Abstract
Modification of proteins by ubiquitin (Ub)-like proteins (UBLs) plays an important role in many cellular processes, including cell cycle progression, nuclear transport, and autophagy. Protein modification occurs via UBL-conjugating and -deconjugating enzymes, which presumably exert a regulatory function by determining the conjugation status of the substrate proteins. To target and identify UBL-modifying enzymes, we produced Nedd8, ISG15, and SUMO-1 in Escherichia coli and equipped them with a C-terminal electrophilic trap (vinyl sulfone [VS]) via an intein-based method. These C-terminally modified UBL probes reacted with purified UBL-activating (E1), -conjugating (E2), and -deconjugating enzymes in a covalent fashion. Modified UBLs were radioiodinated and incubated with cell lysates prepared from mouse cell lines and tissues to allow visualization of polypeptides reactive with individual UBL probes. The cell type- and tissue-specific labeling patterns observed for the UBL probes reflect distinct expression profiles of active enzymes, indicating tissue-specific functions of UBLs. We identify Ub C-terminal hydrolase L1 (UCH-L1) and DEN1/NEDP1/SENP8, in addition to UCH-L3, as proteases with specificity for Nedd8. The Ub-specific protease isopeptidase T/USP5 is shown to react with ISG15-VS. Furthermore, we demonstrate that the desumoylation enzyme SuPr-1 can be modified by SUMO-1-VS, a modification that is dependent on the SuPr-1 active-site cysteine. The UBL probes described here will be valuable tools for the further characterization of the enzymatic pathways that govern modification by UBLs.
Ubiquitin (Ub) is a conserved 76-amino-acid protein attached posttranslationally to substrate proteins. This conjugation occurs through an isopeptide bond between the C-terminal carboxylate of Ub and the ɛ-NH2 of a lysine side chain in the target protein. Conjugation is achieved by the sequential action of an E1 activating enzyme, E2 conjugating enzymes, and E3 ligases (22). The removal of Ub from substrates is carried out by deubiquitinating enzymes. The long-known Ub-specific cysteine protease families of Ub C-terminal hydrolases (UCHs) and Ub-specific processing proteases (UBPs/USPs) were recently joined by a Ub-specific JAMM motif containing metalloprotease and cysteine proteases containing an OTU domain (2, 8, 13, 63, 67).
Ub-like proteins (UBLs) are a set of small proteins that share with Ub the ability to be conjugated to a lysine residue in a substrate protein (26). Many UBLs are related in sequence to Ub, and a three-dimensional fold similar to that in Ub has been reported for Nedd8 and SUMO (3, 66). The UBLs ISG15 (also called UCRP) and FAT10 resemble two Ub moieties fused in tandem (19, 54). UBLs do not generally appear to be assembled into multimeric chains upon conjugation to substrates, with the possible exception of SUMO-2 and SUMO-3 (62). Like Ub, most UBLs are expressed as inactive precursors, with extensions at the C terminus, which prevent direct conjugation (26) (Table 1). These precursors must be processed by specific proteases, which release the mature UBL and the tail. The C termini of the mature forms of most UBLs terminate in a Gly-Gly motif, as does Ub.
TABLE 1.
UBLs to which the UBL-intein-CBD expression approach was applieda
| UBL | % Identity to Ub | % Identity of mouse and human UBLsb | C-terminal amino acidsc | UBL-intein-CBD solubilityd | Radioiodinatione |
|---|---|---|---|---|---|
| Ub | 100 | 100 | LVLRLRGG-XXXXX | + | + |
| Nedd8 | 57 | 100 | LVLALRGG-GGLGQ | + | + |
| ISG15 | 28/34g | 65 | KHLRLRGG-GGDQCA | + | + |
| SUMO-1 | 18 | 100 | VYQEQTGG-HSTV | + | + |
| URM-1 | 13 | 93 | FISTLHGG | + | + |
| Fau | 31 | 95 | VAGRMLGG-KVHGSLARAGKV | + | NDf |
| FAT10 | 28/34g | 68 | LTTHCTGG | − | ND |
| HUB1 | 22 | 100 | GMNLELYY-Q | − | ND |
| Apg12 | 11 | 89 | YCKSQAWG | − | ND |
Characteristics are shown for UBLs from the mouse. The amino acid sequences used are derived from the clones described in Materials and Methods.
Percentages apply to the processed forms of the UBLs.
The dash in the C-terminal sequence indicates the position where processing occurs to generate the mature UBL. URM1, FAT10, and Apg12 are expressed in their mature form. The C-terminal sequence of Fau extends beyond the point shown.
+, soluble; −, insoluble.
+, could be radioiodinated.
ND, not done.
The two numbers indicate homology to Ub within individual domains.
The conjugation pathway of UBLs is analogous to the Ub pathway, involving E1-like and E2-like enzymes and in certain cases also E3-like factors (26). The E1-like enzymes for Nedd8 and SUMO-1, Ula1/Uba3 and Aos1/Uba2, respectively, are dimeric (11, 16, 18, 29, 38). The Uba subunit corresponds to the C terminus of the Ub E1, Uba1, and harbors the active-site cysteine, whereas the other subunits correspond to the Uba1 N terminus. Whereas there are many Ub-specific E2s, each UBL appears to be served by a single E2-like enzyme (18, 27, 38, 45, 53). Several E3-like proteins exist for SUMO (28, 44, 48, 51, 57). Although the SUMO E2 Ubc9 can directly recognize and modify a lysine contained in a sumoylation motif, these E3-like factors facilitate sumoylation of specific substrates. Furthermore, assembled SCF/CBC Ub E3 ligases appear to play a role in the modification of their cullin components by Nedd8 (32). No E3-like factors have been identified yet for any of the other UBLs.
Sumoylation has been implicated in cell cycle progression, nuclear import, the subnuclear localization of target proteins (in particular in relation to polymorphonuclear leukocyte nuclear bodies), and regulation of transcription (5, 12, 37, 47, 51, 56, 58). In these cases SUMO is thought to function as a mediator of protein-protein interactions or to confer conformational changes in the target protein. In addition, sumoylation can antagonize ubiquitination by competing with Ub for modification of specific lysines in target proteins (10, 23). The only known substrates of Nedd8 modification are the cullins (24), which are components of the SCF/CBC Ub E3 ligases. Neddylation of cullins plays a role in the recruitment of E2 to the ligase complex, thereby facilitating Ub conjugation (32, 33, 70). Roles for Nedd8 conjugation in cell cycle progression and cytoskeletal regulation have also been reported (35, 61). Expression of ISG15 is inducible by alpha/beta interferons. ISG15 has multiple targets, of which a limited number, i.e., serpin 2A, phospholipase Cγ1, Jak1, ERK1, and STAT1, have been identified to date (21, 40, 42). In the cell, ISG15 colocalizes with intermediate filaments and ISGylation may be involved in certain aspects of neurological disease or function and may modulate signaling through the JAK-STAT pathway (39, 42, 55).
SUMO deconjugation is catalyzed by a family of cysteine proteases distinct from the Ub-specific UCHs and UBPs. The first member identified was the yeast Ulp1 protein, which catalyzes both SUMO precursor processing and SUMO substrate cleavage (37). In mammals, SENP1/SuPr-2, SENP2/Axam/Supr-1, SMT3IP2/Axam2, SUSP1, SMT3IP1, and an unidentified 30-kDa hydrolase have been described as SUMO proteases (5, 17, 31, 34, 49, 50, 60, 73, 74). The UCH-L3 and USP21 proteases have been described as having dual specificity for Nedd8 and Ub (15, 64). Recently, a subunit of the COP9 signalosome (CSN) harboring a JAMM metalloprotease motif was found to underlie the CSN′s Nedd8 isopeptidase activity (9). In addition, three different groups reported the identification of DEN1/NEDP1/SENP8, a Nedd8-specific protease of the ULP family (14, 43, 71). The only deconjugating protease with specificity for ISG15 identified to date is UBP43/USP18 (41). A mouse deficient in UBP43/USP18 accumulates ISG15 conjugates (but not Ub conjugates) and develops neurological abnormalities, resulting in premature death (55). A ∼100-kDa ISG15 precursor processing enzyme has been described, although its identity remains unknown (52).
Whereas each UBL is apparently served by a single E1-like enzyme and a single E2-like enzyme, multiple UBL proteases likely exist for each UBL (26). After initial identification of some UBL-specific proteases by genetic screens or biochemical assays, further family members were then tentatively assigned to the enzyme family based on sequence similarity (73). However, the specificities of many of these putative proteases for Ub or UBLs have remained undetermined, and more unidentified Ub/UBL proteases are likely to exist.
We have previously described the synthesis of C-terminally modified Ub derivatives that can covalently modify active Ub-deconjugating enzymes (7, 8). By this method, active Ub-specific proteases were targeted and identified based on their specificity for and activity towards Ub. We sought to extend this approach to UBLs. Here we describe the synthesis of Nedd8, ISG15, and SUMO-1 modified with a C-terminal vinyl sulfone (VS) moiety. We characterize their reactivities towards purified UBL-activating (E1), -conjugating (E2), and -deconjugating enzymes. Radiolabeling of the UBL derivatives was used to visualize modified enzymes in cell lysates, and epitope-tagged versions of UBL probes were used to identify proteases with specificity for Nedd8 and ISG15. The approach described here will be valuable for the further characterization of the enzymatic pathways involved in UBL modifications.
MATERIALS AND METHODS
Cloning of Ub-like modifiers into pTYB1.
Sequences of mouse UBLs were found by searching the National Center for Biotechnology Information databases. Clones containing the desired sequences were obtained from Research Genetics and the American Type Culture Collection. The database accession numbers of the sequences used are BE374499 (Nedd8), BF152346 (UCRP), BF100858 (SUMO-1), BG868942 (Fau), BG295603 (URM1), BE624821 (HUB1), BG862330 (FAT10), and AI574302 (Apg12). Open reading frames of the processed forms of the UBLs, omitting the C-terminal codon, were amplified by PCR. The PCR primers used had extensions at their 5′ ends to generate an NdeI restriction site at the N-terminal coding end of the PCR product and a SapI restriction site at the C-terminal coding end of the PCR product. To generate epitope-tagged versions of the UBL fusions, coding sequences for the epitope were included in the 5′ PCR primer, between the NdeI site and the methionine start codon of the UBL sequence. After digestion of the PCR product with NdeI and SapI, the fragment was cloned into the pTYB1 vector (New England Biolabs), generating an in-frame fusion with the intein and chitin binding domain (CBD) (72). All constructs generated were sequence verified.
Synthesis of UBL-VSs.
Expression of UBL-intein-CBD fusion proteins in Escherichia coli was induced at an optical density at 600 nm of ∼0.5 with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) for 2 h at 30°C (8). Cells from a 1-liter bacterial culture were pelleted, resuspended in 30 ml of 50 mM HEPES-100 mM sodium acetate (pH 6.5)-50 μM phenylmethylsulfonyl fluoride (PMSF) and lysed with a French press (1,500 lb/in2). After centrifugation, the supernatant was loaded onto a 15-ml chitin bead column (New England Biolabs) at a flow rate of 0.5 ml/min to allow binding of the UBL-intein-CBD fusion protein. The column was washed with 80 ml of lysis buffer, followed by 50 ml of lysis buffer containing 50 mM β-mercaptoethanesulfonic acid (MESNa) (Sigma). The column, containing buffer with MESNa, was incubated overnight at 37°C to allow on-column cleavage. The UBL-MESNa thioester was eluted with 25 ml of lysis buffer, and the fractions containing UBL-MESNa product were concentrated by using a Centriprep (3,000-molecular-weght cutoff; Millipore). The overall yield was 1 to 5 mg of UBL-MESNa per liter of bacterial culture. The N-terminal methionine of the UBL was removed by processing in certain cases.
UBL-MESNa was converted to the VS derivative in a chemical ligation reaction with a large excess of glycine VS (6). Glycine VS was added to an aliquot of concentrated UBL-MESNa (1 to 2 mg/ml; 500 μl) to a final concentration of 0.25 M, followed by addition of 75 μl of 2 M N-hydroxysuccinimide and 30 μl of 2 M NaOH. The mixture was incubated at 37°C for 1 to 2 h, and the reaction was monitored by liquid chromatography-mass spectrometry (LC-MS) with an LCZ electrospray mass spectrometer (Micromass, Manchester, United Kingdom) coupled with an HP1100 high-pressure liquid chromatography system (Hewlett-Packard, Palo Alto, Calif.). Conversion to the desired product was approximately 30 to 50%, as judged from the LC-MS spectra. The reaction was terminated by addition of 30 μl of 2 M HCl, and the solution was dialyzed against 50 mM sodium acetate (pH 4.5) in a 3.5-ml Slide-a-Lyzer (3,500-molecular-weight cutoff; Pierce). The sample was subsequently aliquoted and stored at −80°C. Probes were used without further purification.
Purification of enzymes.
The purification of recombinant human UCH-L3 from E. coli has been described previously (36). The purification of full-length recombinant human Ubc9 (158 amino acids, 18 kDa) has also been described (4).
Human Aos1 and Uba2 cDNAs were isolated by PCR from a Homo sapiens cDNA library. The large subunit (Uba2) (640 amino acids, 71 kDa) was placed behind a hexahistidine tag by using the pET28b vector (Novagen). The AOS1 gene (encoding 346 amino acids [38 kDa]) was cloned into pET15b (Novagen) without an affinity tag. The two plasmids were transformed into E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene), coexpressed, and purified from lysates by using Ni-nitrilotriacetic acid-agarose resin (Qiagen). The proteins were further purified by gel filtration and anion-exchange chromatography (Superdex 200 and MonoQ; Pharmacia). The resulting peak was concentrated, exchanged into buffer containing 350 mM NaCl-20 mM Tris (pH 8.0)-1 mM β-mercaptoethanol, and frozen for storage at −80°C.
A cDNA encoding the catalytic domain from human SENP2 was amplified by PCR (amino acids 365 to 590, 27 kDa) from an H. sapiens cDNA library, cloned into pET-28b to encode an N-terminal thrombin-cleavable hexahistidine-tagged fusion protein, transformed into E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene), and induced with IPTG. The protein was purified over Ni-nitrilotriacetic acid-agarose resin (Qiagen), dialyzed against 50 mM Tris-HCl (pH 8.0)-200 mM NaCl-2 mM β-mercaptoethanol-bovine thrombin (Sigma), and purified by gel filtration (Superdex 75; Pharmacia). The final fractions were concentrated, exchanged into buffer containing 350 mM NaCl-20 mM Tris (pH 8.0)-1 mM β-mercaptoethanol, and frozen for storage at −80°C.
Preparation of cell lysates.
EL-4 cells (a mouse thymoma cell line) were cultured in RPMI 1640 (Gibco) supplemented with 10% fetal calf serum and 50 U of penicillin-streptomycin per ml. Single-cell suspensions were prepared from mouse tissues by disrupting the organs between glass slides in phosphate-buffered saline. If required, red blood cells were lysed by resuspension in ACK buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM Na2EDTA [pH 7.3]). To lyse cells, cell pellets were resuspended in HR buffer (50 mM Tris [pH 7.4], 5 mM MgCl2, 250 mM sucrose, 2 mM dithiothreitol [DTT], 2 mM ATP) and vortexed vigorously with glass beads. Lysates were centrifuged to remove cell debris, and aliquots were snap frozen in liquid nitrogen and stored at −80°C.
P19 cells were cultured and transiently transfected with pcDNA3.1-HA-SuPr-1 and pcDNA3.1-HA-SuPr-1 C466S as previously described (56). Nuclear extracts were prepared as follows. Cell pellets were resuspended in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT) and incubated on ice for 15 min, and then 0.06% NP-40 was added and samples were vortexed for 10 s. Subsequently, nuclei were spun down, the supernatant was removed, and the nuclei were resuspended in buffer C (20 mM HEPES [pH 7.9], 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT). Samples were shaken for 15 min, and all remaining debris was spun down. The supernatant (nuclear extract) was aliquoted, snap frozen in liquid nitrogen, and stored at −80°C.
Radioiodination of UBL-VSs.
UBL-VSs were radioiodinated as described previously (7). Briefly, 40 μg of UBL-VS was iodinated in 50 mM phosphate buffer (pH 7.5) containing 1 mCi of Na125I, using Iodogen as a catalyst. The reaction was allowed to proceed for 30 min on ice and was quenched with 0.1 mg of tyrosine per ml. Hen egg lysozyme (1 mg/ml) was added as a carrier protein for the subsequent purification over a Sephadex G-25 (Pharmacia) spin column. Iodinated UBL-VSs were stored at −80°C.
Labeling reactions and detection.
Reactions using purified enzymes and UBL-VSs were for 1 to 2 h at 37°C in 75 mM Tris-50 mM NaCl-5 mM MgCl2-2 mM DTT-2 mM ATP. Enzymes were typically used at a 250 nM final concentration, and crude UBL-VS probes were used at approximately 3 to 5 μg per reaction. 125I-UBL-VS (2.5 × 105 to 1 × 106 cpm) was incubated with 20 to 40 μg of cell lysate for 1 h at 37°C. Reactions were terminated by addition of sodium dodecyl sulfate (SDS) sample buffer with β-mercaptoethanol and boiling for 5 min. Where indicated, enzymes or cell lysates were preincubated with 1 mM PMSF or 10 or 20 mM N-ethylmaleimide (NEM) (Sigma) for 15 min on ice prior to addition of the UBL probe.
Polypeptides were resolved by SDS-10 to 12.5% polyacrylamide gel electrophoresis (PAGE). Gels were analyzed by Coomassie brilliant blue staining, silver staining, autoradiography, or immunoblotting, by standard procedures, as indicated. Immunoblot detection of hemagglutinin (HA)-tagged proteins was with the mouse monoclonal anti-HA antibody 12CA5, followed by horseradish peroxidase-conjugated goat anti-mouse secondary antibody and visualization by chemiluminescence according to the manufacturer's instructions.
Isolation of polypeptides reactive with FLAG-Nedd8-VS and HA-ISG15-VS.
EL-4 lysate (20 mg) was incubated for 2 h at 37°C with 300 μg of FLAG-Nedd8-VS, HA-ISG15-VS, or their inactive counterparts (FLAG-Nedd81-75 and HA-ISG151-154). Sepharose beads (150 μl/sample) conjugated to anti-HA antibody 12CA5 or anti-FLAG antibody M2 (100 μg/sample), followed by protein G-Sepharose beads, were added, and the mixtures were incubated overnight at 4°C. Beads were collected by centrifugation at low speed and washed five times with 10 ml of 1× NET buffer (0.5% NP-40, 150 mM NaCl, 5 mM EDTA, 50 mM Tris [pH 7.4]), followed by a final wash with 10 ml of 15 mM Tris (pH 8)-75 mM NaCl. Proteins were eluted with 100 mM glycine (pH 3.0) at 4°C for 45 min and evaporated to dryness, and the pellet was solubilized in 1× reducing SDS sample buffer and loaded on an SDS-polyacrylamide gel.
Protein identification by tandem mass spectrometry.
Relevant bands were excised from Coomassie brilliant blue-stained SDS-polyacrylamide gels and subjected to trypsinolysis. Samples were analyzed by LC-electrospray ionization (ESI)-MS-MS with a QTOF micro-tandem mass spectrometer (Micromass/Waters) as previously described (8).
RESULTS
Synthesis of UBLs with a C-terminal VS.
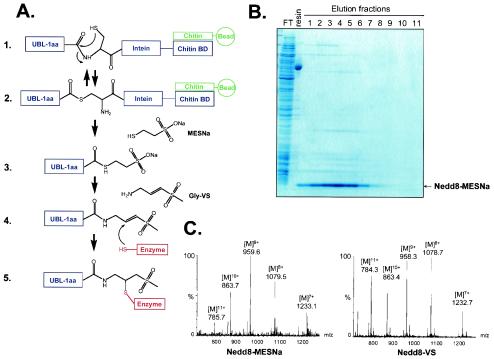
A scheme for the synthesis of UBLs with a C-terminal VS moiety is depicted in Fig. 1A. Open reading frames encoding the processed forms of the UBLs were cloned into the vector pTYB1. The C-terminal residue of the processed form of each UBL was omitted from the expressed version (see below). The pTYB1 vector allows expression in E. coli of the UBLs as a fusion with an intein and a CBD (Fig. 1A, step 1) (72). The soluble fusion proteins were retrieved on a chitin column via their CBDs. The UBLs were then released by a trans-thioesterification reaction induced by inclusion of MESNa sodium salt (Fig. 1A, steps 2 and 3). The UBL-MESNa product released from the chitin column was obtained in good purity, as judged by SDS-PAGE, using this one-step purification method (Fig. 1B). Finally, UBL-MESNa was converted to the VS derivative in a chemical ligation reaction with glycine VS (Fig. 1A, steps 3 and 4). This reaction results in the incorporation of the C-terminal glycine and an electrophilic VS moiety, which may covalently react with nucleophilic active-site residues of UBL-modifying enzymes (Fig. 1A, steps 4 and 5). Conversion reactions were monitored by LC-ESI-MS. The masses determined for UBL-VS derivatives were in good agreement with predicted values (Fig. 1C). This approach was applied to the set of mouse UBLs shown in Table 1. SUMO-1, ISG15, URM-1, and Fau were purified and converted to their VS derivatives at levels comparable to those shown for Nedd8 (Fig. 1B and C and data not shown). For Apg12, FAT10, and HUB1 the UBL-intein-CBD fusion proteins and/or the derived UBL-MESNa products were insoluble under the conditions employed. Purification of soluble forms of these proteins was not further pursued.
FIG. 1.
Synthesis of UBLs with a C-terminal VS. (A) Reaction scheme for UBL-VS synthesis. Step 1, the processed form of a UBL, minus the C-terminal amino acid (−1aa), is expressed as a fusion protein with an intein and a CBD. Soluble fusion protein binds to a chitin affinity column. Step 2, spontaneous N-S acyl rearrangement resulting in an intermediate in which the peptide bond is replaced by a thioester linkage. Step 3, the UBL is released from the column by a transthioesterification reaction induced by incubation with MESNa sodium salt, resulting in the UBL-MESNa product. Step 4, the MESNa group is replaced by glycine-VS in a chemical ligation reaction, producing UBL-VS. Step 5, nucleophilic active-site residues of enzymes can covalently react with the VS group. (B) Nedd8-MESNa purification. FT, flowthrough after loading of lysate on chitin column; resin, chitin resin after Nedd8-MESNa elution. Eluted Nedd8-MESNa fractions were collected after overnight on-column cleavage induced by MESNa. Samples were prepared in SDS sample buffer without β-mercaptoethanol. (C) LC-ESI-MS analysis of Nedd8-MESNa and Nedd8-VS conversion product. The indicated multicharged species of Nedd8-MESNa correspond to a molecular weight of 8,627.6 ± 2.3, in agreement with a predicted molecular weight of 8,628.1. For Nedd8-VS, multicharged species correspond to a molecular weight of 8,620.8 ± 2.5, in agreement with a predicted molecular weight of 8,621.1.
UBL-VSs exhibit specificity in in vitro labeling of deconjugating enzymes.
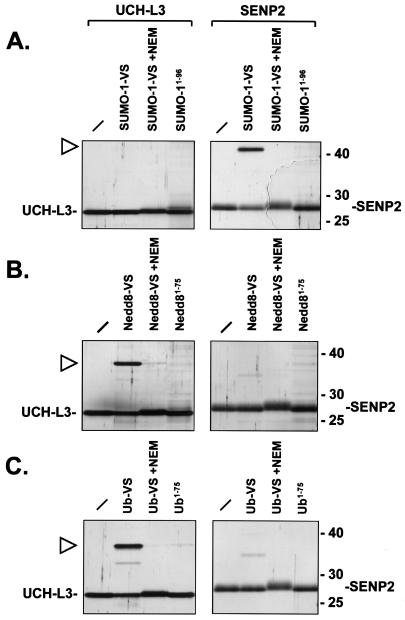
Having prepared the VS-modified UBLs, we wished to establish whether they reacted specifically with the cognate deconjugating enzymes. To this end, Ub C-terminal hydrolase UCH-L3 and the catalytic domain of the desumoylating enzyme SENP2 were expressed in E. coli and purified to apparent homogeneity, as judged by SDS-PAGE. Incubation of the catalytic domain of the desumoylating enzyme SENP2 with SUMO-1-VS led to the appearance of an additional species with a molecular weight consistent with a SENP2-SUMO-1-VS adduct (Fig. 2A). The new species is stable under the reducing conditions used for sample preparation and SDS-PAGE. Formation of this new species was blocked by preincubation of the enzyme with the alkylating agent NEM, consistent with the requirement of the active-site cysteine of SENP2 for catalysis and therefore labeling. Incubation of SENP2 with a SUMO-1 probe lacking the C-terminal glycine VS (SUMO1-96) did not lead to adduct formation, showing that covalent modification of SENP2 is indeed dependent on the C-terminal electrophilic substituent of SUMO-1-VS. We also found that SUMO-1-VS could react with the purified catalytic domain of the yeast ULP1 protease, which is specific for the yeast SUMO-1 homologue SMT3 (reference 46 and data not shown). Incubation of SUMO-1-VS with UCH-L3 did not lead to adduct formation, showing that SUMO-1-VS readily distinguishes between a UCH and a SUMO-specific protease. Nedd8-VS reacted with UCH-L3, an enzyme with reported specificity for both Ub and Nedd8 (64) (Fig. 2B). Again, this reaction was blocked by inclusion of NEM and failed to occur when the VS moiety was omitted (Nedd81-75). Nedd8-VS did not react with SENP2, indicative of its specificity. Reactions with Ub-VS showed a pattern identical to that seen for Nedd8, i.e., formation of an adduct with UCH-L3 but not with SENP2 (Fig. 2C). Radioiodinated Ub-VS and Nedd8-VS also formed adducts with UCH-L3, whereas no adduct formation was seen with radioiodinated SUMO-1-VS or ISG15-VS (see Fig. 5 and data not shown). We therefore conclude that the UBL-VSs produced here react specifically with their cognate deconjugating enzymes through a Michael addition reaction of the active-site cysteine thiol of the enzyme with the VS moiety (Fig. 1A, steps 4 and 5).
FIG. 2.
UBL-VSs react specifically with cognate deconjugating enzymes. (A) Recombinant, purified UCH-L3 enzyme (left panel) and the catalytic domain of SENP2 (right panel) were incubated for 1 h at 37°C with SUMO-1 derivatives. No SUMO-1 probe was added to the first sample in each panel. Where indicated, enzymes were pretreated with 10 mM NEM prior to addition of SUMO-1-VS. SUMO1-96 lacks the C-terminal glycine and VS moiety and therefore is not reactive. The reactions were terminated by addition of SDS sample buffer containing β-mercaptoethanol and boiling for 5 min. Polypeptides were resolved by SDS-11% PAGE and visualized by silver staining. The positions of molecular mass markers (in kilodaltons) are indicated on the right. The positions of UCH-L3 and the catalytic domain of SENP2 are indicated. The triangle marks the position of the SENP2-SUMO-1-VS adduct. (B) Same as panel A but with the Nedd8 derivatives Nedd8-VS and Nedd81-75. Nedd81-75 lacks the C-terminal glycine and VS group. The triangle marks the position of the UCH-L3-Nedd8-VS adduct. (C) Same as panel A but with the Ub derivatives Ub-VS and Ub1-75. Ub1-75 lacks the C-terminal glycine and VS group. The triangle marks the position of the UCH-L3-Ub-VS adduct.
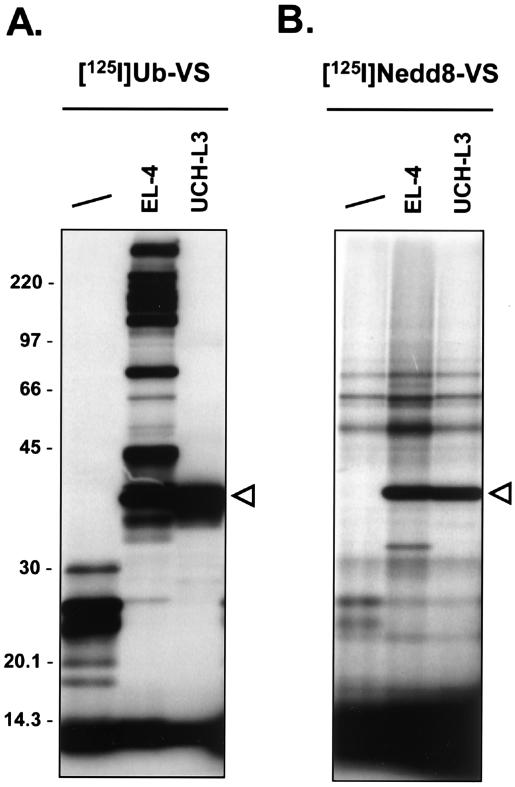
FIG. 5.
UCH-L3 labeling by 125I-Ub-VS and 125I-Nedd8-VS. 125I-Ub-VS (2.5 × 105 cpm) (A) or 125I-Nedd8-VS (2.5 × 105 cpm) (B) was incubated for 1 h at 37°C with lysate buffer (first lane), 20 μg of EL-4 lysate, or 150 ng of recombinant, purified UCH-L3. The reactions were terminated as described for Fig. 2. Polypeptides were resolved by SDS-11% PAGE and visualized by autoradiography. The triangles mark the UCH-L3-Ub-VS (left) and UCH-L3-Nedd8-VS (right) adducts. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
SUMO-1-VS covalently modifies its cognate E1 and E2 enzymes.
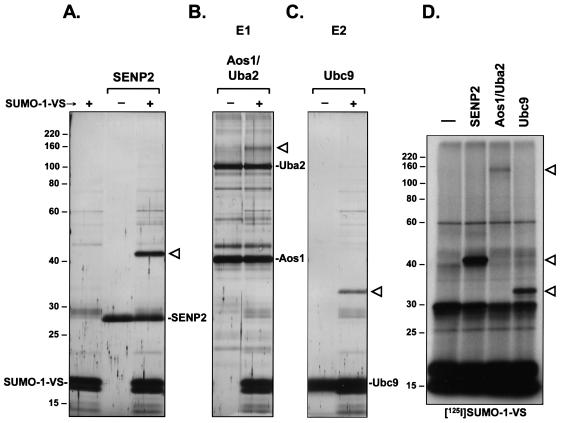
For Ub-VS, the only targets identified in cell lysates were deubiquitinating enzymes (7, 8). Having shown specific labeling of deconjugating enzymes by the UBL-VSs, we asked whether the reactivity of the probes was restricted to deconjugating enzymes or whether enzymes involved in the conjugation pathway were also targets of UBL-VSs. The E1 activating enzyme of SUMO-1 is a heterodimer of Aos1 and Uba2, in which the Uba2 subunit harbors the active-site cysteine (16, 29). The SUMO-1 E2 conjugating enzyme is Ubc9 (27). Aos1/Uba2 and Ubc9 were produced in recombinant form in E. coli and purified. Incubation of SENP2 with SUMO-1-VS led to the formation of a new product, the SENP2-SUMO-1-VS adduct, which was absent from reactions that included either of the two components alone (Fig. 3A). Importantly, incubation of SUMO-1-VS with Aos1/Uba2 led to the appearance of a species with a molecular mass greater than that of Uba2 (Fig. 3B). A species of identical size was observed when radioiodinated SUMO-1-VS was incubated with Aos1/Uba2 (Fig. 3D). As Uba2 contains the E1 active-site cysteine, this species likely represents a Uba2-SUMO-1-VS adduct. In addition, incubation of SUMO-1-VS with the E2 enzyme Ubc9 led to the formation of a species with an apparent molecular mass consistent with a Ubc9-SUMO-1-VS adduct (Fig. 3C). A species of identical size was also observed by autoradiography (Fig. 3D).
FIG. 3.
SUMO-1-VS covalently modifies its cognate E1 and E2 enzymes in vitro. (A) Recombinant, purified SENP2 catalytic domain was incubated for 2 h at 37°C with (+) or without (−) SUMO-1-VS. SUMO-1-VS alone is shown in the first lane. The reactions were terminated as described for Fig. 2. Polypeptides were resolved by SDS-11% PAGE and visualized by silver staining. The triangle marks the position of the SENP2-SUMO-1-VS adduct. The positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) Recombinant, purified SUMO-1-specific E1 activating enzyme, composed of the Aos1/Uba2 heterodimer, was incubated for 2 h at 37°C with (+) or without (−) SUMO-1-VS. Samples were subsequently processed as described for panel A. The triangle marks the position of the Uba2-SUMO-1-VS adduct. (C) Recombinant, purified SUMO-1-specific E2 conjugating enzyme, Ubc9, was incubated for 2 h at 37°C with (+) or without (−) SUMO-1-VS. Samples were subsequently processed as described for panel A. The triangle marks the position of the Ubc9-SUMO-1-VS adduct. (D) 125I-SUMO-1-VS (106 cpm) was incubated for 1 h at 37°C with either no enzyme (first lane) or 12.5 pmol of SENP2, Aos1/Uba2, or Ubc9. Polypeptides were resolved by SDS-11% PAGE and visualized by autoradiography. The triangles mark the positions of the observed SUMO-1-VS adducts (top to bottom) Uba2-SUMO-1-VS, SENP2-SUMO-1-VS, and Ubc9-SUMO-1-VS.
Equimolar amounts of the different enzymes were used in the experiment shown in Fig. 3D. Therefore, the relative intensity of the adducts detected by autoradiography corresponds to the relative amounts of enzyme-SUMO-1-VS adduct formed. It is clear, then, that SENP2 has a higher affinity for and/or greater reactivity towards the SUMO-1 probe than do the SUMO-1-specific E1 and E2 enzymes. Ubc9 (E2) in turn reacts more strongly with SUMO-1-VS than does Aos1/Uba2 (E1). We further found that when all three enzymes were mixed together and incubated with SUMO-1-VS, the SENP2 protease reacted preferentially, with little or no labeling seen for Aos1/Uba2 and Ubc9 (data not shown). We conclude that UBL probes equipped with a C-terminal electrophilic trap can covalently modify purified E1 and E2 enzymes, as well as their deconjugating proteases. However, the affinity or reactivity of the probe appears to be greater for the deconjugation enzyme than for the respective E1 and E2 enzymes.
UBL-VSs label distinct sets of proteins in EL-4 cell lysates.
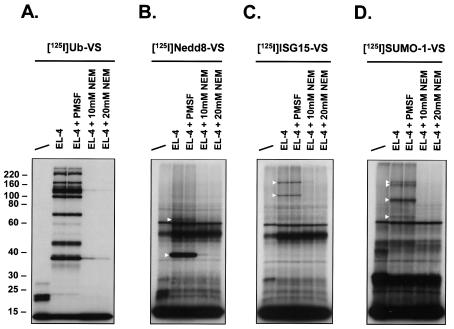
Incubation of radioiodinated Ub-VS with cell lysates allowed detection of specific adducts. These species were identified either genetically or by tandem MS as Ub-VS-adducts with Ub-deconjugating enzymes (7, 8). To examine the labeling profiles of the prepared UBL probes, we incubated EL-4 cell lysates with radioiodinated UBL-VSs (Fig. 4). EL-4 cells were chosen as a model cell line, allowing a direct comparison with Ub-VS labeling profiles, which served as a reference (Fig. 4A) (7, 8). Incubation of EL-4 cell lysates with 125I-Nedd8-VS resulted in strong labeling of a species at ∼38 kDa and another one at ∼65 kDa (Fig. 4B). In addition, polypeptides were detected at ∼32, ∼110, and ∼170 kDa, although their appearance was not always observed (see Fig. 6). The ∼38-kDa band observed with Ub-VS was previously identified as UCH-L3 (8). When recombinant UCH-L3 was incubated with 125I-Ub-VS in parallel with EL-4 lysate, the recombinant UCH-L3 adduct with Ub-VS had a slightly lower apparent molecular weight than the UCH-L3-UbVS adduct formed in cell lysate (Fig. 5A). This small difference in mobility is likely due to the different origin of the recombinant (human) versus the cellular (mouse) UCH-L3. The same experiment performed with 125I-Nedd8-VS yielded results for UCH-L3 identical to those obtained for 125I-Ub-VS (Fig. 5). Given the known dual specificity of UCH-L3 for Ub and Nedd8 (see also Fig. 2B and C) (64), we conclude that the ∼38-kDa band observed for 125I-Nedd8-VS is most likely a UCH-L3-Nedd8-VS adduct (see below). 125I-SUMO-VS and 125I-ISG15-VS did not react with recombinant UCH-L3 (data not shown). Labeling of EL-4 lysates with 125I-ISG15-VS revealed strongly labeled species of ∼110 and ∼210 kDa (Fig. 4C). A weaker adduct at ∼70 kDa was sometimes observed (see Fig. 6C). 125I-SUMO-1-VS labeling produced an abundant adduct at ∼95 kDa and a doublet of ∼160 and ∼180 kDa. A fourth, less abundant, adduct was observed at ∼68 kDa (Fig. 4D). Formation of all observed adducts was resistant to preincubation with the serine protease inhibitor PMSF but was abolished by preincubation with NEM, indicating dependency of adduct formation on active-site cysteine residues (Fig. 4). Fau-VS could not be radioiodinated under the conditions used due to the absence of a tyrosine in the Fau amino acid sequence. We did not observe any labeling of targets in EL-4 lysates when 125I-URM1-VS was used (Table 1 and data not shown).
FIG. 4.
UBL-VSs label distinct sets of proteins in EL-4 lysates. VS derivatives of the indicated UBLs were radiolabeled with Na125I and incubated with EL-4 cell lysates. Per sample, 5 × 105 cpm of 125I-labeled probe and 40 μg of EL-4 lysate were used. First lane in each panel, no lysate added (UBL-VS probe only). EL-4 lysate was pretreated with 1 mM PMSF or with 10 or 20 mM NEM prior to addition of the UBL-VS probes. The reactions were terminated as described for Fig. 2. Polypeptides were resolved by SDS-11% PAGE and visualized by autoradiography. Triangles mark the positions of observed UBL-VS-protein adducts. The radiolabeled UBL-VS probes used were 125I-Ub-VS (A), 125I-Nedd8-VS (B), 125I-UCRP-VS (C), and 125I-SUMO-1-VS (D). The positions of molecular mass markers (in kilodaltons) are indicated on the left.
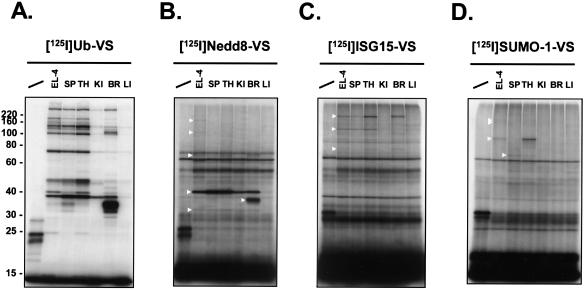
FIG. 6.
UBL-VSs exhibit differential labeling patterns in mouse tissue extracts. Lysates from single-cell suspensions of the indicated mouse tissues were prepared and incubated for 1 h at 37°C with radiolabeled UBL-VSs. Per reaction, 5 × 105 cpm of 125I-UBL-VS and 20 μg of EL-4 or tissue lysate were used. The reactions were terminated as described for Fig. 2. Polypeptides were resolved by SDS-10% PAGE and visualized by autoradiography. First lane in each panel, no lysate added (UBL-VS probe only). SP, spleen; TH, thymus; KI, kidney; BR, brain; LI, liver. Triangles mark the positions of observed UBL-VS-protein adducts. The radiolabeled UBL-VS probes used were125I-Ub-VS (A), 125I-Nedd8-VS (B), 125I-UCRP-VS (C), and 125I-SUMO-1-VS (D). The positions of molecular mass markers (in kilodaltons) are indicated on the left.
UBL-VSs exhibit differential labeling patterns in mouse tissue extracts.
It is possible that EL-4 cells do not express all possible target proteins for the UBL-VSs and that some of these targets are expressed in a tissue-specific fashion. We therefore labeled lysates prepared from various mouse tissues to examine the tissue distribution of active target enzymes (Fig. 6). Differential labeling profiles for the various examined tissues were observed for all probes. 125I-Ub-VS labeling of UCH-L3 (∼38 kDa) was seen in all tissues, although only weakly in liver (Fig. 6A). The intensely labeled 125I-Ub-VS species of ∼36 kDa in brain corresponds to a Ub-VS adduct with UCH-L1, which is known to be abundantly expressed in brain (7, 68). A very similar labeling pattern is seen with 125I-Nedd8-VS for species at ∼38 and ∼36 kDa (Fig. 6B). Together with the previous data (Fig. 4 and 5), it therefore seems likely that these polypeptides represent UCH-L3-Nedd8-VS and UCH-L1-Nedd8-VS adducts, respectively. Reactivity of UCH-L1 towards Nedd8 has not been reported previously. Furthermore, a ∼32-kDa Nedd8-adduct was present in all tissues except liver (Fig. 6B). The ∼65-kDa Nedd8-VS specific adduct was detected only in thymus and brain. Labeling of the ∼110- and ∼170-kDa adducts, as seen for 125I-Nedd8-VS in EL-4 lysates, was not observed in any of the tissues. With 125I-ISG15-VS a ∼210-kDa adduct was detected in spleen, thymus, and brain. The species in the brain sample appeared to have a slightly higher apparent molecular weight than the species in the other samples. The ∼110-kDa polypeptide was present in spleen and thymus only. The ∼70-kDa adduct was detected in thymus and brain (Fig. 6C). 125I-SUMO-1-VS strongly labeled a ∼95-kDa adduct in thymus only. As EL-4 cells are a thymus-derived cell line, this ∼95-kDa adduct may represent a thymus-specific SUMO-1-modifying enzyme. The ∼68-kDa adduct was detected weakly in spleen and thymus. The ∼160/180-kDa doublet was not detected in any of the tissue samples.
SUMO-1-VS modifies SUMO-1-specific protease SuPr-1 via its active-site cysteine.
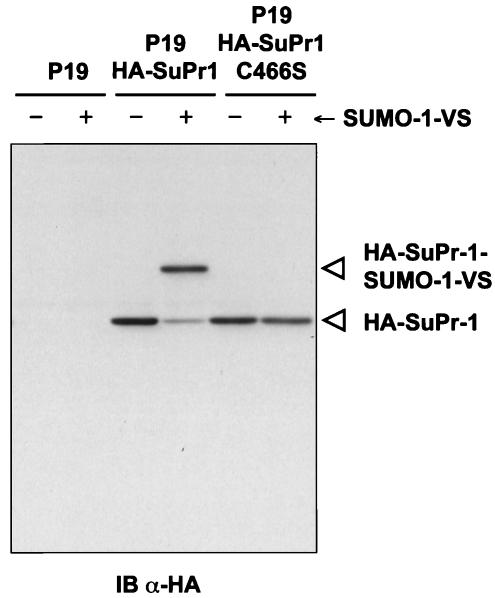
In an initial effort to characterize the proteins labeled in cell lysates by UBL-VSs, we ectopically expressed an HA-tagged version of the recently described mouse SUMO-1-specific protease SuPr-1 (5, 56). Incubation of extracts of HA-SuPr-1-transfected cells with SUMO-1-VS led to the detection of a ∼78-kDa species, in addition to the ∼58-kDa HA-SuPr-1 polypeptide, indicating direct modification of HA-SuPr-1 by SUMO-1-VS (Fig. 7). The formation of this species was strictly dependent on the presence of the active-site cysteine, as a mutant in which the cysteine was replaced by a serine (C466S) was not labeled (Fig. 7). In cell lysates, SUMO-1-VS is therefore capable of targeting SUMO-specific proteases.
FIG. 7.
SUMO-1 modifies SuPr-1 via its active-site cysteine. P19 cells were transiently transfected with a vector expressing HA-SuPr-1 or the catalytically inactive mutant HA-SuPr-1C466S, cells were harvested, and nuclear extracts were prepared. Nuclear extracts (25 μg) were incubated for 1 h at 37°C without (−) or with (+) SUMO-1-VS. The reactions were terminated by addition of SDS sample buffer containing β-mercaptoethanol and boiling for 5 min. Polypeptides were resolved by SDS-10% PAGE and immunoblotted (IB) with an anti-HA (α-HA) antibody.
Identification of UBL-specific proteases.
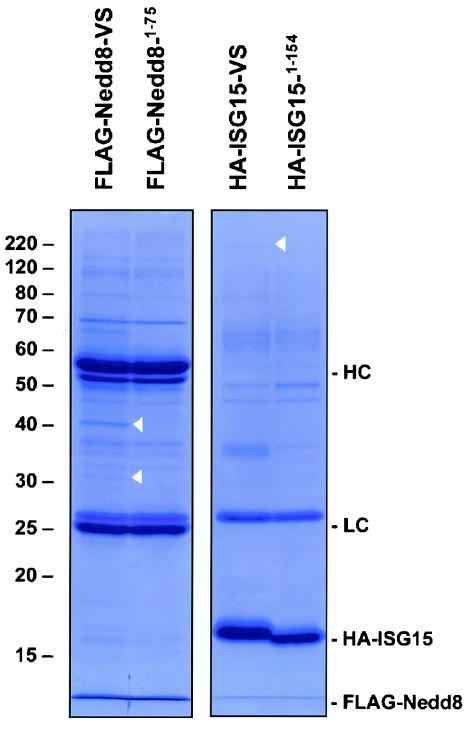
To unambiguously identify the proteins targeted by the UBL probes, we prepared epitope-tagged versions of two of the probes, namely, FLAG-Nedd8-VS and HA-ISG15-VS. Large-scale labeling reactions of these probes with EL-4 cell lysates were performed, followed by immunopurification of the probes and their covalently attached targets. Comparison of the pull-down profiles obtained by using active probes with those obtained by using their inactive counterparts revealed the presence of several specific UBL adducts. Bands were excised from the gel and processed for analysis by tandem MS (Fig. 8). Using FLAG-Nedd8-VS, we identified the ∼39-kDa adduct as the Ub C-terminal hydrolase UCH-L3 and the ∼32-kDa adduct as containing DEN1/NEDP1/SENP8. Further, the 210-kDa adduct observed with HA-ISG15-VS was identified as isopeptidase T/USP5 (Table 2). Identification of targets of SUMO-1-VS is in progress and will be described elsewhere. Epitope-tagged UBL-VS probes were therefore successfully employed in the identification of UBL-specific proteases.
FIG. 8.
Isolation of polypeptides reactive with FLAG-Nedd8-VS and HA-ISG15-VS. EL-4 lysate (20 mg) was incubated for 2 h at 37°C with 300 μg of FLAG-Nedd8-VS, HA-ISG15-VS, or their inactive counterparts (FLAG-Nedd81-75 and HA-ISG151-154). Probes and probe-enzyme adducts were immunopurified by using Sepharose beads conjugated to anti-HA antibody 12CA5 (for HA-ISG15) or anti-FLAG antibody M2 combined with protein G-Sepharose beads (for FLAG-Nedd8). Precipitates were extensively washed, proteins were eluted with 100 mM glycine (pH 3.0) and evaporated to dryness, and the pellet was solubilized in 1× reducing SDS sample buffer and resolved by SDS-11% PAGE. The position of the free probes as well as the antibody light chains (LC) and heavy chains (HC) are indicated on the right. The positions of the bands identified by tandem MS are indicated by triangles on the gels. The positions of molecular mass markers (in kilodaltons) are indicated on the left.
TABLE 2.
UBL-specific proteases identified by tandem MS
| Probe | Protease identified | Molecular mass (kDa)
|
Peptides identifiedc | % Coveraged | |
|---|---|---|---|---|---|
| Calculateda | Observedb | ||||
| FLAG-Nedd8-VS | UCH-L3 | 26.2 | 39 | 6-WLPLEANPEVTNQFLK-21, 75-SQGQDVTSSVYFMK-88, 122-FLEESVSMSPEER-134, 135-AKFLENYDAIR-145, 137-FLENYDAIR-145, 196-TSDETLLEDVIKVCK-210, 196-TSDETLLEDVIKVCKK-211, 212-FMERDPDELR-221 | 34.8 |
| FLAG-Nedd8-VS | DEN1/NEDP1/SENP8 | 25.1 | 32 | 144-GDKLVFVEEK-153, 181-RQPESPLQLLTPTYITKK-198 | 12.2 |
| HA-ISG15-VS | IsoT/USP5 | 95.8 | 210 | 361-IFQNAPTDPTQDFSTQVAK-379, 453-SSENPNEVFR-462, 778-SAAESISESVPVGPK-793 | 5.1 |
Molecular masses of the proteins were calculated with the ProtParam tool from the ExPASy Molecular Biology Server, using the mouse amino acid sequences.
Relevant bands from the gels in Fig. 8 were excised and processed for analysis by tandem MS as described in Materials and Methods.
The individual peptides identified by tandem MS, together with their position in the identified polypeptide, are shown.
Fraction of nonoverlapping peptide sequences identified as part of the entire polypeptide.
DISCUSSION
Here we describe the synthesis of UBLs modified with a C-terminal electrophilic trap, according to a method previously used for Ub. We chose a comprehensive approach in which known mouse UBLs were expressed as fusion proteins containing an intein and a CBD (Table 1). Due to insolubility of fusion proteins and/or UBL-MESNa species or to an inability to radiolabel the probe with 125I, functional probes were obtained only for Nedd8, ISG15, and SUMO-1. The UBL probes could be used either unlabeled, radioiodinated, or equipped with an epitope tag.
UBL-derived probes reacted specifically with purified cognate deconjugating enzymes in vitro (Fig. 2). This reaction was dependent on the presence of the C-terminal glycine VS moiety and was abolished by inclusion of the alkylating agent NEM, consistent with the requirement for the active-site cysteine residue for activity of the enzymes. We conclude that adducts are formed through a Michael addition reaction of the active-site cysteine thiol of the enzyme with the VS moiety. It was important to demonstrate specificity of the probes towards purified proteases to allow a meaningful interpretation of the data obtained with cell lysates (see below).
We further demonstrated that SUMO-1-VS also reacts in vitro with its cognate activating (E1) and conjugating (E2) enzymes, Aos1/Uba2 and Ubc9, respectively (Fig. 3) (16, 27, 29). Although E1s and E2s possess active-site cysteine residues and form thioester intermediates with the UBL C terminus, they do not contain a His/Cys/Asp catalytic triad as do the cysteine proteases of the UCH, UBP, ULP, and OTU families (25, 30, 46). Activation of the C terminus of a UBL by an E1 enzyme involves hydrolysis of ATP and the concomitant formation of an adenylate intermediate. This is followed by transfer of the UBL to the active-site cysteine by formation of a thioester linkage with the UBL C terminus (65). Due to the presence of the VS moiety at the SUMO-1-VS C terminus, an adenylate intermediate cannot be formed and adduct formation with Aos1/Uba2 (Fig. 3) therefore must occur through a direct nucleophilic attack of the active-site cysteine on the incoming SUMO-1-VS. Although the observed formation of an adduct indicates that the SUMO-1-VS C terminus is in sufficient proximity to allow this attack, we consider it likely that the lower efficiency of modification compared to that for Ubc9 and SENP2 is accounted for in part by the inability of Aos1/Uba2 to form an adenylate intermediate, as well as the absence of a general base in the active site (see below).
The crystal structure of Ubc9 in a complex with a physiological substrate showed the absence of other catalytic residues near the essential cysteine residue, in particular a base capable of deprotonating the cysteine (4). Instead, transfer of SUMO appeared to occur merely by the correct positioning of the C terminus of SUMO in proximity to the active-site cysteine. According to our observations, SUMO-1-VS, in the absence of delivery by an E1, can interact with Ubc9 in a way that allows direct nucleophilic attack by the active-site cysteine on the VS moiety (Fig. 3). Based on the mechanistic considerations described above, it is not surprising that the reactivity of Aos1/Uba2 and Ubc9 is lower than that observed for the SENP2 protease. It should be noted that the concentrations of E1 and E2 enzymes used are relatively high compared to those in cell lysates, in which reactivity of the probes with E1 and E2 enzymes is not observed (see below). Nonetheless, starting from purified components, these reactions may be useful in the preparation of stable enzyme-UBL conjugates for structural analysis and may yield further insight into the catalytic mechanisms involved.
We next used radioiodinated UBL-VSs to detect covalent adducts in EL-4 lysates (Fig. 4). Unique labeling patterns were observed for each of the tested UBLs. Four specific adducts were detected with SUMO-1-VS, three adducts were detected with UCRP-VS, and six adducts were detected with Nedd8-VS (Fig. 4 and 6). Labeling of the targets with UBL-VSs was generally less strong than that observed with Ub-VS, possibly reflecting lower expression levels of the target proteins. There are several clues as to the identity of the modified polypeptides. First, labeling is abolished by the inclusion of NEM, consistent with the requirement of active-site cysteines for reactivity (Fig. 4). Second, as the specificity of the probes used is dictated by the UBL domain in question, it is likely that the targets identified are indeed proteins involved in UBL modification. Since the proteases are significantly more reactive with the probe than the relevant E1 and E2 enzymes (Fig. 2 and 3), the adducts detected in cell lysates likely represent UBL-specific proteases. Third, all targets identified so far for Ub-VS are Ub-specific proteases (7, 8). Fourth, a SUMO-1-specific protease (SuPr-1), when expressed ectopically, was modified by SUMO-1-VS via its active-site cysteine (Fig. 7). Fifth, a survey of the reported molecular masses of the E1 and E2 enzymes of SUMO-1, ISG15, and Nedd8 (see the introduction for references) indicated that the observed adducts did not correspond to the masses expected for UBL-E1 and UBL-E2 adducts. Finally, several of the targeted polypeptides were identified as cysteine proteases (Fig. 8; Table 2). Based on these arguments we conclude that the targeted polypeptides most likely represent UBL-specific proteases.
The SUMO-1-VS conjugates had apparent molecular masses of ∼68, ∼95, ∼160, and ∼180 kDa. By ectopically expressing HA-SuPr-1, it became clear that none of these species represented SuPr-1, as a SuPr-1 adduct should have an apparent molecular mass of slightly less than the 78 kDa observed for HA-tagged SuPr-1 (Fig. 7). Our ability to label HA-SuPr-1 with SUMO-1-VS and the failure to detect endogenous SuPr-1 in crude cell lysates illustrates one of the complications in the analysis of UBL-specific proteases. Failure to detect a protease can be due to low expression levels or to the choice of cell or tissue type examined. A survey of the molecular masses of the SUMO proteases reported to date (see the introduction) indicated that SuPr-2/SENP1 (∼70 kDa) is a candidate for the observed ∼95-kDa adduct (5, 17). The other adducts do not correspond in mass to adducts formed by known SUMO proteases and therefore likely represent novel SUMO-specific proteases.
For ISG15, the apparent masses of the adducts were ∼70, ∼110, and ∼210 kDa. The ∼70-kDa adduct could correspond to the ISG15-specific protease UBP43/USP18 (41). The ∼110-kDa adduct likely represents a novel UCRP-specific protease. The ∼210-kDa adduct was identified as isopeptidase T/USP5 (IsoT) by using HA-ISG15-VS combined with tandem MS. IsoT has previously been described as a 93-kDa Ub-specific cysteine protease from the UBP/USP family that is involved in the disassembly of free poly(Ub) chains (1, 20). The large difference in apparent molecular weight between the unmodified IsoT and the IsoT-ISG15 adduct is partly explained by the branched nature of the adduct. IsoT could be further modified by posttranslational modifications, and large differences between predicted and observed molecular weights were also observed for adducts with HA-Ub-VS (8). Based on kinetics studies using Ub derivatives, it has been proposed by several groups that IsoT has two, or perhaps even four, binding sites for Ub (59, 69). ISG15 consists of two Ub-like domains fused in tandem, each around 30% homologous to Ub (Table 1). The multiple Ub binding sites on IsoT may therefore allow binding of ISG15, despite its relatively low sequence similarity with Ub. Furthermore, IsoT may correspond to the unidentified ∼100-kDa cysteine protease that is capable of processing the ISG15 precursor (52). In favor of this hypothesis are the similar molecular weights of the two proteins, their sensitivity to alkylating agents (Fig. 4) (20, 52), their constitutive activity (Fig. 4 and 6) (52), and the observation that their activities can be stimulated by free Ub (59, 69). The accumulation of ISG15 conjugates in UBP43-deficient cells indicates that UBP43 is a major ISG15-deconjugating protease. As conjugation occurs in the absence of UBP43, ISG15 precursor processing must be performed by another protease(s) (55). Based on the above data and considerations, we propose that IsoT may function as an ISG15-processing protease. Whereas UBP43 is exclusively specific for ISG15, IsoT appears to have dual specificity for Ub and ISG15 (Fig. 8) (8).
For Nedd8, the observed masses of the adducts were ∼32, ∼36, ∼38, ∼65, ∼110, and ∼170 kDa. We identified the major species at 38 kDa as UCH-L3 (Fig. 8). We (Fig. 2 and 5) and others (64) have shown that Nedd8 specifically reacts with UCH-L3 in vitro, and the apparent molecular mass of the observed adduct in the EL-4 lysate is consistent with a Nedd8-UCH-L3 adduct. The ∼36-kDa adduct detected with Nedd8-VS in brain extract most likely represents UCH-L1, an enzyme known to be abundantly expressed in this tissue and also reactive with Ub-VS (7, 8, 68). UCH-L1 is the third enzyme (in addition to UCH-L3 and USP21) to have dual specificity for Ub and Nedd8. Dual specificity of enzymes for both Ub and Nedd8 is perhaps not very surprising in view of the homology between Ub and Nedd8 (57% identity), although it raises questions with respect to regulation of these modifications in the cell. The 32-kDa adduct was identified as DEN1/NEDP1/SENP8. This enzyme, belonging to the ULP family of cysteine proteases, appears to be capable of Nedd8 precursor processing, deneddylation of hyperneddylated substrate, deneddylation of mononeddylated cullins in vitro (14, 43, 71). Furthermore, DEN1 is capable of deconjugating Nedd8 from all modified proteins detected in vivo (43). The other observed adducts likely represent novel Nedd8-specific proteases.
EL-4 cell lysates proved to be suitable for the detection of UBL-VS adducts. Apart from Ub/Nedd8-VS detection of UCH-L1 in brain and ISG15-VS detection of a slightly different ∼180-kDa species in brain, all other polypeptides detected in the tissues were also detected in EL-4 cell lysate (Fig. 4 and 6). Nevertheless, the tissue-specific detection of active UBL-modifying enzymes suggests tissue-specific functions of UBLs. Of note was the strong labeling by SUMO-1-VS of a ∼95-kDa adduct in thymus and EL-4 extracts (Fig. 6). Since EL-4 cells are a cell line derived from thymus, this ∼95-kDa adduct may represent a thymus-specific SUMO-1-modifying enzyme.
Taken together, the UBL probes described here have allowed us to identify UBL-specific proteases, based on their reactivity towards the UBL C termini. The proteases identified (UCH-L1 and DEN1/NEDP1/SENP8 for Nedd8 and IsoT for ISG15) belong to protease families (UCH, ULP, and UBP) originally thought to be specific for other UBLs (Ub, SUMO, and Ub, respectively). Two of these proteases, UCH-L1 and IsoT, now appear to have dual specificity for Ub/Nedd8 and Ub/ISG15, respectively. Therefore, the enzyme family to which a UBL-specific protease belongs does not necessarily predict its substrate specificity, and for each enzyme the specificity needs to be determined. For this, as well as for identification of new proteases, the UBL probes described here should be useful. It will be interesting to see whether the as-yet-unidentified proteases present in the variety of distinct adducts reported here belong to any of the known cysteine protease families or to new families.
In addition to UBL protease identification and validation, the UBL probes may have a number of other applications. As the probes described here are mechanism-based inhibitors, the labeling of a target enzyme observed is proportional to the activity of that enzyme. Therefore, these probes can be used to monitor the activity of UBL-specific enzymes under different cellular conditions (7). By equipping the UBLs with different electrophilic groups, it should also be possible to fine-tune the target specificity of the probes, perhaps ultimately leading to the development of probes specific for single proteases (8). An important goal will be to render these probes cell permeative to study the effect of specific inhibition of these enzymes on cellular physiology. Furthermore, the physiological thioester intermediates formed between UBLs and UBL-modifying enzymes are very labile. In contrast, the covalent adducts formed between UBL-VSs and purified enzymes are stable and could therefore facilitate structural analysis of UBL-enzyme complexes. In conclusion, the UBL probes described here are valuable tools for the characterization of the enzymatic pathways that govern the modification by UBLs.
Acknowledgments
We thank LeAnn Williams for expert preparation of samples for MS-MS analysis.
This work was supported by grants from the National Institutes of Health to H.L.P., C.D.L., and D.R. (GM65872) and to K.D.W. (GM30308 and GM66355), as well as by grants from the Harvard Center for Neurodegeneration and Repair and the Alexander and Margaret Stewart Trust Foundation to B.M.K. and H.L.P. G.G. was funded in part by the Hellman Family Fund. T.G.-E. was supported by a grant from the Foggarty Foundation (TW05461). H.O. was financially supported by a fellowship from The Netherlands Organization for Scientific Research (NWO). B.M.K. is a recipient of a Multiple Myeloma Research Foundation (MMRF) Senior Research Award.
REFERENCES
- 1.Amerik, A., S. Swaminathan, B. A. Krantz, K. D. Wilkinson, and M. Hochstrasser. 1997. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 16:4826-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakirev, M. Y., S. O. Tcherniuk, M. Jaquinod, and J. Chroboczek. 2003. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 4:517-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, P., A. Arndt, S. Metzger, R. Mahajan, F. Melchior, R. Jaenicke, and J. Becker. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280:275-286. [DOI] [PubMed] [Google Scholar]
- 4.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. [DOI] [PubMed] [Google Scholar]
- 5.Best, J. L., S. Ganiatsas, S. Agarwal, A. Changou, P. Salomoni, O. Shirihai, P. B. Meluh, P. P. Pandolfi, and L. I. Zon. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell 10:843-855. [DOI] [PubMed] [Google Scholar]
- 6.Bogyo, M., S. Shin, J. S. McMaster, and H. L. Ploegh. 1998. Substrate binding and sequence preference of the proteasome revealed by active-site-directed affinity probes. Chem. Biol. 5:307-320. [DOI] [PubMed] [Google Scholar]
- 7.Borodovsky, A., B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson, and H. L. Ploegh. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20:5187-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borodovsky, A., H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh, and B. M. Kessler. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149-1159. [DOI] [PubMed] [Google Scholar]
- 9.Cope, G. A., G. S. Suh, L. Aravind, S. E. Schwarz, S. L. Zipursky, E. V. Koonin, and R. J. Deshaies. 2002. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298:608-611. [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 11.Desterro, J. M., M. S. Rodriguez, G. D. Kemp, and R. T. Hay. 1999. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274:10618-10624. [DOI] [PubMed] [Google Scholar]
- 12.Dohmen, R. J., R. Stappen, J. P. McGrath, H. Forrova, J. Kolarov, A. Goffeau, and A. Varshavsky. 1995. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270:18099-18109. [DOI] [PubMed] [Google Scholar]
- 13.Evans, P. C., T. S. Smith, M. J. Lai, M. G. Williams, D. F. Burke, K. Heyninck, M. M. Kreike, R. Beyaert, T. L. Blundell, and P. J. Kilshaw. 2003. A novel type of deubiquitinating enzyme. J. Biol. Chem. 278:23180-23186. [DOI] [PubMed] [Google Scholar]
- 14.Gan-Erdene, T., N. Kolli, L. Yin, K. Wu, Z. Q. Pan, and K. D. Wilkinson. 2003. Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 278:28892-28900. [DOI] [PubMed] [Google Scholar]
- 15.Gong, L., T. Kamitani, S. Millas, and E. T. Yeh. 2000. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 275:14212-14216. [DOI] [PubMed] [Google Scholar]
- 16.Gong, L., B. Li, S. Millas, and E. T. Yeh. 1999. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448:185-189. [DOI] [PubMed] [Google Scholar]
- 17.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 18.Gong, L., and E. T. Yeh. 1999. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 274:12036-12042. [DOI] [PubMed] [Google Scholar]
- 19.Haas, A. L., P. Ahrens, P. M. Bright, and H. Ankel. 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 262:11315-11323. [PubMed] [Google Scholar]
- 20.Hadari, T., J. V. Warms, I. A. Rose, and A. Hershko. 1992. A ubiquitin C-terminal isopeptidase that acts on polyubiquitin chains. Role in protein degradation. J. Biol. Chem. 267:719-727. [PubMed] [Google Scholar]
- 21.Hamerman, J. A., F. Hayashi, L. A. Schroeder, S. P. Gygi, A. L. Haas, L. Hampson, P. Coughlin, R. Aebersold, and A. Aderem. 2002. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J. Immunol. 168:2415-2423. [DOI] [PubMed] [Google Scholar]
- 22.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 23.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 24.Hori, T., F. Osaka, T. Chiba, C. Miyamoto, K. Okabayashi, N. Shimbara, S. Kato, and K. Tanaka. 1999. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18:6829-6834. [DOI] [PubMed] [Google Scholar]
- 25.Hu, M., P. Li, M. Li, W. Li, T. Yao, J. W. Wu, W. Gu, R. E. Cohen, and Y. Shi. 2002. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111:1041-1054. [DOI] [PubMed] [Google Scholar]
- 26.Jentsch, S., and G. Pyrowolakis. 2000. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10:335-342. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, E. S., and G. Blobel. 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272:26799-26802. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16:5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston, S. C., S. M. Riddle, R. E. Cohen, and C. P. Hill. 1999. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 18:3877-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadoya, T., H. Yamamoto, T. Suzuki, A. Yukita, A. Fukui, T. Michiue, T. Asahara, K. Tanaka, M. Asashima, and A. Kikuchi. 2002. Desumoylation activity of axam, a novel axin-binding protein, is involved in downregulation of beta-catenin. Mol. Cell. Biol. 22:3803-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway, and J. W. Conaway. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, K. I., S. H. Baek, Y. J. Jeon, S. Nishimori, T. Suzuki, S. Uchida, N. Shimbara, H. Saitoh, K. Tanaka, and C. H. Chung. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102-14106. [DOI] [PubMed] [Google Scholar]
- 35.Kurz, T., L. Pintard, J. H. Willis, D. R. Hamill, P. Gonczy, M. Peter, and B. Bowerman. 2002. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295:1294-1298. [DOI] [PubMed] [Google Scholar]
- 36.Larsen, C. N., J. S. Price, and K. D. Wilkinson. 1996. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues. Biochemistry 35:6735-6744. [DOI] [PubMed] [Google Scholar]
- 37.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 38.Liakopoulos, D., G. Doenges, K. Matuschewski, and S. Jentsch. 1998. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17:2208-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loeb, K. R., and A. L. Haas. 1994. Conjugates of ubiquitin cross-reactive protein distribute in a cytoskeletal pattern. Mol. Cell. Biol. 14:8408-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malakhov, M. P., K. I. Kim, O. A. Malakhova, B. S. Jacobs, E. C. Borden, and D. E. Zhang. 2003. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 278:16608-16613. [DOI] [PubMed] [Google Scholar]
- 41.Malakhov, M. P., O. A. Malakhova, K. I. Kim, K. J. Ritchie, and D. E. Zhang. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976-9981. [DOI] [PubMed] [Google Scholar]
- 42.Malakhova, O. A., M. Yan, M. P. Malakhov, Y. Yuan, K. J. Ritchie, K. I. Kim, L. F. Peterson, K. Shuai, and D. E. Zhang. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendoza, H. M., L. N. Shen, C. Botting, A. Lewis, J. Chen, B. Ink, and R. T. Hay. 2003. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 278:25637-25643. [DOI] [PubMed] [Google Scholar]
- 44.Miyauchi, Y., S. Yogosawa, R. Honda, T. Nishida, and H. Yasuda. 2002. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277:50131-50136. [DOI] [PubMed] [Google Scholar]
- 45.Mizushima, N., T. Yoshimori, and Y. Ohsumi. 2002. Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid method. FEBS Lett. 532:450-454. [DOI] [PubMed] [Google Scholar]
- 46.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865-876. [DOI] [PubMed] [Google Scholar]
- 47.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell. Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 48.Nakagawa, K., and H. Yokosawa. 2002. PIAS3 induces SUMO-1 modification and transcriptional repression of IRF-1. FEBS Lett. 530:204-208. [DOI] [PubMed] [Google Scholar]
- 49.Nishida, T., F. Kaneko, M. Kitagawa, and H. Yasuda. 2001. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting beta-catenin degradation. J. Biol. Chem. 276:39060-39066. [DOI] [PubMed] [Google Scholar]
- 50.Nishida, T., H. Tanaka, and H. Yasuda. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423-6427. [DOI] [PubMed] [Google Scholar]
- 51.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 52.Potter, J. L., J. Narasimhan, L. Mende-Mueller, and A. L. Haas. 1999. Precursor processing of pro-ISG15/UCRP, an interferon-beta-induced ubiquitin-like protein. J. Biol. Chem. 274:25061-25068. [DOI] [PubMed] [Google Scholar]
- 53.Pru, J. K., K. J. Austin, A. L. Haas, and T. R. Hansen. 2001. Pregnancy and interferon-tau upregulate gene expression of members of the 1-8 family in the bovine uterus. Biol. Reprod. 65:1471-1480. [DOI] [PubMed] [Google Scholar]
- 54.Raasi, S., G. Schmidtke, and M. Groettrup. 2001. The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J. Biol. Chem. 276:35334-35343. [DOI] [PubMed] [Google Scholar]
- 55.Ritchie, K. J., M. P. Malakhov, C. J. Hetherington, L. Zhou, M. T. Little, O. A. Malakhova, J. C. Sipe, S. H. Orkin, and D. E. Zhang. 2002. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 16:2207-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 57.Sapetschnig, A., G. Rischitor, H. Braun, A. Doll, M. Schergaut, F. Melchior, and G. Suske. 2002. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seufert, W., B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373:78-81. [DOI] [PubMed] [Google Scholar]
- 59.Stein, R. L., Z. Chen, and F. Melandri. 1995. Kinetic studies of isopeptidase T: modulation of peptidase activity by ubiquitin. Biochemistry 34:12616-12623. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki, T., A. Ichiyama, H. Saitoh, T. Kawakami, M. Omata, C. H. Chung, M. Kimura, N. Shimbara, and K. Tanaka. 1999. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J. Biol. Chem. 274:31131-31134. [DOI] [PubMed] [Google Scholar]
- 61.Tateishi, K., M. Omata, K. Tanaka, and T. Chiba. 2001. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 63.Verma, R., L. Aravind, R. Oania, W. H. McDonald, J. R. Yates III, E. V. Koonin, and R. J. Deshaies. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298:611-615. [DOI] [PubMed] [Google Scholar]
- 64.Wada, H., K. Kito, L. S. Caskey, E. T. Yeh, and T. Kamitani. 1998. Cleavage of the C-terminus of NEDD8 by UCH-L3. Biochem. Biophys. Res. Commun. 251:688-692. [DOI] [PubMed] [Google Scholar]
- 65.Walden, H., M. S. Podgorski, and B. A. Schulman. 2003. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature 422:330-334. [DOI] [PubMed] [Google Scholar]
- 66.Whitby, F. G., G. Xia, C. M. Pickart, and C. P. Hill. 1998. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273:34983-34991. [DOI] [PubMed] [Google Scholar]
- 67.Wilkinson, K. D. 1997. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11:1245-1256. [DOI] [PubMed] [Google Scholar]
- 68.Wilkinson, K. D., K. M. Lee, S. Deshpande, P. Duerksen-Hughes, J. M. Boss, and J. Pohl. 1989. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science 246:670-673. [DOI] [PubMed] [Google Scholar]
- 69.Wilkinson, K. D., V. L. Tashayev, L. B. O'Connor, C. N. Larsen, E. Kasperek, and C. M. Pickart. 1995. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry 34:14535-14546. [DOI] [PubMed] [Google Scholar]
- 70.Wu, K., A. Chen, and Z. Q. Pan. 2000. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275:32317-32324. [DOI] [PubMed] [Google Scholar]
- 71.Wu, K., K. Yamoah, G. Dolios, T. Gan-Erdene, P. Tan, A. Chen, C. G. Lee, N. Wei, K. D. Wilkinson, R. Wang, and Z. Q. Pan. 2003. DEN1 is a dual function protease capable of processing the C-terminus of Nedd8 deconjugating hyper-neddylated CUL1. J. Biol. Chem. 278:28882-28891. [DOI] [PubMed] [Google Scholar]
- 72.Xu, M. Q., and T. C. Evans, Jr. 2001. Intein-mediated ligation and cyclization of expressed proteins. Methods 24:257-277. [DOI] [PubMed] [Google Scholar]
- 73.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, H., H. Saitoh, and M. J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]