Abstract
Congenital heart disease is the most common form of human birth defects, yet much remains to be learned about its underlying causes. Here we report that mice lacking functional ADAM19 (mnemonic for a disintegrin and metalloprotease 19) exhibit severe defects in cardiac morphogenesis, including a ventricular septal defect (VSD), abnormal formation of the aortic and pulmonic valves, leading to valvular stenosis, and abnormalities of the cardiac vasculature. During mouse development, ADAM19 is highly expressed in the conotruncus and the endocardial cushion, structures that give rise to the affected heart valves and the membranous ventricular septum. ADAM19 is also highly expressed in osteoblast-like cells in the bone, yet it does not appear to be essential for bone growth and skeletal development. Most adam19−/− animals die perinatally, likely as a result of their cardiac defects. These findings raise the possibility that mutations in ADAM19 may contribute to human congenital heart valve and septal defects.
ADAMs (mnemonic for a disintegrin and metalloprotease) are membrane-anchored glycoproteins with key roles in fertilization, neurogenesis, angiogenesis, Alzheimer's disease, and the release of proteins such as epidermal growth factor (EGF) receptor ligands and tumor necrosis factor family members from the plasma membrane (3, 4, 17, 37, 39, 41). ADAM19 (also referred to as meltrin β) was initially identified in muscle cells and was later found to be expressed in several other tissues, most prominently in heart, lung, and bone (18, 27, 52), during dendritic cell differentiation (13) and Notch-induced T-cell maturation (9). The catalytic activity of ADAM19 towards candidate substrates has been explored by overexpression in cells and by purifying recombinantly expressed soluble forms of the entire ectodomain or the pro- and metalloprotease domains (7, 42, 49, 53). Overexpressed ADAM19 enhances ectodomain shedding of two of several splice variants of neuregulin I-β (42), a ligand for the ErbB family of receptor tyrosine kinases (11). Furthermore, overexpression of ADAM19 increases ectodomain release of tumor necrosis factor-related activation-induced cytokine (TRANCE, also referred to as osteoprotegerin-ligand [OPGL]) (7), a protein with important roles in osteoclast differentiation, dendritic cell survival, and mammary gland development (12, 25, 28).
In light of the high expression of ADAM19 in heart and bone and its ability to cleave TRANCE as well as splice variants of neuregulin I-β, we were interested in evaluating the function of ADAM19 in mice, with an emphasis on its role in heart and bone development. Here we present an analysis of mice lacking functional ADAM19 (adam19−/− mice).
MATERIALS AND METHODS
Generation of adam19−/− mice.
adam19+/− mice were generated by the Sloan-Kettering Institute transgenic facility by following standard procedures using stem cells with a secretory gene trap insertion in ADAM19 (30). All mice evaluated in this study were of mixed genetic background (129Sv/C57BL6), and morphological and histological comparisons between wild-type and adam19−/− mice were performed with littermates that resulted from matings of heterozygous parents. Genotyping was performed either by Southern blotting (50) or by PCR. For PCR analysis, two sets of primers were used simultaneously to detect both the wild-type and mutant alleles. The primers used to detect the wild-type allele were TTA CCA GGA ACA GTG CCA GC (sense primer) and ATC ACT GCT GTC TTT GAG ATC (antisense primer). The primers specific for the mutant allele were GCC CTG AAT GAA CTG CAG GAC G and CAC GGG TAG CCA ACG CTA TGT C.
Enzymatic deglycosylation and Western blot analysis.
Treatment of cell lysates with endoglycosidase H (EndoH; New England Biolabs) or peptide-N-glycosidase (PNGase; New England Biolabs) and Western blot analysis were performed as previously described (32).
In situ hybridization.
Timed matings were set up to generate embryos at different stages of gestation (embryonic day 11.5 [E11.5], E13.5, and E16.5) for an analysis of ADAM19 expression by mRNA in situ hybridization. Mouse embryos were fixed in 4% paraformaldehyde overnight at 4°C, and graded series of ethanol were subsequently used to dehydrate the fixed embryos. Dehydrated tissues were cleared with Histoclear, embedded in paraffin, sectioned, and mounted on Fisher Superfrost Plus slides. The appropriate linearized plasmids were used to prepare 33P-labeled RNA probes with T7 or Sp6 RNA polymerases by using a ribonucleotide triphosphate mix with 12 μM cold UTP and 4 μM hot UTP. The RNA in situ hybridization procedure was performed essentially as described previously (33).
LacZ staining.
Embryos at E11.5 were fixed in 2% paraformaldehyde for 10 min at 4°C, washed extensively in phosphate-buffered saline (PBS), and stained overnight at 37°C in PBS-10 mM MgCl2-0.2 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)/ml. Embryos were refixed in 4% paraformaldehyde for 30 min and then processed for sectioning as described above.
Whole-mount skeletal preparation.
E18.5 embryos were eviscerated, fixed in 95% ethanol overnight, and then stained overnight with 0.05% alcian blue 8GX in 95% ethanol and 5% acetic acid. After being rinsed and washed with 95% ethanol overnight, they were placed in 2% KOH until the bones became clearly visible. The bones were stained with 0.1% alizarin red in 1% KOH overnight and rinsed in 1% KOH-20% glycerol for 2 days. For storage, specimens were transferred into a 1:1 mixture of 95% ethanol and glycerol.
Immunohistochemistry.
The sections were prepared as described above, postfixed with ice-cold acetone for 10 min, and immersed in 0.1% H2O2 to inactivate the endogenous peroxidase. Following preincubation with PBS-10% normal goat serum-2% bovine serum albumin for 30 min, the slides were incubated for 3 h with antibodies against the platelet endothelial cell adhesion molecule 1 (PECAM-1/CD31; Santa Cruz Biotech, Santa Cruz, Calif.) and then washed and incubated with biotin-conjugated goat anti-rabbit immunoglobulin G. Bound antibodies were visualized by using the avidin-biotin complex detection method according to the manufacturer's instructions (Vector Laboratories, Burlingame, Calif.). After development the sections were counterstained with hematoxylin.
TRANCE, HB-EGF, and neuregulin I-β1 and I-β2 expression constructs.
To facilitate the detection of both precursor and shed forms of TRANCE, heparin-binding (HB)-EGF, and neuregulin I-β1 and I-β2, all proteins were expressed as fusion proteins bearing an alkaline phosphatase (AP) module in their extracellular domain. pAPtag5-TRANCE and the HB-EGF-AP plasmid have been described previously (7, 46). The HB-EGF-AP plasmid was a gift from S. Higashiyama (Osaka University Medical School). pEF-BOS-HA-neuregulin I-β1 (kindly provided by A. Fujisawa-Sehara, Kyoto University [42]) was used as a template to amplify a portion of mouse neuregulin I-β1 sequence (nucleotides 880 to 1210) bearing a 5′ XhoI site and a 3′ XbaI. The digested fragment was then subcloned at the corresponding restriction sites of the pAPtag5 vector (Genhunter Corp.), yielding a protein with the alkaline phosphatase tag attached at the N terminus of the EGF repeat. Human neuregulin I-β2 partial cDNA (nucleotides 977 to 2374; GenBank accession number NM_013957) was obtained from the MDA-MB-231 cell line by reverse transcription-PCR using primers carrying external XbaI sites (5′-GCTCTAGAAACCACTGGGACAAGCCATCTTG-3′ and 5′-GCTCTAGAGTTATACAGCAATAGGGTCTTG-3′). The identity of the neuregulin I-β2 isoform was confirmed by sequencing, and the XbaI-digested fragment was subcloned at the corresponding site into the pAPtag5 vector.
Ectodomain shedding assays.
COS-7 cells were cotransfected with either pAPtag5-TRANCE, HB-EGF-AP, pAPtag5-neuregulin I-β1, or pAPtag5-neuregulin I-β2, together with full-length wild-type (pcDNA3-ADAM19) or E>A mutant (pcDNA3-ADAM19E/A) ADAM19 (7). Mouse embryonic fibroblasts (mEFs) were prepared from 13.5-day-old wild-type or ADAM19-deficient embryos and transfected with the indicated plasmids by using Lipofectamine2000 (Invitrogen) as previously described (7, 50). The day following transfection, each well was washed once in PBS and incubated for 1 h in Opti-MEM I (Invitrogen) (nonstimulated conditioned medium) and then for an additional hour in Opti-MEM I containing 25 ng of phorbol 12-myristate 13-acetate (PMA)/ml (stimulated conditioned medium). Conditioned media were collected, and cells were lysed in PBS containing 1% Triton X-100, 2 μg of leupeptin/ml, 10 μg of soybean trypsin inhibitor/ml, 500 μM iodoacetamide, and 1 mM 1,10-phenanthroline. His-tagged shed forms of TRANCE-AP in the supernatant as well as membrane-anchored TRANCE-AP in cell lysate were concentrated by using Talon metal affinity resin and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as previously described (7). Shed HB-EGF-AP, neuregulin I-β1-AP, and neuregulin I-β2-AP present in the conditioned media were concentrated by using concanavalin A-Sepharose (Amersham Biosciences) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as described previously (7). COS-7 cell lysates were assayed for the expression of wild-type or catalytically inactive (E>A mutant) ADAM19 by Western blotting with anti-ADAM19 polyclonal antibodies as described previously (7).
RESULTS AND DISCUSSION
Mice lacking functional ADAM19 were obtained from ES cells with a secretory gene trap insertion in the ADAM19 gene (Fig. 1 a; see Materials and Methods) (30). The progeny of matings of heterozygous ADAM19 mutant mice had a Mendelian distribution of the targeted allele at birth (28.0% +/+, 47.2% +/−, 24.8% −/−; n = 254). However, ∼80% of homozygous adam19−/− mice died in the first few days after birth, while heterozygous ADAM19 mutant mice were healthy and fertile (distribution of offspring at day 21 after birth [P21], 30.8% +/+, 64.9% +/−, 4.3% −/−; n = 487).
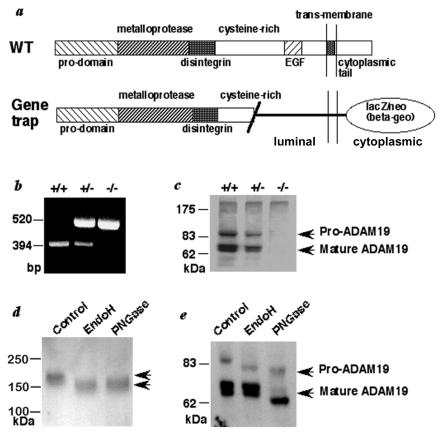
FIG. 1.
ADAM19 gene trap construct. (a) Diagram of wild-type ADAM19 and the mutant form generated by a secretory gene trap insertion (30). (b) Results of PCR genotyping. (c) Western blots of brain extracts of wild-type (+/+), heterozygous (+/−), or homozygous (−/−) ADAM19 mutant mice were analyzed as described previously (50) with an antibody raised against the ectodomain of ADAM19. (d and e) Western blots of brain extracts from adam19−/− mice (d) or wild-type mice (e) treated with EndoH or peptide-N-glycosidase (PNGase) were probed with an antibody against LacZ (d) or against the ectodomain of ADAM19 (e) (see also Fig. 1c). N-linked high mannose glycans are converted into complex carbohydrates by passage through the medial-Golgi network. Complex carbohydrates are usually resistant to treatment with EndoH, whereas treatment with PNGase removes both high-mannose glycans and complex N-linked carbohydrate residues. Acquisition of EndoH resistance therefore indicates that a glycoprotein has emerged from the ER and has passed through the medial-Golgi network. The arrows in panel d mark the position of the mutant ADAM19/LacZ/neo (β-geo) fusion protein.
Western blot analysis of brain extracts confirmed that wild-type ADAM19 is absent in homozygous mutant mice (Fig. 1b and c). Using an anti-β-galactosidase antibody to detect the mutant form of ADAM19 (Fig. 1a), we observed that the N-linked carbohydrate residues never acquire resistance to endoglycosidase H (EndoH) (Fig. 1d), indicating that most or all of the mutant protein never reaches the medial Golgi compartment. In contrast, mature wild-type ADAM19 acquires resistance to EndoH and is cleaved by a proprotein convertase in the trans-Golgi network (Fig. 1e) (22). The gene trap insertion into the cysteine-rich domain of ADAM19 evidently prevents proper protein folding, resulting in retention of mutant ADAM19 in the endoplasmic reticulum (ER) by chaperones and subsequent degradation (45). Therefore, since no detectable amounts of mutant ADAM19 emerge from the ER and adam19+/− mice have no evident pathological phenotype, the abnormal phenotypes in adam19−/− mice likely result from a loss of ADAM19 function and not from a dominant negative effect of the mutant protein.
In situ examination of the thoracic cavity of adam19−/− mice revealed abnormally distended blood-filled vascular structures on the anterior wall of the right ventricle (Fig. 2 b, d, and e). Immunohistochemical evaluation of the right ventricle demonstrated that the walls of these distended vessels are composed of endothelial cells (CD31 positive) (Fig. 2d and e). Furthermore, blood vessels within the myocardium of adam19−/− mice appeared abnormal, with disrupted smooth muscle cell ensheathment (Fig. 2g and i; wild-type controls are shown in Fig. 2f and h). In electron micrographs of myocardial endothelial cells of adam19−/− mice, we frequently observed perivascular edema in capillaries, extensive vacuolization of endothelial cells (Fig. 2k and l; wild-type control is shown in Fig. 2j), and occasional rupture of both capillaries and arterioles with release of erythrocytes into the extravascular space (data not shown). These ultrastructural defects are consistent with the focal intramyocardial hemorrhage observed in some adam19−/− hearts at the light microscopic level (data not shown).
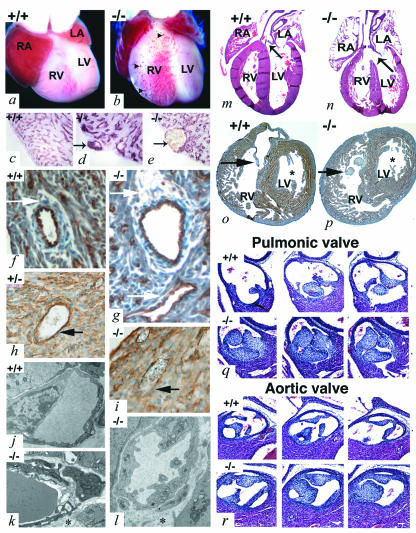
FIG. 2.
Heart defects in adam19−/− mice. Ventral view of hearts from newborn wild-type (a) or adam19−/− mice (b). Distended blood-filled vascular structures are indicated by arrowheads. (c, d, and e) CD31 staining confirms the vascular nature of the abnormal structures (shown by arrows in panels d and e). (f through i) Cross-section through CD31-stained (f and g) and α-smooth muscle actin-stained (h and i) blood vessel within the septal myocardium of wild-type (f), adam19+/− (h), and adam19−/− (g and i) animals. The sections shown in panels f and g are from newborn mice, whereas the sections in panels h and i are from E17.5 embryos. Arrows indicate normal smooth muscle ensheathment in a wild-type mouse (f and h) and abnormal or apparently lacking smooth muscle cell ensheathment of vessels in an adam19−/− mouse (g and i). Note that α-smooth muscle-actin also stains myocardial cells surrounding smooth muscle cells in h and i. (j, k, and l) Electron micrographs of cross-sections through myocardial capillaries of a wild-type (j) or adam19−/− (k and l) heart (the asterisks in k and l indicate perivascular edema). (m and n) Longitudinal section through a heart of a newborn wild-type (m) or adam19−/− (n) mouse stained with hematoxylin and eosin; the aortic valve is indicated by arrows; the aorta is overriding VSD in panel n. (o and p) Cross-sections through a heart of a newborn wild-type (o) or adam19−/− (p) mouse. In panels o and p, an arrow points to the tricuspid valve, which displays a lack of remodeling in the adam19−/− heart (p). An asterisk is placed next to the leaflets of the mitral valve. (q and r) Cross-sections of the pulmonic (q) and aortic valve (r) of a wild-type heart and an adam19−/− heart. Three comparable serial sections of 10 μm in thickness, spaced three sections (30 μm) apart, are shown for each valve. Abbreviations: RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle.
A histological evaluation of adam19−/− hearts revealed membranous ventricular septal defects (VSD) (Fig. 2n) as well as stenotic aortic and pulmonic valves with abnormally thickened leaflets in all hearts examined at birth (12 of 12; Fig. 2q and r). adam19−/− neonates exhibited moderate malrotation of the cardiac outflow tracts, in some cases resulting in an overriding aorta. These defects resemble conotruncal malformations, such as Tetralogy of Fallot and double-outlet right ventricle, which are important forms of human congenital heart disease with high morbidity. Thickened tricuspid valves were found in about one-half of adam19−/− hearts (Fig. 2p), and atrial septal defects were observed in one-third of adam19−/− hearts (data not shown). The mitral valve appeared normal in all adam19−/− hearts examined (Fig. 2n and p), and septation of the outflow tract into pulmonary artery and aorta was also unaffected (Fig. 2q and r).
The membranous aspect of the ventricular septum and the heart valves arising from the endocardial cushions between E10.5 and E13.5 (24, 29) prompted evaluation of ADAM19 expression in these developmental structures. In situ mRNA hybridization revealed prominent ADAM19 expression in the atrioventricular and conotruncal endocardial cushions between E10.5 and E12.5 (Fig. 3 a, b, e, and f; also data not shown). This expression pattern is consistent with the observed defects in adam19−/− mice. The complete penetrance of proximal conotruncal defects (VSD and aortic and pulmonic valve defects) compared to the partial penetrance of atrioventricular defects (ostium primum atrial septal and tricuspid valve defects) in adam19−/− mice demonstrates that the proximal conotruncal endocardial cushion is most sensitive to loss of ADAM19 activity.
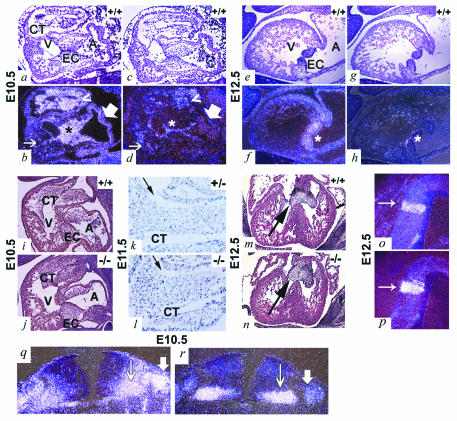
FIG. 3.
Analysis of the expression of ADAM19 and neuregulin during heart and brain development by in situ mRNA hybridization and comparison of endocardial cushion development in wild-type and adam19−/− embryos. Hearts of wild-type E10.5 (a through d) and E 12.5 embryos (e through h), were probed with antisense RNA probes for mouse ADAM19 (a, b, e, and f) or neuregulin (c, d, g, and h) (51). In panels b, d, f, and h, an asterisk marks the atrioventricular endocardial cushion. In panels b and d, a thick arrow points to the atrial endocardium, where ADAM19, but little neuregulin, is expressed. A thin arrow points to the ventricular endocardium, where there is some neuregulin expression, but little ADAM19 expression. The arrowhead (b and d) points towards a limited dorsal area of the conotruncal outflow tract where expression of ADAM19 and neuregulin appear to overlap. Panels a, c, e, and g are bright field; panels b, d, f, and h are dark field. (i through n) Sections of wild-type (i and m), adam19+/− (k), or adam19−/− (j, l, and n) hearts at E10.5 (i and j), E11.5 (k and l), and E12.5 (m and n). Sections in panels k and l are of an E11.5 conotruncal endocardial cushion of a heterozygous (k) heart or homozygous ADAM19 mutant heart (l) stained with X-Gal in order to detect cells expressing lacZ under the ADAM19 promoter. Cells located below the gap indicated by an arrow in panels k and l are part of the conotruncal endocardial cushion. In panels m and n, the endocardial cushion is marked by an arrow. (o through r) Adjacent sections of the neuroepithelium of the fourth ventricle of E12.5 embryos (o and p) or of the developing spinal cord of E10.5 embryos (q and r) were probed with antisense RNA probes for mouse ADAM19 (o and q) or neuregulin (p and r) (51). In panels o and p, an arrow points to a region of enhanced coexpression of ADAM19 and neuregulin in a defined area of the neuroepithelium of the fourth ventricle. In panels q and r, thick arrows mark the dorsal root ganglion and thin arrows mark the ventral horn of the spinal cord. Abbreviations: A, atrium; V, ventricle; CT, conotruncal endocardial cushion; EC, atrioventricular endocardial cushion.
Cardiac neural crest cells make important contributions to the morphogenesis of the conotruncal endocardial cushion, suggesting that ADAM19 may have a role in neural crest migration (24). Furthermore, the related ADAM13 has been implicated in neural crest migration in Xenopus laevis (1). However, in adam19−/− mice, other structures that depend on migration of neural crest cells, such as the thymus, hyoid bone, larynx, and palate, appeared morphologically normal (data not shown). Moreover, adam19−/− and wild-type embryos had endocardial cushions of similar shape and cellularity at E10.5 and E12.5, when neural crest migration to the region is complete (Fig. 3i, j, m, and n). Finally, when we stained heart sections of heterozygous or homozygous mutant ADAM19 embryos with X-Gal to mark cells expressing lacZ under the ADAM19 promoter, we found no difference in the distribution of stained cells at E11.5 (Fig. 3k and l). This result argues against a role for ADAM19 in the migration of ADAM19-expressing cells into the endocardial cushion. Instead, the heart defects seen in newborn adam19−/− mice, in particular the thickened and improperly remodeled aortic, pulmonic, and tricuspid valves, are more consistent with a key role for ADAM19 regulating morphogenesis and remodeling of the endocardial cushion after ADAM19-expressing cells have entered this structure.
Recent studies have demonstrated that mice lacking HB-EGF also have thickened aortic and pulmonic valves (19, 20). Furthermore, mice lacking ADAM17, which has a critical role in processing and presumably also activating HB-EGF (34, 44), phenocopy mice lacking HB-EGF with respect to the thickened aortic and pulmonic valves (20). In order to test whether ADAM19 might also contribute to HB-EGF shedding from the endocardium, we coexpressed both proteins in Cos-7 cells and also compared HB-EGF shedding in adam19+/− and adam19−/− mEFs. In order to confirm that ADAM19 is active in coexpression experiments, we included TRANCE/OPGL, an osteoclast differentiation factor and dendritic cell survival factor that is a known substrate of ADAM19 (7), as a positive control (Fig. 4 a). When HB-EGF was coexpressed with wild-type ADAM19 or a catalytically inactive mutant (E>A), no difference in constitutive or PMA-induced shedding was observed (Fig. 4a). Furthermore, we saw no difference in constitutive or PMA-stimulated shedding of HB-EGF from adam19−/− mEF cells compared to heterozygous controls (Fig. 4b) or wild-type controls (data not shown). Taken together, these experiments argue against a role of ADAM19 in HB-EGF shedding.
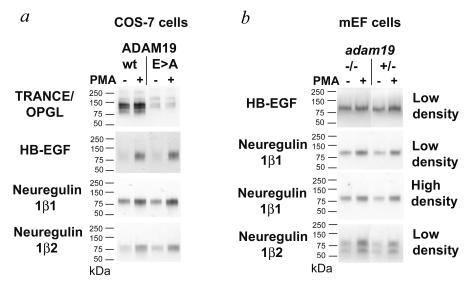
FIG. 4.
Role of ADAM19 in ectodomain shedding of TRANCE, HB-EGF, neuregulin I-β1, and neuregulin I-β2. (a) Coexpression of alkaline phosphatase-tagged candidate substrate proteins with wild-type ADAM19 or a catalytically inactive mutant (HEIGH > HAIGH; E>A) in Cos-7 cells. Coexpression of alkaline phosphatase-tagged TRANCE with wild-type ADAM19 strongly increases constitutive shedding compared to coexpression with the catalytically inactive mutant (E>A). Addition of PMA only leads to a minor further enhancement of TRANCE shedding, suggesting that ADAM19 is not strongly stimulated by PMA in Cos-7 cells. In parallel experiments, coexpression of wild-type ADAM19 with HB-EGF or neuregulin I-β1 or I-β2 did not lead to increased constitutive or PMA-stimulated shedding compared to the inactive mutant. Similar results were obtained in coexpression experiments with hemagglutinin-tagged neuregulin I-β1 constructs kindly provided by Shirakabe et al. (42) (data not shown). In all experiments, the expression levels of the candidate substrates as well as wild-type and mutant forms of ADAM19 in the cell lysates were comparable (data not shown). (b) Ectodomain shedding of candidate substrate proteins in primary mEFs isolated from adam19−/− or adam19+/− mice. Cells were plated at 105 (low density) or 3 × 105 (high density) per well of a six-well plate. No significant difference in constitutive or stimulated ectodomain shedding of these substrates was seen in comparison of adam19−/− and adam19+/− mEFs plated at low or high density (results from HB-EGF- and neuregulin I-β2-expressing mEFs plated at high density are not shown). TRANCE shedding is also not significantly affected in adam19−/− cells, presumably because the expression levels of ADAM19 are relatively low in these cells compared to those of other TRANCE sheddases (40).
ADAM19 has also been implicated in ectodomain shedding of neuregulin I-β, a protein that is essential for proper trabeculation of the heart during early development (26, 36, 42). However, when we coexpressed neuregulin I-β1 or I-β2 with wild-type or mutant ADAM19 in Cos-7 cells, no difference in ectodomain shedding was evident (Fig. 4a). Furthermore, no significant defect was seen in the constitutive or PMA-stimulated shedding of neuregulin I-β1 and I-β2 in sparsely or densely seeded adam19−/− mEFs compared to heterozygous or wild-type controls (Fig. 4b and data not shown). When we compared the expression pattern of ADAM19 and neuregulin in adjacent sections of E10.5 and E12.5 mouse embryos, we found little, if any, coexpression in the endocardial cushion (Fig. 3a through h). Neuregulin has also been implicated in the development of the cardiac conduction system (38), yet we found no differences between the electrocardiograms of newborn adam19−/− mice and those of wild-type mice (data not shown). Taken together, these results suggest that the heart defects seen in adam19−/− mice are not due to defects in processing neuregulin I-β1 or I-β2. It remains to be determined whether ADAM19 has a role in processing other neuregulin isoforms (11) in cells and tissues where both proteins are coexpressed, such as in the endocardium overlying the endocardial cushion and a limited portion of the outflow tract of the developing heart at E10.5 (Fig. 3b and d), a defined area of neuroepithelial cells within the fourth brain ventricle (Fig. 3o and p), and part of the ventral horn of the spinal cord and dorsal root ganglia (Fig. 3q and r; see also reference 27).
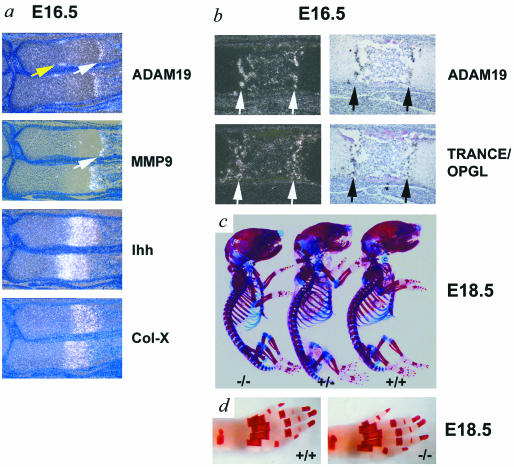
In a previous study, it was demonstrated that ADAM19 is highly expressed in bone (18). Because ADAM19 can cleave the osteoclast differentiation factor TRANCE in Cos-7 cells (see above and reference 7), we evaluated whether ADAM19 may have a role in bone development. In situ hybridization showed that ADAM19 is highly expressed in a subpopulation of cells residing immediately adjacent to the zone of collagen-X-expressing hypertrophic chondrocytes in the growth plate (Fig. 5 a and b). The expression pattern of ADAM19 resembles that of MMP9 (48) (Fig. 5a), which is also found next to cells expressing collagen-X. Furthermore, at E16.5, TRANCE appears to be coexpressed with ADAM19 (Fig. 5b), raising the possibility that ADAM19 could participate in TRANCE shedding in this particular bone area. Finally, a distinct domain of ADAM19 expression is found in a subset of highly proliferating chondrocytes adjacent to the perichondrium and distal to early hypertrophic chondrocytes expressing Indian hedgehog. Whole-mount alizarin red and alcian blue staining as well as morphometric analysis of bone sections from wild-type and adam19−/− mice did not uncover evident histopathological differences in bone development (Fig. 5c and d and data not shown). Further studies will be necessary to evaluate bones of surviving adult adam19−/− mice for more-subtle defects in bone remodeling that may not be apparent during bone development.
FIG. 5.
Evaluation of potential roles of ADAM19 in developing bone. (a and b) Serial sections of wild-type E16.5 forelimbs were hybridized with antisense probes for ADAM19, matrix metalloproteinase 9 (MMP9), Indian hedgehog (Ihh), collagen-X (Col-X) (a) and ADAM19 or TRANCE/OPGL (b). The expression pattern of ADAM19 resembles that of MMP9 (white arrows) immediately adjacent to the zone of hypertrophic cells, which is marked by expression of Col-X. The yellow arrow points to ADAM19 expression in proliferating chondrocytes distal to the Ihh expression domain (a). Furthermore, the expression of ADAM19 is similar or identical to the expression of TRANCE/OPGL (b). (c) Whole-mount alizarin red and alcian blue staining of adam19−/−, adam+/−, and adam+/+ littermates shows comparable bone development in all three genotypes at E18.5. (d) Alizarin red-stained whole-mount hind paws of adam19+/+ and adam19−/− mice also show comparable development at E18.5.
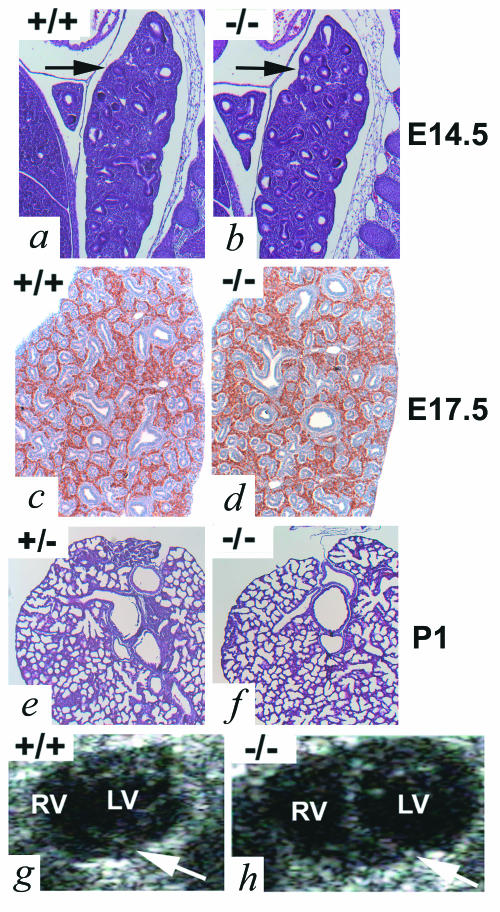
In addition to heart and bone, ADAM19 mRNA is also highly expressed in the lung (18). However, a histopathological analysis of lungs of adam19−/−, adam19+/− and wild-type embryos at E14.5 and E17.5 as well newborn mice of all three genotypes did not reveal any apparent defects in the absence of ADAM19 (Fig. 6 a through f and data not shown).
FIG. 6.
Evaluation of lung development (a through f) and echocardiographic analysis of adult hearts (g and h) in the presence or absence of ADAM19. Sections shown in panels a through f are of lungs from wild-type (a and c) or adam19−/−(b and d) embryos at E14.5 (a and b; sections are stained with hematoxylin and eosin [H&E]), E17.5 (c and d; sections are immunostained with anti-CD34 to mark the vasculature and counterstained with hematoxylin), or of adam19+/− (e) or adam19−/− (f) newborn mice (P1; H&E stained). Arrows in a and b point to lung tissue. No major differences in lung morphology, branching, or alveolarization were detected in adam19−/− compared to wild-type or adam19+/− controls. An echocardiographic analysis of adult hearts of 2-month-old mice (g and h) shows an enlarged right ventricle in an adam19−/− mouse (h) compared to a wild-type control (g). The arrows indicate the position of the ventricular septum in panels h and g. Abbreviations: RV, right ventricle, LV, left ventricle.
The overall analysis of adam19−/− mice suggests that their postnatal lethality is most likely due to the ventricular septal defect combined with aortic and pulmonic valvular stenosis. Following birth, when the right and left circulation are normally separated, a persistent VSD coupled with pulmonic valve stenosis would lead to volume and pressure overload of the right ventricle, resulting in progressive right ventricular dilatation and hypertrophy. Similar malformations (e.g., VSD with pulmonic stenosis and atresia, Tetralogy of Fallot) as well as aortic stenosis are well-recognized causes of heart failure and death in humans. The vascular anomalies observed in adam19−/− mice may further compromise myocardial perfusion. Although some adam19−/− mice survive, echocardiography revealed an enlarged right ventricle in all five adult adam19−/− animals examined (Fig. 6g and h and data not shown). This is consistent with sublethal cardiac defects that are still compatible with long-term but nonstressed survival. Adult adam19−/− mice appear healthy and are fertile, although four of five pregnant adam19−/− mice died late in pregnancy, likely due to increased circulatory demands. Indeed, human adults with similar forms of congenital heart disease represent high-risk obstetrical patients.
Congenital heart disease is the most common form of birth defects in humans, yet little is known about the underlying molecular causes (2, 8, 15, 29, 43, 47). In light of the critical role of mouse ADAM19 in heart development, it will be interesting to determine whether certain types of human congenital heart defects are caused by mutations in ADAM19, which is located on human chromosome 5q33.3. To date, only a few proteins on the cell surface or in the extracellular matrix have been implicated in endocardial cushion development, including neurotrophin-3 (10), tumor growth factor β (5, 6) and the related BMP6 and BMP7 (23), and most recently HB-EGF and ADAM17 (19, 20). It will now be interesting to further explore potential functional connections between ADAM19 and these proteins as well as other molecules that are implicated in endocardial cushion transformation and/or conotruncal defects, such as RXRα, smad6, and TBX1 (14, 16, 21, 31, 35). It is also possible that ADAM19-dependent cell-cell interactions or signaling via its cytoplasmic domain contribute to its role in heart development (39). We anticipate that the discovery of an essential role for ADAM19 in heart development in mice will lead to a better understanding of the causes underlying human congenital heart disease.
Acknowledgments
This work was supported by National Institutes of Health grant RO1 GM64750 (to C.P.B.).
We thank Phil Leighton, Bill Skarnes, and Marc Tessier-Lavigne for generously providing embryonic stem cells carrying a gene trap insertion in ADAM19, Leona Cohen-Gould for providing electron micrographs, Thomas Ludwig, Willie Mark, Liz Lacy, Jay Edelberg, and David Christini for valuable advice, and Thadeous Kacmarczyk, Dan Harrigan, Kevin Curran, Maria Kobi, Conny Kreschel, and members of the Sloan-Kettering Institute transgenic facility for excellent technical assistance.
REFERENCES
- 1.Alfandari, D., H. Cousin, A. Gaultier, K. Smith, J. M. White, T. Darribere, and D. W. DeSimone. 2001. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr. Biol. 11:918-930. [DOI] [PubMed] [Google Scholar]
- 2.Basson, C. T., and C. E. Seidman. 1998. Genetic studies of myocardial and vascular disease, p. 2429-2447. In E. J. Topol (ed.), Textbook of cardiovascular medicine. Lippincott-Raven, Philadelphia, Pa.
- 3.Black, R. A., and J. M. White. 1998. ADAMs: focus on the protease domain. Curr. Opin. Cell Biol. 10:654-659. [DOI] [PubMed] [Google Scholar]
- 4.Blobel, C. P. 2000. Remarkable roles of proteolysis on and beyond the cell surface. Curr. Opin. Cell Biol. 12:606-612. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. B., A. S. Boyer, R. B. Runyan, and J. V. Barnett. 1999. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science 283:2080-2082. [DOI] [PubMed] [Google Scholar]
- 6.Camenisch, T. D., D. G. Molin, A. Person, R. B. Runyan, A. C. Gittenberger-de Groot, J. A. McDonald, and S. E. Klewer. 2002. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev. Biol. 248:170-181. [DOI] [PubMed] [Google Scholar]
- 7.Chesneau, V., D. Becherer, Y. Zheng, H. Erdjument-Bromage, P. Tempst, and C. P. Blobel. 2003. Catalytic properties of ADAM19. J. Biol. Chem. 278:22331-22340. [DOI] [PubMed] [Google Scholar]
- 8.Chien, K. R. 2000. Genomic circuits and the integrative biology of cardiac diseases. Nature 407:227-232. [DOI] [PubMed] [Google Scholar]
- 9.Deftos, M. L., E. Huang, E. W. Ojala, K. A. Forbush, and M. J. Bevan. 2000. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity 13:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donovan, M. J., R. Hahn, L. Tessarollo, and B. L. Hempstead. 1996. Identification of an essential nonneuronal function of neurotrophin 3 in mammalian cardiac development. Nat. Genet. 14:210-213. [DOI] [PubMed] [Google Scholar]
- 11.Falls, D. L. 2003. Neuregulins: functions, forms, and signaling strategies. Exp. Cell Res. 284:14-30. [DOI] [PubMed] [Google Scholar]
- 12.Fata, J. E., Y. Y. Kong, J. Li, T. Sasaki, J. Irie-Sasaki, R. A. Moorehead, R. Elliott, S. Scully, E. B. Voura, D. L. Lacey, W. J. Boyle, R. Khokha, and J. M. Penninger. 2000. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103:41-50. [DOI] [PubMed] [Google Scholar]
- 13.Fritsche, J., M. Moser, S. Faust, A. Peuker, R. Buttner, R. Andreesen, and M. Kreutz. 2000. Molecular cloning and characterization of a human metalloprotease disintegrin-a novel marker for dendritic cell differentiation. Blood 96:732-739. [PubMed] [Google Scholar]
- 14.Galvin, K. M., M. J. Donovan, C. A. Lynch, R. I. Meyer, R. J. Paul, J. N. Lorenz, V. Fairchild-Huntress, K. L. Dixon, J. H. Dunmore, M. A. Gimbrone, Jr., D. Falb, and D. Huszar. 2000. A role for smad6 in development and homeostasis of the cardiovascular system. Nat. Genet. 24:171-174. [DOI] [PubMed] [Google Scholar]
- 15.Goldmuntz, E., and B. S. Emanuel. 1997. Genetic disorders of cardiac morphogenesis. The DiGeorge and velocardiofacial syndromes. Circ. Res. 80:437-443. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, P. J., S. W. Kubalak, T. Pexieder, H. M. Sucov, R. M. Evans, and K. R. Chien. 1996. RXR alpha deficiency confers genetic susceptibility for aortic sac, conotruncal, atrioventricular cushion, and ventricular muscle defects in mice. J. Clin. Investig. 98:1332-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horiuchi, K., G. Weskamp, L. Lum, H. P. Hammes, H. Cai, T. A. Brodie, T. Ludwig, R. Chiusaroli, R. Baron, K. T. Preissner, K. Manova, and C. P. Blobel. 2003. Potential role for ADAM15 in pathological neovascularization in mice. Mol. Cell. Biol. 23:5614-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue, D., M. Reid, L. Lum, J. Krätzschmar, G. Weskamp, Y. M. Myung, R. Baron, and C. P. Blobel. 1998. Cloning and initial characterization of mouse meltrin beta and analysis of the expression of four metalloprotease-disintegrins in bone cells. J. Biol. Chem. 273:4180-4187. [DOI] [PubMed] [Google Scholar]
- 19.Iwamoto, R., S. Yamazaki, M. Asakura, S. Takashima, H. Hasuwa, K. Miyado, S. Adachi, M. Kitakaze, K. Hashimoto, G. Raab, D. Nanba, S. Higashiyama, M. Hori, M. Klagsbrun, and E. Mekada. 2003. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc. Natl. Acad. Sci. USA 100:3221-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson, L. F., T. H. Qiu, S. W. Sunnarborg, A. Chang, C. Zhang, C. Patterson, and D. C. Lee. 2003. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 22:2704-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jerome, L. A., and V. E. Papaioannou. 2001. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat. Genet. 27:286-291. [DOI] [PubMed] [Google Scholar]
- 22.Kang, T., Y. G. Zhao, D. Pei, J. F. Sucic, and Q. X. Sang. 2002. Intracellular activation of human adamalysin 19/disintegrin and metalloproteinase 19 by furin occurs via one of the two consecutive recognition sites. J. Biol. Chem. 277:25583-25591. [DOI] [PubMed] [Google Scholar]
- 23.Kim, R. Y., E. J. Robertson, and M. J. Solloway. 2001. Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev. Biol. 235:449-466. [DOI] [PubMed] [Google Scholar]
- 24.Kirby, M. L. 1999. Contribution of neural crest to heart and vessel morphology, p. 179-193. In R. P. Harvey and N. Rosenthal (ed.), Heart development. Academic Press, London, United Kingdom.
- 25.Kong, Y. Y., H. Yoshida, I. Sarosi, H. L. Tan, E. Timms, C. Capparelli, S. Morony, A. J. Oliveira-dos-Santos, G. Van, A. Itie, W. Khoo, A. Wakeham, C. R. Dunstan, D. L. Lacey, T. W. Mak, W. J. Boyle, and J. M. Penninger. 1999. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature 397:315-323. [DOI] [PubMed] [Google Scholar]
- 26.Kramer, R., N. Bucay, D. J. Kane, L. E. Martin, J. E. Tarpley, and L. E. Theill. 1996. Neuregulins with an Ig-like domain are essential for mouse myocardial and neuronal development. Proc. Natl. Acad. Sci. USA 93:4833-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurisaki, T., A. Masuda, N. Osumi, Y. Nabeshima, and A. Fujisawa-Sehara. 1998. Spatially- and temporally-restricted expression of meltrin alpha (ADAM12) and beta (ADAM19) in mouse embryo. Mech. Dev. 73:211-215. [DOI] [PubMed] [Google Scholar]
- 28.Lacey, D. L., E. Timms, H. L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliott, A. Colombero, G. Elliott, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y. X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 29.Lamers, W. H., and A. F. Moorman. 2002. Cardiac septation: a late contribution of the embryonic primary myocardium to heart morphogenesis. Circ. Res. 91:93-103. [DOI] [PubMed] [Google Scholar]
- 30.Leighton, P. A., K. J. Mitchell, L. V. Goodrich, X. Lu, K. Pinson, P. Scherz, W. C. Skarnes, and M. Tessier-Lavigne. 2001. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature 410:174-179. [DOI] [PubMed] [Google Scholar]
- 31.Lindsay, E. A., F. Vitelli, H. Su, M. Morishima, T. Huynh, T. Pramparo, V. Jurecic, G. Ogunrinu, H. F. Sutherland, P. J. Scambler, A. Bradley, and A. Baldini. 2001. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410:97-101. [DOI] [PubMed] [Google Scholar]
- 32.Lum, L., and C. P. Blobel. 1997. Evidence for distinct serine protease activities with a potential role in processing the sperm protein fertilin. Dev. Biol. 191:131-145. [DOI] [PubMed] [Google Scholar]
- 33.Manova, K., K. Nocka, P. Besmer, and R. F. Bachvarova. 1990. Gonadal expression of c-kit encoded at the W locus of the mouse. Development 110:1057-1069. [DOI] [PubMed] [Google Scholar]
- 34.Merlos-Suarez, A., S. Ruiz-Paz, J. Baselga, and J. Arribas. 2001. Metalloprotease-dependent protransforming growth factor-alpha ectodomain shedding in the absence of tumor necrosis factor-alpha-converting enzyme. J. Biol. Chem. 276:48510-48517. [DOI] [PubMed] [Google Scholar]
- 35.Merscher, S., B. Funke, J. A. Epstein, J. Heyer, A. Puech, M. M. Lu, R. J. Xavier, M. B. Demay, R. G. Russell, S. Factor, K. Tokooya, B. S. Jore, M. Lopez, R. K. Pandita, M. Lia, D. Carrion, H. Xu, H. Schorle, J. B. Kobler, P. Scambler, A. Wynshaw-Boris, A. I. Skoultchi, B. E. Morrow, and R. Kucherlapati. 2001. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104:619-629. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, D., and C. Birchmeier. 1995. Multiple essential functions of neuregulin in development. Nature 378:386-390. [DOI] [PubMed] [Google Scholar]
- 37.Primakoff, P., and D. G. Myles. 2000. The ADAM gene family: surface proteins with an adhesion and protease activity packed into a single molecule. Trends Genet. 16:83-87. [DOI] [PubMed] [Google Scholar]
- 38.Rentschler, S., J. Zander, K. Meyers, D. France, R. Levine, G. Porter, S. A. Rivkees, G. E. Morley, and G. I. Fishman. 2002. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc. Natl. Acad. Sci. USA 99:10464-10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlöndorff, J., and C. P. Blobel. 1999. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein ectodomain shedding. J. Cell Sci. 112:3603-3617. [DOI] [PubMed] [Google Scholar]
- 40.Schlöndorff, J. S., L. Lum, and C. P. Blobel. 2001. Biochemical and pharmacological criteria define two shedding activities for TRANCE/OPGL that are distinct from the TNF alpha convertase (TACE). J. Biol. Chem. 276:14665-14674. [DOI] [PubMed] [Google Scholar]
- 41.Seals, D. F., and S. A. Courtneidge. 2003. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 17:7-30. [DOI] [PubMed] [Google Scholar]
- 42.Shirakabe, K., S. Wakatsuki, T. Kurisaki, and A. Fujisawa-Sehara. 2001. Roles of meltrin beta/ADAM19 in the processing of neuregulin. J. Biol. Chem. 276:9352-9358. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava, D., and E. N. Olson. 2000. A genetic blueprint for cardiac development. Nature 407:221-226. [DOI] [PubMed] [Google Scholar]
- 44.Sunnarborg, S. W., C. L. Hinkle, M. Stevenson, W. E. Russell, C. S. Raska, J. J. Peschon, B. J. Castner, M. J. Gerhart, R. J. Paxton, R. A. Black, and D. C. Lee. 2002. Tumor necrosis factor-alpha converting enzyme (TACE) regulates epidermal growth factor receptor ligand availability. J. Biol. Chem. 277:12838-12845. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, T., Q. Yan, and W. J. Lennarz. 1998. Complex, two-way traffic of molecules across the membrane of the endoplasmic reticulum. J. Biol. Chem. 273:10083-10086. [DOI] [PubMed] [Google Scholar]
- 46.Tokumaru, S., S. Higashiyama, T. Endo, T. Nakagawa, J. Miyagawa, K. Yamamori, Y. Hanakawa, H. Ohmoto, K. Yoshino, Y. Shirakata, Y. Matsuzawa, K. Hashimoto, and N. Taniguchi. 2000. Ectodomain shedding of epidermal growth factor receptor ligands is required for keratinocyte migration in cutaneous wound healing. J. Cell Biol. 151:209-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaughan, C. J., and C. T. Basson. 2000. Molecular determinants of atrial and ventricular septal defects and patent ductus arteriosus. Am. J. Med. Genet. 97:304-309. [DOI] [PubMed] [Google Scholar]
- 48.Vu, T. H., J. M. Shipley, G. Bergers, J. E. Berger, J. A. Helms, D. Hanahan, S. D. Shapiro, R. M. Senior, and Z. Werb. 1998. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell 93:411-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei, P., Y. G. Zhao, L. Zhuang, S. Ruben, and Q. X. Sang. 2001. Expression and enzymatic activity of human disintegrin and metalloproteinase ADAM19/meltrin beta. Biochem. Biophys. Res. Commun. 280:744-755. [DOI] [PubMed] [Google Scholar]
- 50.Weskamp, G., H. Cai, T. A. Brodie, S. Higashyama, K. Manova, T. Ludwig, and C. P. Blobel. 2002. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol. Cell. Biol. 22:1537-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson, D. G., J. A. Bailes, J. E. Champion, and A. P. McMahon. 1987. A molecular analysis of mouse development from 8 to 10 days post coitum detects changes only in embryonic globin expression. Development 99:493-500. [DOI] [PubMed] [Google Scholar]
- 52.Yagami-Hiromasa, T., T. Sato, T. Kurisaki, K. Kamijo, Y. Nabeshima, and A. Fujisawa-Sehara. 1995. A metalloprotease-disintegrin participating in myoblast fusion. Nature 377:652-656. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, Y. G., P. Wei, and Q. X. Sang. 2001. Inhibitory antibodies against endopeptidase activity of human adamalysin 19. Biochem. Biophys. Res. Commun. 289:288-294. [DOI] [PubMed] [Google Scholar]