Abstract
The strength and duration of mitogen-activated protein kinase (MAPK) signaling have been shown to regulate cell fate in different cell types. In this study, a general mechanism is described that explains how subtle differences in signaling kinetics are translated into a specific biological outcome. In fibroblasts, the expression of immediate early gene (IEG)-encoded Fos, Jun, Myc, and early growth response gene 1 (Egr-1) transcription factors is significantly extended by sustained extracellular signal-regulated kinase 1 and 2 (ERK1 and -2) signaling. Several of these proteins contain functional docking site for ERK, FXFP (DEF) domains that serve to locally concentrate the active kinase, thus showing that they can function as ERK sensors. Sustained ERK signaling regulates the posttranslational modifications of these IEG-encoded sensors, which contributes to their sustained expression during the G1-S transition. DEF domain-containing sensors can also interpret the small changes in ERK signal strength that arise from less than a threefold reduction in agonist concentration. As a result, downstream target gene expression and cell cycle progression are significantly changed.
The induction of immediate early genes (IEGs) following exposure to extracellular stimuli represents the first major transcriptional program that precedes changes in a variety of cellular responses. The extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), and p38 mitogen-activated protein kinase (MAPK) pathways, as well as phosphatidylinositol 3-kinase (PI3-kinase) signaling (3, 11), are known to positively regulate IEGs, which results in various cellular outcomes, such as cell proliferation, differentiation, and oncogenic transformation. Specifically, the MEK1/2-ERK signaling pathway plays a crucial role in IEG induction by directly activating IEG promoter-bound transcription factors (40). This results in transient transcription of the IEGs (12) and after a further 30 to 45 min, the appearance of gene products such as the Fos, Jun, and Myc family of transcription factors, as well as other transcription factors, such as Egr-1.
One puzzling feature associated with the physiological role of IEG induction is that their global expression is associated with agents as diverse as mitogenic growth factors, differentiating factors, and environmental insults, such as osmotic shock and ionizing radiation. Therefore, if the same panel of IEGs are induced by different extracellular agents, how does this lead to the generation of an appropriate biological outcome? One possibility is that different signaling pathways induce a specific subset of IEGs, and this promotes a specific outcome. Substantial experimental evidence suggests that this is not the case (2, 12, 24, 35). What is known, however, is that the kinetics and amplitude of ERK signaling can regulate various cell fate decisions (21, 39). This laboratory has recently illustrated how the c-Fos IEG product functions as a sensor for the duration of ERK signaling in fibroblasts (26). The expression of c-Fos is posttranslationally regulated by differences in signal kinetics, and this controls c-Fos-dependent proliferation and transformation. The fundamental significance of this is demonstrated by the observation that changes in the duration of ERK signaling dictate whether or not c-Fos protein is expressed transiently (30 min) or in a more sustained manner (up to 4 h). This suggests that differences in signal properties, such as duration and strength, can modulate an IEG expression program that is initially generic at the transcriptional level to one that is fine-tuned in a signal-specific manner at the posttranslational level.
The process whereby c-Fos activity is regulated during sustained ERK signaling occurs in two stages (26). First, nuclear localization of ERK and the 90-kDa ribosomal S6 kinase (RSK) leads to the phosphorylation of Ser374 and Ser362 in the COOH terminus of c-Fos, respectively, and this reduces the rate of c-Fos degradation. Under conditions of transient ERK signaling, the c-fos gene is induced, but c-Fos protein, appearing after the initial stimulus, is not modified and therefore is unstable. Second, phosphorylation of Ser374 and Ser362 primes the additional phosphorylation of Thr325 and, to a lesser extent, Thr331. Efficient phosphorylation of these latter two residues is dependent on an ERK docking DEF (docking site for ERK, FXFP) domain found between amino acids (aa) 343 and 346 (Phe-Thr-Tyr-Pro). Importantly, mutation of the c-Fos DEF domain reduces AP-1 transcription and completely prevents the ability of c-Fos to mediate cellular transformation (26). The essential role for the DEF domain in c-Fos function prompted us to visually examine other IEG products for the presence of DEF domains. This revealed that in addition to c-Fos, other IEG products from the Fos, Jun, and Myc family contain putative DEF domains, suggesting that they may also function as ERK sensors. The majority of these ERK sensors are encoded by proto-oncogenes and are therefore predicted to have an important role in promoting cellular transformation. In this study, we show that several of these putative sensors, (Fra-2, Fra-1, and c-Myc) indeed have functional DEF domains and that ERK docking controls the phosphorylation of physiologically relevant sites. Interestingly, the transcriptional induction of one of these sensors, Fra-1, is itself under the direct control of the earlier-expressed c-Fos sensor, indicating that the products of growth factor-inducible genes can provide specific long-term temporal information about the properties of ERK signaling. ERK sensors also exhibit the ability to distinguish between small differences in the strength of ERK signals that result from minute changes in agonist concentration. This ultimately results in substantial changes in downstream gene expression and the efficiency with which cells enter S phase.
MATERIALS AND METHODS
Plasmids.
The pCDNA3 Fra-1 and Fra-2 constructs were generated by subcloning mouse fra-1 or fra-2 cDNAs from pCMVSport (18) to pCDNA3 by using the HindIII-ApaI and HindIII-EcoRI restriction sites, respectively. pCDNA3 c-Myc was constructed by subcloning the human c-myc cDNA from pBabe c-Myc (K. Ciekowski, Harvard Medical School) by using the BamHI-EcoRI sites. pCDNA3 Flag-c-Fos was described previously (26). All point mutations in cDNAs were generated by PCR and verified by DNA sequencing.
Cell culture.
NIH 3T3 cells were cultured in Dulbecco's minimal essential medium (DME) containing 10% fetal calf serum (FCS), and Swiss 3T3 cells were cultured in DME containing 10% heat-inactivated fetal bovine serum (FBS). Plasmid DNA was transfected into NIH 3T3 cells (1.8 × 105 per 35-mm-diameter dish) by using Lipofectamine reagent (Invitrogen Corporation) for 6 h, and then cells were deprived of serum growth factors for a further 20 h. Swiss 3T3 cells (0.8 × 106) were cultured in 100-mm dishes for 30 h and then deprived of serum growth factors for 40 h. Epidermal growth factor (EGF; 25 ng/ml) and platelet-derived growth factor BB (PDGF-BB; 20 ng/ml) were obtained from Invitrogen Corp. In certain experiments, cells were preincubated with 5 μM UO126 or vehicle control (0.1% dimethyl sulfoxide [DMSO]) for 30 min prior to addition of growth factors.
Cell lysis and Western blotting.
Following treatment of cells with growth factors, lysis was performed as described previously (31). Cell extracts were either directly subjected to Western analysis or in some cases used for immunoprecipitation. The expression of c-Myc in Swiss 3T3 cells was measured by immunoprecipitating endogenous c-Myc from cell extracts with a mouse anti-c-Myc monoclonal antibody (sc-42) from Santa Cruz Biotechnology, Inc., followed by Western analysis with an anti-c-Myc antibody (06-340) from Upstate Biotechnology, Inc. To measure the phosphorylation of Ser62, c-Myc was overexpressed in NIH 3T3 cells, immunoprecipitated from cell extracts, and then subjected to Western analysis with an anti-phospho-Ser62 antibody (34) or a mouse anti-human c-Myc monoclonal antibody (9E10) to control for variation in c-Myc levels. The anti-ERK and anti-phospho-Thr325 c-Fos antibodies were used as previously described (26). The mouse anti-phospho-ERK1/2 antibody was obtained from Sigma-Aldrich; anti-human c-Fos was obtained from Upstate Biotechnology, Inc.; anti-c-Jun was obtained from Cell Signaling Technologies; and anti-Fra-1 (sc-605), anti-Fra-2 (sc-604), anti-JunB (sc-73), anti-Egr-1 (sc-110), and anti-cyclin D1 (sc-753) antibodies were obtained from Santa Cruz Biotechnology, Inc. For each Western blot, nitrocellulose membrane was rinsed briefly in purified water prior to incubation in blocking solution (phosphate-buffered saline [PBS] containing 0.02% [vol/vol] Tween 20 and 3% [wt/vol] nonfat dried milk) for 20 min. The primary antibodies were diluted 1:1,000 in blocking solution and incubated with the membrane for 12 h at 4°C. The nitrocellulose was rinsed three times with purified water and then incubated with secondary antibody in PBS containing 3% (wt/vol) nonfat dried milk (90 min at room temperature). Finally, the membrane was rinsed three times in water, once in PBS-0.02% (vol/vol) Tween 20 (5 min), and a further five times in water before enhanced chemiluminescence with the ECL system was performed.
Kinase assay.
ERK1 was immunoprecipitated from cell extracts, and its kinase activity was determined by suspending the immune complex in kinase buffer (7) in the presence of 10 μCi of [γ-32P]ATP and 2 μg of glutathione S-transferase (GST)-RSKD2/K464R at 30°C for 10 min. Kinase reactions were stopped by adding sodium dodecyl sulfate sample buffer and boiling for 3 min. Samples were resolved using sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the phosphorylation of GST-RSKD2/K464R was quantitated with a phosphorimager.
BrdU analysis.
Indirect immunofluorescence analysis was performed as described previously (26), except cells were incubated with bromodeoxyuridine (BrdU) for 15 min prior to fixation. For quantitation, cells were harvested with trypsin, washed in PBS, and fixed in ice-cold 70% ethanol (1 h). To extract histones and denature cellular DNA, cells were incubated with 0.1 N HCl with 0.5% Triton X-100 (10 min), resuspended in double-distilled water, incubated at 100°C (10 min), and washed in PBS containing 0.5% Triton X-100. For direct immunofluorescence staining, cells were incubated for 30 min with anti-BrdU fluorescein isothiocyanate-conjugated antibody (Becton Dickinson), washed in 0.5% Tween 20 plus 1% bovine serum albumin in PBS, resuspended in propidium iodide stain buffer (0.5% Tween 20, 250 μg of DNase-free RNase A per ml, 20 μg of propidium iodide per ml). After staining, samples were analyzed by fluorescence-activated cell sorting (FACS) with a FACScan flow cytometer (Becton Dickinson), and data were acquired and analyzed with CellQuest software (Becton Dickinson Immunocytometry Systems).
RESULTS
Functional DEF domains in Fra-1, Fra-2, and c-Myc.
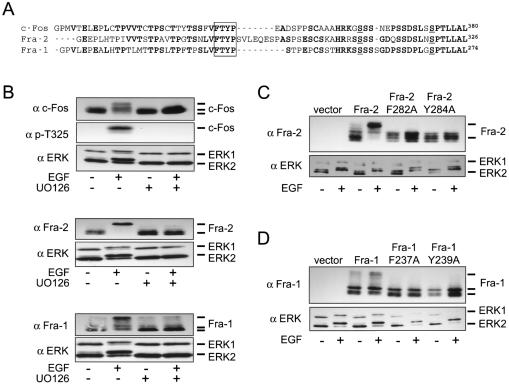
The COOH termini of c-Fos, Fra-1, and Fra-2 show significant amino acid identity (Fig. 1A, boldface residues). Specifically, the priming phosphorylation sites (underlined residues) and the DEF domain (boxed residues) found in c-Fos are conserved in Fra-1 and Fra-2, suggesting that Fra-1 and Fra-2 respond to ERK signaling like c-Fos. The electrophoretic mobility of c-Fos can be used as an indication of the phosphorylation state (Fig. 1B), and this mobility shift is partly due to an increase in the phosphorylation of Thr325 (Fig. 1B) as described previously (26). Like c-Fos, Fra-1 and Fra-2 also undergo a growth factor-regulated mobility shift that is inhibited when cells are pretreated with the MEK1/2 inhibitor UO126 (Fig. 1B). In each case, the growth factor-regulated mobility shift of c-Fos, Fra-2, and Fra-1 was accompanied by a mobility shift of endogenous ERK1/2 indicating the phosphorylation and activation of ERK signaling. To test the role of the putative DEF domains in regulating the Fra-2 and Fra-1 mobility shift, alanine substitution of Phe282 or Tyr284 in Fra-2 and Phe237 or Tyr239 in Fra-1 was performed. These mutations completely inhibited the growth factor-regulated mobility shift of ectopically expressed Fra-2 (Fig. 1C) and Fra-1 (Fig. 1D), suggesting that the DEF domain controls Fra hyperphosphorylation. The DEF domain has been found to exclusively direct ERK to phosphorylate phosphoacceptor sites lying just NH2 terminal to the binding motif (15, 26). Putative phosphoacceptor sites (Thr-Pro) are indeed located NH2 terminal to the DEF domains in Fra-2 and Fra-1 (Fig. 1A), and some of these are known to be phosphorylated in vivo (25, 43). Interestingly, in the case of Fra-1, one of these sites (Thr231) controls transcriptional transactivation (43).
FIG. 1.
DEF domain-dependent regulation of Fra proteins. (A) Alignment of the c-Fos, Fra-1, and Fra-2 COOH termini showing amino acid identity (boldface letters), ERK and RSK priming phosphorylation sites (underlined), and DEF domains (box). The sequences are from rat (c-Fos) and mouse (Fra-1 and Fra-2) proteins, and the numbers indicate amino acid positions. (B) NIH 3T3 cells were transfected with the indicated Fos alleles, serum deprived, and treated with EGF (50 ng/ml) for 5 min. Where indicated, cells were pretreated with UO126 (5 μM) for 30 min before treatment with EGF for 5 min. Cell extracts were then subjected to immunoblotting with anti-c-Fos, anti-phospho-Thr325 c-Fos, and anti-ERK antibodies. (C and D) NIH 3T3 cells were transfected with the indicated DEF domain mutant Fra-2 and Fra-1 alleles and treated as described for panel B. Immunoblotting was performed with the anti-Fra-2, anti-Fra-1, and anti-ERK antibodies.
In addition to Fos family proteins, we have noted that Myc proteins also contain putative DEF domains (26). The DEF domain in c-Myc is located between aa 196 and 199 and is COOH terminal to the growth factor-regulated phosphorylation sites Ser62 and Thr58 (Fig. 2A). ERK-mediated phosphorylation of Ser62 results in increased c-Myc stability, but when PI3-kinase signaling is low, Ser62 phosphorylation can also prime the phosphorylation of Thr58, which promotes ubiquitination and degradation of c-Myc (34). Given the pivotal role that Ser62 phosphorylation plays in c-Myc, a potent inducer of transformation and cell death, understanding how efficient phosphorylation of this site is achieved is of great importance. Phosphorylation of Ser62 is regulated by the MEK-ERK pathway (Fig. 2B), in agreement with previous studies (34). Replacement of the critical DEF domain residue Phe196 or Tyr198 with alanine reduced the growth factor-regulated increase in Ser62 phosphorylation (Fig. 2C). This observation indicates that aa 196 to 199 comprise a DEF domain and that this is required for the regulation of c-Myc Ser62 phosphorylation. The above observations show that in addition to c-Fos, the IEG products Fra-1, Fra-2, and c-Myc also contain functional DEF domains, indicating that these can also serve as molecular sensors for ERK signaling.
FIG. 2.
DEF domain-mediated regulation of c-Myc phosphorylation. (A) The illustration details the putative DEF domain (FPYP), NH2-terminal phosphorylation site Thr58 and Ser62, Myc box II (MbII), basic DNA binding domain (B), and helix-loop-helix (HLH)-leucine zipper (zip) domains. (B) NIH 3T3 cells were transfected with c-Myc or control vector and treated as described in the legend to Fig. 1B. c-Myc was immunoprecipitated (IP) from cell lysates, and each sample was analyzed with anti-c-Myc or anti-phospho-specific Ser62 (pS62) antibodies. Western blot analysis of ERK activation in the cell lysate was measured with the anti-phospho-ERK1/2 antibody. IgG, immunoglobulin G. (C) Quiescent cells expressing the indicated c-Myc proteins were treated with EGF for 5 min and lysed, and c-Myc immunoprecipitations were performed. Western blot analysis of immunoprecipitates used anti-c-Myc or anti-phospho-Ser62 antibodies, and analysis of cell lysate used anti-c-Myc or anti-ERK antibodies. The data shown are representative of three experiments.
Kinetics of EGF- and PDGF-stimulated IEG induction.
Differences in the duration of ERK signaling have profound effects on the phosphorylation state and stability of c-Fos (8, 26, 27). To determine if this response to signal duration is restricted to c-Fos alone, the expression kinetics of additional IEG products containing DEF domains was examined. Treatment of quiescent Swiss 3T3 cells with EGF induced transient activation of ERK (Fig. 3A) and transient accumulation of c-Fos (Fig. 3A, lanes 1 to 8). In contrast, PDGF treatment was associated with sustained ERK activity and c-Fos expression (Fig. 3A, lanes 9 to 15). Note that after 45 min of induction with EGF or PDGF, similar levels of c-Fos are expressed. In the case of transient ERK signaling, c-Fos is unstable and disappears, whereas sustained ERK activation is required for c-Fos stabilization, as shown previously (26). The expression of c-Jun, a c-Fos dimerization partner, was identical to that of c-Fos (Fig. 3A). Low levels of Fra-2 were reproducibly detected when signaling was transiently activated (Fig. 3A, lane 3) (data not shown), which might indicate either its reduced stability when hypophosphorylated or weak transcriptional induction by EGF. In contrast, higher levels of Fra-2 were observed up to 4 h after PDGF treatment (Fig. 3A, lanes 9 to 15). Fra-1 was only detected when ERK signaling was sustained, and its expression was delayed with respect to the other Fos proteins and c-Jun (Fig. 3A, lanes 9 to 15). In addition to these observations, the abundance and/or electrophoretic mobility of c-Fos, c-Jun, and Fra-2 significantly changed 3 to 4 h after PDGF induction (Fig. 3A, lanes 14 and 15). This may reflect reduced ERK-mediated phosphorylation of these proteins, which in turn affects protein stability and/or mobility. Fra-1 levels remained elevated at these later time points, and no change in electrophoretic mobility was observed (Fig. 3A, lanes 14 and 15).
FIG. 3.
Global induction of IEGs. (A) Quiescent Swiss 3T3 cells were treated with EGF (25 ng/ml) or PDGF-BB (20 ng/ml) for the indicated times and lysed. Western blotting was used to visualize the expression of Fos family proteins and c-Jun, as well as ERK activation kinetics (lower panel). (B) Cells were treated as described for panel A, and lysates were analyzed with anti-Egr-1, anti-JunB, or anti-ERK antibodies. (C) The induction of c-Myc was analyzed by initially immunoprecipitating c-Myc from extracts followed by Western analysis. The activation of ERK was analyzed with an anti-phospho-ERK1/2 (αpERK) antibody. All data shown are representative of at least four experiments.
The expression of two other DEF domain-containing IEG products, Egr-1 and JunB, was also similar to that of c-Fos. Expression of both was transient in cells treated with EGF but was sustained in PDGF-treated cells (Fig. 3B, lanes 1 to 13). Although the initial levels of EGF or PDGF induction of Egr-1 and JunB expression after 60 min were identical, levels of both proteins changed dramatically at later time points (Fig. 3B, compare lane 11 with lane 5). The electrophoretic mobility of JunB always appeared as a triplet when PDGF was used (Fig. 3B, lanes 9 to 13), whereas EGF induction was associated with a doublet JunB species (Fig. 3B, lane 3). This suggests that although both growth factors initially induce JunB expression, PDGF-regulated posttranslational modification of JunB leads to its stabilization.
When c-Myc expression was analyzed, a transient and weak induction was observed in EGF-treated cells (Fig. 3C, lanes 1 to 7), which likely reflects the short t1/2 (∼7.5 min) for hypophosphorylated c-Myc (34). Under sustained ERK signaling conditions with PDGF, levels of c-Myc were higher and were maintained for the duration of the experiment (Fig. 3C, lanes 8 to 14).
Sustained ERK signaling is required for extended expression of IEG products.
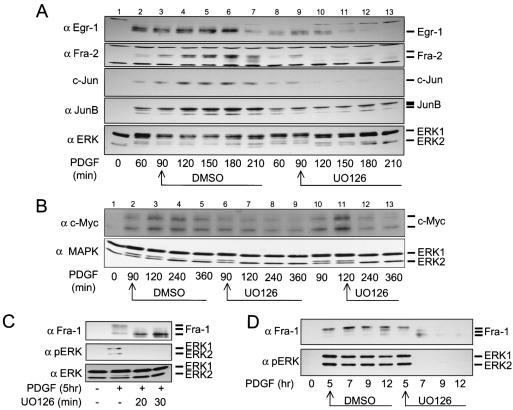
The results from the experiments described above illustrate a strong correlation between sustained ERK signaling and sustained expression of several IEG products containing DEF domains. To extend this further, we induced IEG product expression with PDGF and examined the requirement for sustained ERK activity for maintenance of IEG protein expression. If sustained ERK signaling is required for the extended expression of several of these IEG products, inhibition of ERK signaling with the MEK inhibitor should result in their rapid turnover. Thus, UO126 was added to cells already induced with PDGF (90 min) in order to convert the sustained signal into a more transient signal (Fig. 4A, lower panel, compare lanes 3 to 7 with lanes 9 to 13). Manipulating ERK signaling in this way had a strong effect on the extended expression of all IEG products examined. The expression of Egr-1, Fra-2, c-Jun, and JunB was reduced when UO126 was added to PDGF-treated cells (Fig. 4A). UO126 also reduced the levels of c-Myc detected in cells treated with PDGF for 2, 4, or 6 h (Fig. 4B, compare lanes 3 to 5 with 7 to 9). Delaying the addition of UO126 until after 2 h of PDGF treatment allowed c-Myc to reach its maximum expression level (Fig. 4B, lane 11), but levels then subsequently declined after a further 2 and 4 h of culture in the presence of both PDGF and UO126 (Fig. 4B, lanes 12 and 13). This is in contrast to maximal c-Myc levels being maintained in control cells (Fig. 4B, lanes 3 to 5, and Fig. 3C, lanes 12 to 14). Since PDGF does not significantly induce ERK5 activation in Swiss 3T3 cells (L.O. Murphy and J. Blenis, unpublished observations), the effect of UO126 on IEG expression (Fig. 4) can be fully attributed to inhibition of the MEK1/2-ERK pathway and not the MEK5-ERK5 pathway, the activation of which is also UO126 sensitive (17).
FIG. 4.
Sustained expression of IEG products requires the ERK pathway. Swiss 3T3 cells were induced with PDGF for 90 min and then treated with UO126 (5 μM) or DMSO (0.1%) for a further 120 min. (A) The levels of Egr-1, Fra-2, c-Jun, and JunB and the ERK activation kinetics were assayed by Western blotting. (B) After initially treating the cells with PDGF for 90 (lanes 2 to 10) or 120 min (lanes 11 to 13), DMSO or UO126 was added to cells. c-Myc was immunoprecipitated from cell lysates and subjected to Western analysis. (C) Swiss 3T3 cells were induced for 5 h with PDGF and then treated with UO126 for 20 or 30 min before cell lysis. Control cells (−) were treated with DMSO for 30 min. α pERK, anti-phospho-ERK. (D) Cells induced with PDGF for 5 h were incubated in the presence and absence (DMSO) of UO126 for a further 7 h. Cell extracts were prepared at the indicated times and subjected to Western analysis to detect levels of Fra-1 and the activation kinetics of ERK. The data shown are representative of three experiments.
The delayed induction of Fra-1 relative to other IEG products in 3T3 fibroblasts is partly due to the fact that c-Fos expression is required for the initial induction of fra-1 transcription (33). Therefore, to determine if sustained ERK signaling regulates Fra-1 protein expression during the G1-S transition, UO126 was added after 5 h of PDGF treatment, which allows Fra-1 to reach its maximum level of expression (Fig. 4C). Note that at this time point, c-Fos is relatively hypophosphorylated (26), and its levels are relatively low (Fig. 3A, lane 15). The presence of UO126 for only 20 or 30 min was sufficient to completely inhibit the mobility shift of Fra-1, indicating an important role for ERK signaling in posttranslational control of Fra-1 (Fig. 4C). Furthermore, this change in electrophoretic mobility preceded the complete disappearance of Fra-1, indicating an additional role for ERK in Fra-1 stability (Fig. 4D). In the absence of UO126, the levels of hyperphosphorylated Fra-1 are maintained after 9 to 12 h of induction (Fig. 4D), which corresponds to the G1-S boundary in Swiss 3T3 cells. Taken together, these data show that several IEG products (Fra-2, c-Jun, JunB, c-Myc, and Egr-1) and Fra-1 exhibit the same biochemical response to ERK signal duration that was previously described for c-Fos (26), providing strong evidence that these additional proteins are also ERK signal sensors.
Reduced ERK signal strength results in loss of IEG hyperphosphorylation, stability, and subsequent target gene expression.
The mechanisms that regulate ERK signal duration are not fully understood but may involve differential regulation at the level of phosphatases (29, 32, 36, 37), receptor internalization, and trafficking (30) and possibly alternative modes of Raf activation by Ras or Rap1 (42). Differences in signal amplitude and duration may also be simply a consequence of receptor density. Low receptor numbers would result in transient pathway activation, whereas 10- to 100-fold more receptors would result in more persistent activation (9, 38). We hypothesized that by reducing the amount of PDGF used to treat quiescent Swiss 3T3 cells, the sustained activation of ERK signaling would be converted into a more transient activation. If true, then IEG product phosphorylation and stability would be adversely affected. Indeed, reducing the PDGF concentration from 10 to 4 ng/ml converted the expression profile of c-Fos and Egr-1 from sustained to transient (Fig. 5A). Furthermore, while the initial activation of ERK after 10 min of induction was not significantly altered by reducing the amount of PDGF, the sustained phase of ERK signaling was (Fig. 5C). As anticipated, Thr325 phosphorylation in c-Fos was greatly reduced in cells treated with 4 ng of PDGF per ml compared to cells treated with 10 ng of PDGF per ml, despite the fact that c-Fos levels were nearly identical in both cases (Fig. 5B). Thus, the response associated with sustained ERK signaling (hyperphosphorylation and stabilization of IEG product) is changed to a transient response (hypophosphorylation and transient expression of IEG product) by a surprisingly small decrease in agonist concentration.
FIG. 5.
Incremental decreases in ERK signal strength and duration have large effects on IEG product phosphorylation and sustained expression. (A) Quiescent Swiss 3T3 cells were treated with different amounts of PDGF for the indicated times and lysed. The levels of endogenous c-Fos and Egr-1 and the activation kinetics of ERK were measured by Western blotting.α pERK, anti-phospho-ERK. (B) Quiescent Swiss 3T3 cells were treated with different doses of PDGF and lysed. The phosphorylation of Thr325 in c-Fos and total levels of c-Fos were measured by Western blotting. (C) ERK1 was immunoprecipitated from PDGF-treated cells, and its kinase activity was quantitated with an immune complex kinase assay. The data shown are the mean ± standard error from three independent experiments. (D and E) Cells were treated as described for panel A, and the levels of Fra-2 and Fra-1 were analyzed by Western analysis. The data are representative of three to four experiments.
When ERK1 activity was examined in cells treated with 10 or 4 ng of PDGF per ml, a significant 30 to 40% reduction in kinase activity was observed 1 to 2 h after induction (Fig. 5C). Since RSK activation is dependent on ERK signaling, the reduction in ERK activity associated with 4 ng/ml is also accompanied by a similar reduction in RSK phosphotransferase activity (data not shown). Therefore, differences inThr325 phosphorylation (Fig. 5B) likely reflects priming efficiency (RSK and ERK mediated) as well as DEF-dependent regulation of Thr325 (ERK mediated). These observations show that small decreases in ERK activity result in a substantial reduction in the phosphorylation state and stability of c-Fos. Finally, the expression of Fra-2 and Fra-1 was also examined under conditions of limiting PDGF (Fig. 5D). Fra-2 expression was induced and sustained for 4 h in cells treated with 10 ng of PDGF per ml (Fig. 5D, lanes 2 to 4), and as noted above (Fig. 3A), the electrophoretic mobility of Fra-2 changes 3 to 4 h after induction (Fig. 5D, lane 4). While the initial levels of Fra-2 in cells treated with 6, 5, or 4 ng of PDGF per ml were the same as those in cells treated with 10 ng/ml (compare lane 2 with lanes 5, 8, and 11), the electrophoretic mobility of Fra-2 changed after only 2 h in these cells (compare lane 3 with lanes 6, 9, and 12), indicating a more rapid decrease in phosphorylation state. Furthermore, decreasing the amount of PDGF from 10 to 4 ng/ml also converted Fra-2 expression from sustained to transient (Fig. 5D, compare lanes 4 and 13). Fra-1 was also equally induced after 2 h by all concentrations of PDGF used (Fig. 5D, compare lanes 2, 5, 8, and 11), but the electrophoretic mobility of Fra-1 was faster at the lower doses (compare lane 2 with lanes 5, 8, and 11), indicating differential phosphorylation of Fra-1. Furthermore, the levels of Fra-1 remaining after 4, 8, and 12 h were consistently lower in cells treated with 4 ng of PDGF per ml than those in cells treated with 10 ng/ml (Fig. 5D and E).
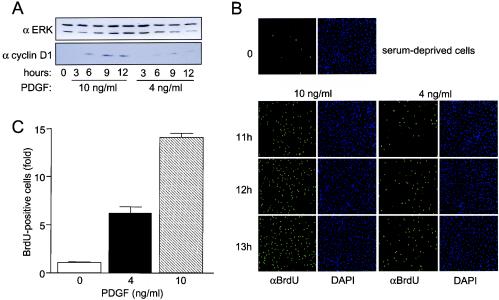
If the effects of ERK signal strength and duration on cell cycle control are mediated via IEG-encoded transcription factors, then sensor-regulated downstream gene expression should also be affected by signal kinetics. Since the cyclin D1 gene is under the direct control of the AP-1 transcription complex (5), cyclin D1 expression could represent a critical link between ERK signaling kinetics, IEG sensors, and cell cycle progression. If this is the case, small decreases in ERK signal strength that convert sustained c-Fos expression to more transient expression (Fig. 5A) would also be predicted to affect cyclin D levels. To examine this, quiescent Swiss 3T3 cells were cultured in the presence of 10 or 4 ng of PDGF per ml for 3, 6, 9, or 12 h, and cyclin D1 expression was measured by immunoblotting. Although the two doses of PDGF induced cyclin D1 expression (i.e., no delayed induction with 4 ng/ml), the levels were significantly reduced in cells treated with 4 ng/ml (Fig. 6A). Finally, to determine whether the small decrease in ERK signal strength differentially regulates G1-S progression, BrdU incorporation was performed, and quantitated by fluorescence-activated cell sorter (FACS) analysis. Both doses induced BrdU incorporation after 11 to 13 h; however, at each time point, the number of BrdU-positive cells was significantly reduced when 4 ng/ml was used (Fig. 6B). Quantitation showed that after 12 h in the presence of 10 ng of PDGF per ml, there was more than a 6-fold increase in BrdU incorporation, whereas 4 ng/ml was associated with only a 2.5-fold increase (Fig. 6C). These observations show that a two- to threefold change in agonist concentration is associated with subtle differences in ERK signal strength and duration. These apparently modest changes in ERK signal timing, however, are propagated into substantial differences in the phosphorylation state and expression of multiple IEG products, downstream sensor-regulated gene expression, and cell cycle progression.
FIG. 6.
ERK signal strength affects AP-1 target gene expression and S-phase entry. (A) Quiescent Swiss 3T3 cells were treated with PDGF (10 or 4 ng/ml), and cyclin D1 levels in cell extracts were measured with an anti-cyclin D antibody. (B) Quiescent Swiss 3T3 cells cultured on glass coverslips were treated with PDGF, and BrdU incorporation was assayed as described in Materials and Methods. The data shown are representative of three independent experiments. DAPI, 4′,6′-diamidino-2-phenylindole. (C) Quiescent Swiss 3T3 cells were induced with PDGF for 11.75 h, treated with BrdU (20 μM) for 15 min, and then fixed. The number of BrdU-positive cells in each treatment group was quantitated by FACS analysis as described in Materials and Methods. The data shown are the mean ± standard error from triplicate determinations and are representative of three independent experiments.
DISCUSSION
The findings presented in this study show how the strength and overall duration of ERK signaling control the global expression of several DEF domain-containing IEG products and how this relates to the expression of second-tier genes and cell cycle progression. First, we show that the putative DEF domains in Fra-1, Fra-2, and c-Myc are indeed functional. Like c-Fos, the DEF domains found in these transcription factors regulate their hyperphosphorylation and are predicted to control their biological function. Second, all DEF domain-containing IEG products examined display the same differential response to the duration of ERK signals, and prolonged ERK signaling is required for the sustained expression of these IEG products through the G1 phase of the cell cycle. Third, a small reduction in ERK signal strength results in significant loss of hyperphosphorylation and stability of these IEG products. Fourth, expression of the AP-1 target gene, cyclin D1, is significantly reduced when ERK signal strength and duration are decreased. Fifth, the IEG expression response to signal duration is tightly correlated with quantitative differences in cell cycle progression. Taken together, these observations argue that the posttranslational regulation of IEG products provides the cell with a mechanism to respond to subtle changes in ERK signal strength and duration, which in turn leads to significant changes in cellular function.
Understanding how sustained ERK and RSK signaling regulates c-Fos has provided a molecular framework to explain how differences in signal duration lead to the generation of a specific biological outcome (26). During sustained signaling, ERK- and RSK-mediated phosphorylation of the c-Fos COOH terminus primes hyperphosphorylation at Thr325 and Thr331 (26). Hyperphosphorylation also requires an intact DEF domain, and when mutated, Fos-mediated transformation is inhibited (26). In the present study, we have specifically shown that Fra-2, Fra-1, and c-Myc also have functional DEF domains that are required for their hyperphosphorylation. Experimental and sequence analyses show that posttranslational control of Fra-2 and Fra-1 is likely very similar to that of c-Fos due to the presence of priming phosphorylation sites and DEF domain-mediated phosphorylation sites in their COOH termini. Interestingly, the transactivation potential of Fra-1 is completely inhibited when Thr231 is mutated to alanine (43), and our findings indicate that this residue is under the regulation of the DEF domain. In the case of c-Myc, the DEF domain regulates the ERK-dependent phosphorylation of Ser62. Since phosphorylation of Ser62 promotes c-Myc stability, DEF-dependent docking of ERK to c-Myc would lead to c-Myc stabilization. Interestingly, the DEF domain and NH2-terminal phosphorylation sites Ser62 and Thr58 both flank a region known as Myc box II (MbII). MbII is required for the assembly of a high-molecular-weight complex that contains the transformation-transactivation domain-associated protein (TRRAP) and TIP49/TIP48 (23, 41). This complex is required for Myc-dependent transformation and may be involved in chromatin modification of Myc target genes (10). Apart from directly phosphorylating Ser62, ERK docking to c-Myc may also have a role in regulating the recruitment and/or activity of the TRRAP complex.
Several other IEG products have expression patterns that are characteristic of the Fos and Myc family sensors. For example, differences in signal duration can regulate c-Jun expression just like c-Fos. Based on sequence analysis, however, it is predicted that c-Jun does not contain a functional DEF domain (26). Nevertheless, it is known that the ERK pathway can control the phosphorylation of Ser63 and Ser73 during sustained signaling in PC12 cells (19). How can c-Jun expression be regulated by sustained ERK signaling in the absence of a DEF domain? One possibility is that transphosphorylation of Ser63 and Ser73 by Fos-associated ERK would result in increased c-Jun stabilization and transcriptional activation. In support of this, JNK-mediated transphosphorylation of c-Jun dimerization partners has been demonstrated (16). Another possibility is that the stabilization of Fos family proteins indirectly controls the stability of Jun proteins, because Jun/Fos dimers are more stable than Jun/Jun dimers (1).
The phosphorylation of Thr102 and Thr104 in JunB is thought to be predominantly regulated by JNK and not p38 or ERK (20). Interestingly, these amino acids, which are required for JunB-driven interleukin-4 expression in Th2 lymphocytes (20), are located NH2 terminal to the putative DEF domain in JunB (26). It is possible that in different cell types and in the presence of an appropriate partner protein, the DEF domain facilitates the ERK-dependent phosphorylation of JunB, and future work is required to investigate this possibility. Finally, ERK-mediated differentiation of PC12 cells requires Egr-1 transcription activity to increase the expression of p35, an Egr-1 target gene (14). While the role of the Egr-1 DEF domain is presently unknown in this process, the possibility exists that ERK docking controls the stability and/or transcriptional activity of Egr-1. Interestingly, several putative ERK phosphorylation sites (Ser-Pro) are located NH2 terminal to this DEF domain (26), and Egr-1 itself is phosphorylated in vivo (6).
The observations in this study illuminate a global mechanism that is responsible for dictating cellular responses to ERK signal duration. This mechanism takes into account the effect of both signal kinetics and signal strength. We believe that the presence of DEF domains in several growth factor-induced sensors enables the cell to respond to small increases or decreases in signal strength. This concept is best exemplified by the response of Fra-1 to sustained ERK signals. In Swiss 3T3 fibroblasts, although ERK activity at the G1-S boundary is only 10 to 20% of that measured after 10 min of initial stimulation, it is nevertheless sufficient to promote hyperphosphorylation of Fra-1. This and other evidence strongly suggests that DEF domains locally concentrate the active ERK to Fos, Jun, Myc, and Egr-1 sensors during sustained signaling, and this results in efficient phosphorylation of target phosphoacceptor sites.
The transcriptional induction of fra-1 can be directly regulated by c-Fos (4) and as a result is compromised in c-fos−/− osteoclasts and fibroblasts (22, 33). This suggests that in addition to the role of ERK signaling directly on Fra-1 protein, the effect of signal duration on the c-Fos sensor can also determine the transcriptional induction of fra-1. Although Fra-1 expression occurs only when ERK signaling is sustained (i.e., cells treated with PDGF but not EGF), c-Fos is still transiently expressed and is hypophosphorylated in EGF-treated cells but is not sufficient to induce Fra-1. This implies that growth factor-induced fra-1 expression requires prior ERK-mediated phosphorylation and/or stabilization of c-Fos. This important observation discriminates between an absolute requirement of c-fos for the induction of fra-1 expression, as indicated by the behavior of c-fos−/− cells, and an essential posttranslational role for ERK signaling in this process. Further support for the latter mechanism comes from experiments in which signal strength was reduced. With low doses of PDGF, although the expression of c-Fos was relatively transient, it was nonetheless phosphorylated to a greater extent than that normally associated with EGF treatment, and critically, this preceded Fra-1 expression in Swiss 3T3 cells. We believe that after the initial c-Fos-dependent induction of fra-1 (33), Fra-1 protein is hyperphosphorylated by ERK, and this primarily contributes to sustained Fra-1 expression in G1. Thus, the c-Fos-Fra-1 pathway is an example of how two sensors that have overlapping expression profiles provide the cell with temporal information about the amplitude of ERK signaling.
The ability of a single cell to respond to minute changes in the concentration of extracellular stimuli underlies processes that control embryonic development, cell morphology, migration, differentiation, and proliferation. In fibroblasts, a two- to threefold change in PDGF concentration can result in a modest reduction in ERK signal strength. This small change in ERK signaling, however, leads to large differences in the expression kinetics and phosphorylation of IEG-encoded sensors and cell cycle progression. This response to agonist concentration resembles the behavior of Drosophila and Xenopus embryos to morphogens. In these systems, as little as a two- to fourfold change in morphogen concentration can result in quantitative differences in gene expression and cell fate outcome (13). Many morphogens, such as Screw and Activin, control rapid activation of “early” target genes independent of ongoing protein synthesis (13), which resembles the IEG response to extracellular stimuli. Morphogens are also associated with the delayed expression of “late” target genes, and it has been proposed that the combination of intracellular signaling and the presence of early proteins can regulate late gene expression (28). During embryonic development, an IEG sensor-like mechanism could allow individual cells to interpret their position in a morphogen gradient and therefore would be required to generate a specific cell fate.
In conclusion, the uniform response shown by DEF domain-containing IEG products to differences in ERK signal duration and strength demonstrates that these gene products can collectively act as sensors for ERK signaling. Since IEG products are induced by a diverse range of extracellular stimuli, these proteins may have the general capacity to interpret differences in the kinetics of signaling pathways other than ERK. Therefore, this model in part may help explain how complex intracellular signals can be decoded and ultimately transduced into a phenotypic change in cell behavior.
Acknowledgments
We are grateful to Y. Taya and J. Nevins for generously supplying the phospho-Ser62 c-Myc antibody and E. Tulchinsky and K. Ciekowski for plasmids. We wish to acknowledge members of the Blenis laboratory for their critical feedback.
L.O.M. is a Leukemia & Lymphoma Society Special Fellow. This work was supported by NIH grant RO1CA46595 (J.B.) and a Special Grant for Research from the American Cancer Society, New England Division (L.O.M.).
REFERENCES
- 1.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 2.Balmanno, K., and S. J. Cook. 1999. Sustained MAP kinase activation is required for the expression of cyclin D1, p21Cip1 and a subset of AP-1 proteins in CCL39 cells. Oncogene 18:3085-3097. [DOI] [PubMed] [Google Scholar]
- 3.Bebien, M., S. Salinas, C. Becamel, V. Richard, L. Linares, and R. A. Hipskind. 2003. Immediate-early gene induction by the stresses anisomycin and arsenite in human osteosarcoma cells involves MAPK cascade signaling to Elk-1, CREB and SRF. Oncogene 22:1836-1847. [DOI] [PubMed] [Google Scholar]
- 4.Bergers, G., P. Graninger, S. Braselmann, C. Wrighton, and M. Busslinger. 1995. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Mol. Cell. Biol. 15:3748-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. R., E. Nigh, R. J. Lee, H. Ye, M. A. Thompson, F. Saudou, R. G. Pestell, and M. E. Greenberg. 1998. Fos family members induce cell cycle entry by activating cyclin D1. Mol. Cell. Biol. 18:5609-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, X., R. A. Koski, A. Gashler, M. McKiernan, C. F. Morris, R. Gaffney, R. V. Hay, and V. P. Sukhatme. 1990. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol. Cell. Biol. 10:1931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, R.-H., and J. Blenis. 1990. Identification of Xenopus S6 protein kinase homologs (pp90rsk) in somatic cells: phosphorylation and activation during initiation of cell proliferation. Mol. Cell. Biol. 10:3204-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, R. H., P. C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12:1493-1502. [PubMed] [Google Scholar]
- 9.Duckworth, B. C., and L. C. Cantley. 1997. Conditional inhibition of the mitogen-activated protein kinase cascade by wortmannin. Dependence on signal strength. J. Biol. Chem. 272:27665-27670. [DOI] [PubMed] [Google Scholar]
- 10.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 11.Fambrough, D., K. McClure, A. Kazlauskas, and E. S. Lander. 1999. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 97:727-741. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, M. E., L. A. Greene, and E. B. Ziff. 1985. Nerve growth factor and epidermal growth factor induce rapid transient changes in proto-oncogene transcription in PC12 cells. J. Biol. Chem. 260:14101-14110. [PubMed] [Google Scholar]
- 13.Gurdon, J. B., and P. Y. Bourillot. 2001. Morphogen gradient interpretation. Nature 413:797-803. [DOI] [PubMed] [Google Scholar]
- 14.Harada, T., T. Morooka, S. Ogawa, and E. Nishida. 2001. ERK induces p35, a neuron-specific activator of Cdk5, through induction of Egr1. Nat. Cell Biol. 3:453-459. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, D., D. Glossip, H. Xing, A. J. Muslin, and K. Kornfeld. 1999. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 13:163-175. [PMC free article] [PubMed] [Google Scholar]
- 16.Kallunki, T., T. Deng, M. Hibi, and M. Karin. 1996. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87:929-939. [DOI] [PubMed] [Google Scholar]
- 17.Kamakura, S., T. Moriguchi, and E. Nishida. 1999. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 274:26563-26571. [DOI] [PubMed] [Google Scholar]
- 18.Kustikova, O., D. Kramerov, M. Grigorian, V. Berezin, E. Bock, E. Lukanidin, and E. Tulchinsky. 1998. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol. Cell. Biol. 18:7095-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppa, S., R. Saffrich, W. Ansorge, and D. Bohmann. 1998. Differential regulation of c-Jun by ERK and JNK during PC12 cell differentiation. EMBO J. 17:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, B., C. Tournier, R. J. Davis, and R. A. Flavell. 1999. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 18:420-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179-185. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo, K., J. M. Owens, M. Tonko, C. Elliott, T. J. Chambers, and E. F. Wagner. 2000. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet. 24:184-187. [DOI] [PubMed] [Google Scholar]
- 23.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 24.Milbrandt, J. 1986. Nerve growth factor rapidly induces c-fos mRNA in PC12 rat pheochromocytoma cells. Proc. Natl. Acad. Sci. USA 83:4789-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami, M., M. Ui, and H. Iba. 1999. Fra-2-positive autoregulatory loop triggered by mitogen-activated protein kinase (MAPK) and Fra-2 phosphorylation sites by MAPK. Cell Growth Differ. 10:333-342. [PubMed] [Google Scholar]
- 26.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4:556-564. [DOI] [PubMed] [Google Scholar]
- 27.Okazaki, K., and N. Sagata. 1995. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 14:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pages, F., and S. Kerridge. 2000. Morphogen gradients. A question of time or concentration? Trends Genet. 16:40-44. [DOI] [PubMed] [Google Scholar]
- 29.Paul, S., A. C. Nairn, P. Wang, and P. J. Lombroso. 2003. NMDA-mediated activation of the tyrosine phosphatase STEP regulates the duration of ERK signaling. Nat. Neurosci. 6:34-42. [DOI] [PubMed] [Google Scholar]
- 30.Pierce, K. L., S. Maudsley, Y. Daaka, L. M. Luttrell, and R. J. Lefkowitz. 2000. Role of endocytosis in the activation of the extracellular signal-regulated kinase cascade by sequestering and nonsequestering G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 97:1489-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards, S. A., J. Fu, A. Romanelli, A. Shimamura, and J. Blenis. 1999. Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr. Biol. 9:810-820. [DOI] [PubMed] [Google Scholar]
- 32.Saxena, M., S. Williams, K. Tasken, and T. Mustelin. 1999. Crosstalk between cAMP-dependent kinase and MAP kinase through a protein tyrosine phosphatase. Nat. Cell Biol. 1:305-311. [DOI] [PubMed] [Google Scholar]
- 33.Schreiber, M., C. Poirier, A. Franchi, R. Kurzbauer, J. L. Guenet, G. F. Carle, and E. F. Wagner. 1997. Structure and chromosomal assignment of the mouse fra-1 gene, and its exclusion as a candidate gene for oc (osteosclerosis). Oncogene 15:1171-1178. [DOI] [PubMed] [Google Scholar]
- 34.Sears, R., F. Nuckolls, E. Haura, Y. Taya, K. Tamai, and J. R. Nevins. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein, R., S. Orit, and D. J. Anderson. 1988. The induction of a neural-specific gene, SCG10, by nerve growth factor in PC12 cells is transcriptional, protein synthesis dependent, and glucocorticoid inhibitable. Dev. Biol. 127:316-325. [DOI] [PubMed] [Google Scholar]
- 36.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2:110-116. [DOI] [PubMed] [Google Scholar]
- 37.Theodosiou, A., and A. Ashworth. 26June2002, posting date. MAP kinase phosphatases. Genome Biol. 3:REVIEWS3009. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- 38.Traverse, S., K. Seedorf, H. Paterson, C. J. Marshall, P. Cohen, and A. Ullrich. 1994. EGF triggers neuronal differentiation of PC12 cells that overexpress the EGF receptor. Curr. Biol. 4:694-701. [DOI] [PubMed] [Google Scholar]
- 39.Werlen, G., B. Hausmann, D. Naeher, and E. Palmer. 2003. Signaling life and death in the thymus: timing is everything. Science 299:1859-1863. [DOI] [PubMed] [Google Scholar]
- 40.Whitmarsh, A. J., P. Shore, A. D. Sharrocks, and R. J. Davis. 1995. Integration of MAP kinase signal transduction pathways at the serum response element. Science 269:403-407. [DOI] [PubMed] [Google Scholar]
- 41.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321-330. [DOI] [PubMed] [Google Scholar]
- 42.York, R. D., H. Yao, T. Dillon, C. L. Ellig, S. P. Eckert, E. W. McCleskey, and P. J. Stork. 1998. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature 392:622-626. [DOI] [PubMed] [Google Scholar]
- 43.Young, M. R., R. Nair, N. Bucheimer, P. Tulsian, N. Brown, C. Chapp, T.-C. Hsu, and N. H. Colburn. 2002. Transactivation of Fra-1 and consequent activation of AP-1 occur extracellular signal-regulated kinase dependently. Mol. Cell. Biol. 22:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]