression is down-regulated 36. The association of C/EBP transcription factors with mitotic chromosomes and their direct repression of ribosomal RNA genes suggest that these phenotype-specific regulatory proteins mediate lineage commitment and maintenance through their bookmarking of target gene loci on metaphase chromosomes during the mitotic clonal expansion of pre-adipocytes 36, 37(Figure 2).
Figure 2. Coordination of cell growth, proliferation and differentiation of mesenchymal stem cells into three distinct lineages.
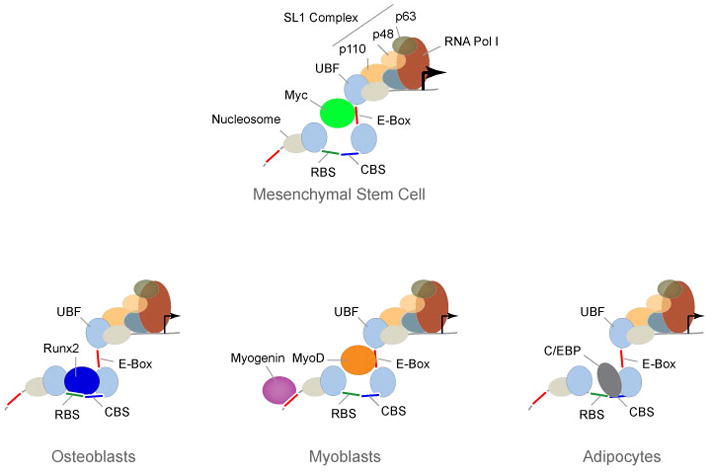
Mesenchymal stem cells (MSCs) can be differentiated into osteoblasts, myoblasts, or adipocytes, depending upon the available cohort of regulatory proteins, providing an example of the developmental relevance of gene bookmarking. A ribosomal DNA (rDNA) repeat that is occupied by the proliferation-promoting Myc transcription factor is illustrated. Myc upregulates rRNA transcription in non-committed, proliferating MSCs and contributes to growth potential. In response to extracellular signals, MSCs express either Runx2 when differentiated into osteoblasts, muscle regulatory factors (i.e., MyoD and myogenin) when differentiated into myoblasts, or C/EBP upon adipocyte differentiation. These phenotypic proteins occupy rDNA repeats as cells differentiate into their respective lineages and down regulate rRNA expression, concomitant with the exit of MSCs from the cell cycle, reduced cell growth and initiation of a differentiation.
Cellular response to stress
The inducible heat shock protein 70 (hsp70i) gene is primarily upregulated upon an increase in temperature through transcriptional activity of the heat shock factor 1 (HSF1) protein 63. Extensive studies from the laboratory of Kevin Sarge 19 show that unlike many genes, the hsp70i gene maintains open chromatin conformation in the promoter region as demonstrated by DNase I accessibility experiments20. Mechanistically, HSF2, which is a family member of HSF1 and contains binding sites within the DNase hypersensitivity sites, occupies the hsp70i promoter during mitosis and interacts with the CAP-G subunit of condensin complexes 38. Concomitantly, HSF2 also recruits the serine/threonine phosphatase PP2A to the promoter, which deactivates the condensin complexes by dephosphorylation of CAP-G subunit. As a result, the region remains in open chromatin conformation and accessible to DNase I. Blocking the activity of HSF2 and the consequent bookmarking of the hsp70i gene compromises cell survival in the G1 phase 38.
Compromised Gene Bookmarking in Disease
Recently, evidence has also emerged that deregulation of gene bookmarking may have an important role in disease 51, 64–66, suggesting that understanding this mechanism better may have clinical utility. An example of the clinical relevance of gene bookmarking is provided by the ischemic-reperfusion injury related reactivation of the hepatocyte nuclear factor beta 1 (HNFβ1), a necessary step in the pathology of the disease. HNFβ remains associated with mitotic chromosomes 65. While cells lacking HNFβ1 are quiescent and maintain the transcriptional state of crucial cytogenic target genes, the expression of these genes is lost when cells are forced to proliferate by an ischemic-reperfusion injury. In addition, the chromatin of target genes acquires a condensed state. The authors propose that because HNFβ1 remains associated with mitotic chromosomes, it may be functioning as a bookmarking factor that controls the transcriptional regulation of target genes following mitosis.
Another example is leukemic fusion protein, AML1-ETO. AML1-ETO is generated by the translocation of a portion of chromosome 8 carrying the ETO gene in frame with the DNA binding domain of the Runx1 (also designated AML1) gene located on chromosome 21. Expression of AML1-ETO in myeloid progenitor cells results in a differentiation block and enhanced proliferative potential (reviewed in 47). Association of the leukemic AML1-ETO protein with mitotic chromosomes upregulates ribosomal RNA (rRNA) synthesis when compared to wild type Runx1, which downregulates the expression of rRNA genes 51. Upregulated rRNA synthesis is correlated with enhanced cell proliferation. Thus, AML1-ETO association with rRNA genes during mitosis and their upregulation post-mitotically provides a novel epigenetic mechanism for continued cell proliferation, a hallmark of sustained tumor phenotype 51.
Recently, it has also been shown that the mixed lineage leukemia protein (MLL), a chromatin remodeling factor that is associated with leukemia and regulates transcription by recruiting chromatin modifying machinery to target genes, remains associated with mitotic chromatin 66. This retention favors rapid reactivation of target genes, for example Meis1, Hoxc11, and EEF1A1, all of which have been implicated in the onset and progression of mixed lineage leukemia, in the G1 phase of the cell cycle by recruiting other chromatin modifying factors (i.e., Menin, RbBP5 and ASH2L) and by retaining the methylation of lysine 4 of histone H3.
Conclusions and Future Prospects
Here, we have provided examples that offer mechanistic insight into gene bookmarking and its role in post-mitotic gene regulation. We propose that epigenetic inheritance through mitosis plays a vital role in maintenance of cellular identity as well as coordinates lineage commitment. In our model, in addition to inheritable histone marks and non-genetic DNA modifications, phenotypic transcription factors and components of the basal transcriptional machinery are retained on mitotic chromosomes to convey necessary information to initiate and sustain lineage commitment (Figure 3). It is clear that all genes are not bookmarked during mitosis and those that are bookmarked, are not regulated by a single epigenetic mechanism. A likely scenario is that a series of epigenetic mechanisms operate on each gene that is mitotically bookmarked. For example, during mitosis a gene that is required for lineage maintenance in post-mitotic cells may carry a unique pattern of histone modifications, an accessible chromatin conformation and be occupied by a sequence-specific transcription factor at its promoter; together these distinct, yet functionally overlapping epigenetic mechanisms provide a mitotic epigenetic signature for a gene that is bookmarked for post-mitotic expression.
Figure 3. Mitotic retention of sequence-specific transcription factors as an epigenetic mechanism for lineage maintenance in normal cells and for sustained tumor phenotype in cancer cells.
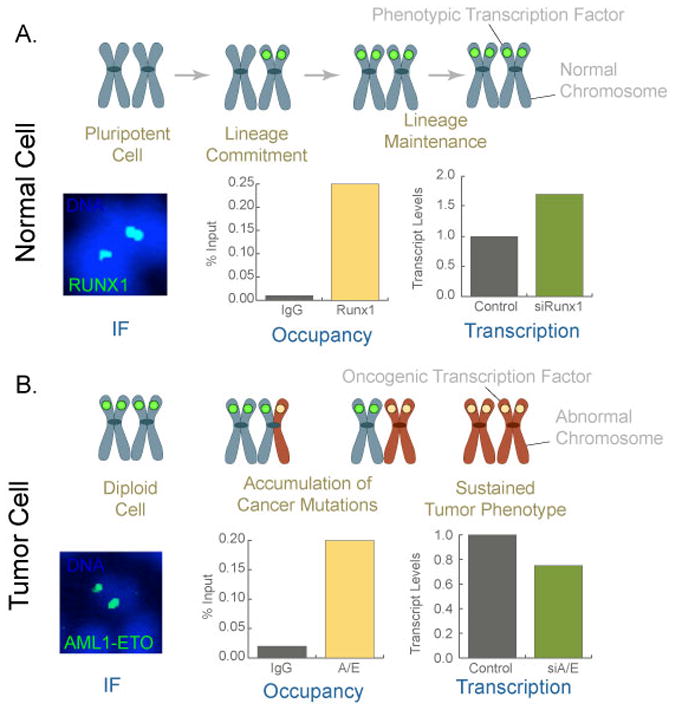
Gene loci on metaphase chromosomes (blue color) are occupied by phenotypic transcription factors (green circle) for lineage maintenance through successive cell divisions in normal cells. This possibility is shown by Runx1, a hematopoietic master regulator, which associates with mitotic chromosomes (immunofluorescence; IF), occupies target genes during mitosis (Occupancy as shown by chromatin immunoprecipitation assays), and regulate their expression in interphase (as represented by Transcription, where transcripts are quantitatively measured from cells with (control) or without Runx1 (siRunx1)). In tumors, as cells undergo genomic instability and accumulate cancer mutations, it is possible that one allele (shown here as red chromosome arm) is occupied by oncogenic transcription factor (which can be a phenotypic protein, ectopically expressed in cancer cells). Successive cell divisions and acquisition of additional cancer mutations then lead to the replacement of phenotypic regulatory proteins with oncogenic transcription factors. Occupancy of target gene loci with oncogenic proteins results in a sustained tumor phenotype. This possibility is reflected by the chimeric AML1-ETO (A/E) protein that is encoded by the translocated Runx1 gene in frame with ETO gene. The oncogenic Runx1 translocation associates with mitotic chromosomes (IF), occupies target genes during mitosis (Occupancy), and regulate their expression in interphase (Transcription). Images and data are modified from Reference 51.
Key Terms
Architectural epigenetics
Composite of inherited structural and functional regulatory information during mitosis that includes the cohort of regulatory and co-regulatory factors that mediate transcription and the chromatin organization of cognate gene loci.
The functions of gene bookmarking during mitosis may include, but are not restricted to, the poising of gene expression for those genes that are involved in progression of the G1 phase of the cell cycle, maintenance of cellular identity, and commitment of progenitor cells to a specific lineage. Mitotic association of phenotype-specific regulatory proteins with genes in an allele-specific manner in pluripotent cells might facilitate the asymmetric distribution of transcription factors to cells that are destined to commit to a particular lineage. In committed cells, both alleles can be accessible to phenotypic transcription factors during mitosis thus leading to equal partitioning into progeny cells for the maintenance of the lineage.
The implications of gene bookmarking remain to be fully explored. Key questions include: do genes that do not retain cognate regulatory factors have other mitotic bookmarks for activation or suppression in progeny cells? When do mitotically displaced or degraded transcription factors resume occupancy of target genes? Why only a select set of genes maintains mitotic bookmarks? Are genes epigenetically regulated in a selective manner during asymmetric cell division? What are the identities of the transcriptional regulatory proteins and their co-factors that occupy mitotic gene loci? What is the dynamics of transcription factor interaction with mitotic chromosomes in live cells? While complex mechanistically and technically challenging, these dimensions are experimentally approachable. From recent studies, we already know that same gene (e.g., hsp70i) can be book marked by persistence of a histone modifications as well as retention of transcription factors during mitosis, indicating that these mechanisms are not mutually exclusive. Similarly, chromatin immunoprecipitation experiments have shown that the osteogenic Runx transcription factor carries TLE1, a co-repressor, through mitosis 15–18. Lower organisms, such as yeast and Drosophila, offer examples of asymmetric cell division, with implications in lineage commitment. However, it remains challenging to comprehensively address the bookmarking of many genes due to the availability of specific antibodies and accessibility of regulatory factors by antibodies during mitosis. Advances in live cell imaging, high-resolution immunofluorescence microscopy and genome-wide proteomics now allow us to address some of these fundamental biological questions.
Despite these open questions and a requirement for in-depth studies that firmly link gene bookmarking with the onset and progression of disease, there is evidence to suggest that gene bookmarking may be clinically relevant. Specific targeting of gene-bookmarking mechanisms could offer novel options for targeted therapy, with enhanced specificity and reduced off-target activity compared with global inhibition of chromatin modifications or DNA methylation. Importantly, the mitotic association of regulatory proteins results in focal concentrations of factors that can favorably influence pharmacological kinetics, i.e., a minimal drug concentration will be required. In addition, the assessment of genes that are bookmarked, combined with global, genome-wide evaluation of histone modifications and DNA methylation, may provide a more complete epigenetic signature for the diagnosis and prognosis of cancer and for monitoring its progression and the efficacy of therapy.
Acknowledgments
The authors thank Patricia Jamieson for her assistance in preparation of this article. This work was supported by National Institutes of Health grants P01 AR048818 and P01 CA082834. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Reference List
- 1.Urnov FD, Wolffe AP. Above and within the genome: epigenetics past and present. J Mammary Gland Biol Neoplasia. 2001;6:153–167. doi: 10.1023/a:1011304606604. [DOI] [PubMed] [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 4.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 5.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Nathan D, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shilatifard A. CHROMATIN MODIFICATIONS BY METHYLATION AND UBIQUITINATION: Implications in the Regulation of Gene Expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 8.Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 14.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 15.Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow CM, et al. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saurin AJ, et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke LJ, et al. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J. 2005;24:3291–3300. doi: 10.1038/sj.emboj.7600793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle. 2009;8:818–823. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- 21.Groudine M, Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982;30:131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- 22.Zeng C, et al. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma H, Siegel AJ, Berezney R. Association of chromosome territories with the nuclear matrix. Disruption of human chromosome territories correlates with the release of a subset of nuclear matrix proteins. J Cell Biol. 1999;146:531–542. doi: 10.1083/jcb.146.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Spatial relationship between transcription sites and chromosome territories. J Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaich AK, et al. Subnuclear trafficking and gene targeting by steroid receptors. Ann N Y Acad Sci. 2004;1024:213–220. doi: 10.1196/annals.1321.002. [DOI] [PubMed] [Google Scholar]
- 26.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner S, Chiosea S, Nickerson JA. The spatial targeting and nuclear matrix binding domains of SRm160. Proc Natl Acad Sci U S A. 2003;100:3269–3274. doi: 10.1073/pnas.0438055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi JY, et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci, USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.North T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 30.Spencer CA, Kruhlak MJ, Jenkins HL, Sun X, Bazett-Jones DP. Mitotic transcription repression in vivo in the absence of nucleosomal chromatin condensation. J Cell Biol. 2000;150:13–26. doi: 10.1083/jcb.150.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He S, Davie JR. Sp1 and Sp3 foci distribution throughout mitosis. J Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- 32.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Young DW, et al. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- 34.Young DW, et al. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weintraub H, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 36.Ali SA, et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci U S A. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xing H, et al. Mechanism of hsp70i gene bookmarking. Science. 2005;307:421–423. doi: 10.1126/science.1106478. [DOI] [PubMed] [Google Scholar]
- 39.Michelotti EF, Sanford S, Levens D. Marking of active genes on mitotic chromosomes. Nature. 1997;388:895–899. doi: 10.1038/42282. [DOI] [PubMed] [Google Scholar]
- 40.Duncan R, et al. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 41.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michelotti GA, et al. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaidi SK, et al. Nuclear microenvironments in biological control and cancer. Nat Rev Cancer. 2007;7:454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- 44.Lian JB, et al. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- 45.Cameron ER, Neil JC. The Runx genes: lineage-specific oncogenes and tumor suppressors. Oncogene. 2004;23:4308–4314. doi: 10.1038/sj.onc.1207130. [DOI] [PubMed] [Google Scholar]
- 46.Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-beta superfamily signaling. Curr Opin Genet Dev. 2003;13:43–47. doi: 10.1016/s0959-437x(03)00007-8. [DOI] [PubMed] [Google Scholar]
- 47.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 48.Vradii D, et al. Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype. Proc Natl Acad Sci, USA. 2005;102:7174–7179. doi: 10.1073/pnas.0502130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Javed A, et al. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci, USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pande S, et al. Subnuclear targeting of the Runx3 tumor suppressor and its epigenetic association with mitotic chromosomes. J Cell Physiol. 2009;218:473–479. doi: 10.1002/jcp.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakshi R, et al. The leukemogenic t(8;21) fusion protein AML1-ETO controls ribosomal RNA genes and associates with nucleaolar organizing regions at mitotic chromosomes. J Cell Sci. 2008;21:3981–3990. doi: 10.1242/jcs.033431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaidi SK, et al. Mitotic partitioning and selective reorganization of tissue specific transcription factors in progeny cells. Proc Natl Acad Sci USA. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao A, Moi P. Regulation of the globin genes. Pediatr Res. 2002;51:415–421. doi: 10.1203/00006450-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 54.McGhee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G. A 200 base pair region at the 5′ end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- 55.Xin L, et al. Exploring cellular memory molecules marking competent and active transcriptions. BMC Mol Biol. 2007;8:31. doi: 10.1186/1471-2199-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 57.Whitelaw E, Tsai SF, Hogben P, Orkin SH. Regulated expression of globin chains and the erythroid transcription factor GATA-1 during erythropoiesis in the developing mouse. Mol Cell Biol. 1990;10:6596–6606. doi: 10.1128/mcb.10.12.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohun T. Muscle differentiation. Curr Opin Cell Biol. 1992;4:923–928. doi: 10.1016/0955-0674(92)90119-w. [DOI] [PubMed] [Google Scholar]
- 59.Dias P, Dilling M, Houghton P. The molecular basis of skeletal muscle differentiation. Semin Diagn Pathol. 1994;11:3–14. [PubMed] [Google Scholar]
- 60.Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 61.Rudnicki MA, Jaenisch R. The MyoD family of transcription factors and skeletal myogenesis. BioEssays. 1995;17:203–209. doi: 10.1002/bies.950170306. [DOI] [PubMed] [Google Scholar]
- 62.MacDougald OA, Lane MD. Transcriptional regulation of gene expression during adipocyte differentiation. Annu Rev Biochem. 1995;64:345–373. doi: 10.1146/annurev.bi.64.070195.002021. [DOI] [PubMed] [Google Scholar]
- 63.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9:122–133. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quyn AJ, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–181. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 65.Verdeguer F, et al. A mitotic transcriptional switch in polycystic kidney disease. Nat Med. 2010;16:106–110. doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Blobel GA, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 68.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 69.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 70.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]


